Abstract
The role of cholesterol in the etiology of Alzheimer’s disease (AD) is still controversial. Some studies aiming to explore the association between lipids and/or lipid lowering treatment and AD indicate a harmful effect of dyslipidemia and a beneficial effect of statin therapy on AD risk. The findings are supported by genetic linkage and association studies that have clearly identified several genes involved in cholesterol metabolism or transport as AD susceptibility genes, including Apolipoprotein E (APOE), Apolipoprotein J (APOJ, CLU) and the sortilin-related receptor (SORL1). Functional cell biology studies support a critical involvement of lipid raft cholesterol in the modulation of AbetaPP processing by β- and γ-secretase resulting in altered Aβ production. Contradictory evidence comes from epidemiological studies showing no or controversial association between dyslipidemia and AD risk, cell biology studies suggesting that there is little exchange between circulating and brain cholesterol, that increased membrane cholesterol is protective by inhibiting loss of membrane integrity through amyloid cytotoxicity, and that cellular cholesterol inhibits co-localization of BACE1 and AbetaPP in non-raft membrane domains and thereby increasing generation of plasmin, an Aβ-degrading enzyme. The aim of this review is to summarize the findings of epidemiologic and cell biologic studies aiming to elucidate the role of cholesterol in AD etiology.
Keywords: Alzheimer’s disease, cholesterol, Aβ peptides, AbetaPP, neurodegeneration, amyloid, genetics
INTRODUCTION
Alzheimer’s disease (AD) is the most common cause of dementia and the fourth leading cause of death in industrialized nations. About 25 million people worldwide are affected and its incidence and costs are predicted to double every 20 years due to increasing life expectancy.[1,2,3] These figures emphasize the role of AD as a major health problem which will even impose a higher burden in the near future.
Clinical hallmarks of AD are progressive loss of cognitive function in particular the memory domain, ultimately leading to complete dependency and death. The main histopathological changes are extracellular neuritic plaques composed of β-amyloid peptide (Aβ) and intracellular neurofibrillary tangles consisting of an abnormally phosphorylated form of the protein tau.
Extensive research has been done to explore the role of dyslipidemia, one of the most common syndromes in Western societies, in etiology of dementia, but the results are controversial. The aim of this review is to summarize the findings of epidemiologic and cell biological studies aiming to elucidate the role of cholesterol in AD etiology.
DATA COLLECTION
The papers were identified by PubMed (MEDLINE) search, using cholesterol, AD, dementia, statins, lipid-lowering agents, gene and genetics as key words in various combinations. Inclusion criteria: time period between 1985 and 2011, original articles only, English language. All suitable articles were evaluated taking into account their internal validity (e.g. population type, drop-out rate, diagnostic procedure, confounding control, presence of bias, statistical power) and causal criteria (strength of association and temporality). For epidemiological studies, cross-sectional studies were excluded, as they do not allow conclusions regarding causal relationships.
EPIDEMIOLOGICAL STUDIES RELATING PLASMA CHOLESTEROL LEVELS WITH AD
The main longitudinal studies on the association between serum lipid levels and development of AD later in life are summarized in Table 1. Several studies reported on the effect that high levels of cholesterol had on risk.[4,5,6] Four articles reported that high cholesterol had no effect.[7,8,9,10] Others reported that high total cholesterol levels increased risk significantly.[6,11,12] An additional paper from the CAIDE study also looked at high midlife cholesterol effects and reported that a high midlife total cholesterol of greater than or equal to 251 mg/dL increased risk significantly.[5] In contrast, three articles reported an association between higher total cholesterol levels in late life and a reduced risk of dementia.[13,14,15]
Table 1.
