Abstract
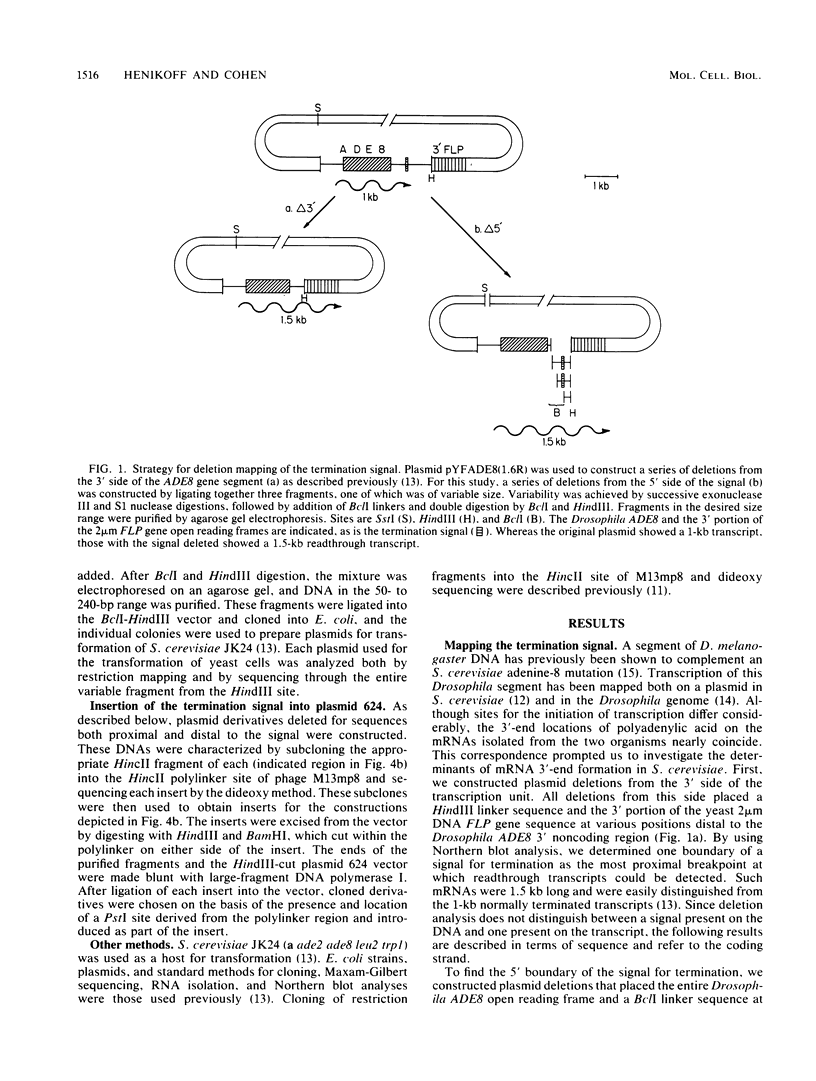
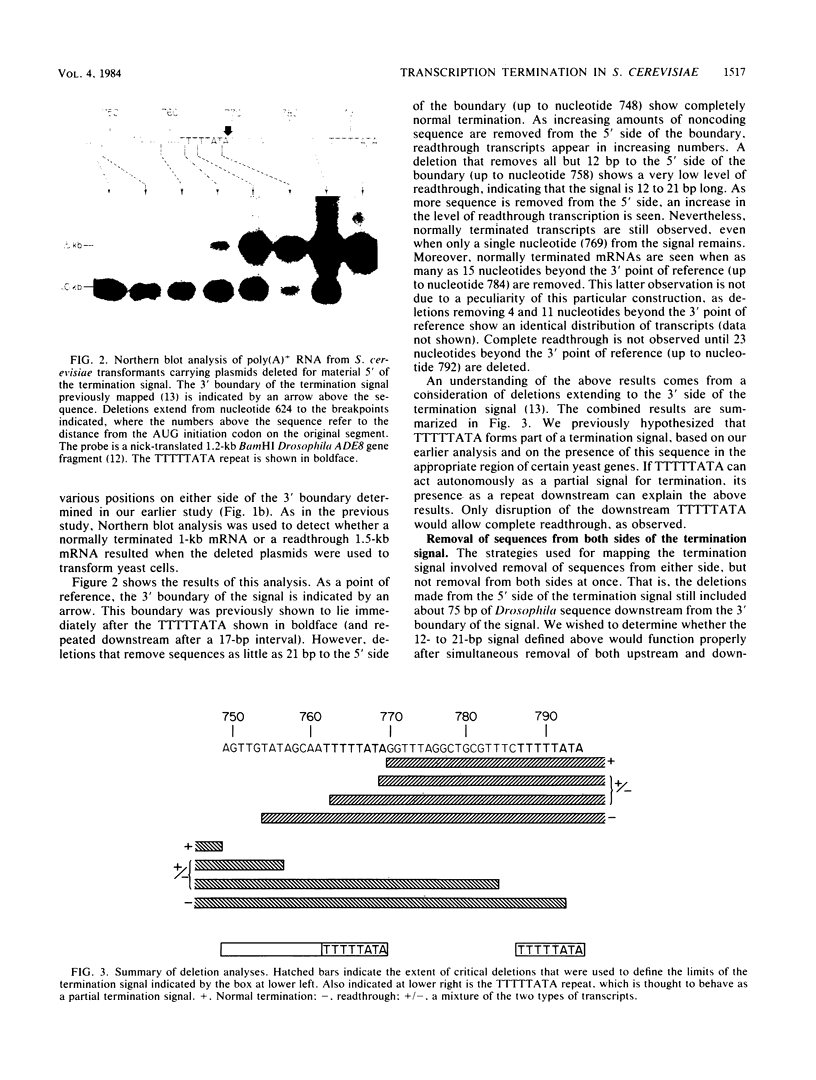
We have mapped a signal sequence for mRNA 3'-end formation in Saccharomyces cerevisiae by using a Drosophila melanogaster DNA segment that complements a yeast adenine-8 mutation. That the 3' end of the transcript in S. cerevisiae nearly coincides with that in D. melanogaster is consistent with the possibility that mRNA termini are similarly determined in both organisms. Deletion analysis reveals that the complete signal is no more than 21 base pairs long. Part of the signal is the sequence TTTTTATA, which is seen in the termination region of several yeast genes. TTTTTATA appears to be able to act autonomously as a partial termination signal. The efficiency of the complete signal is affected by substitution of sequences downstream from it. This modulation of the effect of a signal is consistent with termination in S. cerevisiae, resembling rho-dependent termination in bacteria.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alber T., Kawasaki G. Nucleotide sequence of the triose phosphate isomerase gene of Saccharomyces cerevisiae. J Mol Appl Genet. 1982;1(5):419–434. [PubMed] [Google Scholar]
- Birchmeier C., Folk W., Birnstiel M. L. The terminal RNA stem-loop structure and 80 bp of spacer DNA are required for the formation of 3' termini of sea urchin H2A mRNA. Cell. 1983 Dec;35(2 Pt 1):433–440. doi: 10.1016/0092-8674(83)90176-9. [DOI] [PubMed] [Google Scholar]
- Birchmeier C., Grosschedl R., Birnstiel M. L. Generation of authentic 3' termini of an H2A mRNA in vivo is dependent on a short inverted DNA repeat and on spacer sequences. Cell. 1982 Apr;28(4):739–745. doi: 10.1016/0092-8674(82)90053-8. [DOI] [PubMed] [Google Scholar]
- Bourouis M., Jarry B. Vectors containing a prokaryotic dihydrofolate reductase gene transform Drosophila cells to methotrexate-resistance. EMBO J. 1983;2(7):1099–1104. doi: 10.1002/j.1460-2075.1983.tb01552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke R. L., Tekamp-Olson P., Najarian R. The isolation, characterization, and sequence of the pyruvate kinase gene of Saccharomyces cerevisiae. J Biol Chem. 1983 Feb 25;258(4):2193–2201. [PubMed] [Google Scholar]
- Darnell J. E., Jr Variety in the level of gene control in eukaryotic cells. Nature. 1982 Jun 3;297(5865):365–371. doi: 10.1038/297365a0. [DOI] [PubMed] [Google Scholar]
