Abstract
In order to obtain lipid producing strain with high-yield, the wild type stain Rhodotorula glutinis was treated by low ion implantation, and optimization of fermentation medium for higher lipid yield was carried out using mutant strain. It was found that the strain had a higher positive mutation rate when the output power was 10 keV and the dose of N+ implantation was 80 × 2.6 × 1013 ions/cm2. Then a high-yield mutant strain D30 was obtained through cid-heating coupling ultrasonic method and lipid yield was 3.10 g/L. Additionally, the surface response method was used to optimize fermentation medium. The three significant factors (glucose, peptone, KH2PO4) were optimized using response surface methodology (RSM), and the optimized parameters of fermentation medium were as follows: glucose 73.40 g/L, peptone 1.06 g/L and KH2PO4 3.56 g/L. Finally the fermentation characteristic of high-yield mutation strain D30 was studied, when fermentation time was 10 days, which lipid yield increased to 7.81 g/L. Fatty acid composition of the lipid was determined by GC, and the most represented fatty acids of mutant D30 were C16:0 (11.4 %), C16:1 (5.66 %), C18:1 (49.3 %), and C18:2 (27.0 %).
Keywords: Lipid, Rhodotorula glutinis, Ion implantation, Response surface methodology
Introduction
Natural sources of lipid include plants, animals and microorganisms [1]. Traditionally, lipid is produced from plant oils or animal fats. With the increasing population and continued demand for industrial utilization, the plant and animal sources alone cannot meet the total requirement [2]. Therefore, there is currently increased interest in identifying potential new lipid sources for use [3].
Microbial lipids, known as single cell oils (SCOs) and produced by oleaginous microorganisms including algae, yeast, bacteria and fungi have potential application as new lipid sources. Usually, the fatty acid composition of microbial lipid is similar to that of plant oils [4]. In addition, microbial lipids contain high fractions of polyunsaturated fatty acids and have the potential to serve as a source of significant quantities of transportation fuels [5]. Also, microbial lipid have many advantages, such as short life cycle, less labor required, less affection by venue, season and climate [6]. Thus microbial lipid is a potential alternative lipid resource for production.
As a new mutagenesis technique, low-energy ion implantation was first explored as a new agent for mutation induction in rice. Through 26 years of development, the technique has made much progress and has been widely applied in the breeding of crops, plants and microbes [7]. Ion implanted is characterized by a higher mutation rate and wider mutational spectrum with less damage for the implanted organism, and suggested that a combination of energy deposition, mass deposition and charge exchange of energetic ions will result in genetic damage [8].
The response surface methodology (RSM) is a combination of mathematical and statistical approaches used for modeling different phenomena and optimizing the experimental results as a function of various parameters [9]. The main advantage of RSM is the reduced number of experimental runs needed to provide sufficient information for statistically acceptable result. It is a faster and less expensive method for gathering research results than the classical method [10]. Therefore, the most extensive applications of RSM are in situations where several input variables potentially influence some performance measure or quality characteristic of the product or process [11].
The aim of the present study was improving lipid yield of Rhodotorula glutinis. The wild type strain was treated by low-energy nitrogen ions to induce mutations, and obtaining a high-yield mutant strain. Then RSM were employed to optimize fermentation medium components of the mutant strain.
Materials and Methods
Microorganisms
Rhodotorula glutinis 31596, a lipid-producing microorganism, was obtained from China Center of Industrial Culture Collection (CICC).
Media and Cultivation
The compositions of media were listed as follows: medium I, as slant medium, 10 % wort with 2 % agar and 2 % peptone, pH 5.8, sterilized for 30 min at 115 °C. Medium II, used for seed culture, 10 % wort with 2 % peptone, pH 5.8, sterilized for 30 min at 115 °C. Medium III (g/L), used for lipid fermentation, glucose: 80; peptone: 1.8; (NH4)2SO4: 1.1; KH2PO4: 2.5; pH 5.8, sterilized for 30 min at 115 °C. Seed culture of the strain was grown aerobically in a 250 mL. Erlenmeyer flask containing 30 mL of the seed medium with shaking at 190 rpm, 28 °C for 24 h. Fermentation were performed in 250 mL Erlenmeyer flasks containing 30 mL medium III, with shaking at 190 rpm, 28 °C for 96 h.
Ion Implantation Equipment
Implantation sources were produced by ion beam bioengineering instrument (Patent No. CN 93103361.6, Yu Zengliang et al., 2000, People’s Republic of China) devised by ASIPP (Chinese Academy of Sciences, Institute of plasma physics).