Longitudinal observational studies relating lipid levels with risk of dementia
| Study | Cohort | Population size/follow-up time | Diagnostic criteria | Findings |
|---|---|---|---|---|
| Kuusisto et al. (1997)[14] | Kuopio | n = 980 (46 AD) Age range: 66–75 years Follow-up: 3.5 years |
DSM-IIIR | Low total cholesterol associated with increased AD risk |
| Notkola et al. (1998)[11] | Seven Countries Study (Finland) | n = 444 men (47 dementia, 27 AD) Baseline age: 40–59 years Follow-up: 15–25 years |
DSM-IIIR | High total cholesterol associated with increased AD risk |
| Moroney et al. (1999)[26] | WHICAP | n = 987 (126 AD) Mean age: 73 years Follow-up: 2.5 years |
NINCDS-ADRDA | Low total cholesterol associated with increased AD risk |
| Romas et al. (1999)[9] | WHICAP | N=1,449 75.8±6.4 years Cross-sectional study |
NINCDS-ADRDA | No association |
| Kalmijn et al. (2000)[22] | HAAS | n = 3,734 men (251 dementia, 82 AD, 73 VaD) Mean baseline age: 53 years Mean follow-up: 25 years |
DSM-IIIR, NINCDS-ADRDA | Tryglicerides positively associated with AD risk |
| Kivipelto et al. (2001)[5] | CAIDE Study | n = 1,449 (57 dementia, 48 AD) Mean baseline age: 50 years Mean follow-up: 21 years |
DSM-IV, NINCDS-ADRDA | High total cholesterol associated with increased AD risk |
| Tan et al. (2003)[10] | Framingham Study | n =1,026 subjects who had undergone biennial evaluation for cardiovascular risk factors since 1950 and who were alive and free of stroke and dementia at examination cycle 20 (1988–1989); 77 developed AD afterwards | DSM-IV, NINCDS-ADRDA | No association |
| Reitz et al. (2004)[14] | WHICAP | n = 1,168 (119 AD, 54 VaD) Mean age: 78.4 years Follow-up: 4.8 years |
DSM-IIIR, NINCDS-ADRDA | Higher total cholesterol associated with decreased AD risk. Higher LDL-C and non-HDL-C associated with increased VaD risk |
| Dufouil et al. (2005)[30] | Three-City Study | n=9,294 Age: 74.2±5.5 Cross-sectional analyses |
DSM-IV, NINCDS-ADRDA | Hyperlipidemia associated with increased risk of non-AD dementia, lipid- lowering treatment associated with decreased dementia risk |
| Whitmer et al. (2005)[12] | Kaiser Permanent Medical Care Program of Northern California) |
n = 8,845 (721 dementia) Baseline age: 40–44 years Mean follow-up: 27 years |
ICD9CM | Positive association |
| Li et al. (2005)[7] | ACT Study | n = 2,141 (152 AD) Mean age: 74.9 years Follow-up: 5.6 years |
DSM-IV, NINCDS-ADRDA | No association |
| Mainous et al (2005)[8] | NHEFS | n=6,558 Age: 40–74 years at baseline Follow-up: 20 years |
ICD-9 | No association |
| Mielke et al (2005)[13] | Göteborg | n=382 (93 dementia) Baseline Age: 70 years Follow-up: 18 years |
DSM-IIIR, NINCDS-ADRDA | High total cholesterol levels associated with reduced risk of dementia |
| Kivipelto et al. (2005)[6] | Caide Study | n = 1,449 (61 dementia, 48 AD) Mean baseline age: 50 years Mean follow-up: 21 years |
DSM-IV, NINCDS-ADRDA | High total cholesterol associated with increased AD risk |
| Hayden et al (2006)[4] | Cache County Study | n=3,264 (185 dementia) Mean Age: 73.7 Follow-up: 3.2 years |
DSM-IIIR, NINCDS-ADRDA | High total cholesterol associated with increased AD risk |
| Stewart (2007)[16] | HAAS | 1027 Japanese American men (56 AD) Mean Age: 80.2 Follow-up: 26 years |
DSM-IIIR, NINCDS-ADRDA | decline in cholesterol levels in men at least 15 years before dementia diagnosis |
| Reitz et al (2010)[19] | WHICAP | n=1,130 (101 AD) Mean Age: 75.7 Follow-up: 4 years |
DSM-IIIR, NINCDS-ADRDA | High HDL levels associated with reduced AD risk |
| Beydoun et al (2010)[17] | BLSA | n=1,604 (259 dementia, 182 AD) Mean Age: 57.6 Follow-up: 25 years |
DSM-IIIR, NINCDS-ADRDA | decline in total cholesterol associated with increased dementia risk |
| Solfrizzi et al (2010)[20] | ILSA | n=2,097 Age: 72.9±5.6 Follow-up: 3.5 years |
DSM-IIIR, NINCDS-ADRDA | Metabolic syndrome associated with higher risk of VAD |
| Reynolds et al. (2010)[21] | Swedish Adoption Twin Study of Aging | N=819 twins Age: 50–96 years Follow-up: 16 years |
