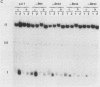
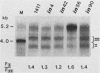
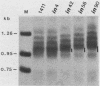
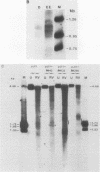
Abstract
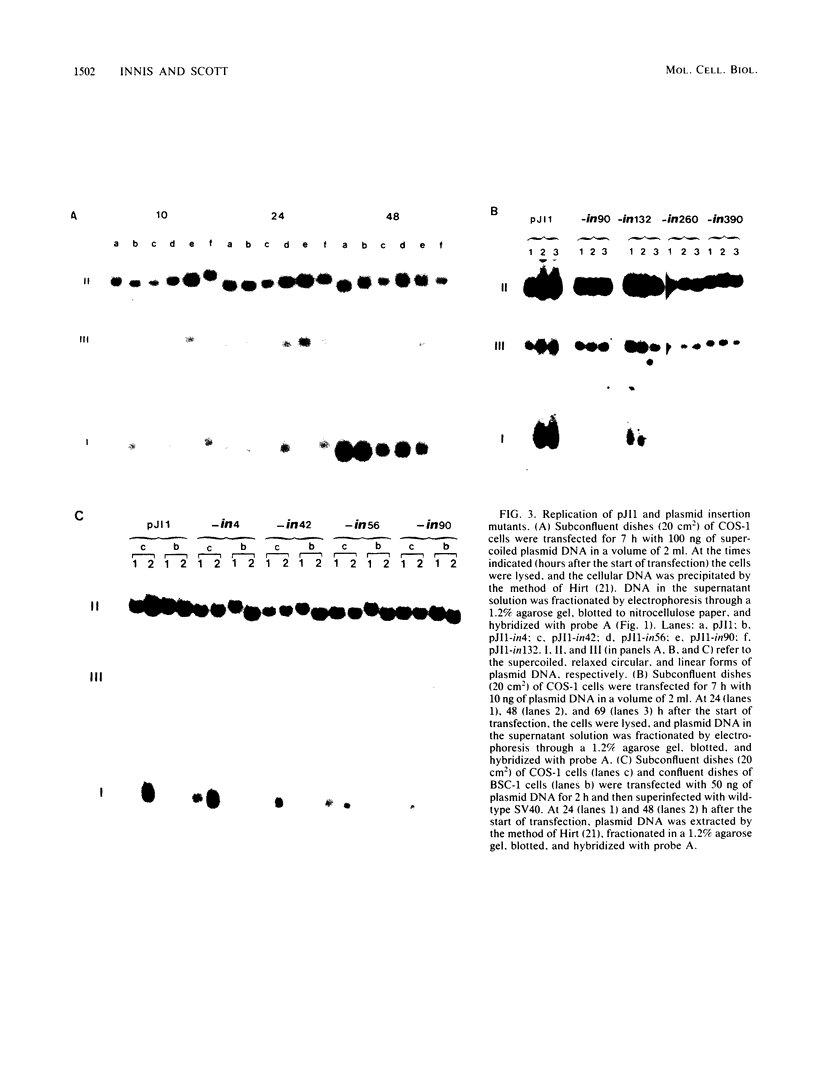
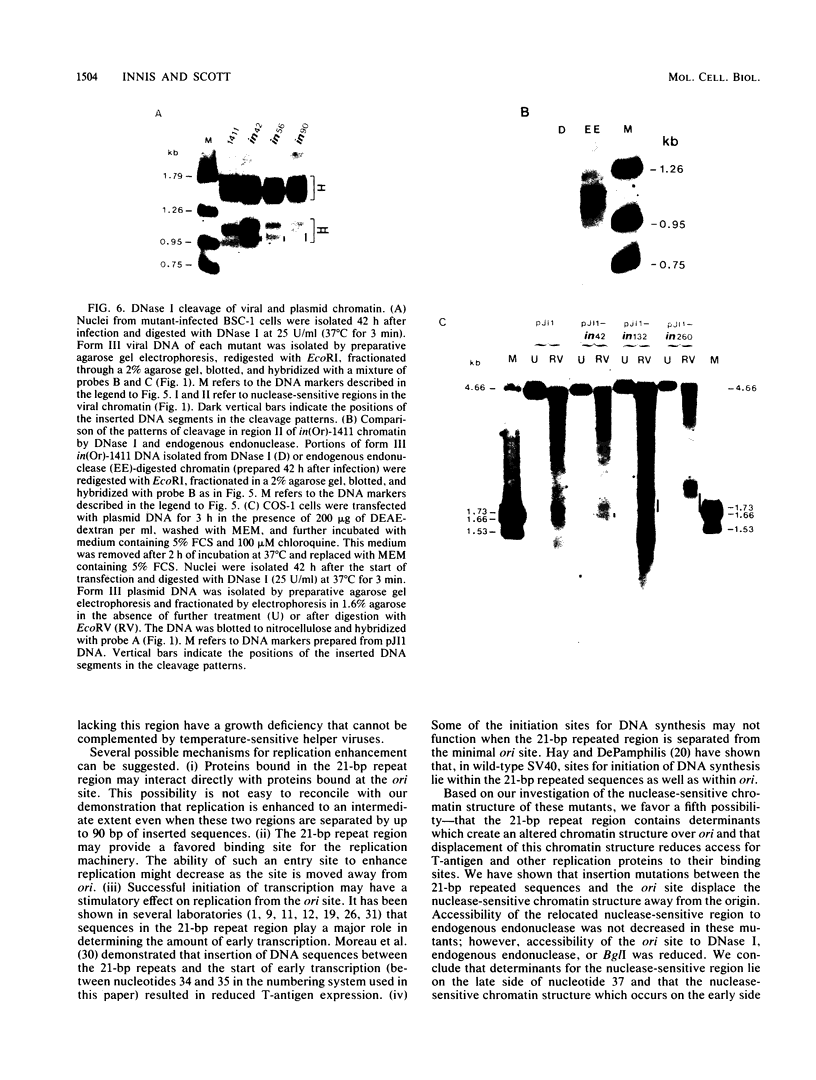
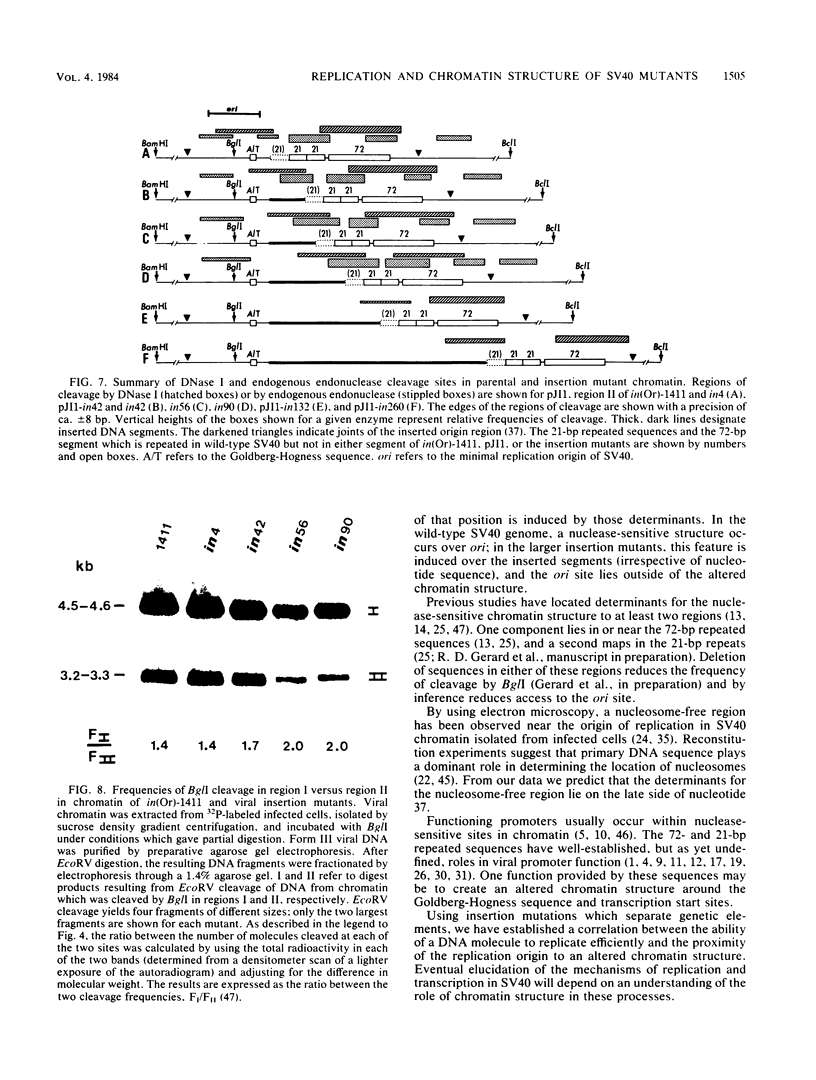
Insertion of DNA segments into the nuclease-sensitive region of simian virus 40 alters both replication efficiency and chromatin structure. Mutants containing large insertions between the simian virus 40 origin of replication (ori site) and the 21-base-pair repeated sequences replicated poorly when assayed by transfection into COS-1 cells. Replication of mutants with shorter insertions was moderately reduced. This effect was cis-acting and independent of the nucleotide sequence of the insert. The nuclease-sensitive chromatin structure was retained in these mutants, but the pattern of cleavage sites was displaced in the late direction from the ori site. New cleavage sites appeared within the inserted sequences, suggesting that information specifying the nuclease-sensitive chromatin structure is located on the late side of the inserts. Accessibility to BglI (which cleaves within the ori site) was reduced in the larger insertion mutants. These results support the conclusion that efficient function of the viral origin of replication is correlated with its proximity to an altered chromatin structure.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Bergsma D. J., Olive D. M., Hartzell S. W., Subramanian K. N. Territorial limits and functional anatomy of the simian virus 40 replication origin. Proc Natl Acad Sci U S A. 1982 Jan;79(2):381–385. doi: 10.1073/pnas.79.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne B. J., Davis M. S., Yamaguchi J., Bergsma D. J., Subramanian K. N. Definition of the simian virus 40 early promoter region and demonstration of a host range bias in the enhancement effect of the simian virus 40 72-base-pair repeat. Proc Natl Acad Sci U S A. 1983 Feb;80(3):721–725. doi: 10.1073/pnas.80.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright I. L., Abmayr S. M., Fleischmann G., Lowenhaupt K., Elgin S. C., Keene M. A., Howard G. C. Chromatin structure and gene activity: the role of nonhistone chromosomal proteins. CRC Crit Rev Biochem. 1982;13(1):1–86. doi: 10.3109/10409238209108709. [DOI] [PubMed] [Google Scholar]
