Abstract
The coagulase negative staphylococci (CNS) are the most prevalent mastitis pathogen group yet their virulence characteristics have not been well described. We investigated the presence of 19 classical and newly described staphylococcal superantigen (SAg) genes in CNS isolates from bovine intramammary infections (IMI). A total of 263 CNS representing 11 different Staphylococcus spp. were examined, and 31.2% (n = 82) of CNS isolates had one or more SAg genes; there were 21 different SAg gene combinations. The most prevalent combination of SAg genes (seb, seln, and selq; n = 45) was found in S. chromogenes, S. xylosus, S. haemolyticus, S. sciuri subsp. carnaticus, S. simulans and S. succinus. The genes for SAgs appear to be widely distributed amongst CNS isolated from bovine IMI.
Keywords: coagulase-negative staphylococci, staphylococcal superantigens, multiplex PCR
1. INTRODUCTION
Staphylococcal superantigens (SAgs) including staphylococcal enterotoxins (SEs) and toxic shock syndrome toxin-1 (TSST-1) were originally identified in S. aureus. Staphylococcal enterotoxins were named according to their emetic activities following oral administration in a primate model. Several SEs were designated as SE-like (SEl) since they either lack emetic properties or their emetic activities have not been tested in this model (Lina et al., 2004). There are 5 different types of classical SEs, SEA through SEE, which are antigenically distinct (Bergdoll, 1989). Recently, new types of SEs (SEG, SEH, SEI, SElJ, SElK, SElL, SElM, SElN, SElO, SElP, SElQ, SElR, SES, SET, SElU, and SElV) have been described in S. aureus (Ono et al., 2008; Seo and Bohach, 2007; Thomas et al., 2006).
Compared to S. aureus, a relatively limited numbers of studies have examined the prevalence of SAg genes in Coagulase negative staphylococci (CNS) isolates. Crass and Bergdoll (1986) first identified CNS that produced TSST-1, SEA, or both from patients with toxic shock syndrome. Some studies also demonstrated that classical SEs and TSST-1 were produced by CNS species isolated from human clinical samples and intramammary infections of cattle and other ruminants (Bautista et al., 1988; Kuroishi et al., 2003; Valle et al., 1991; Cunha et al., 2007). Conversely, several larger surveys failed to detect genes encoding SAgs in CNS from human and veterinary specimens (Becker et al., 2001a; Becker et al., 2001b; Jaulhac et al., 1991; Kreiswirth et al., 1987; Nemati et al., 2008).
These observations collectively suggest that CNS isolates from bovine IMI could be a possible source of SAgs. Survey studies examining genetic constructs for SAg in CNS heretofore have been limited in scope, with either a limited number of samples collected or a limited number of SAg genes probed. To build upon these previous investigations we collected milk from more than 1000 cows on 6 dairy farms to gain a better insight into the prevalence of CNS with genes for SAgs. The focus of the current study was to investigate the prevalence and distribution of CNS bovine IMI isolates that possess the genes for classical and/or newly described SAgs.
2. MATERIALS AND METHODS
2.1. Bacterial strains
Milk samples were aseptically collected in duplicate from functional mammary quarters on six dairies in Idaho and Washington. A mammary quarter was considered infected when a single type of CNS was presumptively identified on sheep blood agar after 48 h incubation at 37 °C and 5% CO2, from duplicate milk samples, and the number of colony-forming unit/ml of milk was greater than 200 (National Mastitis Council, 1987). A total of 263 CNS isolates which caused IMI were identified and speciation was determined by partial 16S rRNA and/or rpoB gene sequence analyses as previously described (Park et al., 2009). The species of CNS from bovine IMI used in the present study included: S. chromogenes (n = 190), S. xylosus (n = 24), S. haemolyticus (n = 16), S. sciuri subsp. carnaticus (n = 9), S. hyicus (n = 8), S. simulans (n = 7), S. caprae (n = 3), S. epidermidis (n = 2), S. succinus (n = 2), S. capitis (n = 1), and S. hominis (n = 1). Six S. aureus reference strains (Table 1) were used to establish multiplex PCR for 19 different SAg genes. All bacterial strains were stored in 20% glycerol solution at −80 °C.
Table 1.