DSM-IV, NINCDS-ADRDA | In women but not men, higher HDL-C and lower triglyceride levels predict better maintenance of cognitive abilities |
Five articles from 4 studies reported on the affect interactions between cholesterol and other risk factors have on risk for AD. One paper from the NHEFS examined the interaction between high total cholesterol and high transferrin saturation and reported that the interaction between cholesterol greater than 261 mg/dL and transferrin saturation greater than 34.9% did not affect risk, whereas cholesterol greater than 268 mg/dL in combination with a transferrin saturation greater than 37% increased risk.[8] A paper from the CAIDE study reported an interaction between high midlife serum cholesterol greater than or equal to 251 mg/dL and high midlife systolic blood pressure (>=160 mm Hg) that significantly increased risk.[5] Two articles, from the CAIDE and ACT studies, reported no interaction between total cholesterol and APOEε4.[5,7] Others reported a significant decline in cholesterol levels before onset of dementia.[16,17] Studies examining the association between high-density-lipoprotein (HDL-C) levels, low-density lipoprotein (LDL-C) levels or triglyceride levels and risk of dementia also remained controversial, although some reported associations between low HDL-C levels or increased LDL-C or triglyceride levels and an increased risk of dementia or cognitive decline.[18,19,20,21,22,23,24,25,26] Of note, low HDL-C levels and high triglyceride levels also occur as part of the metabolic syndrome, therefore these findings would be in line with the hypothesis of an association between metabolic syndrome and a higher risk of dementia[20]. Studies exploring the effect of triglyceride levels on dementia risk were also inconclusive.
STATIN TREATMENT AND ALZHEIMER’S DISEASE
The main effect of statins is to lower low-density lipoprotein cholesterol (LDL-C) by blocking cholesterol synthesis in the liver. As a group, statins are thought to prevent cardiovascular disease by improving endothelial function, reducing inflammatory responses, maintaining stability of atherosclerotic plaques, and preventing the formation of thrombi,[27] even in those with normal cholesterol levels. These pleiotropic effects of statins have led to interest in using them to treat noncardiovascular conditions including AD. Several well-performed, population-based observational studies[28,29,30,31,32] show a protective effect of statin therapy on the risk of AD and dementia, even after adjustment for demographic and vascular confounders. The results from these five studies are quite consistent, ranging from a 38% to 43% lower risk of AD or dementia with an average follow up time of 5 to 6 years. However, in contrast, several controlled randomized trials exploring the effect of statin on progression of AD have found no benefit.[33,34,35,36,37,38] One explanation for the dramatic differences between observational studies and clinical trials may be that treatment trials tend to be short term and include patients with advanced disease who are typically older. Observational cohort studies normally include a wider age range of people with normal cognition to more-advanced disease and are usually longer in duration. Li and colleagues report greater benefits from statin therapy for those younger than 80, as did a previous study.[28,39] Consistent with these findings, in one of the smaller trials of atorvastatin, the authors noted that an earlier stage of progression, higher starting cholesterol, and APOE 34 status could modify the effects of statin on change in cognitive function in patients with AD.[37]
Another issue emerging from clinical trials is confounding by indication (indication bias) meaning that the indication for a drug being prescribed or not prescribed confounds the relationship between the drug and the disease. People with dementia are likely to be denied statins, reflecting a trend to be less aggressive in their medical care including the treatment of vascular risk factors.