Mutagenesis Method
After being aerobic cultured in seed medium. The fresh cultured cells of Rhodotorula glutinis were diluted in sterilized physiological salt solution, and dilution rate is 10−5 0.1 mL suspension was spread on an empty sterilized Petri dish (90 mm) and desiccated by sterilized air to make a dry membrane of cells. Then, the dishes were put into the sample holder and implanted by nitrogen ion beam under dry and vacuum condition with energy of 10 keV. The dose for implantation ranged from 20 × 2.6 × 1013 to 120 × 2.6 × 1013 ions/cm2 with interval dose 20 × 2.6 × 1013 ions/cm2. At the same time, in order to evaluate the effects of vacuum on mutation, the bacteria of control group without N+ beam implantation were also put into the target chamber.
Mutant Selection [12]
After ion implantation, strain of Rhodotorula glutinis were washed out from the plates with sterilized physiological salt solution, then was spread on the plates to isolate high-yield lipid producing mutants. The number of colonies was counted to determine the survival ratio. The colonies were transferred to test tubes, then fermentation after seed culture. The positive and negative mutants were calculated, those mutant with high-yield were selected.
Determination of Fatty Acid Composition by GC
The lipid extracted was esterified with KOH–MeOH. Fatty acid composition of the sample was determined using agilent 6890N gas chromatography(GC) equipped with a flame ionization detector and quartz capillary columns DB-WAX(30 m × 0.25 mm × 0.25 μm). The carrier gas was nitrogen. The flow rates of air, nitrogen (carrier gas), hydrogen and make-up gas were 400, 1, 40, 30 mL/min, respectively. The split ratio was 1:100. The detector temperature was programmed at 270 °C. The injector temperature was set at 250 °C. The column temperature was programmed for three stages: 90 °C initial temperature, increased to 190 °C at 7 °C/min and increased to 215 °C at 3 °C/min, it was then held at 215 °C for 10 min. Finally, temperature was increased to 230 °C at 20 °C/min and held for 5 min.
Results and Discussion
Effect of N+ Implantation on Survival Ratio of Rhodotorula glutinis
Low-energy ions was the atoms deprived of some or all electrons, whose energy were lower than 100 keV and the protonic number was more than two. It was well known that the mutagen of N+ has the ability to cause higher mutation frequency and wider mutations spectra than other ions such as Ar+, H+, He+, and it was the most common ion used in ion beam implantation [13]. For ion implantation, mutation frequency and screen efficiency are closely related to the energy and the doses of ions [14]. The mechanism including energy absorption, mass deposition, and charge exchange has been proposed by accumulated evidences in breeding, gene transfer, and cell modification [15].
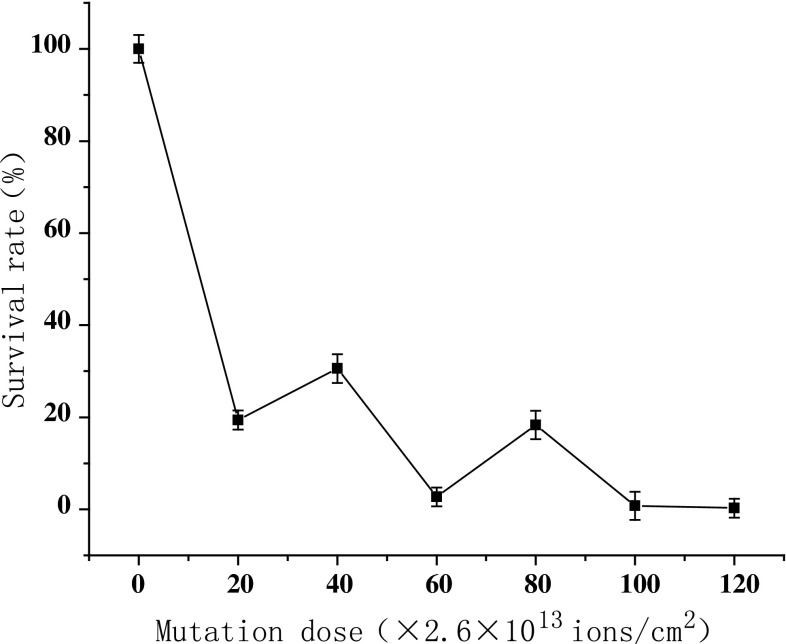
As was shown in Fig. 1, Firstly, the survival rate decreased sharply, then decreased slowly with doses in the range of 20 × 2.6 × 1013 ions/cm2 and 60 × 2.6 × 1013 ions/cm2. However, the survival rate increased slightly with dose from 60 × 2.6 × 1013 to 80 × 2.6 × 1013 ions/cm2, and decreased again as the dose exceeded 80 × 2.6 × 1013 ions/cm2. Such a peculiar survival rate curve, indicating the abnormal radiation damage, was regarded as the typical and important mutation effect of ion beam implantation. The shape of survival rate was quite different from the results of other traditional irradiation, such as UV, X-ray or γ-ray [16]. This phenomenon could be explained by a reasonable mechanism based on the energy and momentum deposition effect at lower doses and by the charge stimulating effect at higher doses [17].The low dosage ions only damage and etching of the cell surface, so the survival rate is higher. With the increase in dosage, the superficial injury to the cell becomes serious, and massive free OH radicals and radials cause the survival rate to drop. When they drop to a certain extent (40 × 2.6 × 1013, 80 × 2.6 × 1013 ions/cm2), some kind of internal repair mechanism of the cell such as SOS (error-prone repair) is activated, and the survival rate have a temporary rise. But when the dose is further increased, the cell damage becomes irreparable [18].
