Abstract
Pseudomonas is a highly versatile bacterium at the species level with great ecological significance. These genetically and metabolically diverse species have undergone repeated taxonomic revisions. We propose a strategy to identify Pseudomonas up to species level, based on the unique features of their 16S rDNA (rrs) gene sequence, such as the frame work of sequences, sequence motifs and restriction endonuclease (RE) digestion patterns. A species specific phylogenetic framework composed of 31 different rrs sequences, allowed us to segregate 1,367 out of 2,985 rrs sequences of this genus, which have been classified at present only up to genus (Pseudomonas) level, as follows: P. aeruginosa (219 sequences), P. fluorescens (463 sequences), P. putida (347 sequences), P. stutzeri (197 sequences), and P. syringae (141 sequences). These segregations were validated by unique 30–50 nucleotide long motifs and RE digestion patterns in their rrs. A single gene thus provides multiple makers for identification and surveillance of Pseudomonas.
Electronic supplementary material
The online version of this article (doi:10.1007/s12088-013-0412-1) contains supplementary material, which is available to authorized users.
Keywords: Pseudomonas, Diversity, Evolution, Framework, Phylogeny, Restriction endonuclease
Introduction
Gene and genome sequencing has tremendously increased our knowledge of the microbes. The most extensively studied among these is 16S rDNA (rrs) gene [1] (http://rdp.cme.msu.edu). Most molecular techniques allow identification of organism up to genus level [2]. An alternative to full length rrs sequence is the multi-locus sequence analysis or multi locus sequence typing, which cannot correctly interpret phylogenetic differences among closely related species within Acinetobacter and Pseudomonas [2–4]. Pseudomonas is a genetically and metabolic diverse genus that has undergone repeated taxonomic revisions [5, 6]. Phylogenetic analysis based on concatenated sequences of four core HKGs (housekeeping genes—rrs, gyrB, rpoB and rpoD), allowed segregation of different groups as (i) intrageneric cluster I composed of P. aeruginosa, P. flavescens, P. mendocina, P. resinovorans, [3] and (ii) P. fluorescens intrageneric cluster II consisting of P. chlororaphis, P. fluorescens, P. syringae and P. putida [7] Studies based on the nucleotide sequences of the genes rrs, gyrB and rpoD [8–10], revealed that P. aeruginosa and P. stutzeri form a group quite distinct from that constituted by P. syringae, P. fluorescens and P. putida. In spite of these extensive analyses, a definitive lineage could not be established and hence needs re-evaluation [3, 