CHOLESTEROL AND ALZHEIMER’S DISEASE
One indication that lipids may play an important role in AbetaPP processing and Aβ production are given by the common feature that all proteins involved in AbetaPP processing are integral membrane proteins. Moreover, the Aβ producing γ-secretase cleavage takes place in the middle of the membrane suggesting that the lipid environment of the cleavage enzymes influences Aβ production and hence AD pathogenesis.[40]
The brain contains ~25–30% of the total body cholesterol and is thus the cholesterol-richest organ in the body. Cholesterol is an essential component of cell membranes and plays a crucial role in the development and maintenance of neuronal plasticity and function.[41]
Biosynthesis
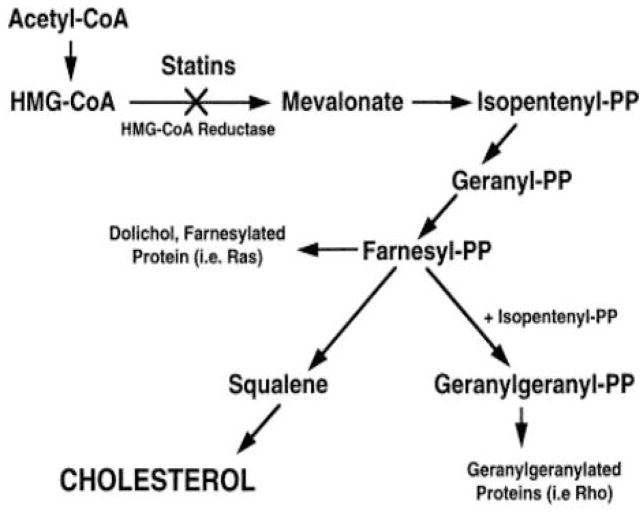
The cholesterol homoeostasis in the brain is regulated by several processes that include synthesis, storage, transport, and removal. While glial cells and neurons can synthesize cholesterol de novo, cholesterol can also be recycled from extracellular locations within the CNS. However, essentially all the cholesterol used in the brain is synthesized within the CNS. The biosynthesis of cholesterol is a lengthy process requiring more than 20 reactions and intermediates (Figure 1). The rate limiting step of cholesterol biosynthesis is the catalysation of the formation of mevalonate by 3-hydroxy-3-methylglutaryl-CoA reductase (HMG-CoAR), this step is therefore the prominent pharmacological target. However, interruption at this step by statins may decrease the generation of downstream intermediate products with important metabolic functions such as isoprenoids (including farnesyl phosphates) which are important precursors for molecules involved in cell signalling and inflammatory responses.[42]
Fig. 1.
Cholesterol Synthesis
Transport and Storage
In the CNS, cholesterol is transported and stored through several pathways. Astrocytes synthesise cholesterol and in addition recycle cholesterol that has been released from degenerating nerve terminals, or excess cholesterol which transport out of cells is mediated by ABCA1, a membrane protein that facilitates the formation of APOE–cholesterol–phospholipid (ApoE-Chol-PL) complex and requires hydrolysis of ATP for its activity.[43] APOE binds this synthesized or recycled cholesterol and acts as a ligand for cell-surface-lipoprotein receptors, such as LDL-receptor-related proteins (LRP). In addition, it regulates lipid transport into neurons and clearing cholesterol from the extracellular matrix. The CNS contains the second greatest concentration of APOE mRNA after the liver, making APOE the most prevalent lipoprotein in the CNS.[44] Neurons and glial cells contain large endocytic membranic LRP-receptors which bind and internalise LDL.[45] The endosome that results from the internalised APOE–cholesterol–phospholipid complex then fuses with lysosomes, which contain the necessary hydrolytic enzymes to allow the intracellular release of free cholesterol. The free cholesterol released intracellularly from the lipoprotein complex provides negative feedback to HMG-CoAR to reduce endogenous synthesis of cholesterol, while esterification by acyl-coenzyme-A cholesterol acyltransferase (ACAT) allows for more efficient storage.[45] This intracellular pool of cholesterol serves as the source for synaptic and dendritic formation and remodelling.[41]
Efflux of cholesterol from CNS
The blood–brain barrier (BBB) prevents direct transport of cholesterol between peripheral circulation and the CNS. Due to neurodegeneration, in AD there is increased release of cholesterol from degenerating neurons and synapses, as well as from impaired transport of cholesterol in APOEε4 carriers.[46,47] A high cholesterol environment may lead to low cytofacial to exofacial cholesterol ratio, and favours activity of β- and γ-secretases, which increases Aβ production from amyloid-beta protein precursor (AbetaPP). Because there is no degradation mechanism for cholesterol, excess cholesterol must be transported from the brain into the circulation. APOE-dependent mechanisms involving HDL-like lipoprotein allow the clearance of 1–2 mg of cholesterol from the CNS each day but 6–7 mg of cholesterol are eliminated from the brain in the form of 24S-hydroxylcholesterol, which is lipophilic and can freely cross the blood–brain barrier.[48] Over 90% of circulating 24S-hydroxycholesterol originates in the brain. Consequently, blood concentrations of 24S-hydroxycholesterol reflect CNS cholesterol turnover.[49] In addition to cholesterol-24 hydroxylase-associated mechanisms, the efflux of cholesterol from the CNS might also be mediated by members of the superfamily of ATP-binding cassette membrane transport proteins (ABCA). These proteins have a channel-like topology and are able to transport various solutes—including ions, drugs, peptides, proteins, sugars, and lipids—across cell membranes by the coupling the transport to ATP hydrolysis.