Nucleotide sequences of primers and expected size of PCR products (bp) of staphylococcal superantigenic toxins used in this study
| Gene | Primer | Oligonucleotides sequence (5′to 3′) | PCR product (bp) |
PCR set |
References |
|---|---|---|---|---|---|
| sea | SEA-F | CAGCATACTATATTGTTTAAAGGC | 400 | 1 | This study |
| SEA-R | CCTCTGAACCTTCCCATC | ||||
| seb | SEB-F | GTATGGTGGTGTAACTGAGCA | 351 | 1 | This study |
| SEB-R | TCAATCTTCACATCTTTAGAATCA | ||||
| sec | SEC-F | CTCAAGAACTAGACATAAAAGCTAG G |
271 | 1 | (Becker et al., 1998) |
| SEC-R | TCAAAATCGGATTAACATTATCC | ||||
| sed | SED-F | CTAGTTTGGTAATATCTCCTTTAAAC G |
319 | 1 | (Becker et al., 1998) |
| SED-R | TTAATGCTATATCTTATAGGGTAAAC ATC |
||||
| see | SEE-F | CAGTACCTATAGATAAAGTTAAAAC AAGC |
178 | 1 | (Becker et al., 1998) |
| SEE-R | TAACTTACCGTGGACCCTTC | ||||
| seg | SEG-F | AAGTAGACATTTTTGGCGTTCC | 287 | 2 | (Omoe et al., 2002) |
| SEG-R | AGAACCATCAAACTCGTATAGC | ||||
| seh | SEH-F | GTCTATATGGAGGTACAACACT | 213 | 2 | (Omoe et al., 2002) |
| SEH-R | GACCTTTACTTATTTCGCTGTC | ||||
| sei | SEI-F | GGTGATATTGGTGTAGGTAAC | 454 | 2 | (Omoe et al., 2002) |
| SEI-R | ATCCATATTCTTTGCCTTTACCAG | ||||
| selj | SEJ-F | ATAGCATCAGAACTGTTGTTCCG | 152 | 2 | (Omoe et al., 2005) |
| SEJ-R | CTTTCTGAATTTTACCACCAAAGG | ||||
| selk | SEK-F | TAGGTGTCTCTAATAATGCCA | 293 | 3 | (Omoe et al., 2005) |
| SEK-R | TAGATATTCGTTAGTAGCTG | ||||
| sell | SEL-F | TAACGGCGATGTAGGTCCAGG | 383 | 4 | (Omoe et al., 2005) |
| SEL-R | CATCTATTTCTTGTGCGGTAAC | ||||
| selm | SEM-F | GGATAATTCGACAGTAACAG | 379 | 3 | (Omoe et al., 2005) |
| SEM-R | TCCTGCATTAAATCCAGAAC | ||||
| seln | SEN-F | CATCATGCTTATACGGAGGAG | 301 | 4 | This study |
| SEN-R | CCCACTGAACCTTTTACGTT | ||||
| selo | SEO-F | TGTGTAAGAAGTCAAGTGTAG | 214 | 3 | (Omoe et al., 2005) |
| SEO-R | TCTTTAGAAATCGCTGATGA | ||||
| selp | SEP-F | TGATTTATTAGTAGACCTTGG | 381 | 2 | (Omoe et al., 2005) |
| SEP-R | ATAACCAACCGAATCACCAG | ||||
| selq | SEQ-F | TCAAGGAGTTAGTTCTGGAAATT | 251 | 4 | This study |
| SEQ-R | GCTTACCATTGACCCAGAGA | ||||
| selr | SER-F | GGATAAAGCGGTAATAGCAG | 166 | 4 | (Omoe et al., 2005) |
| SER-R | GTATTCCAAACACATCTAAC | ||||
| selu | SEU-F | ATCAGAAACAAACATTAAAGCCCA | 500 | 4 | This study |
| SEU-R | TGACCATTTCCTTCGATAAACTTTAT | ||||
| tst1 | TST-F | AAGCCCTTTGTTGCTTGCG | 447 | 3 | (Becker et al., 1998) |
| TST-R | ATCGAACTTTGGCCCATACTTT |
2.2. Genomic DNA purification
Genomic DNA was purified as previously described (Pitcher et al., 1989). Briefly, bacteria were cultured in 5 ml of Todd Hewitt broth (THB, Becton Dickinson Diagnostic Systems, Sparks, MD) at 37 °C for 18 h and then harvested by centrifugation at 12,000 g for 5 min. The cell pellet was resuspended in TE buffer (50 mM Tris-HCl pH 7.0, 10 mM EDTA), digested by treatment with 10 µl of lysostaphin (1 mg/ml, Sigma-Aldrich Co., St. Louis, MO) and 100 µl of lysozyme (100 mg/ml, Sigma-Aldrich Co.) at 37 °C for 1 h. Cell lysates were treated with 500 µl of lysis buffer (5 M guanidine thiocyanate, 5 mM EDTA, 0.5% v/v sarkosyl) followed by 250 µl of ammonium acetate solution (7.5 M, Sigma-Aldrich Co.). After mixing with an equal volume of chloroform:isoamyl alcohol (24:1), the sample was centrifuged at 18,000 g for 10 min. The upper aqueous phase containing DNA was collected into a fresh microtube and precipitated with isopropanol, followed by washing with 70% ethanol. The DNA concentration was adjusted to100 ng/µl by the addition of deionized water to achieve an OD = 2.0 at 260 nm (Nanodrop Technologies, Wilmington, DE).