Fig. 1.
Survival rate curve of N+ iron implantation. The survival fraction can be calculated from the formula: Survival fraction = S1/S0, where S1 was the number of clones scored on treated plates and S0 was the number of clones scored on control plates without ion implantation
Effect of N+ Implantation on Mutation Rate of Rhodotorula glutinis
According to current microbic breeding theory, the highest positive mutation ratio was obtained when the lethal ratio of microorganism ranges from 70 to 80 %.
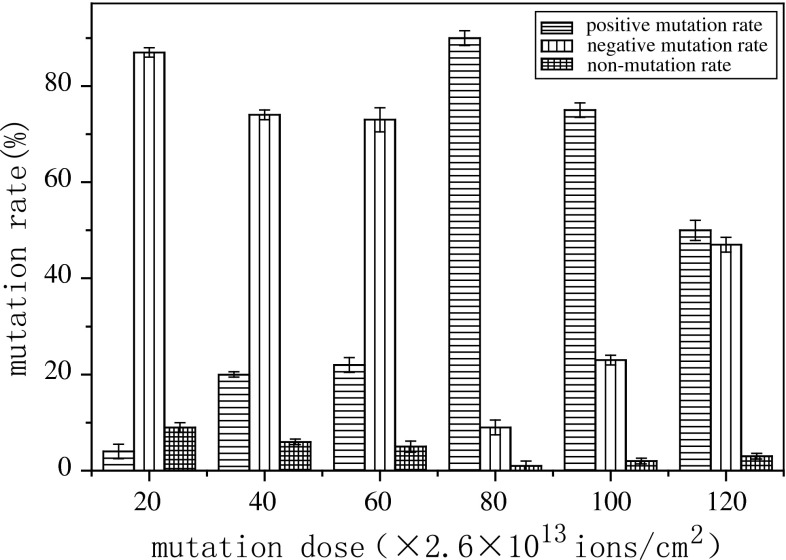
Mutation ratio of Rhodotorula glutinis related to different doses of N+ implantation are shown in Fig. 2. In a range of 80 × 2.6 × 1013–100 × 2.6 × 1013 ions/cm2, the positive mutation ratio was observed to be higher than the negative mutation ratio, especially when the dose was 80 × 2.6 × 1013 ions/cm2. Out of this range, negative mutation was dominant. Considering both lethal rate and positive mutation rate, 10 keV, 80 × 2.6 × 1013 ions/cm2 were chosen as optimal mutation parameters.
Fig. 2.
Effect of Implantation dose on mutation rate. The positive mutants/the negative mutants were defined as those in which lipid yield was increased/decreased by more than 5 % when compared with the original strain. Between +5 % and −5 % is defined as non-mutation. The mutation ratio was calculated as the number of either positive or negative mutants divided by the total number of screened mutants
As a result, D30 was screened. The yield of lipid was as high as 3.10 g/L which increased by 33.05 % (the original strain is 2.33 g/L) when fermentation time was 96 h.
Response Surface Methodology
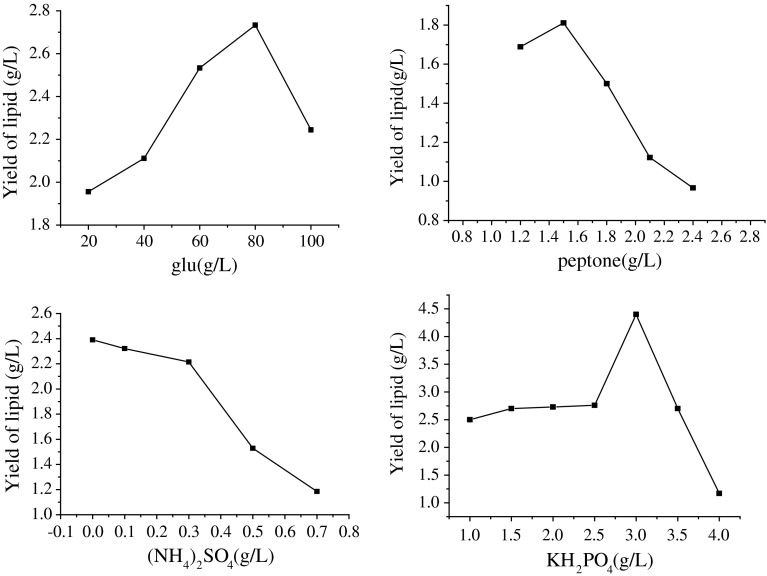
Response surface methodology (RSM) consists of a group of empirical techniques devoted to the evaluation of relations existing between a cluster of controlled experimental factors and the measured responses, according to one or more selected criteria [19]. Based on the results obtained in preliminary experiments (Fig. 3), Fig. 3 indicated glucose, peptone and KH2PO4 were major variables in the lipid production, but (NH4)2SO4 did not need to add. The carbon source provided energy and necessary carbon skeleton for the growth of strains, the nitrogen source was material (nucleic acids, amino acids, protein, etc.) to constitute cell and nitrogen metabolites, phosphorus participated in sugar metabolism and energy metabolism. Hence, these variables were selected to find the optimized conditions for higher lipid production using response surface methodology.