ROLE OF CHOLESTEROL IN AD PATHOGENESIS
Aβ production
Cholesterol modulates the activity of the enzymes involved in AbetaPP processing and the production of Aβ. The cleavage of AbetaPP by α-secretase results in non-amyloidogenic or soluble AbetaPP. However, cleavage by the membrane associated β- and γ-secretases results in amyloidogenic forms that aggregate as extracellular plaques In animal studies, dietary cholesterol accelerates Aβ deposition in the brain whereas cholesterol-lowering drugs lower it.[50,51] Other in-vitro studies have shown that a high cholesterol environment results in reduced production of soluble amyloid precursor protein.[52,53] The mechanism by which cholesterol affects Aβ production and metabolism remains unclear. It is possible that a change in membrane properties, including stiffness and fluidity, modulates the activities of membrane-bound enzymes such as the secretases. The high cholesterol content in lipid rafts, the membrane regions where these enzymes are located, seems to facilitate the clustering of the β and γ-secretases with their substrates, thereby promoting the pathogenetic cleavage of amyloid precursor protein into amyloidogenic forms.[54] However, the significance of this in humans is unknown.
Neurofibrillary tangles and phosphorylation of tau
The role of cholesterol homoeostasis in tau phosphorylation and formation of neurofibrillary tangles also remains unclear. The injection of Aβ42 into rat cortices caused a significant increase in hyperphosphorylation of tau protein.[55] Because membrane cholesterol modulates Aβ fibrillogenesis and upregulates Aβ42 formation, it is has been suggested that excess membrane cholesterol may indirectly promote production of neurofibrillary tangles.[56] The notion underlying this hypothesis is, that neurofibrillary tangles (NFTs) are composed primarily of hyperphosphorylated tau, and that Aβ can induce tau phosphorylation.
Cerebrovascular Disease
Another pathway through which dyslipidemia may influence the risk of AD is cerebrovascular disease. It is clear that both cerebral micro- and macrovascular disease increases AD risk. However, whether dyslipidemia in fact affects risk of cerebrovascular changes remains controversial.
Exocytosis, and apoptosis
Finally, there is evidence that high concentrations of oxysterols are able to provoke neuronal apoptosis and exocytosis.[57,58] Cholesterol oxidation product concentrations increase as a consequence to neuroinflammation associated with brain injury. However, they in turn enhance exocytosis and neurotransmitter release, thereby aggravating excitotoxicity.[58] Thus, alterations in levels and metabolism of cholesterol can be seen as both initiating point and consequence in neurodegeneration.
It is important to emphasize that alterations in cholesterol metabolism are not specific for AD. Several other neurodegenerative diseases also show changes in cholesterol and its metabolites in the brain, including Parkinson’s disease, Huntington’s disease, Niemann-Pick type C, and multiple sclerosis.
GENES INVOLVED IN CHOLESTEROL METABOLISM AND RISK OF AD
Only ~2.9% (666 out of ~ 23,000) of all genes have been investigated for association with AD in hypothesis-driven approaches so far. Interestingly, among these the cholesterol metabolic pathway is highly represented as ~40% of the cholesterol-related genes have been investigated. Promising, robust associations have been reported in particular for five genes (APOE, CLU, SORL1, LDLR, CH25H). Most other genes have not been replicated consistently. However, it is important to keep in mind that for most genes the investigated markers do not capture the entire genetic variation, and that the sample size of most studies performed was clearly too small to detect loci with modest effect sizes.