2.3. Multiplex PCR for SAg genes
A multiplex PCR method described by Omoe et al. (2005) was used to amplify 19 different SAg genes (18 SE and TSST-1 genes). It appeared that primers for sea, seb, seln, and selq genes used in a previous study generated non-specific amplification in some species of CNS isolates, particularly S. chromogenes, the most prevalent species in bovine IMI. Thus primers for sea, seb, seln and selq genes were replaced with newly designed ones. Additionally, one more primer for the selu gene was included in this study. Primers and the expected size of PCR products for each SAg gene are described in Table 1. Multiplex PCR was performed with 4 different sets of primer mixtures (Set 1: sea, seb, sec, sed, see; Set 2: seg, seh, sei, selj, selp; Set 3: selk, selm, selo, tst1; Set 4: sell, seln, selq, selr, selu). Each reaction mixture (50 µl) consisted of 10×PCR buffer (Applied Biosystems Inc., Foster City, CA), 1U Taq DNA polymerase (Applied Biosystems Inc.), 0.4mM of dNTP mix (Applied Biosystems Inc.), 0.3 µM of each primer, 100 ng of template DNA and sterile deionized water. Multiplex PCR for SAg genes was carried out with the following thermal cycling conditions: an initial denaturation of DNA at 95°C for 10 min was followed by 35 cycles of amplification (95 °C for 30 s, 53 °C for 45 s, and 72 °C for 90 s), ending with a final extension at 72 °C for 10 min. The mixture of genomic DNA from S. aureus reference strains were used for multiplex PCR of each set as a positive control. All PCR products were analyzed by 1% agarose gel electrophoresis in 0.5× Tris-borate-EDTA buffer. Amplification of non-template controls was attempted with each reaction to determine if DNA contamination occurred.
3. RESULTS
3.1. Modification of multiplex PCR for SAg genes