Fig. 3.
Effect of variables on oil production
The range and the levels of the experimental variables investigated in this study are given in the Table 1. The central values (zero level) chosen for experiment design were glucose 80 g/L, peptone 1.5 g/L and KH2PO4 3 g/L. Once the variables having the greatest influence on the responses were identified, a 15-run Box-Behnken Design was used to optimize the levels of these variables. The exact values of the three significant factors effecting lipid yield were determined using surface response methodology by Box–Behnken design as given in Table 2, and after the responses were obtained, they were subjected to multiple nonlinear regression and optimization using the statistical analysis software (SAS9.0). The second-order polynomial models were applied to correlate these variables. The Prob > |t| was used to evaluate the significance of the models.
Table 1.
Factors and levels of the experimental variables
| Code | Factors | −1 | 0 | 1 |
|---|---|---|---|---|
| X1 | Glu (g/L) | 53.3 | 80 | 120 |
| X2 | Peptone (g/L) | 1 | 1.5 | 2.25 |
| X3 | kH2PO4 (g/L) | 2 | 3 | 4.5 |
Table 2.
The Box–Behnken design for the three independent variables and predicted values
| Runs | X1 | X2 | X3 | Response Value/g/L |
|---|---|---|---|---|
| 1 | −1 | −1 | 0 | 2.78 |
| 2 | −1 | 1 | 0 | 2.45 |
| 3 | 1 | −1 | 0 | 2.85 |
| 4 | 1 | 1 | 0 | 1.96 |
| 5 | 0 | −1 | −1 | 1.55 |
| 6 | 0 | –1 | 1 | 2.82 |
| 7 | 0 | 1 | −1 | 1.20 |
| 8 | 0 | 1 | 1 | 2.02 |
| 9 | −1 | 0 | −1 | 1.20 |
| 10 | 1 | 0 | −1 | 1.55 |
| 11 | −1 | 0 | 1 | 2.75 |
| 12 | 1 | 0 | 1 | 2.20 |
| 13 | 0 | 0 | 0 | 2.99 |
| 14 | 0 | 0 | 0 | 2.96 |
| 15 | 0 | 0 | 0 | 3.11 |
The responses in Table 2 were correlated by nonlinear regression using the following full quadratic polynomial model:
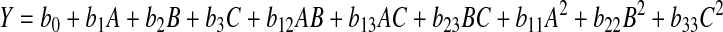 |
3 |
Where, Y was the desired response, bi were the coefficients, A, B and C were the three factors. Table 3 lists the associated significant levels obtained from statistical analysis software (SAS9.0).
Table 3.
Analysis of variance
| Source | DF | MS | SS | F | Prob > |t| | Significance |
|---|---|---|---|---|---|---|
| X1 | 1 | 0.04805 | 0.04805 | 11.56438 | 0.019238 | * |
| X2 | 1 | 0.702112 | 0.702112 | 168.9801 | 0.0001 | ** |
| X3 | 1 | 2.300513 | 2.300513 | 553.6733 | 0.0001 | ** |
| X1*X1 | 1 | 0.214898 | 0.214898 | 51.72036 | 0.000809 | ** |
| X1*X2 | 1 | 0.0784 | 0.0784 | 18.86883 | 0.007402 | ** |
| X1*X3 | 1 | 0.2025 | 0.2025 | 48.73646 | 0.000928 | ** |
| X2*X2 | 1 | 0.266683 | 0.266683 | 64.18356 | 0.00049 | ** |
| X2*X3 | 1 | 0.050625 | 0.050625 | 12.18412 | 0.01746 | * |
| X3*X3 | 1 | 2.691283 | 2.691283 | 647.7215 | 0.0001 | ** |
| Model | 9 | 6.323518 | 0.702613 | 169.1006 | 0.0001 | ** |
| Lack of fit | 3 | 0.008175 | 0.002725 | 0.43254 | 0.753157 | |
| Pure Error | 2 | 0.0126 | 0.0063 | |||
| Total | 14 | 6.34498 |
* As significance (p < 0.05); ** as top-significance (p < 0.01)
The application of response surface methodology yielded the following regression equation which is an empirical relationship between the logarithmic values of lipid yields and test variables in coded unit:
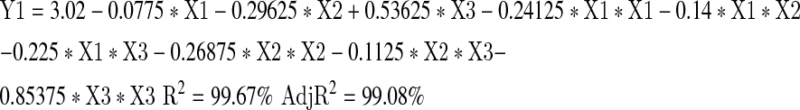 |
4 |
In the test above, Y is the response, that is, the lipid yields expressed in logarithmic values, and X1, X2 and X3 are the coded values of the test variables (glucose, peptone and KH2PO4 respectively). The goodness of fit of the model was checked by the determination coefficient (R2). In this case, the value of the determination coefficient (R2 = 99.67 %) indicates that only 0.33 % of total variations are not explained by the model. The value of the adjusted determination coefficient (AdjR2 = 99.08 %) is also very high, which indicates a high significance of the model. A higher value of the correlation coefficient (R = 99.83 %) signifies an excellent correlation between the independent variables. At the same time a relatively lower value of the coefficient of variation (CV = 2.81) indicates improved precision and reliability of the conducted experiments.
