Abstract
This communication reports the increase in fluorescence resonance energy transfer (FRET) efficiency between two laser dyes in the presence of deoxyribonucleic acid (DNA). Two types of molecular logic gates have been designed where DNA acts as input signal and fluorescence intensity of different bands are taken as output signal. Use of these logic gates as a DNA sensor has been demonstrated.
Keywords: FRET, DNA, Molecular logic gate, Sensor
Introduction
Recent research on molecular logic gate was initiated after the pioneering work of de Silva [1] using the idea that molecules can be used for processing information just like the electronic logic gates. Remarkable progress has been made since then in the development of molecular logic gates [2–14]. The change in fluorescence characteristics of a dye due to the introduction of some external agent can be considered to be analogous to the digital responses in electronic logic gates. Molecules can undergo changes in ground or exited state due to the interference of some external chemical or biological materials [15]. In most cases, these kinds of changes can be realized in terms of the basic operations of logic gates using the familiar Boolean logic [16]. The development of molecular systems such as logic gates and circuits is of immense interest among the researchers that pursue advanced technologies. Basic logic operation of a molecular logic gate is the same as that of the electronic one, the only difference is in the input and output signal. Nowadays, these molecular logic gates can be used as simple as well as more complex devices [17–19] such as adders and subtractors [2–4], multiplexers/demultiplexers [5–7], encoders/decoders [8, 9], keypad locks [10–14] etc. Among many applications of molecular logic gates, one of the most interesting is the investigation of the inside components of a cell, where silicon-based analogues are not expected to reach [2]. To have some structural and functional ideas about different biological materials like DNA, RNA, and proteins, etc., the development of techniques for sensing and monitoring them is in great demand [20]. A homogeneous sensing ensemble, based on DNA quadruplexes, was reported by Margulies et al. [21] where the sensing ensemble can provide a direct analysis of the properties of the target proteins.
In designing nanosized devices, the issue of connectivity plays an important role. The strength of semiconductor devices depends on this connectivity where the output from one nanosized device is used to control the input of another. Whereas in the case of the molecular logic gate, it is not easy to pass the output of one gate to serve as an input to the next due to the difference in nature and properties of output-input characteristics. From this point of view, it must be stressed that the molecular logic gate or computation need not follow the conventional semiconductor blue print.
Using some basic logic, molecules can process and manipulate information like electronic computers and the human brain. There are many molecular logic gates where chemicals are used as inputs and optical signals are the outputs [22]. For the sensing of different organic [23], inorganic [24], and biological [21, 25] materials, these molecular logic gates are now being extensively used. To better understand the outputs of the sensors, they are compared with some well-known digital logic gates and from different outputs of those logic gates we can have some idea about the different structural features of the test sample.
Fujimoto et al. reported the detection of target DNAs by excimer-monomer switching of pyrene using the fluorescence resonance energy transfer (FRET) process [26]. A DNA-based nanomachine was reported by Liu et al. using the FRET phenomenon [20]. Also for encrypting messages on DNA strands, various methods have been accomplished [27–29].
The present communication reports the effect of DNA on fluorescence resonance energy transfer (FRET) between two dyes acriflavine (Acf) and rhodamine B (RhB). Acf and RhB are in principle suitable for energy transfer. Both the dyes are highly fluorescent. Fluorescence spectrum of Acf overlaps with absorption spectrum of RhB. By using this FRET process, we are able to construct a photo-regulated fluorescence switch. The output of the switch is mimicking the electronic NOT and YES/NOT logic gates. This kind of “ON–OFF” switching of fluorescence intensity can be varied by the introduction of photochemically active biomolecule DNA. It has been observed that the incorporation of DNA in the FRET pair modulates the FRET efficiency. This has been used to design the molecular logic gate, which is capable of sensing the presence of DNA.
Experimental section
Both acriflavine (Acf) and rhodamine B (RhB) were purchased from Sigma Chemical Co., USA and were used as received. Ultrapure Milli-Q water (resistivity 18.2 MΩ-cm) was used as solvent. The DNA used is sheared Salmon sperm DNA having a size of nearly about 2,000 bp with approximate GC content 41.2%, purchased from SRL India and was used as received. The purity of DNA was checked by UV-Vis absorption and fluorescence spectroscopy before use. UV-Vis absorption and fluorescence spectra of the solutions were recorded by a Perkin Elmer Lambda-25 absorption spectrophotometer and Perkin Elmer LS-55 fluorescence spectrophotometer, respectively. For fluorescence measurement, the excitation wavelength was 420 nm. The concentration of the individual dye in aqueous solution was 10 − 6 M. In order to create the mixed dye solution, the dye solutions were mixed with 1:1 volume ratio.