11]. Recent works on exploring the latent features of rrs have revealed reliable framework sequences, unique motifs and in silico restriction mapping, allowing species level identification of organisms in the cases of Bacillus, Clostridium, Stenotrophomonas and Streptococcus [6, 12–14]. The genomic tools developed in these studies can be extended to discriminate novel taxa and may prove helpful to classify an organism, when it encounters a never-seen-before situation [15]. In this work, we have employed around 1,350 rrs gene sequences (each >1,200 nucleotides, nts) of five species of Pseudomonas sensu stricto to select unique molecular markers within rrs sequences, to be used as phylogenetic frameworks for identifying Pseudomonas up to species level. The results were validated through the presence of molecular markers: unique motif(s) and restriction endonuclease (RE) digestion patterns.
Materials and Methods
Pseudomonas rrs sequence were retrieved from RDP/NCBI sites: http://rdp.cme.msu.edu/; http://www.ncbi.nlm.nih.gov/ (Supplementary Tables 1, 2). The phylogenetic, restriction enzyme sites and Pseudomonas species-specific motif analysis among of rrs sequences has been detailed as a Supplementary Material 2.
Results
Phylogenetic Frame Work for Pseudomonas Species
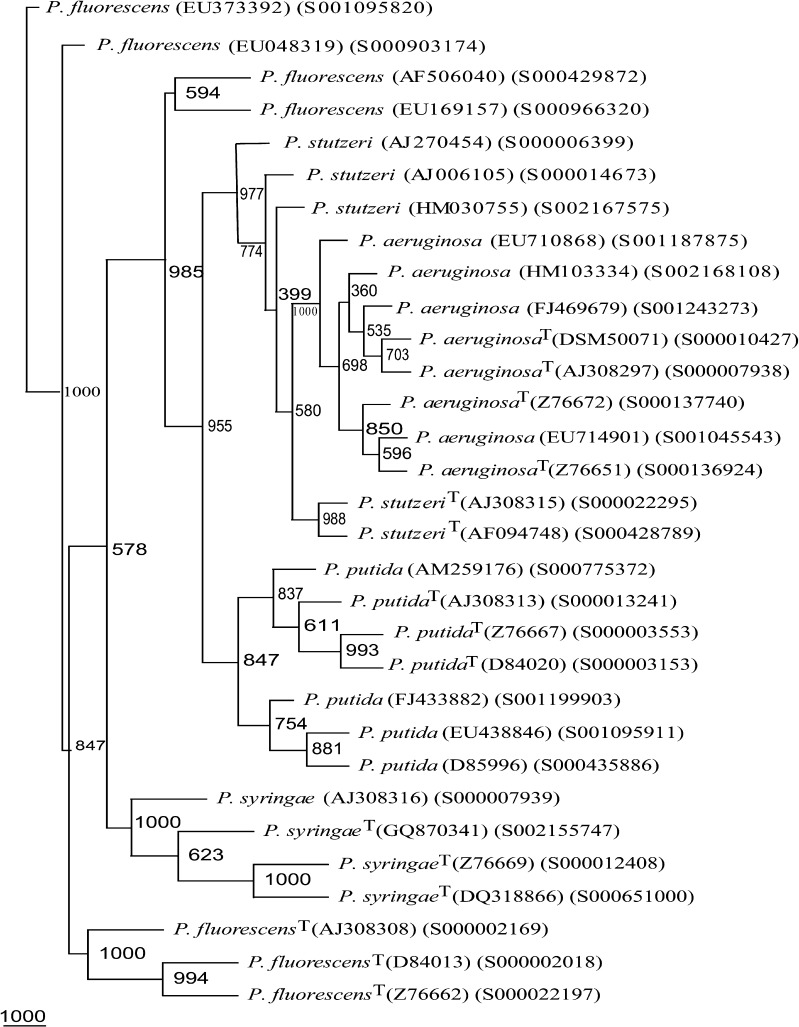
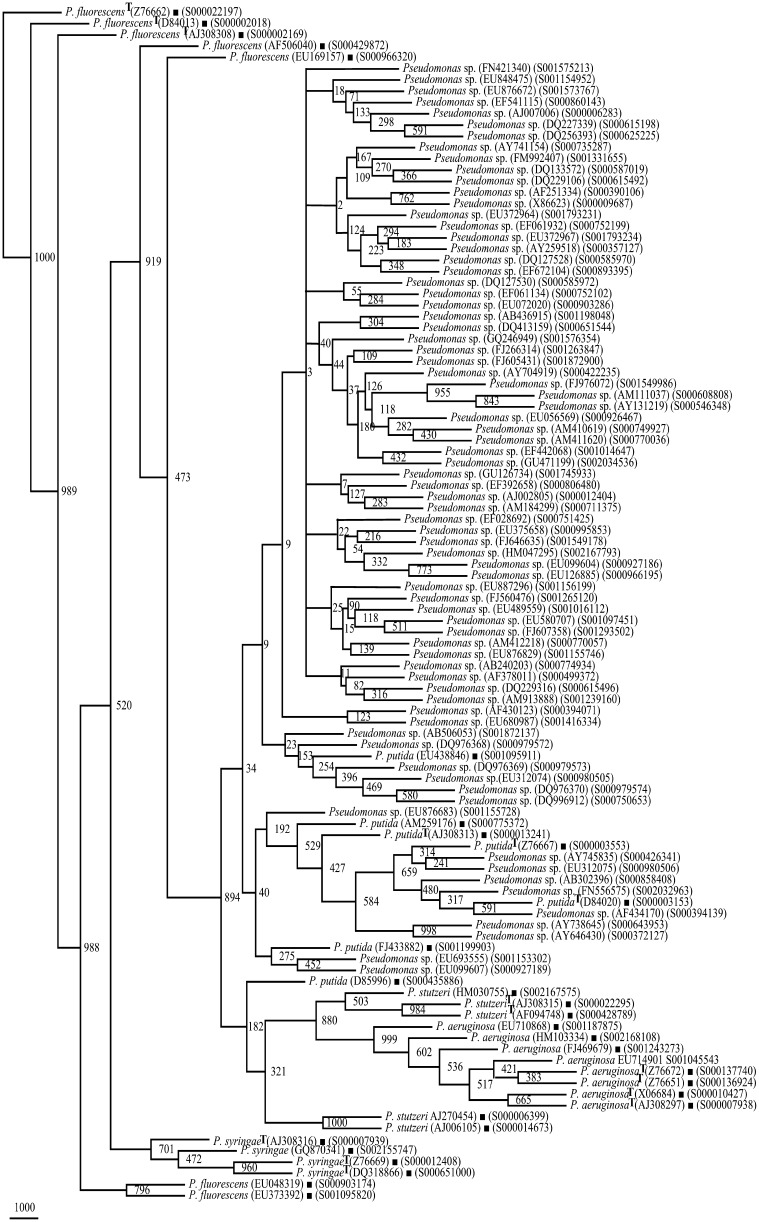
On the basis of 1,350 rrs sequences (>1,200 nts each) belonging to group I Pseudomonas spp.: P. aeruginosa (375 sequences), P. fluorescens (273 sequences), P. putida (414 sequences), P. stutzeri (199 sequences), and P. syringae (89 sequences) (Supplementary Tables 1, 2), phylogenetic frame was developed. It was employed to identify 2,985 rrs sequences belonging to organisms designated at present only up to genus level (Supplementary Table 2). For P. aeruginosa, four phylogenetic trees based on 375 rrs sequences enabled us to select eight frame work sequences (FWS) (Supplementary Table 3; Supplementary Fig. 1). Similarly, we could select seven FWS each of P. fluorescens and P. putida, five FWS in the case of P. stutzeri and four FWS for P. syringae (Supplementary Table 3; Supplementary Figs. 1–3). The total genetic diversity of 1,350 rrs sequences belonging to five Pseudomonas spp. could be represented by 31 as FWS including 15 type strains. A phylogenetic tree based on these 31 FWS (Fig. 1; Table 1) showed high heterogeneity (low Bootstrap values) within each Pseudomonas species (Figs. 1, 2, 3), enabling easy separation into distinct groups.
Fig. 1.
Phylogenetic tree of 31 rrs framework sequences of P. aeruginosa, P. fluorescens, P. putida, P. stutzeri, and P. syringae. T type strain
Table 1.