- Cremisi C. The appearance of DNase I hypersensitive sites at the 5' end of the late SV40 genes is correlated with the transcriptional switch. Nucleic Acids Res. 1981 Nov 25;9(22):5949–5964. doi: 10.1093/nar/9.22.5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danna K., Nathans D. Specific cleavage of simian virus 40 DNA by restriction endonuclease of Hemophilus influenzae. Proc Natl Acad Sci U S A. 1971 Dec;68(12):2913–2917. doi: 10.1073/pnas.68.12.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio D., Nathans D. Cold-sensitive regulatory mutants of simian virus 40. J Mol Biol. 1980 Jun 15;140(1):129–142. doi: 10.1016/0022-2836(80)90359-9. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983 Nov;35(1):79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. DNAase I-hypersensitive sites of chromatin. Cell. 1981 Dec;27(3 Pt 2):413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- Everett R. D., Baty D., Chambon P. The repeated GC-rich motifs upstream from the TATA box are important elements of the SV40 early promoter. Nucleic Acids Res. 1983 Apr 25;11(8):2447–2464. doi: 10.1093/nar/11.8.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm M., Berg P. Deletion mapping of DNA regions required for SV40 early region promoter function in vivo. J Mol Appl Genet. 1982;1(5):457–481. [PubMed] [Google Scholar]
- Fromm M., Berg P. Simian virus 40 early- and late-region promoter functions are enhanced by the 72-base-pair repeat inserted at distant locations and inverted orientations. Mol Cell Biol. 1983 Jun;3(6):991–999. doi: 10.1128/mcb.3.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard R. D., Woodworth-Gutai M., Scott W. A. Deletion mutants which affect the nuclease-sensitive site in simian virus 40 chromatin. Mol Cell Biol. 1982 Jul;2(7):782–788. doi: 10.1128/mcb.2.7.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Green M. H., Miller H. I., Hendler S. Isolation of a polyoma-nucleoprotein complex from infected mouse-cell cultures. Proc Natl Acad Sci U S A. 1971 May;68(5):1032–1036. doi: 10.1073/pnas.68.5.