Results and discussion
FRET between Acf and RhB in aqueous solution
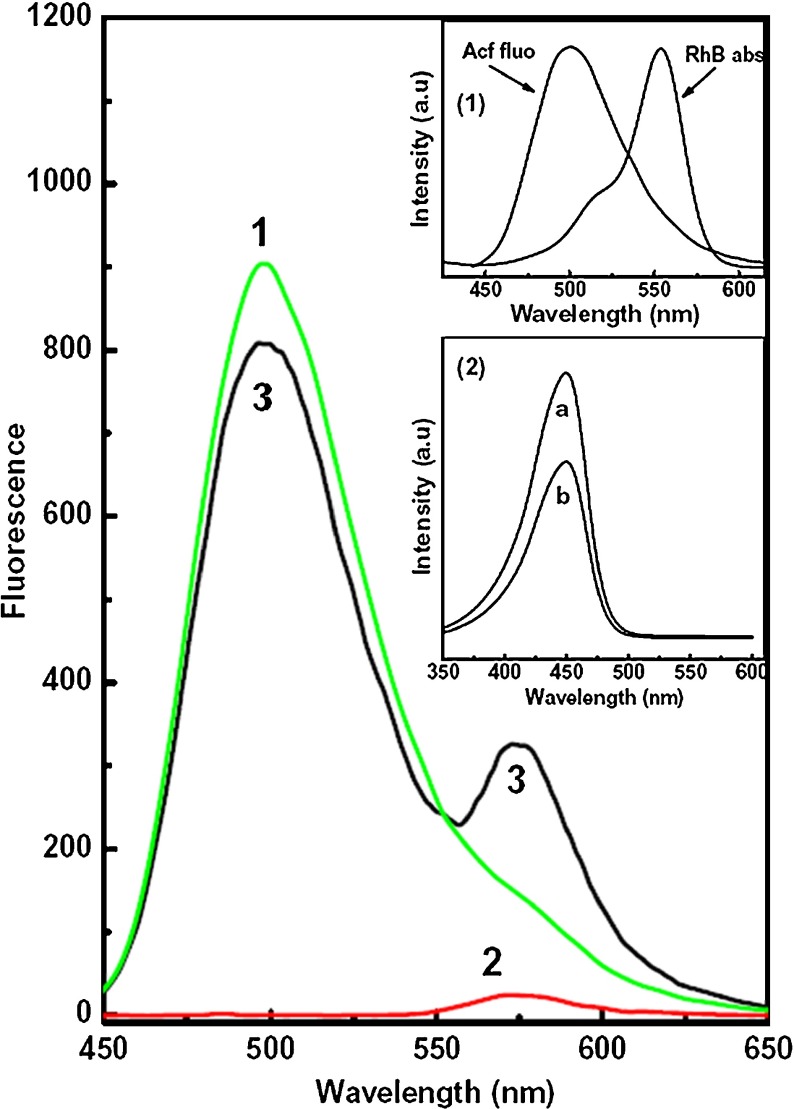
Figure 1 shows the fluorescence spectra of pure Acf (curve 1), pure RhB (curve 2), and their mixture of 1:1 volume ratio (curve 3) in aqueous solution. Spectra shown in Fig. 1 were recorded with excitation wavelength 420 nm (close to the monomer absorption of Acf). This excitation wavelength was selected in order to avoid the direct excitation of the RhB molecules. With this excitation wavelength, Acf shows prominent fluorescence with peak at 500 nm (curve-1 of Fig. 1), whereas RhB fluorescence intensity is almost negligible with a very weak peak at 578 nm (curve-2 of Fig. 1). From the spectral characteristics, it has been observed that both the Acf and RhB are mainly present as monomers in aqueous solution. However, for the fluorescence spectra of Acf-RhB mixed solution (curve 3), the RhB fluorescence intensity increases even with this excitation wavelength (420 nm) as well as Acf fluorescence decreases compared to their pure counterpart. This may be due to the transfer of energy from Acf to RhB. This transferred energy excites more RhB molecules followed by light emission from RhB, which is added to the original RhB fluorescence. As a result, the RhB fluorescence intensity gets sensitized. In order to confirm this, we measure the excitation spectra with emission wavelength fixed at Acf (500 nm) and RhB (578 nm) fluorescence maximum in case of Acf-RhB mixed aqueous solution (inset 2 of Fig. 1). Interestingly, both of the excitation spectra are almost similar and possess characteristic absorption bands of Acf monomers. This confirms that the RhB fluorescence in case of Acf–RhB mixed solution is mainly due to the light absorption by Acf and corresponding transfer to RhB monomer. Thus, FRET between Acf to RhB has been confirmed.