Accession numbers of rrs sequences of Pseudomonas species used for generating phylogenetic framework (http://www.ncbi.nlm.nih.gov/ and http://rdp.cme.msu.edu/)
| Organism | Reference sequence(s) |
|---|---|
| P. aeruginosa | EU714901, FJ469679, EU710868, HM103334, X06684(T), AJ308297(T), Z76651(T), Z76672(T) |
| P. fluorescens | AF506040, EU169157, EU048319, EU373392, D84013(T), AJ308308(T), Z76662(T) |
| P. putida | D85996, AM259176, EU438846, FJ433882, D84020(T), Z76667(T), AJ308313(T) |
| P. stutzeri | AJ270454, AJ006105, HM030755, AJ308315(T), AF094748(T) |
| P. syringae | GQ870341, AJ308316(T), Z76669(T), DQ318866(T) |
| Total | 31 strains |
Type strain designated as (T)
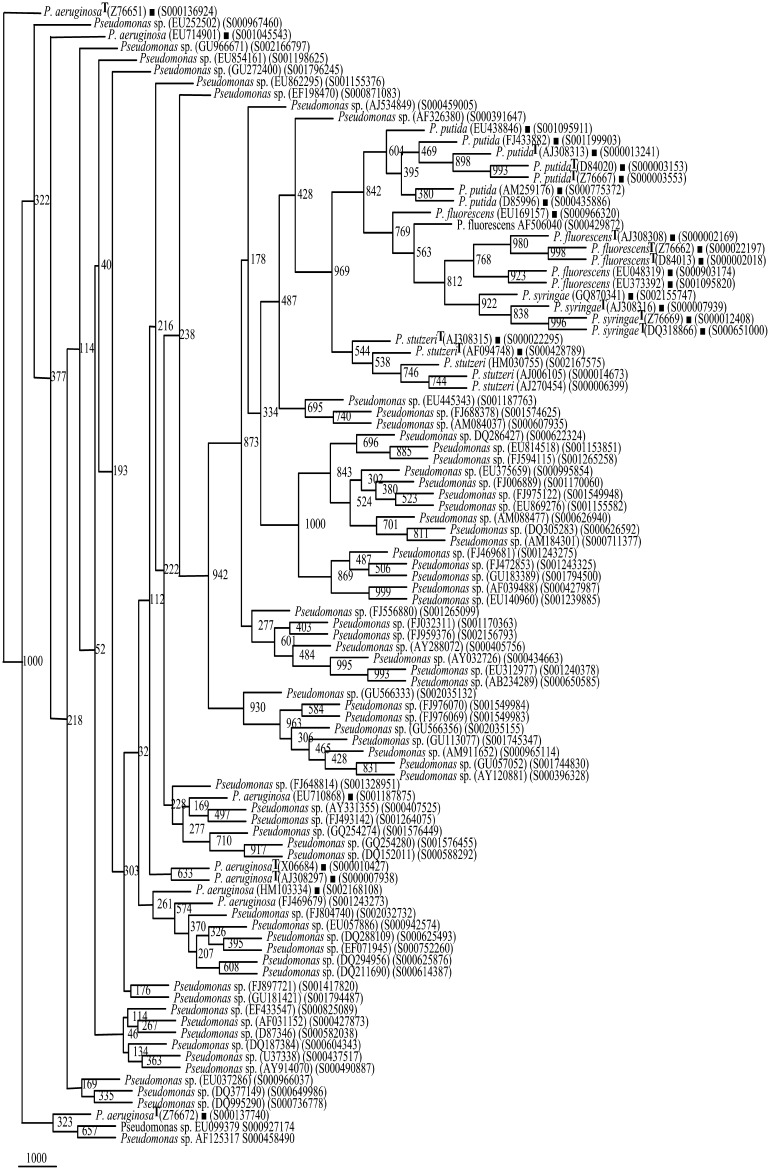
Fig. 2.
Phylogenetic tree of rrs of Pseudomonas sp. (66 segregated as P. aeruginosa, Supplementary Table 4) and Framework sequences (Filled squares) (Fig. 1)
Fig. 3.
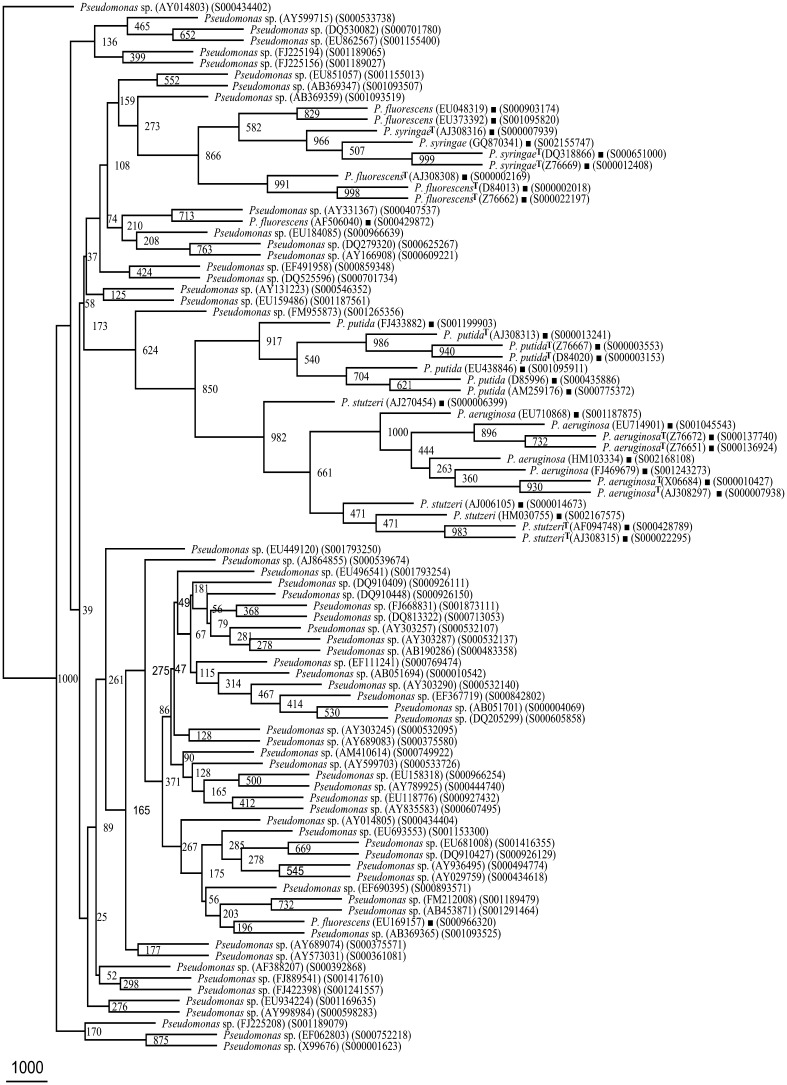
Phylogenetic tree of rrs of Pseudomonas sp. (61 segregated as P. fluorescens, Supplementary Table 4) and Framework sequences (Fig. 1). 61/129 have been presented here. The rest 68 rrs sequences have been presented in Fig. 4
Validation of Species-Specific Phylogenetic FWS