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss P., Dhar R., Khoury G. Simian virus 40 tandem repeated sequences as an element of the early promoter. Proc Natl Acad Sci U S A. 1981 Feb;78(2):943–947. doi: 10.1073/pnas.78.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutai M. W., Nathans D. Evolutionary variants of simian virus 40: Nucleotide sequence of a conserved SV40 DNA segment containing the origin of viral DNA replication as an inverted repetition. J Mol Biol. 1978 Dec 5;126(2):259–274. doi: 10.1016/0022-2836(78)90362-5. [DOI] [PubMed] [Google Scholar]
- Hartzell S. W., Yamaguchi J., Subramanian K. N. SV40 deletion mutants lacking the 21-bp repeated sequences are viable, but have noncomplementable deficiencies. Nucleic Acids Res. 1983 Mar 11;11(5):1601–1616. doi: 10.1093/nar/11.5.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay R. T., DePamphilis M. L. Initiation of SV40 DNA replication in vivo: location and structure of 5' ends of DNA synthesized in the ori region. Cell. 1982 Apr;28(4):767–779. doi: 10.1016/0092-8674(82)90056-3. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Hiwasa T., Segawa M., Yamaguchi N., Oda K. Phasing of nucleosomes in SV40 chromatin reconstituted in vitro. J Biochem. 1981 May;89(5):1375–1389. doi: 10.1093/oxfordjournals.jbchem.a133329. [DOI] [PubMed] [Google Scholar]
- Innis J. W., Scott W. A. Chromatin structure of simian virus 40-pBR322 recombinant plasmids in COS-1 cells. Mol Cell Biol. 1983 Dec;3(12):2203–2210. doi: 10.1128/mcb.3.12.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobovits E. B., Bratosin S., Aloni Y. Formation of a nucleosome-free region in SV40 minichromosomes is dependent upon a restricted segment of DNA. Virology. 1982 Jul 30;120(2):340–348. doi: 10.1016/0042-6822(82)90035-6. [DOI] [PubMed] [Google Scholar]
- Jongstra J., Reudelhuber T. L., Oudet P., Benoist C., Chae C. B., Jeltsch J. M., Mathis D. J., Chambon P. Induction of altered chromatin structures by simian virus 40 enhancer and promoter elements. Nature. 1984 Feb 23;307(5953):708–714. doi: 10.1038/307708a0. [DOI] [PubMed] [Google Scholar]