The variance analysis (Table 3) indicated that the regression model was significant. Based on the estimated value of partial effects, the interaction s between X1 and X2, X1 and X3, X2 and X3 had significant effects on mass of lipid, especially X1 and X3. The quadratic regression model demonstrated the relation among independent variances and response with accuracy. And according to Table 3, the linear/quadratic and cross product are all significant that indicated three factors had interaction with each other.
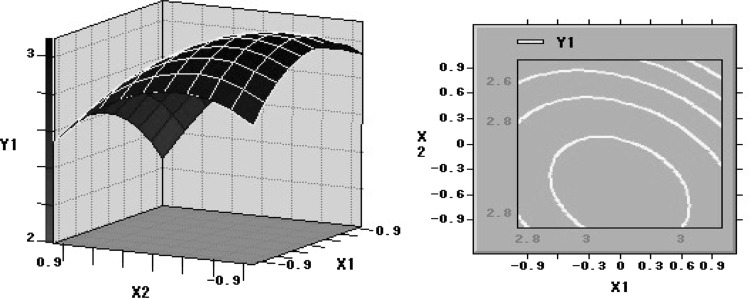
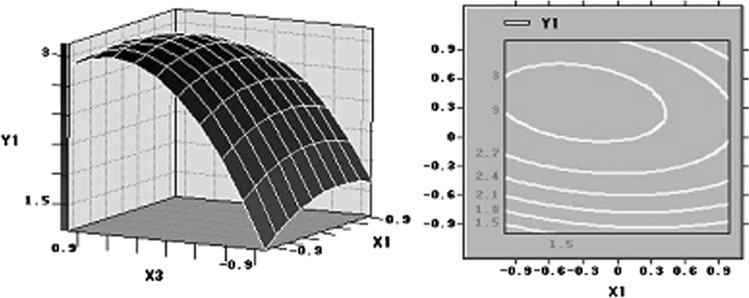
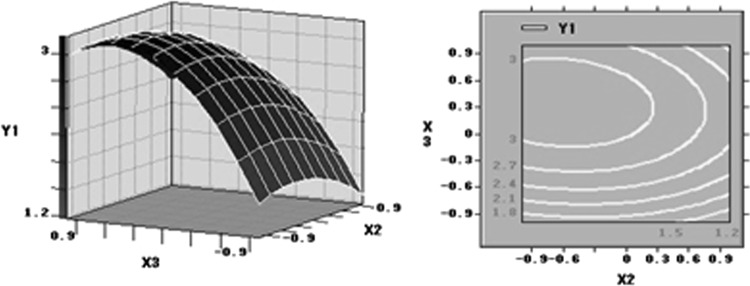
Response surface plots as a function of two factors at a time, maintaining all other at zero levels, are more helpful in understanding both the main and the interaction effects of these two factors. The optimal value of each factor was clearly showed in the three-dimension surface plots (Figs 4, 5, 6). Figure 4 showed the response surface plot obtained as a function of glucose concentration versus peptone concentration, while all other variables are maintained at zero level, mass of lipid reached the maximum level around 73.40 g/L glucose and 1.06 g/L peptone. Figure 5 showed the function of glucose concentration and KH2PO4 concentration. According to the plot, the optimal value of glucose concentration and KH2PO4 concentration were near 73.40 and 3.56 g/L, separately. Further numerical analysis of the respond surface using the statistical analysis software (SAS9.0) revealed that the optimal levels of three main factors were: glucose 763.40 g/L, peptone 1.06 g/L and KH2PO4 3.56 g/L.
Fig. 4.
Response surface plot showing the effect on glucose concentration, peptone concentration and their mutual effect on the production of lipid. Other variables are held at zero level
Fig. 5.
Response surface plot showing the effect on glucose concentration, KH2PO4 concentration and their mutual effect on the production of lipid. Other variables are held at zero level
Fig. 6.