Phylogenetic trees (29) of rrs sequences each belonging to Pseudomonas sp. along with species specific phylogenetic framework composed of 31 rrs sequences, revealed that 1,367 rrs sequences belonged to P. aeruginosa (219 sequences), P. fluorescens (463 sequences), P. putida (347 sequences), P. stutzeri (197 sequences), and P. syringae (141 sequences) (Supplementary Table 4). Final trees to demonstrate that these Pseudomonas spp. fall well within their respective Pseudomonas groups., a small subset was selected from each species as follows: 66 sequences for P. aeruginosa (Fig. 2; Supplementary Table 4), 129 sequences for P. fluorescens (Figs. 3, 4), 75 sequences for P. putida (Fig. 5; Supplementary Table 4), 66 sequences for P. stutzeri (Supplementary Fig. 4; Supplementary Table 4), and 65 sequences P. syringae (Supplementary Fig. 5; Supplementary Table 4). At this rate about 42.41 % of the presently unclassified Pseudomonas sp. could be identified up to species level.
Fig. 4.
Phylogenetic tree of rrs of Pseudomonas sp. (68 segregated as P. fluorescens, Supplementary Table 4) and Framework sequences (Fig. 1)
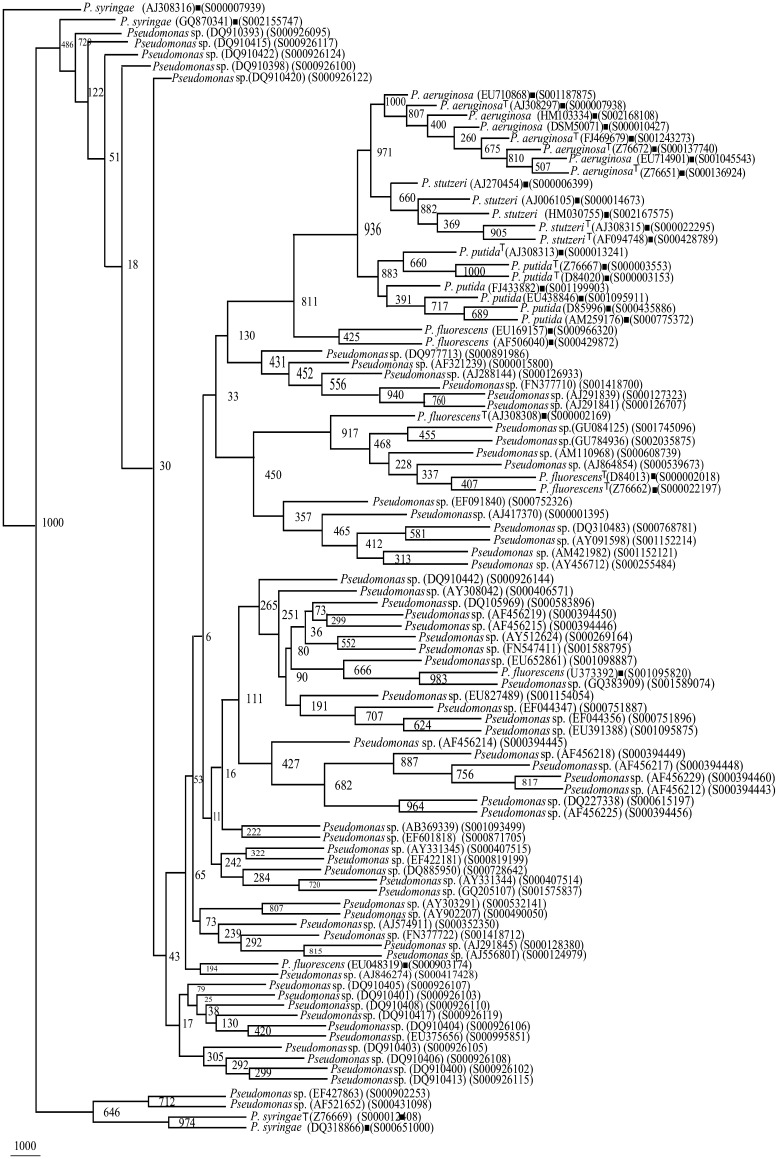
Fig. 5.
Phylogenetic tree of rrs of Pseudomonas sp. (75 segregated as P. putida, Supplementary Table 4) and Framework sequences
In silico RE Digestion Patterns
In the present study, six REs viz AluI, BfaI, DpnII, HaeIII, RsaI and Tru9I were found to be effective in drawing meaningful conclusions (Table 2; Supplementary Table 5).
Table 2.