- Lebowitz P., Ghosh P. K. Initiation and regulation of simian virus 40 early transcription in vitro. J Virol. 1982 Feb;41(2):449–461. doi: 10.1128/jvi.41.2.449-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusky M., Botchan M. Inhibition of SV40 replication in simian cells by specific pBR322 DNA sequences. Nature. 1981 Sep 3;293(5827):79–81. doi: 10.1038/293079a0. [DOI] [PubMed] [Google Scholar]
- Luthman H., Magnusson G. High efficiency polyoma DNA transfection of chloroquine treated cells. Nucleic Acids Res. 1983 Mar 11;11(5):1295–1308. doi: 10.1093/nar/11.5.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Moreau P., Hen R., Wasylyk B., Everett R., Gaub M. P., Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981 Nov 25;9(22):6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers R. M., Rio D. C., Robbins A. K., Tjian R. SV40 gene expression is modulated by the cooperative binding of T antigen to DNA. Cell. 1981 Aug;25(2):373–384. doi: 10.1016/0092-8674(81)90056-8. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Tjian R. Construction and analysis of simian virus 40 origins defective in tumor antigen binding and DNA replication. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6491–6495. doi: 10.1073/pnas.77.11.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Saragosti S., Cereghini S., Yaniv M. Fine structure of the regulatory region of simian virus 40 minichromosomes revealed by DNAase I digestion. J Mol Biol. 1982 Sep 15;160(2):133–146. doi: 10.1016/0022-2836(82)90171-1. [DOI] [PubMed] [Google Scholar]
- Saragosti S., Moyne G., Yaniv M. Absence of nucleosomes in a fraction of SV40 chromatin between the origin of replication and the region coding for the late leader RNA. Cell. 1980 May;20(1):65–73. doi: 10.1016/0092-8674(80)90235-4. [DOI] [PubMed] [Google Scholar]
- Scott W. A., Wigmore D. J. Sites in simian virus 40 chromatin which are preferentially cleaved by endonucleases. Cell. 1978 Dec;15(4):1511–1518. doi: 10.1016/0092-8674(78)90073-9. [DOI] [PubMed] [Google Scholar]
- Shenk T. Construction of a viable SV40 variant containing two functional origins of DNA replication. Cell. 1978 Apr;13(4):791–798. doi: 10.1016/0092-8674(78)90229-5. [DOI] [PubMed] [Google Scholar]