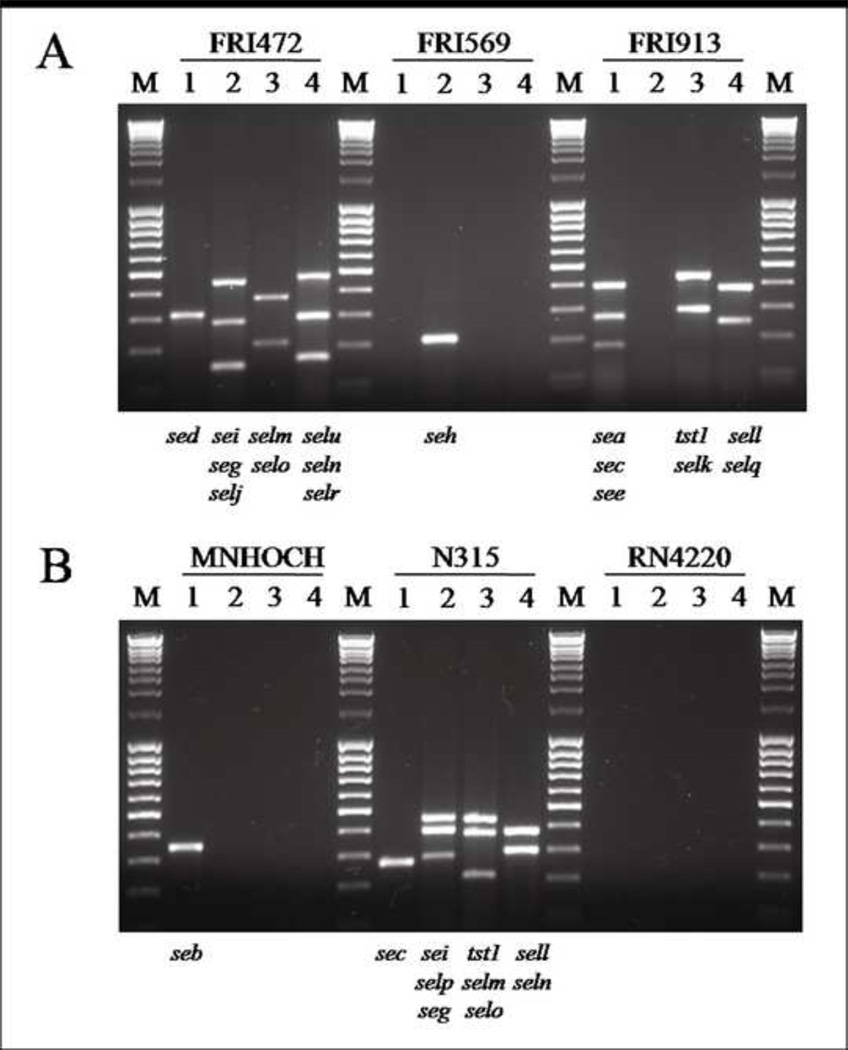
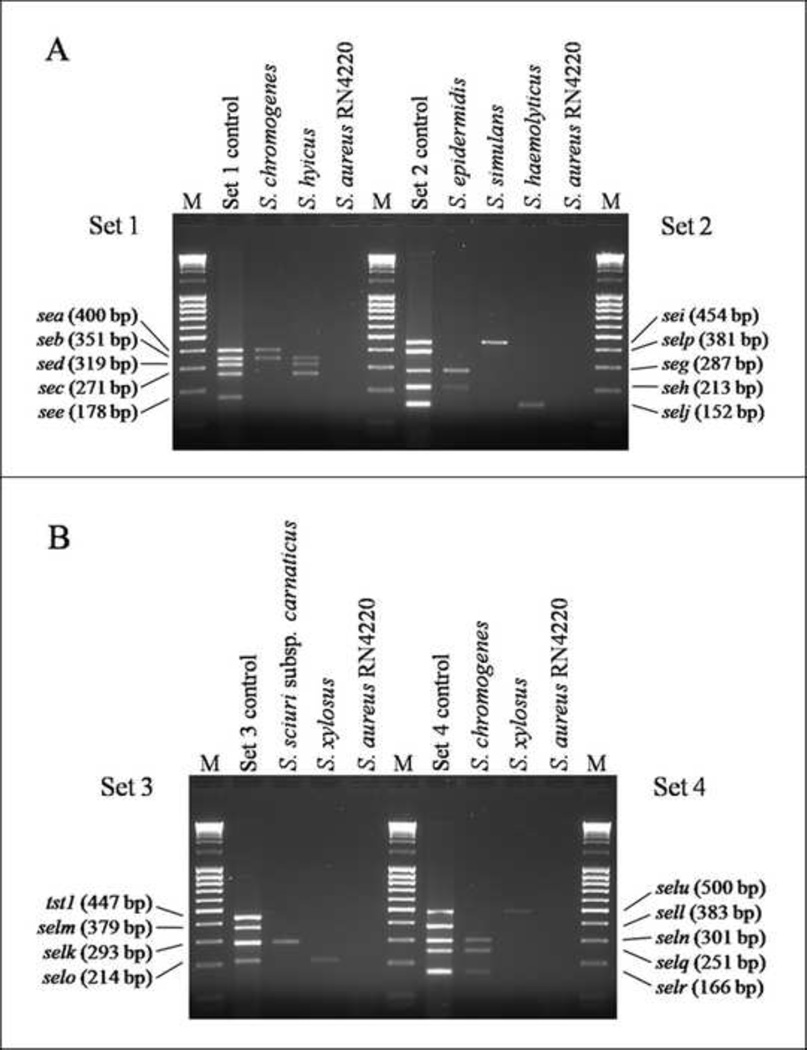
The modified multiplex PCR method in this study successfully amplified 19 different SAg genes from S. aureus reference strains (Fig. 1A and 1B). The sizes of the PCR products obtained from the positive control reference strains corresponded to their predicted sizes (Fig.1 and Table 1). The SAg genes of all reference strains determined by a modified multiplex PCR were exactly identical to those previously described in other studies (Table 2). Non-specific reactions were not observed. An amplicon was not observed in the negative control, S. aureus RN4220. In addition, this modified multiplex PCR specifically amplified SAg genes from S. chromogenes as well as other CNS species of isolates without non-specific amplifications as shown in Fig. 2A and 2B, demonstrating representative multiplex PCR results observed in various CNS species isolated from bovine IMI.
FIG. 1.
Detection of staphylococcal superantigen genes from S. aureus reference strains by multiplex PCR. The SAg genes detected from S. aureus reference strains by multiplex PCR of each primer set were indicated at the bottom of each lane. Lanes; M,100 bp DNA ladder; 1,.primer set 1; 2, primer set 2; 3, primer set 3; 4, primer set 4.
Table 2.
S. aureus reference strains used in this study
| Strains | Superantigen gene(s) | Reference |
|---|---|---|
| FRI 472 | sed, seg, sei, selj, selm, seln, selo, selr, selu | (Monday and Bohach, 1999) |
| FRI 569 | seh | (Su and Wong, 1995) |
| FRI 913 | sea, sec, see, selk, sell, selq, tst1 | (Bania et al., 2006) |
| MNHOCH | seb | (Monday and Bohach, 1999) |
| N315 | sec, seg, sei, sell, selm, seln, selo, selp, tst1 | (Kuroda et al., 2001) |
| RN4220 | None | (Monday and Bohach, 1999) |
FIG. 2.