APOE
The Apolipoprotein E (APOE) gene is 3.6 kb long and located on chromosome 19. It is a 299 amino-acid protein with three common isoforms. APOEε4 differs respectively from APOEε3 and APOEε2 by having arginine residues instead of cysteine at 112 and 158. The amino-acid substitutions have a critical role in determining the three-dimensional structure of APOE leading to changes with its protein binding properties. In the circulation ApoE is a major constituent of chylomicrons. It is the most abundant apolipoprotein in the CNS where it occurs in HDL particles and is involved in cholesterol transfer from the astroglial to the neuronal compartment.[59,60] In addition, APOE act as a pathological chaperone of Aβ, promoting its fibril formation from soluble Aβ by binding interaction between carboxy-terminal domain of apoE and residues 12–28 of full-length Aβ. It has been firmly implicated as the top-ranked susceptibility for AD. In general, the ε2 isoform is associated with lower plasma cholesterol and lower risk of AD, while the ε4 allele is associated with higher plasma concentrations of total and LDL cholesterol, a higher risk of atherosclerosis and higher risk of AD.[61,62] The risks of AD are three and eight times greater in individuals with one or two copies of the ε4 gene respectively, compared with people homozygous for ε3.[63] In most studies, 40–50% of patients with AD have at least one ε4 allele, compared with 10–15% of healthy controls.[63,64,65] Individuals who are homozygous for the ε4 allele and live to age 80 years will almost invariably develop AD, but about 10% of heterozygous ε4 carriers will remain free of AD well into their 80s.[63,66] In addition, the ε4 allele lowers the age at onset of dementia in a gene-dose-dependent manner by as much as 7–9 years per allele,[65] and is associated with AD endophenotypes including disease progression, brain hypometabolism, amyloid deposition in the parenchyma and vasculature (CAA), intraneuronal accumulation of Aβ, and low Aβ42 in CSF.[67,68,69,70] In addition to the APOE ε2/3/4 haplotype a promoter polymorphism is independently associated with AD risk, indicating that not only qualitative differences between the three isoforms but also quantitative variability of APOE levels may modulate the risk for AD.[71]
CLU
Clusterin (CLU), which is located on chromosome 8p21-p12, is 36.3 kb long and encodes the second major Apolipoprotein in the CNS (Apolipoprotein J). In the brain it is predominantly expressed by astrocytes. As of March 1, 2011, CLU was ranked the second strongest susceptibility gene for late-onset AD (www.alzgene.org), based on significant association of genetic variation in this gene and AD in several of the major GWAS performed to date (www.alzgene.org).[72,73] CLU has several functional similarities with apolipoprotein E. It is involved in reverse cholesterol transport as a constituent of HDL particles,[74] and additive to APOE, it also acts as an Aβ chaperone, regulating the conversion of Aβ to insoluble forms as well as its toxicity thereby promoting amyloid plaque formation.[75] Moreover, it may be involved in the transport of Aβ across the blood brain barrier and in its uptake by glial cells and brain macrophages.[76] These effects are influenced by the molar ratios of APOJ and Aβ and by the aggregation state of the latter.[77,78]
SORL1
The SORL1 gene is 177.5 kb long and maps to 11q23.2–q24.2. The encoded sortilin-related receptor containing LDLR class A repeats (SorLA-1) is a multifunctional receptor that binds lipoproteins including APOE-containing particles and mediates their intracellular trafficking. SORL1 belongs to the family of VPS10 receptors, a group of five type I membrane homologues (SORL1, SORT1, SorCS1, SorCS2, SorCS3), that are all characterized by a luminal, extracellular VPS10 domain and are strongly expressed in the central nervous system. In the original study by Rogaeva et al, that included >6,000 subjects from 4 different ethnic groups two clusters of SNPs in SORL1 were identified that are associated with familial and sporadic forms of AD in various ethnic groups including Caucasians, African Americans, Caribbean Hispanics Israeli-Arabs. That study also demonstrated through functional cell biology analyses that SORL1 modulates trafficking of AbetaPP from the cell surface to the Golgi endoplasmic reticulum complex, and that underexpression of SORL1 leads to overexpression of Aβ and an increased risk of AD.[79] In various subsequent studies derived from different ethnic groups the associations of clusters of SNPs in the same two SORL1 regions with AD were replicated.[80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97] The haplotypes were further validated by a collaborative, unbiased meta-analysis of all published Caucasian and Asian datasets (12,464 cases, 17,929 controls; 0.7<OR <1.2, p≤0.001).[98] Finally, the same alleles have also been associated with various AD endophenotypes including age-of-onset of AD, abstract reasoning, white matter lesions, hippocampal atrophy, Aβ42 CSF measures and full-length-SORL1 expression in brain, further validating this gene in AD.[98,99,100,101] Of note, in 2011, also the SORL1 homologue SORCS1 was firmly implicated as a risk gene for AD by demonstrating genetic association of SORCS1 and AD, and by showing that also SORCS1 alters processing of AbetaPP and thereby accumulation of Aβ.[102] Currently, SORL1 is ranked on position 7 in the AlzGene database.