- Shortle D., Nathans D. Local mutagenesis: a method for generating viral mutants with base substitutions in preselected regions of the viral genome. Proc Natl Acad Sci U S A. 1978 May;75(5):2170–2174. doi: 10.1073/pnas.75.5.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D., Nathans D. Regulatory mutants of simian virus 40: constructed mutants with base substitutions at the origin of DNA replication. J Mol Biol. 1979 Jul 15;131(4):801–817. doi: 10.1016/0022-2836(79)90202-x. [DOI] [PubMed] [Google Scholar]
- Subramanian K. N., Shenk T. Definition of the boundaries of the origin of DNA replication in simian virus 40. Nucleic Acids Res. 1978 Oct;5(10):3635–3642. doi: 10.1093/nar/5.10.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. J., Sundin O. H., Bohn M. J. SV40 viral minichromosome: preferential exposure of the origin of replication as probed by restriction endonucleases. Nucleic Acids Res. 1978 Oct;5(10):3469–3477. doi: 10.1093/nar/5.10.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. J., Sundin O., Bohn M. A stretch of "late" SV40 viral DNA about 400 bp long which includes the origin of replication is specifically exposed in SV40 minichromosomes. Cell. 1979 Feb;16(2):453–466. doi: 10.1016/0092-8674(79)90021-7. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldeck W., Föhring B., Chowdhury K., Gruss P., Sauer G. Origin of DNA replication in papovavirus chromatin is recognized by endogenous endonuclease. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5964–5968. doi: 10.1073/pnas.75.12.5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk B., Oudet P., Chambon P. Preferential in vitro assembly of nucleosome cores on some AT-rich regions of SV40 DNA. Nucleic Acids Res. 1979 Oct 10;7(3):705–713. doi: 10.1093/nar/7.3.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Wigmore D. J., Eaton R. W., Scott W. A. Endonuclease-sensitive regions in SV40 chromatin from cells infected with duplicated mutants. Virology. 1980 Jul 30;104(2):462–473. doi: 10.1016/0042-6822(80)90348-7. [DOI] [PubMed] [Google Scholar]
- Wu C., Bingham P. M., Livak K. J., Holmgren R., Elgin S. C. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell. 1979 Apr;16(4):797–806. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]