- Galli G., Hofstetter H., Stunnenberg H. G., Birnstiel M. L. Biochemical complementation with RNA in the Xenopus oocyte: a small RNA is required for the generation of 3' histone mRNA termini. Cell. 1983 Oct;34(3):823–828. doi: 10.1016/0092-8674(83)90539-1. [DOI] [PubMed] [Google Scholar]
- Hatfield G. W., Sharp J. A., Rosenberg M. Pausing and termination of human RNA polymerase II transcription at a procaryotic terminator. Mol Cell Biol. 1983 Oct;3(10):1687–1693. doi: 10.1128/mcb.3.10.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N., Aloni Y. Attenuation in SV40 as a mechanism of transcription-termination by RNA polymerase B. Nucleic Acids Res. 1984 Feb 10;12(3):1401–1414. doi: 10.1093/nar/12.3.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N., Skolnik-David H., Aloni Y. Attenuation in the control of SV40 gene expression. Cell. 1982 May;29(1):183–193. doi: 10.1016/0092-8674(82)90102-7. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Cloning exons of mapping of transcription: characterization of the Drosophila melanogaster alcohol dehydrogenase gene. Nucleic Acids Res. 1983 Jul 25;11(14):4735–4752. doi: 10.1093/nar/11.14.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Furlong C. E. Sequence of a Drosophila DNA segment that functions in Saccharomyces cerevisiae and its regulation by a yeast promoter. Nucleic Acids Res. 1983 Feb 11;11(3):789–800. doi: 10.1093/nar/11.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Kelly J. D., Cohen E. H. Transcription terminates in yeast distal to a control sequence. Cell. 1983 Jun;33(2):607–614. doi: 10.1016/0092-8674(83)90441-5. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Sloan J. S., Kelly J. D. A Drosophila metabolic gene transcript is alternatively processed. Cell. 1983 Sep;34(2):405–414. doi: 10.1016/0092-8674(83)90374-4. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Tatchell K., Hall B. D., Nasmyth K. A. Isolation of a gene from Drosophila by complementation in yeast. Nature. 1981 Jan 1;289(5793):33–37. doi: 10.1038/289033a0. [DOI] [PubMed] [Google Scholar]
- Holland J. P., Holland M. J. The primary structure of a glyceraldehyde-3-phosphate dehydrogenase gene from Saccharomyces cerevisiae. J Biol Chem. 1979 Oct 10;254(19):9839–9845. [PubMed] [Google Scholar]
- Holmes W. M., Platt T., Rosenberg M. Termination of transcription in E. coli. Cell. 1983 Apr;32(4):1029–1032. doi: 10.1016/0092-8674(83)90287-8. [DOI] [PubMed] [Google Scholar]
- Kadesch T. R., Chamberlin M. J. Studies of in vitro transcription by calf thymus RNA polymerase II using a novel duplex DNA template. J Biol Chem. 1982 May 10;257(9):5286–5295. [PubMed] [Google Scholar]