Response surface plot showing the effect on peptone concentration, KH2PO4 concentration and their mutual effect on the production of lipid. Other variables are held at zero level
Validation Experiment
Validation experiment was conducted separately under the optimal conditions. By validation experiment, the maximum yield of lipid production reached 3.41 g/L, and this result was very close to the predicted value of 3.43 g/L, increasing by 10.00 % than original mutant. Therefore, the optimal parameters obtained from the Response surface methodology were valid.
The Fermentation Kinetics of Mutant D30
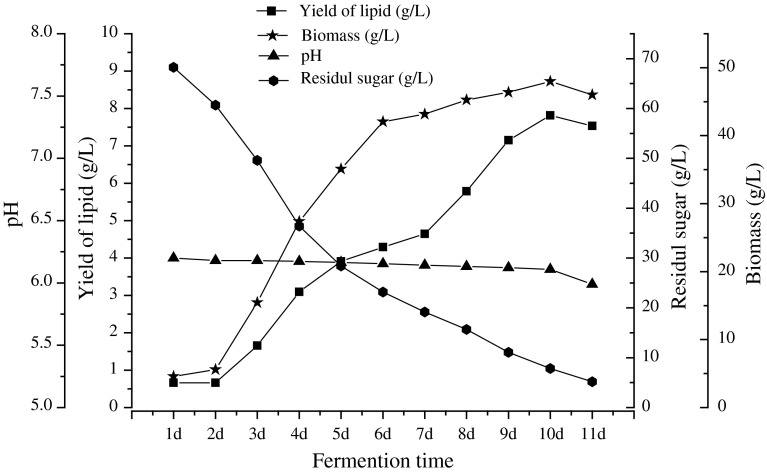
Strain D30 was cultured under above culture condition in an agitated flask shaking at 190 rpm at 28 °C for 11 days observation was carried on per 24 h, three replicates were employed (Fig. 7).
Fig. 7.
Fermentation curve of D30
In this paper, Fig. 7 showed that the lipid was accumulating with cell growing from 1 to 10 days. When the culture time was 10 days, the maximum of lipid yield and the biomass was obtained and they were 7.81 and 18.04 g/L respectively, When fermentation time was 10–11 days, pH, lipid yield and biomass were decreased simultaneously. Strain began to autolysis, glucose was almost consumed absolutely, lipid was used as carbon source again.
Fatty Acid Composition of Rhodotorula glutinis 31596 and Mutant D30
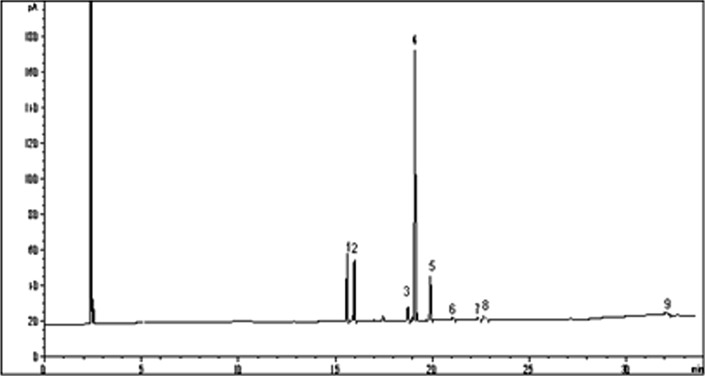
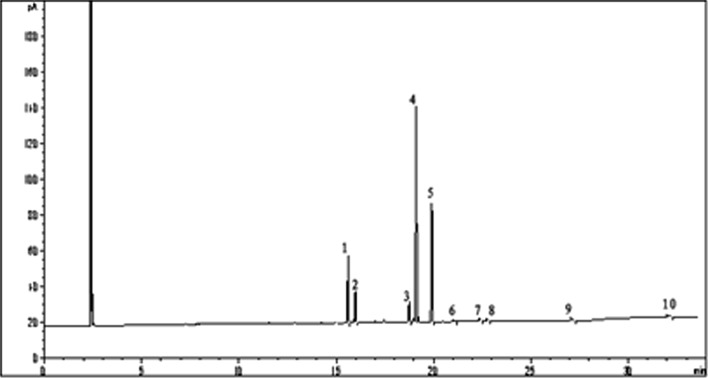
The fatty acid composition of Rhodotorula glutinis 31596 and mutant D30 is shown in Figs. 8 and 9; and Tables 4 and 5, separately, that expressed as percentage of total fatty acids. The analyzed lipid sample of mutant D30 showed a concentration of approximately 17.06 % of saturated fatty acids, 55.26 % of monounsaturated, and 28.07 % of polyunsaturated, increasing by 16.27 % than Rhodotorula glutinis 31596. The most represented fatty acids of mutant D30 were C16:0 (11.4 %), C16:1 (5.66 %), C18:1 (49.3 %), and C18:2 (27.0 %) (Fig. 9).