Detection of staphylococcal superantigen genes from CNS isolates from bovine intramammary infections by multiplex PCR. The mixtures of genomic DNAs from 5 S. aureus reference strains were used for multiplex PCR as a positive control for each primer sets. Genomic DNA extracted from S. aureus RN4220 was used as a negative control for multiplex PCR. Results shown are representative data obtained from various species of bovine IMI CNS isolates harboring SAg genes. (A) primer set 1 and 2, (B) primer set 3 and 4; Lane M: 100 bp DNA ladder.
3.2. Prevalence and distribution of SAg genes
Of the 263 CNS IMI isolates tested, 82 CNS isolates (31.2%) were found to have one or more SAg genes (Table 3). Isolates with the selq gene (22.4%, 59/263) were most prevalent followed by those with the seb (20.9%, 55/263) or seln gene (20.9%, 55/263) (Table 3). The see, sell, selm, selp, or tst1 gene was not detected in any of isolates in this study (Table 3). The SAg genes were widely distributed among 9 different CNS species including S. chromogenes, S. xylosus, S. hyicus, S. haemolyticus, S. sciuri subsp. carnaticus, S. simulans, S. capitis, S. epidermidis, and S. succinus (Table 3). Of the S. chromogenes, 22.1 % (42/190) of the isolates were SAg gene-positive (Table 3). Staphylococcus xylosus was the second most prevalent CNS species in bovine IMI and 45.8% (11/24) of the isolates were SAg gene-positive (Table 3). All S. hyicus (n = 8) and S. capitis (n = 1) were found to have SAg genes. No SAg genes were detected amongst S. caprae and S. hominis isolates (Table 3).
Table 3.
Staphylococcal superantigen (SAg) genes observed in coagulase-negative staphylococcal isolates from bovine intramammary infections
| Speciesa | No. of isolates |
No. of SAg gene-positive isolates |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SAg genesb |
sea | seb | sec | sed | see | seg | seh | sei | selj | selk | sell | selm | seln | selo | selp | selq | selr | selu | tst1 | ||
| S. chromogenes | 190 | 42 | 4 | 33 | 1 | 1 | 0 | 0 | 0 | 0 | 2 | 4 | 0 | 0 | 33 | 0 | 0 | 37 | 1 | 3 | 0 |
| S. xylosus | 24 | 11 | 0 | 9 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 1 | 0 | 9 | 0 | 1 | 0 |
| S. haemolyticus | 16 | 6 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 3 | 0 | 0 | 0 |
| S. sciuri subsp. | 9 | 7 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| carnaticus | |||||||||||||||||||||
| S. hyicus | 8 | 8 | 0 | 4 | 2 | 3 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 4 | 0 | 0 | 4 | 0 | 0 | 0 |
| S. simulans | 7 | 5 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 3 | 0 | 0 | 0 |
| S. caprae | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| S. epidermidis | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| S. succinus | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| S. capitis | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| S. hominis | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 263 | 82 | 4 | 55 | 3 | 6 | 0 | 2 | 2 | 5 | 5 | 12 | 0 | 0 | 55 | 1 | 0 | 59 | 1 | 4 | 0 |
The species was determined by partial 16S rRNA gene sequence analysis except S. caprae and S. capitis which were determined by partial rpoB gene sequence analysis.
No. of isolates that were positive for at least one SAg gene.
A total of 21 different SAg gene combinations were observed among the 82 SAg gene-positive isolates (Table 4). The most common SAg gene combination was seb, seln, and selq, which was present in 54.9% (45/82) of SAg gene-positive CNS isolates (Table 4). Most of SAg gene-positive isolates had both classical and newly described SAg genes (73.2%, 60/82) (Table 4). Nineteen isolates had only newly described SAg genes whereas 3 isolates had classical SAg genes only (Table 4). Of the 42 SAg gene-positive S. chromogenes isolates, the combination of seb, seln, and selq genes was most prevalent (69.0%, 29/42) and the same combination also was most commonly observed in SAg gene-positive S. xylosus isolates (Table 4).
Table 4.