In silico RE activity in rrs of Pseudomonas spp.: AluI and DpnII
| Pseudomonas spp. | Fa | RE digestion fragments (nucleotides, nts) | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AluI | ||||||||||||||||||||
| P. putida | 254/414 | * | 162 | * | 403 | * | 211 | * | 207 | * | 209 | * | 162 | * | ||||||
| P. putida | 54/414 | * | 162 | * | 403 | * | 195 | * | 15 | * | 207 | * | 209 | * | 162 | * | ||||
| P. putida | 61/414 | * | 162 | * | 215 | * | 188 | * | 211 | * | 207 | * | 209 | * | 163 | * | ||||
| P. aeruginosa | 321/375 | * | 162 | * | 403 | * | 211 | * | 206 | * | 208 | * | 162 | * | ||||||
| P. aeruginosa | 19/375 | * | 158 | * | 403 | * | 418 | * | 208 | * | 162 | * | ||||||||
| P. stutzeri | 143/199 | * | 162 | * | 403 | * | 211 | * | 207 | * | 209 | * | 162 | * | ||||||
| P. stutzeri | 13/199 | * | 162 | * | 137 | * | 264 | * | 211 | * | 207 | * | 208 | * | 162 | * | ||||
| P. stutzeri | 16/199 | * | 162 | * | 402 | * | 211 | * | 175 | * | 32 | * | 209 | * | 162 | * | ||||
| P. syringae | 79/89 | * | 403 | * | 196 | * | 15 | * | 207 | * | 209 | * | 162 | * | ||||||
| P. fluorescens | 116/273 | * | 160 | * | 599 | * | 15 | * | 207 | * | 209 | * | 162 | * | ||||||
| P. fluorescens | 47/273 | * | 162 | * | 402 | * | 196 | * | 15 | * | 207 | * | 209 | * | 162 | * | ||||
| P. fluorescens | 55/273 | * | 403 | * | 196 | * | 15 | * | 207 | * | 209 | * | 162 | * | ||||||
| P. fluorescens | 30/273 | * | 403 | * | 196 | * | 15 | * | 207 | * | 209 | * | ||||||||
| DpnII | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. aeruginosa | 123/375 | * | 183 | * | 77 | * | 24 | * | 83 | * | 449 | * | 9 | * | 447 | * | 12 | * | 225 | * | ||
| P. aeruginosa | 200/375 | * | 77 | * | 24 | * | 83 | * | 449 | * | 9 | * | 447 | * | 12 | * | ||||||
| P. aeruginosa | 13/375 | * | 77 | * | 24 | * | 83 | * | 449 | * | 9 | * | 447 | * | 12 | * | 170 | * | ||||
| P. stutzeri | 29/199 | * | 260 | * | 24 | * | 83 | * | 449 | * | 9 | * | 448 | * | 12 | * | 225 | * | ||||
| P. stutzeri | 118/199 | * | 77 | * | 24 | * | 83 | * | 449 | * | 9 | * | 448 | * | 12 | * | ||||||
| P. stutzeri | 31/199 | * | 183 | * | 77 | * | 24 | * | 83 | * | 449 | * | 9 | * | 448 | * | 12 | * | 225 | * | ||
| P. syringae | 88/89 | * | 262 | * | 24 | * | 83 | * | 906 | * | 12 | * | 225 | * | ||||||||
| P. fluorescens | 148/273 | * | 24 | * | 83 | * | 906 | * | 12 | * | ||||||||||||
| P. fluorescens | 23/273 | * | 24 | * | 83 | * | 906 | * | 12 | * | 225 | * | ||||||||||
| P. fluorescens | 20/273 | * | 77 | * | 24 | * | 83 | * | 906 | * | 12 | * | ||||||||||
| P. fluorescens | 51/273 | * | 260 | * | 24 | * | 83 | * | 906 | * | 12 | * | 225 | * | ||||||||
| P. putida | 250/414 | * | 24 | * | 83 | * | 906 | * | 12 | * | ||||||||||||
| P. putida | 17/414 | * | 24 | * | 83 | * | 906 | * | 12 | * | 225 | * | ||||||||||
| P. putida | 103/414 | * | 260 | * | 24 | * | 83 | * | 906 | * | 12 | * | ||||||||||
| P. putida | 31/414 | * | 260 | * | 24 | * | 83 | * | 905 | * | 12 | * | 207 | * | ||||||||
aFrequency of organisms showing this RE digestion pattern. Asterisk indicates RE site in the rrs sequences
AluI
In silico digestion of rrs sequences of five Pseudomonas spp. (Table 2) with AluI resulted in digestion patterns, which could distinguish them into three major groups. Group 1 with a AluI pattern of 162-403-211-207-209-162 nts fragments occurred among P. aeruginosa (321/375 sequences), P. putida (254/414 sequences) and P. stutzeri (143/199 sequences). Since, these three Pseudomonas spp. were indistinguishable, we may propose them to have a common ancestor. Group 2 was composed of rrs sequences belonging to P. fluorescens. Here two different genetic events seem to have happened simultaneously: (1) shift in the AluI site between the fragments 403 and 211 nts, leading to the appearance of two new fragments of 559 and 55 nts, and (2) appearance of an additional RE site in the 211 nts fragment leading to two fragments of 196 and 15 nts. Further evolution among P. fluorescens seems to have happened at 5′ end in 55/273 sequences such that a distinct 162 nts fragment is no longer evident. Another subpopulation of P. fluorescens (30/273 sequences) seems to have evolved by the disappearance of AluI sites as 5′ and 3′ ends. Organisms belonging to P. syringae had a AluI digestion pattern of 403-196-15-207-209-162 nts in their rrs at a frequency of 79/89 sequences. This pattern in P. syringae was indistinguishable from that observed in a small population (55/273 sequences) of P. fluorescens. In conclusion, we may state that P. aeruginosa, P. putida and P. stutzeri cannot be distinguished among themselves on the basis of AluI digestion of their rrs. P. fluorescens has evolved into different populations and that P. syringae resembles one of the subgroups of P. fluorescens.