- Kotval J., Zaret K. S., Consaul S., Sherman F. Revertants of a transcription termination mutant of yeast contain diverse genetic alterations. Genetics. 1983 Mar;103(3):367–388. doi: 10.1093/genetics/103.3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leer R. J., van Raamsdonk-Duin M. M., Molenaar C. M., Cohen L. H., Mager W. H., Planta R. J. The structure of the gene coding for the phosphorylated ribosomal protein S10 in yeast. Nucleic Acids Res. 1982 Oct 11;10(19):5869–5878. doi: 10.1093/nar/10.19.5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., Berk A. J. Inhibition of RNA cleavage but not polyadenylation by a point mutation in mRNA 3' consensus sequence AAUAAA. Nature. 1983 Oct 13;305(5935):600–605. doi: 10.1038/305600a0. [DOI] [PubMed] [Google Scholar]
- Montgomery D. L., Leung D. W., Smith M., Shalit P., Faye G., Hall B. D. Isolation and sequence of the gene for iso-2-cytochrome c in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jan;77(1):541–545. doi: 10.1073/pnas.77.1.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. L., Sharp P. A. Site-specific polyadenylation in a cell-free reaction. Cell. 1984 Mar;36(3):581–591. doi: 10.1016/0092-8674(84)90337-4. [DOI] [PubMed] [Google Scholar]
- Platt T. Termination of transcription and its regulation in the tryptophan operon of E. coli. Cell. 1981 Apr;24(1):10–23. doi: 10.1016/0092-8674(81)90496-7. [DOI] [PubMed] [Google Scholar]
- Richardson J. P. Activation of rho protein ATPase requires simultaneous interaction at two kinds of nucleic acid-binding sites. J Biol Chem. 1982 May 25;257(10):5760–5766. [PubMed] [Google Scholar]
- Rubin G. M., Spradling A. C. Genetic transformation of Drosophila with transposable element vectors. Science. 1982 Oct 22;218(4570):348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Salditt-Georgieff M., Darnell J. E., Jr A precise termination site in the mouse beta major-globin transcription unit. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4694–4698. doi: 10.1073/pnas.80.15.4694. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Skolnik-David H., Hay N., Aloni Y. Site of premature termination of late transcription of simian virus 40 DNA: enhancement by 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole. Proc Natl Acad Sci U S A. 1982 May;79(9):2743–2747. doi: 10.1073/pnas.79.9.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]