Combinations of staphylococcal superantigen (SAg) genes observed in 82 SAg gene-positive coagulase-negative staphylococcal isolates from bovine intramammary infections
| Speciesa | Combinations of SAg genes | No. of isolates |
|---|---|---|
|
S. chromogenes (n = 42) |
sea, seb, selj, selk, seln, selq | 1 |
| sea, seb, selj, selk, seln, selq, selr | 1 | |
| sea, seb, selk, seln, selq | 1 | |
| sea, selk | 1 | |
| seb, seln, selq | 29 | |
| seb, seln, selq, selu | 1 | |
| sec | 1 | |
| sed | 1 | |
| selq | 4 | |
| selu | 2 | |
|
S. xylosus (n = 11) |
seb, seln, selq | 8 |
| seb, seln, selq, selo | 1 | |
| seg | 1 | |
| selu | 1 | |
|
S. hyicus (n = 8) |
seb, sec, sed, selk, seln, selq | 1 |
| seb, sed, seln, selq | 1 | |
| seb, selk, seln, selq | 2 | |
| sec, sed, selk | 1 | |
| selk | 3 | |
|
S. sciuri subsp. carnaticus (n = 7) |
seb, seln, selq | 2 |
| sei | 4 | |
| selk | 1 | |
|
S. haemolyticus (n = 6) |
seb, sed, seln, selq | 1 |
| seb, seln, selq | 2 | |
| selj | 3 | |
|
S. simulans (n = 5) |
seb, seln, selq | 3 |
| sed | 1 | |
| sei | 1 | |
|
S. capitis (n = 1) |
seh | 1 |
|
S. epidermidis (n = 1) |
seh, seg | 1 |
|
S. succinus (n = 1) |
seb, seln, selq | 1 |
The species was determined by partial 16S rRNA gene sequence analysis except S. caprae and S. capitis which were determined by partial rpoB gene sequence analysis.
4. DISCUSSION
Staphylococcal SAgs are typically associated with S. aureus as virulence factors and have profound effects on the host immune system through the subversion of immune responses and delays in the establishment of pathogen-specific immunity (McCormick et al., 2001; Seo et al., 2007). Several previous studies (Bautista et al., 1988; Kuroishi et al., 2003; Valle et al., 1991) have suggested the possibility that various CNS species isolated from ruminant mastitis possess the genes to produce some of the SAgs. We found that one or more classical and/or newly described SAg genes were observed in 31.2% of CNS isolates and these SAg genes were widely distributed in 9 different CNS species. In this study, a group of seb, seln, and selq genes was the most prevalent SAg gene combination (n = 45) in CNS IMI isolates and was observed in various CNS species (Table 4). The current study demonstrated that the seb gene was the most common classical SAg while only 3 isolates harbored the sec gene. This was not consistent with other findings (Cunha et al., 2007; Kuroishi et al., 2003; Valle et al., 1991) that SEC was the most prevalent classical SAg in CNS isolates regardless of the origin of isolates.
Still other investigators reported that no SAg genes were detected in human and veterinary CNS isolates and thus, SAgs, were not associated with CNS isolates (Becker et al., 2001a; Becker et al., 2001b; Nemati et al., 2008). Nemati et al. (2008) investigated the presence of 18 different SAg genes in CNS isolated from clinical or subclinical bovine IMI using PCR as similar to our study and showed that none of the CNS isolates were positive for SAg genes. They raised a possibility that the failure of SAg gene detection could be due to the low sequence similarity of SAg genes between CNS and S. aureus. However, we note in their study genomic DNA was prepared by a boiling method routinely used in PCR technique for Gram-negative microorganism. In our study we based our DNA extraction on cell lysis using lysostaphin and lysozyme rather than boiling cells. We conjecture that our use of enzymes to lyse cells may result in a better yield and quality of DNA, leading to fewer false-negative detection results, than might be found with boiling cells which could have denatured the DNA before extraction.
In this study, we noticed a particular phenomenon that SAg genes detected in CNS IMI isolates seemed to be unstable. Generally, the amplification intensity of SAg genes from CNS IMI isolates was much weaker than those from S. aureus reference strains (the positive control) (Fig. 2A and 2B) although an equal amount of genomic DNA was used for multiplex PCR. Furthermore, the amplicon intensity became weaker or even disappeared when genomic DNA was extracted from the culture following several passages of the organisms under laboratory conditions. Thus it could be hypothesized that genetic elements harboring SAg genes in CNS from bovine IMI are unstable, with stability losses with passing generations. Evidence in support of this hypothesis can be found in a recent report (Ubeda et al., 2007) where SAg genes in some S. aureus, chromosomally integrated in a staphylococcal pathogenic island, could be replicated as multimeric plasmids and transferred to other staphylococci, both S. aureus and CNS. However, replication of multimeric plasmids was unstable such that some daughter cells in the passage of culture lost multimeric plasmid harboring SAg genes. Clearly further study is necessary to define the genetic elements harboring SAg genes in CNS and to identify culture conditions which are needed to stabilize the maintenance of genetic elements.