DpnII
DpnII digestion patterns of rrs among Pseudomonas spp. (Table 2), lead to three groups. P. fluorescens and P. putida were indistinguishable due to a similar RE pattern: 24-83-906-12 nts. P. aeruginosa (200/375 sequences) and P. stutzeri (118/199 sequences) had similar DpnII pattern in their rrs: 77-24-83-449-9-448-12. Among the five Pseudomonas spp., P. syringae had a unique DpnII pattern: 262-24-83-906-12-225, intermediate to the previous two groups. Here certain RE sites seem to have disappeared resulting in the merger of small fragments such as 185-77 into 262 nts and 449-9-448 into a single fragment and 906 nts. Incidentally, this unique pattern of P. syringae (88/89 rrs sequences) matches with a small population of P. fluorescens (51/273 rrs sequences). Each of the Pseudomonas spp., except P. syringae showed subgroups, where they show resemblance to those from other subgroups. Unlike AluI digestion pattern of rrs of Pseudomonas spp., where P. aeruginosa, P. putida and P. stutzeri were indistinguishable, here, with DpnII, P. putida could be separated out of this group. The observations made with DpnII, once again supported that P. syringae is likely to have evolved as a sub population of P. fluorescens as was also observed with AluI.
The information on the digestion patterns generated by REs—HaeIII, RsaI, Tru9I and BfaI have been presented as Supplementary Material 1 and Tables (Supplementary Table 5).
In brief, though not very effective as a tool to distinguish Pseudomonas spp. with authenticity, it does provide an opportunity to conclude that P. fluorescens has at least three subpopulations, of which two can be easily distinguished from all other Pseudomonas spp. being considered here.
Validation of Framework Sequences by In Silico RE Activity on rrs Sequences of Organisms Identified as Pseudomonas spp.
After the initial segregation of Pseudomonas spp. (1,367 isolates) (Supplementary Table 4) on the basis of phylogenetic FWS analysis (represented by 31 sequences including 15 type strains), it is important to validate them. Here, unique RE digestion patterns deduced from the rrs sequences of five known Pseudomonas spp. provided the necessary evidences. The details of the validation process have been presented as Supplementary Material 1 (Supplementary Tables 6–10).
Nucleotide Motif Analysis for the Validation of Framework Sequences
Additional evidences to further validate the segregation of Pseudomonas spp. done on the basis of phylogenetic FWS analysis were collected by the presence of nucleotide motifs (30–50 nts) deduced from isolates of the five known Pseudomonas spp. The sequences of 89–414 data sets submitted group-wise to MEME (Multiple EM for Motif Elicitation) program (http://meme.nbcr.net/meme3/meme.html) revealed 45 motifs (15 each of 30, 40 and 50 nts) for each species. To validate the categorization and classification of 1,367 rrs sequences belonging to five Pseudomonas spp. (Table 3), motifs unique to a particular Pseudomonas sp. were identified. A 30 nts unique motif (M15) was found to occur with a very high frequency of (355/375) in P. aeruginosarrs sequences. The 50 nts motifs (M9, M12 and M15) were also found to occur with higher frequency among P. aeruginosa rrs sequences (341–352/375). These motifs were found to validate the rrs sequences identified as P. aeruginosa on the basis of FWS analysis. A search for unique motifs in rrs of P. stutzeri revealed four motifs viz. M12 (30 nts), M11 (40 nts), M12 (40 nts) and M9 (50 nts), which occurred with high frequency and could be validated on P. stutzeri identified on the basis of FWS. Only one motifs M15 (40 nts) could be categorized as unique to P. putida and appeared with a frequency of 194/414. Similarly, only one—M12 (40 nts) out of 45 motifs was found to be unique among 89 P. syringae rrs sequences. None of the 45 motifs among rrs sequences of P. fluoresens could be identified as unique (Table 3). However, by exclusion principle, we can separate it from other four Pseudomonas spp. used in this study.
Table 3.
Phylogenetic relationships of Pseudomonas sp. with frame work organisms supported by unique motifs
| Motif | Frequency of signature | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide sequence | Unique no. | Size (nts) | Pseudomonas spp. | Pseudomonas sp.a | ||||||||
| PAE (375) | PST (199) | PFL (273) | PPT (414) | PSY (89) | PAE (219) | PST (197) | PFL (463) | PPT (347) | PSY (141) | |||
| P. aeruginosa | ||||||||||||
| GCTAATACCGCATACGTCCTGAGGGAGAAA | M15 | 30 | 355 | 0 | 0 | 1 | 0 | 80 | 1 | 0 | 0 | 0 |
| ATCAGATGAGCCTAGGTCGGATTAGCTAGTTGGTGGGGTAAAGGCCTACC | M9 | 50 | 341 | 1 | 0 | 1 | 0 | 119 | 5 | 0 | 0 | 0 |
| AAACGGGCGCTAATACCGCATACGTCCTGAGGGAGAAAGTGGGGGATCTT | M12 | 50 | 347 | 0 | 0 | 1 | 0 | 79 | 1 | 0 | 0 | 0 |
| TGGCCTTGACATGCTGAGAACTTTCCAGAGATGGATTGGTGCCTTCGGGA | M15 | 50 | 352 | 1 | 0 | 2 | 0 | 121 | 13 | 0 | 1 | 0 |
| P. stutzeri | ||||||||||||
| TAACGCATTAAGTCGACCGCCTGGGGAGTA | M12 | 30 | 3 | 190 | 0 | 6 | 0 | 8 | 171 | 0 | 0 | 0 |
| TTTTGACGTTACCGACAGAATAAGCACCGGCTAACTTCGT | M11 | 40 | 1 | 134 | 0 | 2 | 0 | 16 | 138 | 0 | 0 | 0 |
| TATGGCAGAGGGTGGTGGAATTTCCTGTGTAGCGGTGAAA | M12 | 40 | 0 | 185 | 1 | 6 | 0 | 4 | 165 | 0 | 0 | 0 |
| TGTTTTGACGTTACCGACAGAATAAGCACCGGCTAACTTCGTGCCAGCAG | M9 | 50 | 1 | 133 | 0 | 2 | 0 | 11 | 134 | 0 | 0 | 0 |
| P. putida | ||||||||||||
| AACCTGGGAACTGCATCCAAAACTGGCAAGCTAGAGTACG | M15 | 40 | 0 | 0 | 2 | 194 | 0 | 0 | 0 | 0 | 182 | 0 |
| P. syringae | ||||||||||||
| TAAAGCGCGCGTAGGTGGTTTGTTAAGTTGAATGTGAAAT | M12 | 40 | 0 | 0 | 3 | 3 | 68 | 0 | 0 | 14 | 0 | 23 |
| P. fluorescens | No motif could be designated as unique | |||||||||||
aIsolates designated as Pseudomonas sp. on the basis of FWS (Supplementary Table 3)
PAE: Pseudomonas aeruginosa; PFL: P. fluorescens; PPT: P. putida; PST: P. stutzeri; PSY: P. syringae
Based on two criteria, ten motifs were unique to four Pseudomonas spp., and could be validated among rrs sequences as follows: 121/219 of P. aeruginosa, 171/197 of P. stutzeri, 182/347 of P. putida and 23/141 of P. syringae, identified above on the basis of FWS.