Fig. 1.
Fluorescence spectra of Acf (1), RhB (2), and Acf + RhB (3) (1:1 volume ratio) in water solution. Excitation wavelength was 420 nm (Acf absorption maximum) and concentration of individual dye (pure Acf and RhB) 10 − 6 M. Inset (1) shows the normalized absorption spectrum of rhodamine B and fluorescence spectrum of acriflavine in water solution and inset (2) shows the excitation spectra for Acf+RhB mixture with emission wavelengths at 500 nm (a) and 578 nm (b)
FRET between Acf and RhB in the presence of DNA
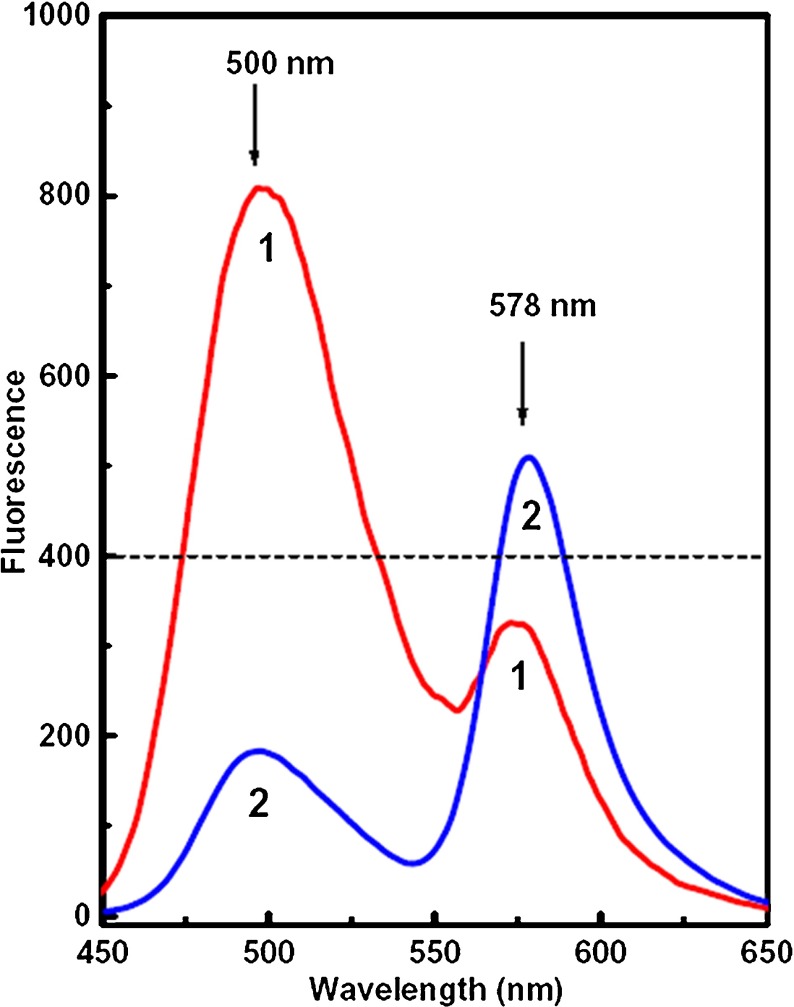
Figure 2 shows the fluorescence spectra of Acf–RhB mixed aqueous solution (1:1 volume ratio) in the presence (curve 2) and in absence (curve 1) of DNA. The DNA concentration was 1 μg/ml. It is interesting to observe that in the presence of DNA, the RhB fluorescence intensity increases and the Acf fluorescence intensity decreases further compared to that in the absence of DNA. This indicates that the presence of DNA influences the extent of energy transfer.
Fig. 2.