To the best of our knowledge, this is the first study that investigated extensively and showed the presence of classical and/or newly described staphylococcal SAg genes in various species of CNS isolates from bovine IMI. We acknowledge the need to determine under what conditions those detected SAg genes in CNS isolates are maintained and expressed at the protein level. Our findings support the conjecture that CNS isolates from bovine IMI could serve as a possible reservoir of classical and newly described SAgs typically associated with the S. aureus pathogen.
ACKNOWLEDGMENT
We are grateful to Dorothy Newkirk and Claudia Deobald for excellent technical assistance. This work was supported in part by the National Institutes of Health Grant P20 RR15587, P20 RR016454, and U54AI57141, and the Idaho Agricultural Experimental Station, and Idaho Dairymen’s Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bania J, Dabrowska A, Bystron J, Korzekwa K, Chrzanowska J, Molenda J. Distribution of newly described enterotoxin-like genes in Staphylococcus aureus from food. Int. J. Food. Microbiol. 2006;108:36–41. doi: 10.1016/j.ijfoodmicro.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Bautista L, Gaya P, Medina M, Nunez M. A quantitative study of enterotoxin production by sheep milk staphylococci. Appl. Environ. Microbiol. 1988;54:566–569. doi: 10.1128/aem.54.2.566-569.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K, Haverkamper G, von Eiff C, Roth R, Peters G. Survey of staphylococcal enterotoxin genes, exfoliative toxin genes, and toxic shock syndrome toxin 1 gene in non-Staphylococcus aureus species. Eur. J. Clin. Microbiol. Infect. Dis. 2001a;20:407–409. doi: 10.1007/pl00011281. [DOI] [PubMed] [Google Scholar]
- Becker K, Keller B, von Eiff C, Bruck M, Lubritz G, Etienne J, Peters G. Enterotoxigenic potential of Staphylococcus intermedius. Appl. Environ. Microbiol. 2001b;67:5551–5557. doi: 10.1128/AEM.67.12.5551-5557.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K, Roth R, Peters G. Rapid and specific detection of toxigenic Staphylococcus aureus: use of two multiplex PCR enzyme immunoassays for amplification and hybridization of staphylococcal enterotoxin genes, exfoliative toxin genes, and toxic shock syndrome toxin 1 gene. J. Clin. Microbiol. 1998;36:2548–2553. doi: 10.1128/jcm.36.9.2548-2553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergdoll MS. Staphylococcus aureus. New York, NY: Foodborne Bacterial Pathogens. Marcel Dekker, Inc.; 1989. pp. 463–523. [Google Scholar]
- Crass BA, Bergdoll MS. Involvement of coagulase-negative staphylococci in toxic shock syndrome. J. Clin. Microbiol. 1986;23:43–45. doi: 10.1128/jcm.23.1.43-45.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha MLRS, Calsolari RAO, Junior JPA. Detection of enterotoxin and toxic shock syndrome toxin 1 genes in Staphylococcus, with emphasis on coagulase-negative staphylococci. Microbiol. Immunol. 2007;51:381–390. doi: 10.1111/j.1348-0421.2007.tb03925.x. [DOI] [PubMed] [Google Scholar]
- Jaulhac B, De Buyser ML, Dilasser F, Prevost G, Piedmont Y. Screening of staphylococci for the toxic shock syndrome toxin-1 (TSST-1) gene. Lett. Appl. Microbiol. 1991;13:90–92. doi: 10.1111/j.1472-765x.1991.tb00578.x. [DOI] [PubMed] [Google Scholar]
- Kreiswirth BN, Schlievert PM, Novick RP. Evaluation of coagulase-negative staphylococci for ability to produce toxic shock syndrome toxin 1. J. Clin. Microbiol. 1987;25:2028–2029. doi: 10.1128/jcm.25.10.2028-2029.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, Lian J, Ito T, Kanamori M, Matsumaru H, Maruyama A, Murakami H, Hosoyama A, Mizutani-Ui Y, Takahashi NK, Sawano T, Inoue R, Kaito C, Sekimizu K, Hirakawa H, Kuhara S, Goto S, Yabuzaki J, Kanehisa M, Yamashita A, Oshima K, Furuya K, Yoshino C, Shiba T, Hattori M, Ogasawara N, Hayashi H, Hiramatsu K. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- Kuroishi T, Komine K, Kai K, Itagaki M, Kobayashi J, Ohta M, Kamata S, Kumagai K. Concentrations and specific antibodies to staphylococcal enterotoxin-C and toxic shock syndrome toxin-1 in bovine mammary gland secretions, and inflammatory response to the intramammary inoculation of these toxins. J. Vet. Med. Sci. 2003;65:899–906. doi: 10.1292/jvms.65.899. [DOI] [PubMed] [Google Scholar]