LDLR
The LDLR gene, currently ranked on position 11 in the AlzGene database, is located on chromosome 19p13.3 and comprises 44 kb. Its gene product, the low density lipoprotein receptor is a major APOE receptor in the brain. Mutations in this gene cause autosomal dominant familial hypercholesterolemia. Mouse experiments show that LDLR has a beneficial effect on AD pathology via enhancement of Aβ clearance.[103] There are several small genetic association studies with controversial results.[104,105,106,107,108] However, a small meta-analysis in the AlzGene database on the exon 10 polymorphism rs5930 is positive with an OR of 0.85 (95% CI 0.72–0.99) for the rare A allele.
CH25H
CH25H is a small 1.6 kb long 686 bases-long, intronless gene located at 10q23. As described above, the gene product cholesterol 25-hydroxylase catalyses the formation of 25-hydroxycholesterol from cholesterol. Association between genetic variants in CH25H and AD have been explored in 10 case-control studies and 2 family-based studies. Out of these, two case-controls studies, which were overlapping, were positive.[109,110] In addition, the AlzGene meta-analysis of SNP rs13500 with about 2700 individuals is positive (www.alzgene.org). Based on these data, this gene is ranked on position 28 in the AlzGene database. However, it has to be noted that there are several negative studies on CH25H that have not been included in the meta-analysis.
CETP
The Cholesteryl ester transfer protein (CETP) is a key player in lipid metabolism that catalyses the transfer of cholesteryl esters from HDL particles to triglyceride-rich lipoproteins in exchange for triglycerides. CETP, a protein composed of 439 amino acid residues, is coded by the CETP gene, which is located on chromosome 16q21 and contains 14 exons. The 405V allele of the CETP I405V polymorphism (rs5882) in exon 14 was previously associated with lower levels of CETP protein, higher levels of circulating HDL,[111] a lower incidence of cardiovascular disease,[111] and longer survival.[112] Ten case-control studies have evaluated the role of the CETP gene in AD. Out of these, two reported an association,[113,114] while eight studies were negative.
ABCA1
The ABCA1 gene is 147kb long and is located at 9q31.1, a chromosomal region linked to AD in several family-based whole genome scans. As described above, it encodes the ATP-binding cassette transporter A1 a central regulator of reverse cholesterol transport. ABCA1 mediates efflux of cellular cholesterol to lipid-poor HDL particles. Loss-of-function mutations in ABCA1 cause Tangier disease, characterized by low HDL cholesterol levels leading to arteriosclerosis and premature coronary artery disease. A potential role of ABCA1 in AD is supported by some experiments in which ABCA1 deficient mice were cross-bred with amyloid-beta precursor protein- (AbetaPP)-transgenic mice modelling the amyloid pathology of AD. These double-mutant mice exhibit higher amyloid load in their brains compared to AbetaPP -transgenic mice with physiological ABCA1 levels.[115,116] As ABCA1 is a regulator of apolipoprotein E (ApoE) levels and lipidation and of the ApoE-mediated transfer of cholesterol from the glial to the neuronal compartment potential ABCA1 effects on AD may be mediated by mechanisms involving ApoE.[117]Variants in ABCA1 have been associated with AD risk and AD endophenotypes including age of onset and amyloid deposition in the brain in case-control studies[118,119,120] and several family-based studies[121,122,123] derived from different ethnic groups. However, there have been also several negative studies. All in all more than 60 single nucleotide polymorphisms (SNPs) have been investigated in up to about 5000 individuals. The most extensively investigated ABCA1 variant is the rs2230806 SNP that predicts a R219K amino acid exchange. Both protective and risk effects have been observed for the A allele of this polymorphism and the AlzGene meta-analyses of studies investigating the association of this and three other variants with AD risk are negative.