Fig. 8.
GC chromatogram of the FAMES of Rhodotorula glutinis 31596. 1 C16:0; 2 C16:1; 3 C18:0; 4 C18:1; 5 C18:2; 6 α-C18:3; 7 C20:0; 8 C20:1; 9 C22:6
Fig. 9.
GC chromatogram of the FAMES of mutant D30. 1 C16:0; 2 C16:1; 3 C18:0; 4 C18:1; 5 C18:2; 6 α-C18:3; 7 C20:0; 8 C20:1; 9 C22:0; 10 C22:6
Table 4.
Contents of fatty acids in Rhodotorula glutinis 31596
| Fatty acids | Content (%) |
|---|---|
| C16:0 | 11.5 |
| C16:1 | 11.0 |
| C18:0 | 3.33 |
| C18:1 | 61.8 |
| C18:2 | 9.93 |
| α-C18:3 | 0.53 |
| C20:0 | 0.33 |
| C20:1 | 0.28 |
| C22:6 | 1.34 |
Table 5.
Contents of fatty acids in mutant D30
| Fatty acids | Content (%) |
|---|---|
| C16:0 | 11.4 |
| C16:1 | 5.66 |
| C18:0 | 4.54 |
| C18:1 | 49.3 |
| C18:2 | 27.0 |
| α-C18:3 | 0.32 |
| C20:0 | 0.60 |
| C20:1 | 0.30 |
| C22:0 | 0.52 |
| C22:6 | 0.75 |
Conclusion
Microbial lipids contain high fractions of polyunsaturated fatty acids that was essential fatty acid to human, and derived from food,. Polyunsaturated fatty acids had the function of losing weight and prevention of cardiovascular disease, anti-inflammatory, anti-tumor [20, 21]. At present, polyunsaturated fatty acids were obtained from plant seeds and marine fish, but not met market demand. In order to obtain lipid producing strain with high-yield, the wild stain Rhodotorula glutinis 31596 was treated by low ion implantation, and studied on optimization of fermentation medium.
By studying, it was found that the original strain had a higher positive mutation rate when the low ion implantation power was 10 keV and the dose of N+ implantation was 80 × 2.6 × 1013 ions/cm2. The mutant D30 was selected through Acid-heating coupling ultrasonic method. And its yield was as high as 3.10 g/L, increasing by 33.05 % than that of the original. It proved once again that ion beam implantation had higher mutation frequency and was a new mutagenesis technique for mutation induction in oleaginous microorganisms.
Based on preliminary experiments, glucose, peptone and KH2PO4 were found to be major variables in the lipid fermentation production. Response surface methodology was employed to obtain the optimal conditions of lipid production and explain the interaction of independent variances. The model of independent variance and response was established. On the basis of the results of statistical analysis, it has been found that the reduced quadratic models were reasonably accurate (R2 = 99.67 %). The optimized parameters of lipid production were as follows: glucose 73.40 g/L, peptone 1.06 g/L and KH2PO4 3.56 g/L. Through optimization, the lipid yield of D30 reached to 3.43 g/L, increased by 10.65 % further.
The fermentation kinetics of mutant D30 was also studied, and the biomass was 18.04 g/L and the yield of lipid was 7.81 g/L when fermentation time was 10 days. Furthermore, mutant D30 contained a considerable amount of unsaturated fatty acids.
Acknowledgments
This study was supported by the opening foundation of the Key Laboratory of Ion Beam Bioengineering, Institute of Plasma Physics, Chinese Academy of Science, and the Key Laboratory of Ion Beam Bioengineering, Anhui Province, grant NO. 2008B004.