- Lina G, Bohach GA, Nair SP, Hiramatsu K, Jouvin-Marche E, Mariuzza R. Standard nomenclature for the superantigens expressed by Staphylococcus. J. Infect. Dis. 2004;189:2334–2336. doi: 10.1086/420852. [DOI] [PubMed] [Google Scholar]
- McCormick JK, Yarwood JM, Schlievert PM. Toxic shock syndrome and bacterial superantigens: an update. Annu. Rev. Microbiol. 2001;55:77–104. doi: 10.1146/annurev.micro.55.1.77. [DOI] [PubMed] [Google Scholar]
- Monday SR, Bohach GA. Use of multiplex PCR to detect classical and newly described pyrogenic toxin genes in staphylococcal isolates. J. Clin. Microbiol. 1999;37:3411–3414. doi: 10.1128/jcm.37.10.3411-3414.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Mastitis Council. Laboratory and Field Handbook on Bovine Mastitis. Fort Atkinson, WI: Hoard and Sons; 1987. [Google Scholar]
- Nemati M, Hermans K, Vancraeynest D, De Vliegher S, Sampimon OC, Baele M, De Graef EM, Pasmans F, Haesebrouck F. Screening of bovine coagulase-negative staphylococci from milk for superantigen-encoding genes. Vet. Rec. 2008;163:740–743. [PubMed] [Google Scholar]
- Omoe K, Hu DL, Takahashi-Omoe H, Nakane A, Shinagawa K. Comprehensive analysis of classical and newly described staphylococcal superantigenic toxin genes in Staphylococcus aureus isolates. FEMS Microbiol. Lett. 2005;246:191–198. doi: 10.1016/j.femsle.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Omoe K, Ishikawa M, Shimoda Y, Hu DL, Ueda S, Shinagawa K. Detection of seg seh, and sei genes in Staphylococcus aureus isolates and determination of the enterotoxin productivities of S. aureus isolates harboring seg, seh, or sei genes. J. Clin. Microbiol. 2002;40:857–862. doi: 10.1128/JCM.40.3.857-862.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono HK, Omoe K, Imanishi K, Iwakabe Y, Hu DL, Kato H, Saito N, Nakane A, Uchiyama T, Shinagawa K. Identification and characterization of two novel staphylococcal enterotoxins, types S and T. Infect. Immun. 2008;76:4999–5005. doi: 10.1128/IAI.00045-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Fox LK, Seo KS, McGuire MA, Park YH, Rurangirwa FR, Sischo WM, Bohach GA. Comparison of phenotypic and genotypic methods for the speciation of coagulase-negative staphylococcal isolates from bovine intramammary infections. Vet. Microbiol. 2009 doi: 10.1016/j.vetmic.2010.06.020. Submitted to. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher DG, Saunders NA, Owen RJ. Rapid extraction of bacterial genomic DNA with guanidine thiocyanate. Lett. Appl. Microbiol. 1989;8:151–156. [Google Scholar]
- Seo KS, Bohach GA. In: Staphylococcus aureus. Doyle M, Montville T, editors. Washington, D.C: ASM Press; 2007. pp. 493–518. Food Microbiology, Fundamentals and Frontiers. [Google Scholar]
- Seo KS, Lee SU, Park YH, Davis WC, Fox LK, Bohach GA. Long-term staphylococcal enterotoxin C1 exposure induces soluble factor-mediated immunosuppression by bovine CD4+ and CD8+ T cells. Infect. Immun. 2007;75:260–269. doi: 10.1128/IAI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YC, Wong AC. Identification and purification of a new staphylococcal enterotoxin. H. Appl. Environ. Microbiol. 1995;61:1438–1443. doi: 10.1128/aem.61.4.1438-1443.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DY, Jarraud S, Lemercier B, Cozon G, Echasserieau K, Etienne J, Gougeon ML, Lina G, Vandenesch F. Staphylococcal enterotoxin-like toxins U2 and V, two new staphylococcal superantigens arising from recombination within the enterotoxin gene cluster. Infect. Immun. 2006;74:4724–4734. doi: 10.1128/IAI.00132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda C, Barry P, Penades JR, Novick RP. A pathogenicity island replicon in Staphylococcus aureus replicates as an unstable plasmid. Proc. Natl. Acad. Sci. U S A. 2007;104:14182–14188. doi: 10.1073/pnas.0705994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle J, Vadillo S, Piriz S, Gomez-Lucia E. Detection of antibodies to staphylococcal enterotoxins in the serum and milk of healthy goats. FEMS Microbiol. Immunol. 1991;3:53–58. doi: 10.1111/j.1574-6968.1991.tb04163.x. [DOI] [PubMed] [Google Scholar]