Fluorescence spectra of Acf–RhB (1:1 volume ratio) mixed aqueous solution in absence of DNA (1) and in presence of DNA (2). Excitation wavelength was 420 nm (Acf absorption maximum) and concentration of individual dye (pure Acf and RhB) 10 − 6 M. DNA concentration was 1 μg/ml
Based on the fluorescence spectra of Figs. 1 and 2, the fluorescence energy transfer efficiency has been calculated using the following equation [30]
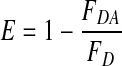 |
where FDA is the relative fluorescence intensity of the donor in the presence of acceptor and FD is the fluorescence intensity of the donor in the absence of the acceptor.
It has been observed that the FRET efficiency of the dye pair increases from 11.37% (absence of DNA) to 79.1% (presence of DNA). These data support the increase in energy transfer between Acf and RhB in the presence of DNA. It is interesting to mention in this context that the FRET process is distance-dependent and if the intermolecular distance between donor and acceptor decreases, then the transfer of energy from donor to acceptor becomes very efficient. FRET is effective over a distance ranging between 1 and 10 nm [31]. Also, an increase in spectral overlap integral enhances the energy transfer [32, 33].
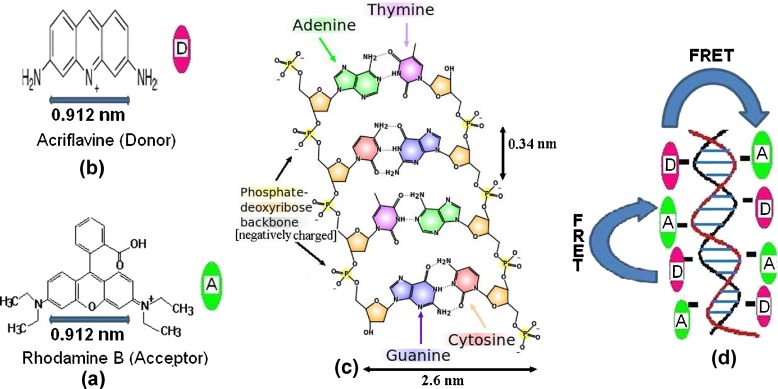
In DNA, the nucleotide bases lie horizontally between the two spiraling polymer strands with negatively charged phosphate backbones attached on either side of the base pair [34, 35]. The distance between two consecutive base pairs is 0.34 nm [36]. In the present case, both of the dyes Acf and RhB used are cationic. In the presence of DNA, they are attached with the DNA strands through the electrostatic attraction with the negatively charged phosphate backbone of DNA. As a result, both of the dyes come close to each other, resulting in a favorable condition for energy transfer. Accordingly, the energy transfer efficiency increases in the presence of DNA. Attachment of the dyes onto the phosphate backbone of DNA has been shown schematically in Fig. 3. It may be mentioned in this context that Shu Wang et al. reported that the negatively charged DNA bring a close electrostatic interaction with the cationic water soluble conjugated polymer backbone referring to an efficient FRET [37]. DNA strands have also been used in FRET-based biosensors, where they are used as spacers between FRET dye pairs.
Fig. 3.
a Molecular structure of rhodamine B, b molecular structure of acriflavine, c structure of DNA showing the negatively charged phosphate deoxyribose backbone, d schematic diagram showing the attachment of Acf & RhB onto phosphate backbone of DNA
Design of molecular logic gates
Based on the efficiency of FRET between Acf and RhB in the presence and absence of DNA, two types of molecular logic gates have been proposed, namely NOT and YES/NOT gates. These molecular logic gates, unlike digital counterparts, sense the presence of a biological material DNA, which acts as an input signal. The output signal is the fluorescence intensity of a particular band (500 and 578 nm). Using these logic gates, it is possible to detect the DNA in aqueous solution up to a very low concentration of 1 μg/ml.
Design of NOT gate as DNA sensor
Based on the spectral characteristic in Fig. 2, we have designed the logic gates. Here we consider the fluorescence intensity of the 500-nm band during FRET between Acf and RhB as the output signal and presence of DNA as input. Fluorescence intensity of 400 units (shown in Fig. 2) has been chosen as the reference level. Table 1 shows the logic of the NOT gate. In the absence of DNA (input = 0), fluorescence intensity at the 500-nm band is greater than the reference level (output = 1). In the presence of DNA (input = 1), the 500-nm fluorescence band intensity is less than the reference level (output = 0). Thus, an effective NOT gate can be developed that can sense the presence of DNA in aqueous solution having a concentration as low as 1 μg/ml. Thus, by observing the fluorescence intensity of the 500-nm band, it is possible to detect the presence of DNA.