References
- 1.Gouda MK, Omar SH, Aouad LM. Single cell oil production by Gordonia sp. DG using agro-industrial wastes. Word J Microbiol Biotechnol. 2008;24:1703–1711. doi: 10.1007/s11274-008-9664-z. [DOI] [Google Scholar]
- 2.Chisti Y. Biodiesel from microalgae. Biotechnology Adv. 2007;25:294–306. doi: 10.1016/j.biotechadv.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Matsui T, Otsuka K, Sato S. Microbial oil production from carbohydrates using Sporobolomyces carnicolor strain O33. Ann Microbiol. 2011;62:861–864. doi: 10.1007/s13213-011-0316-4. [DOI] [Google Scholar]
- 4.Chen HF, Huang C, Xiong L, Chen XD, Ma LL. Microbial oil production from corncob acid hydrolysate by Trichosporon cutaneum. Biotechnol Lett. 2012;34:1025–1028. doi: 10.1007/s10529-012-0869-8. [DOI] [PubMed] [Google Scholar]
- 5.Subramaniam R, Dufreche S, Zappi M, Bajpai R. Microbial lipids from renewable resources: production and characterization. J Ind Microbiol Biotechnol. 2010;37:1271–1287. doi: 10.1007/s10295-010-0884-5. [DOI] [PubMed] [Google Scholar]
- 6.Li Q, Du W, Liu DL. Perspectives of microbial oils for biodiesel production. Appl Microbiol Biotechnol. 2008;80:749–756. doi: 10.1007/s00253-008-1625-9. [DOI] [PubMed] [Google Scholar]
- 7.Gu SB, Yao JM, Yuan QP, Xue PJ, Zheng ZM, Wang L, Yu ZL. A novel approach for improving the productivity of ubiquinone-10 producing strain by low-energy ion beam irradiation. Appl Microbiol Biotechnol. 2005;72:456–461. doi: 10.1007/s00253-005-0283-4. [DOI] [PubMed] [Google Scholar]
- 8.Wu LF, Yu ZL. Radiobiological effects of a low-energy ion beam on wheat. Radiat Environ Biophys. 2001;40:53–57. doi: 10.1007/s004110000078. [DOI] [PubMed] [Google Scholar]
- 9.Kalali A, Ebadi T, Rabbani A, Sadri Moghaddam S. Respone surface methodology approach to the optimization of oil hydrocarbon polluted soil remediation using enhanced soil washing. Int. J. Environ. Sci. Tech. 2011;8:389–400. [Google Scholar]
- 10.Sinkuniene D, Bendikinene V, Juodka B. Respone surface methodology-based optimization of lipase-catalyzed triolein hydrolysis in hexane. Roman Biotechnol Lett. 2011;16:5891–5901. [Google Scholar]
- 11.Myers RH, Montgomery DC. Response surface methodology: process and product optimization using designed experiments. 2. New York: John Wiley and Sons, Inc.; 2002. [Google Scholar]
- 12.Sh Ch Li, HX Liu, Sh B Gu, Wang M, Zh Y Zhu, Wang DD (2010) Screening of lipid high producing mutant from Rhodotorula glutinis by low ion implantation and study on lipid extraction technique. CMBB2010 3:311–316
- 13.Xu A, Yao J, Yu L, Lv S, Wang J, Yan B, Yu Z. Mutation of Gluconobacter oxydans and Bacillus megaterium in a two-step process of L-ascorbic acid manufacture by ion beam. J Appl Microbiol. 2004;96:1317–1323. doi: 10.1111/j.1365-2672.2004.02270.x. [DOI] [PubMed] [Google Scholar]
- 14.Wu M, Li Sh Ch, Yao JM, Pan RR, Yu ZL. Mutant of a xylanase-producing strain of Aspergillus niger in solid state fermentation by low energy ion implantation. World J Microbiol Biotechnol. 2005;21:1045–1049. doi: 10.1007/s11274-004-7870-x. [DOI] [Google Scholar]
- 15.Chen HL, Wan HG, Zhang J, Zeng XX. Mutation induction of Pleurotus ferulae by low-energy N+ ion implantation and characters of the selected mutant. Nucl Sci Tech. 2008;19:13–16. doi: 10.1016/S1001-8042(08)60015-9. [DOI] [Google Scholar]
- 16.Vilaithong T, Yu LD, Alisi C, Phanchaisri B, Apavatjrut P, Anuntalabhochai S. A study of low-energy ion beam effects on outer plant cell structure for exogenous macromolecule transferring. Surf Coat Technol. 2000;128:133–138. doi: 10.1016/S0257-8972(00)00650-2. [DOI] [Google Scholar]
- 17.Song DJ, Yao JM, Shao CL, Yu Z. A possible mechanism of dose related survival of microorganism implanted by N+ ions. Nuclear Tech. 1999;22:129–132. [Google Scholar]
- 18.Xu TT, Bai ZZ, Wang LJ, He BF. Breeding of D(-)-lactic acid high producing strain by low-energy ion implantation and preliminary analysis of related metabolism. Appl Biochem Biotechnol. 2010;160:314–321. doi: 10.1007/s12010-008-8274-4. [DOI] [PubMed] [Google Scholar]
- 19.Adinarayana K, Ellaiah P. Response surface optimization of the critical medium components for the production of alkaline protease by a newly isolated Bacillus sp. J Pharm Pharmaceut Sci. 2002;5:272–278. [PubMed] [Google Scholar]
- 20.Li XC, Jacob MR, Elsohly HN. Acetylenic acids inhibiting azole-resistant Candida albicans from Pentagonia gigantifolia. J Nat Prod. 2003;66:1132–1135. doi: 10.1021/np030196r. [DOI] [PubMed] [Google Scholar]
- 21.Terry PD, Rohan TE, Wolk A. Intakes of fish and marine fatty acids and risks of cancers of the breast and prostate and of other hormone-related cancers: a review of the epidemiologic evidence. Am J Clin Nutr. 2003;77:532–543. doi: 10.1093/ajcn/77.3.532. [DOI] [PubMed] [Google Scholar]