Discussion
Economically important Pseudomonas spp. have been equated to a “dumping ground” [8]. It has been suggested to redefine true diversity of Pseudomonas [16]. The use of rpoD as an alternative to rrs, gives poor resolution of P. fluorescens, P. syringae, P. entomophila and P. putida [17]. Similarly, oprD gene revealed extensive genetic mosaicism [18]. Certain strains of P. fluorescens and P. syringae yielded low DNA–DNA hybridization (DDH) values between 25 and 39 %, demanding their segregation into the different species. Through studies aimed at re-classifying Pseudomonas spp., quite a few have been renamed as: (i) Comamonas, (ii) Acidovorax, (iii) Burkholderia and later as Ralstonia, (iv) Brevundimonas, and (v) Stenotrophomonas [7]. The big issue is the eroding confidence in using rrs. Efforts have been made to explore and exploit the hidden features of rrs of Bacillus, Clostridium, Stentrophomonas and Streptococcus spp. [6, 12–14]. In comparison to these genera, Pseudomonas posed a much bigger challenge as around 3,000 of rrs sequencing entries, were not classified beyond the genus status.
Restriction Enzyme Sites
A survey of different works reveals that certain REs sites are effective in distinguishing even closely related rrs sequences. Around seven different REs—AluI, BfaI, DpnII, HaeIII, RsaI and Tru9I, have been quite instrumental in distinguishing around 2,000 strains belonging to ten species of Bacillus spp., 750 strains of 15 species of Clostridium spp. and a few strains of Stenotrophomonas spp. [6, 12, 13]. In the present study, unique digestion patterns were observed with HaeIII for P. aeruginosa, and with AluI and BfaI for P. fluorescens. P. stutzeri could be distinguished on the basis of unique digestion pattern achievable with Tru9I.
The True Pseudomonas
It is worth noting that a large proportion of isolates deposited in the RDP database as P. aeruginosa, P. stutzeri and P. syringae have been properly identified (Table 3). Here, 355/375 rrs sequences of P. aeruginosa, 190/199 rrs sequences of P. stutzeri and 68/89 of those belonging to P. syringae were found to be supported by FWS and motif analyses. The problem seems to lie with P. putida and P. fluorescens, where only around 47 % (194/414) of the rrs sequences of the former could be validated through motif analysis. This prompts one to conclude that bacterial populations may be reclassified as novel species. The demand for more stringent identification of P. syringae is evident as only 23/141 were found to possess unique motifs. Previous studies have shown that P. fluorescens has closest affiliation to P. rhodesiae and P. gessardii [19], such that those rrs sequences which fall within the FWS but did not have any motif unique to P. fluorescens may actually belong to different species. P. fluorescens Pf-5 is so similar to P. syringae such that it might be mistaken for the later [17]. P. syringae with its high genetic diversity has been proposed to be split into around 17 different species [20]. Significance of our work lies in its potential as a tool for the identification of Pseudomonas isolated from diverse geographic locations [21].
The molecular tools developed here can be used to identify Pseudomonas spp. and provide multiple makers within a single gene (rrs). It may be implied that there is a need to carry out a mega study based on all the genera for which rrs sequences have been deposited in the RDP database. This strategy is likely to provide clues and authentic evidences on the location of candidatus phyla whose members are yet to be cultured or identified through metagenomic studies. It will thus fill the gaps in the evolutionary scale.
Electronic supplementary material
Acknowledgments
This work was supported by the Council of Scientific and Industrial Research, India: CSIR-IGIB and CSIR-NEERI.