Table 1.
Function table of NOT gate using fluorescence intensity
| Input DNA | Output (Fluorescence intensity of 500-nm band) |
|---|---|
| 0 (Absence of DNA) | 1 (Fluorescence intensity greater than reference level) |
| 1 (Presence of DNA) | 0 (Fluorescence intensity less than reference level) |
Design of YES-NOT gate as DNA sensor
In this case, the input is similar to that of the NOT gate, where the output signals are the fluorescence intensities of 500- and 578-nm bands. When the input signal is zero (absence of DNA), the intensity of the 500-nm band is greater than the reference level (output = 1) and for the 578-nm band, the fluorescence intensity is less than the reference level (output = 0). When the input signal is 1 (presence of DNA), the output of the 500-nm band is 0 whereas the 578-nm band is 1. In this case, the YES-NOT gate strongly confirms the presence and absence of DNA in the aqueous solution. Table 2 shows the logic of the YES-NOT gate. Here, by comparing the intensity at 500 nm with the reference level it is possible to detect the presence or absence of DNA.
Table 2.
Function table of YES-NOT gate using fluorescence intensity
| Input DNA | Output (Fluorescence intensity of 500-nm band) | Output (Fluorescence intensity of 578-nm band) |
|---|---|---|
| 0 (Absence of DNA) | 1 (Fluorescence intensity greater than reference level) | 0 (Fluorescence intensity less than reference level) |
| 1 (Presence of DNA) | 0 (Fluorescence intensity less than reference level) | 1 (Fluorescence intensity greater than reference level) |
It is worthwhile to mention in this context that in the present manuscript the experiments have been done with sheared salmon sperm DNA having a size of nearly about 2,000 bp with approximate GC content 41.2%. The actual size of the genomic DNA is approximately 3 × 109 bp, which is sheared to 2,000 bp. Therefore, the sheared DNA in solution contains a huge number of different kinds of sequences. In order to check the dependence of experimental results on the specific sequences of DNA, we have also tested all of the experiments with isolated human DNA (GC content 40%) and found similar results (result not shown). Therefore, the working principle of the designed logic gate depends on the interaction of Acf-RhB with phosphate moiety of DNA and is independent of any specific sequences of DNA. Using this designed logic gate, only the presence or absence of DNA can be detected.
Conclusions
Based on the experimental observation that the presence of DNA increases the fluorescence resonance energy transfer (FRET) between the two laser dyes, acriflavine (Acf) and rhodamine B (RhB), two types of molecular logic gates, namely a NOT and a YES-NOT gate, have been designed. These two molecular logic gates have been found to be efficient in detecting the presence of DNA in aqueous solution having concentrations as low as 1 μg/ml.
Acknowledgements
The author SAH is grateful to DST, CSIR, and DAE for financial support to carry out this research work through DST Fast-Track project Ref. No. SE/FTP/PS-54/2007, CSIR project Ref. 03(1146)/09/EMR-II, and DAE Young Scientist Research Award (No. 2009/20/37/8/BRNS/3328).
References
- 1.de Silva PA, Gunaratne NHQ, Mccoy CP. A molecular photoionic and gate based on fluorescent signaling. Nature. 1993;364:42–44. doi: 10.1038/364042a0. [DOI] [Google Scholar]
- 2.Margulies D, Melman G, Shanzer A. A molecular full-adder and full-subtractor, an additional step toward a moleculator. J. Am. Chem. Soc. 2006;128:4865–4871. doi: 10.1021/ja058564w. [DOI] [PubMed] [Google Scholar]
- 3.Pischel U. Chemical approaches to molecular logic elements for addition and subtraction. Angew. Chem. Int. Ed. 2007;46:4026–4040. doi: 10.1002/anie.200603990. [DOI] [PubMed] [Google Scholar]
- 4.Bozdemir OA, Sozmen F, Büyükcakir O, Guliyev R, Cakmak Y, Akkaya EU. Reaction-based sensing of fluoride ions using built-in triggers for intramolecular charge transfer (ICT) and photoinduced electron transfer (PET) Org. Lett. 2010;12:1400–1403. doi: 10.1021/ol100172w. [DOI] [PubMed] [Google Scholar]
- 5.Andreasson J, Straight SD, Bandyopadhyay S, Mitchell RH, Moore TA, Moore AL, Gust D. Molecular 2:1 digital multiplexer. Angew. Chem. Int. Ed. 2007;46:958–961. doi: 10.1002/anie.200603856. [DOI] [PubMed] [Google Scholar]
- 6.Amelia M, Baroncini M, Credi A. A simple unimolecular multiplexer/demultiplexer. Angew. Chem. Int. Ed. 2008;47:6240–6243. doi: 10.1002/anie.200802018. [DOI] [PubMed] [Google Scholar]
- 7.Perez-Inestrosa E, Montenegro JM, Collado D, Suau R. Molecular 1:2 demultiplexer. Chem. Commun. A. 2008;9:1085–1087. doi: 10.1039/b717690b. [DOI] [PubMed] [Google Scholar]
- 8.Andreasson J, Straight SD, Moore TA, Moore AL, Gust D. Molecular all-photonic encoder-decoder. J. Am. Chem. Soc. 2008;130:11122–11128. doi: 10.1021/ja802845z. [DOI] [PubMed] [Google Scholar]
- 9.Ceroni P, Bergamini G, Balzani V. Old molecules, new concepts: [Ru(bpy)3]2+ as a molecular encoder-decoder. Angew. Chem. Int. Ed. 2009;48:8516–8518. doi: 10.1002/anie.200904764. [DOI] [PubMed] [Google Scholar]
- 10.Margulies D, Felder CE, Melman G, Shanzer A. A molecular keypad lock: a photochemical device capable of authorizing password entries. J. Am. Chem. Soc. 2007;129:347–354. doi: 10.1021/ja065317z. [DOI] [PubMed] [Google Scholar]
- 11.Strack G, Ornatska M, Pita M, Katz E. Biocomputing security system: concatenated enzyme-based logic gates operating as a biomolecular keypad lock. J. Am. Chem. Soc. 2008;130:4234–4235. doi: 10.1021/ja7114713. [DOI] [PubMed] [Google Scholar]
- 12.Sun W, Zhou C, Xu CH, Fang CJ, Zhang C, Li ZX, Yan CH. A fluorescent-switch-based computing platform in defending information risk. Chem. Eur. J. 2008;14:6342–6351. doi: 10.1002/chem.200800576. [DOI] [PubMed] [Google Scholar]
- 13.Suresh, M., Ghosh, A., Das, A.: A simple chemosensor for Hg2 + and Cu2 + that works as a molecular keypad lock. Chem. Commun. 3906–3908 (2008) [DOI] [PubMed]
- 14.Andreasson J, Straight SD, Moore TA, Moore AL, Gust D. An all-photonic molecular keypad lock. Chem. Eur. J. 2009;15:3936–3939. doi: 10.1002/chem.200900043. [DOI] [PubMed] [Google Scholar]
- 15.Bozdemir OA, Guliyev R, Büyükcakir O, Selcuk S, Kolemen S, Gulseren G, Nalbantoglu T, Boyaci H, Akkaya EU. Selective manipulation of ICT and PET processes in styryl-bodipy derivatives: applications in molecular logic and fluorescence sensing of metal ions. J. Am. Chem. Soc. 2010;132:8029–8036. doi: 10.1021/ja1008163. [DOI] [PubMed] [Google Scholar]
- 16.Hayes JP. Introduction to Digital Logic Design. Addison- Company: Wesley Publishing; 1993. [Google Scholar]
- 17.de Silva PA, Uchiyama SN. Molecular logic and computing. Nanotechnology. 2007;2:399–410. doi: 10.1038/nnano.2007.188. [DOI] [PubMed] [Google Scholar]
- 18.Szacizowski K. Digital information processing in molecular systems. Chem. Rev. 2008;108:3481–3548. doi: 10.1021/cr068403q. [DOI] [PubMed] [Google Scholar]