References
- 1.Jeraldo P, Chia N, Goldenfeld N. On the suitability of short reads of 16S rRNA for phylogeny-based analyses in environmental surveys. Environ Microbiol. 2011;13:3000–3009. doi: 10.1111/j.1462-2920.2011.02577.x. [DOI] [PubMed] [Google Scholar]
- 2.Kämpfer P, Glaeser SP. Prokaryotic taxonomy in the sequencing era—the polyphasic approach revisited. Environ Microbiol. 2012;14:291–317. doi: 10.1111/j.1462-2920.2011.02615.x. [DOI] [PubMed] [Google Scholar]
- 3.Hilario E, Buckley TR, Young JM. Improved resolution on the phylogenetic relationships among Pseudomonas by combined analysis of atpD, carA, recA and 16S rDNA. Antonie Van Leeuwenhoek. 2004;86:51–64. doi: 10.1023/B:ANTO.0000024910.57117.16. [DOI] [PubMed] [Google Scholar]
- 4.Mazzaglia A, Studholme DJ, Taratufolo MC, Cai R, Almeida NF, Goodman T, Guttman DS, Vinatzer BA, Balestra GM. Pseudomonassyringae pv. actinidiae (PSA) isolates from recent bacterial canker of Kiwifruit outbreaks belong to the same genetic lineage. PLoS One. 2012;7:e36518. doi: 10.1371/journal.pone.0036518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lalucat J, Bennasar A, Bosch R, Garcia-Valdés E, Palleroni NJ. Biology of Pseudomonas stutzeri. Microbiol Mol Biol Rev. 2006;70:510–547. doi: 10.1128/MMBR.00047-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peix A, Ramírez-Bahena MH, Velázquez E. Historical evolution and current status of the taxonomy of genus Pseudomonas. Infect Genet Evol. 2009;9:1132–1147. doi: 10.1016/j.meegid.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Verma V, Raju SC, Kapley A, Kalia VC, Purohit HJ. Evaluation of genetic and functional diversity of Stenotrophomonas isolates from diverse effluent treatment plants. Bioresour Technol. 2011;101:7744–7753. doi: 10.1016/j.biortech.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Mulet M, Lalucat J, Garcia-Valdes E. DNA sequence-based analysis of the Pseudomonas species. Environ Microbiol. 2010;12:1513–1530. doi: 10.1111/j.1462-2920.2010.02181.x. [DOI] [PubMed] [Google Scholar]
- 9.Anzai Y, Kim H, Park JY, Wakabayashi H, Oyaizu H. Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int J Syst Evol Microbiol. 2000;50:1563–1589. doi: 10.1099/00207713-50-4-1563. [DOI] [PubMed] [Google Scholar]
- 10.Moore ERB, Mau M, Arnscheidt A, Bottger EC, Hutson RA, Collins MD, Van De Peer Y, De Wachter R, Timmis KN. The determination and comparison of the 16S rRNA gene sequences of species of the genus Pseudomonas sensu stricto and estimation of the natural intrageneric relationships. Syst Appl Microbiol. 1996;19:478–492. doi: 10.1016/S0723-2020(96)80021-X. [DOI] [Google Scholar]
- 11.Spiers AJ, Buckling A, Rainey PB. The causes of Pseudomonas diversity. Microbiology. 2000;146:2345–2350. doi: 10.1099/00221287-146-10-2345. [DOI] [PubMed] [Google Scholar]
- 12.Kalia VC, Mukherjee T, Bhushan A, Joshi J, Shankar P, Huma N. Analysis of the unexplored features of rrs (16S rDNA) of the genus Clostridium. BMC Genomics. 2011;12:18. doi: 10.1186/1471-2164-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porwal S, Lal S, Cheema S, Kalia VC. Phylogeny in aid of the present and novel microbial lineages: diversity in Bacillus. PLoS One. 2009;4:e4438. doi: 10.1371/journal.pone.0004438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lal D, Verma M, Lal R. Exploring internal features of 16S rRNA gene for identification of clinically relevant species of the genus Streptococcus. Ann Clin Microbiol Antimicrob. 2011;10:28. doi: 10.1186/1476-0711-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lan Y, Wang Q, Cole JR, Rosen GL. Using the RDP classifier to predict taxonomic novelty and reduce the search space for finding novel organisms. PLoS One. 2012;7:e32491. doi: 10.1371/journal.pone.0032491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silby MW, Cerdeño-Tárraga AM, Vernikos GS, Giddens RS, Jackson RW, Preston GM, Zhang X-X, Moon CD, Gehrig SM, Godfrey SAC, Knight CG, Malone JG, Robinson Z, Spiers AJ, Harris S, Challis GL, Yaxley AM, Harris D, Seeger K, Murphy L, Rutter S, Squares R, Quail MA, Saunders E, Mavromatis K, Brettin TS, Bentley D, Shothersall J, Stephens E, Thomas CM, Parkhill J, Levy SB, Rainey PB, Thomson NR. Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens. Genome Biol. 2009;10:R51. doi: 10.1186/gb-2009-10-5-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiil K, Binnewies TT, Willenbrock H, Hansen SK, Yang L, Jelsbak L, Ussery DW, Friis C. Comparative genomics of Pseudomonas. In: Rehm BHA, editor. Pseudomonas: model organism, pathogen, cell factory. Weinheim: Wiley-VCH; 2008. pp. 1–24. [Google Scholar]
- 18.Pirnay JP, De Vos D, Cochez C, Bilocq F, Vanderkelen A, Zizi M, Ghysels B, Cornelis P. Pseudomonas aeruginosa displays an epidemic population structure. Environ Microbiol. 2002;4:898–911. doi: 10.1046/j.1462-2920.2002.00321.x. [DOI] [PubMed] [Google Scholar]
- 19.Costa R, Salles JF, Berg G, Smalla K. Cultivation-independent analysis of Pseudomonas species in soil and in the rhizoshpere of field-grown Verticillium dahliae host plants. Environ Microbiol. 2006;8:2136–2149. doi: 10.1111/j.1462-2920.2006.01096.x. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto S, Kasai H, Arnold DL, Jackson RW, Vivian A, Harayama S. Phylogeny of the genus Pseudomonas: intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology. 2000;146:2385–2394. doi: 10.1099/00221287-146-10-2385. [DOI] [PubMed] [Google Scholar]
- 21.Morris CE, Sands DC, Vanneste JL, Montarry J, Oakley B, Guibaud Glaux C. Inferring the evolutionary history of the plant pathogen Pseudomonas syringae from its biogeography in headwaters of rivers in North America, Europe, and New Zealand. Mol Biol. 2010;1:e00107–e00110. doi: 10.1128/mBio.00107-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.