- 19.Andreasson J, Pischel U. Smart molecules at work—mimicking advanced logic operations. Rev. Chem. Soc. 2010;39:174–188. doi: 10.1039/b820280j. [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Zhou Y, Yang Y, Wang W, Qu L, Chen C, Liu D, Zhang D, Zhu D. Photo-pH dually modulated fluorescence switch based on DNA spatial nanodevice. J. Phys. Chem., B. 2008;112:6893. doi: 10.1021/jp8020485. [DOI] [PubMed] [Google Scholar]
- 21.Margulies D, Hamilton AD. Digital analysis of protein properties by an ensemble of DNA quadruplexes. J. Am. Chem. Soc. 2009;131:9142. doi: 10.1021/ja900848t. [DOI] [PubMed] [Google Scholar]
- 22.Strack G, Ornatska M, Pita M, Katz E. Biocomputing security system: concatenated enzyme-based logic gates operating as a biomolecular keypad lock. J. Am. Chem. Soc. 2008;130:4234–4235. doi: 10.1021/ja7114713. [DOI] [PubMed] [Google Scholar]
- 23.Pais VF, Remón P, Collado D, Andréasson J, Inestrosa EP, Pischel U. OFF-ON-OFF fluorescence switch with T-Latch function. Org. Lett. 2011;13:5572–5575. doi: 10.1021/ol202312n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar M, Kumar R, Bhalla V. Optical chemosensor for Ag+, Fe3+, and cysteine: information processing at molecular level. Org. Lett. 2011;13:366–369. doi: 10.1021/ol102543e. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Katz E. Digital biosensors with built-in logic for biomedical applications—biosensors based on a biocomputing concept. Anal. Bioanal. Chem. 2010;398:1591–1603. doi: 10.1007/s00216-010-3746-0. [DOI] [PubMed] [Google Scholar]
- 26.Fujimoto K, Shimizu H, Inouye M. Unambiguous detection of target DNAs by excimer-monomer switching molecular beacons. J. Org. Chem. 2004;69:3271–3275. doi: 10.1021/jo049824f. [DOI] [PubMed] [Google Scholar]
- 27.Clelland CT, Risca V, Bancroft C. Hiding messages in DNA microdots. Nature. 1999;399:533–534. doi: 10.1038/21092. [DOI] [PubMed] [Google Scholar]
- 28.Mao C, LaBean TH, Reif JH, Seeman NC. Logical computation using self assembly of DNA triple cross-over molecules. Nature. 2000;407:493–496. doi: 10.1038/35035038. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka K, Okamoto A, Saito I. Public-key system using DNA as a one-way function for key distribution. Biosystems. 2005;81:25–29. doi: 10.1016/j.biosystems.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 30.Seth D, Chakrabarty D, Chakraborty A, Sarkar NS. Study of energy transfer from 7-amino coumarin donors to rhodamine 6G acceptor in non-aqueous reverse micelles. Chem. Phys. Lett. 2005;401:546–552. doi: 10.1016/j.cplett.2004.11.119. [DOI] [PubMed] [Google Scholar]
- 31.Förster TH. Experimentelle und theoretische Untersuchung des Zwischenmolekularen übergangs von Elektrinenanregungsenergie. Z. Naturforsch. 1949;4A:321–327. [Google Scholar]
- 32.Hussain SA, Chakraborty S, Bhattacharjee D, Schoonheydt RA. Fluorescence resonance energy transfer between organic dyes adsorbed onto nano-clay and Langmuir–Blodgett (LB) films. Spectrochim. Acta, Part A. 2010;75:664–670. doi: 10.1016/j.saa.2009.11.037. [DOI] [PubMed] [Google Scholar]
- 33.Förster TH. Modern Quantum Chemistry, Istanbul Lectures, Part III: Action of Light and Organic Crystals. New York: Academic Press; 1965. [Google Scholar]
- 34.Malicka J, Gryczynski I, Lakowicz JR. DNA hybridization assays using metal-enhanced fluorescence. Biochem. Biophys. Res. Commun. 2003;306:213–218. doi: 10.1016/S0006-291X(03)00935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mathur N, Aneja A, Bhatnagar PK, Mathur PC. A new FRET-based sensitive DNA sensor for medical diagnostics using PNA probe and water-soluble blue light emitting polymer. J. Sens. 2008;2008:1–6. doi: 10.1155/2008/270475. [DOI] [Google Scholar]
- 36.Watson JD, Crick FHC. A structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 37.Wang S, Gaylord BS, Bazan GC. Fluorescein provides a resonance gate for FRET from conjugated polymers to DNA intercalated dyes. J. Am. Chem. Soc. 2004;126:5446–5451. doi: 10.1021/ja035550m. [DOI] [PubMed] [Google Scholar]