ABCA2
The gene encoding the ATP-binding cassette transporter A2 (ABCA2) is 21kb long, is located on chromosome 9q34, and is expressed predominantly in brain regions prone to develop AD pathology, specifically in oligodendrocytes. Simmilar to ABCA1 it may regulate intracellular lipid transport and plays a part in myelination. Out of three case-control studies that explored the association between variation in ABCA2 and AD, two reported associations of the rare T allele of rs908832 with AD,[124,125] while one was study did not find an association.[126]
ABCG1
The ABCG1 gene is 97kb long and maps to 21q22.3. The ATP-binding cassette transporter G1 is a half-transporter that either homo- or hetero-dimerizes with ABCG4 to form a functional transporter. It mediates cholesterol efflux to lipidated lipoprotein complexes including ApoE discs. In the CNS ABCG1 is broadly expressed in neuronal, glial, and microglial cells, predominantly in the hippocampus. Evidence for an effect of the genetic risk for AD is sparse. Association of ABCG1 variants with AD was observed in two samples in one study.[127] Partly conflicting effects of ABCG1 on AbetaPP metabolism have been observed in vitro.[128,129] In vivo over-expression of ABCG1 in a transgenic mouse model of AD did not alter amyloid pathology.[130]
APOA1
The apolipoprotein A-1 (APOA1) gene is with 1,9 kb small and is located at 11q23-q24. Apolipoprotein A-I is mainly expressed in the liver but is also present in the CNS. It is the major component of plasma HDL particles and mediates reverse cholesterol transport to the liver. Only three studies have explored genetic associations between these genes and AD, out of which one reported an asspociation between the A allele of the APOA1 -75bp G/A polymorphism and an increased risk for AD in subjects with an age at onset of 66 years or younger.[131] However, this association was not replicated. APOA1 inhibits the aggregation and toxicity of Aβ in vitro.[132] However, amyloid pathology is unaltered in APOA1-deficient AbetaPP-transgenic mice.[133]
APOA4
The APOA4 gene is 2,5kb long and located on the cytogenetic band 11q23. Apolipoprotein A-IV is primarily expressed in the intestine and is a component of chylomicrons and HDL particles. Genetic association of APOA4 has been observed, but the four association studies that have been performed were contradictory.[110,134,135,136]
APOC1
The APOC1 gene is 5028 bases long and is located on chromosome 19q13.2 about 5000 bases downstream the APOE locus. Its gene product the apolipoprotein C-I is primarily expressed in the liver. As part of the APOE/APOC1/APOC4/APOC2 cluster it has been extensively investigated for association with AD (www.alzgene.org). Most studies were positive and the AlzGene meta-analysis of the most extensively studied rs11568822 (ins/del) polymorphism shows a robust association of the deletion allele with an increased AD risk with an OR of 2.07 (95% CI 1.67–2.57). It is possible that this association may be explained by linkage disequilibrium (LD) with the APOE locus (http://hapmap.ncbi.nlm.nih.gov). However, it cannot be excluded, that more loci than APOE alone, including APOC1 may contribute to the strong connection of this chromosomal region with AD.
APOC2
APOC2 is 7.3 kb long and is located on chromosome 19q13.2 as part of the APOE/APOC1/APOC4/APOC2 cluster. A study showing genetic association of APOC2 with AD is the first that links chromosome 19q13.2 with the disease and, given that this association may be owing to LD with APOE, may be the first to catch a glimpse of the one strong genetic risk factor for AD.[137] Subsequent studies on APOC2 yielded positive and negative results.
APOC3
The APOC3 gene is 3.2 kb long and resides on chromosome 11q23.1-q23.2 closely linked to the APOA1 and the APOA4 gene. There is no study showing a significant association.
CAV1
The CAV1 gene is 36,4 kb long and is located at 7q31.1. Cavelolin 1 is the main proteinacious component of the cholesterol enriched caveola membrane domains. In the only genetic association study that has been published to date, haplotypes of a polymorphic purin complex about 1.5 kb upstream the CAV1 gene were associated with AD.[138] Support for this comes from studies that have observed regulatory effects of caveolin 1 on AbetaPP processing as well as increased caveolin expression in the brains of AD patients have been described.[139,140]
CD36
The CD36 gene is 74,8 kb long and resides on 7q11.2. Its gene product is a scavenger receptor with multiple functions including the uptake of oxidized LDL particles. Genetic association with AD was described in one study[141] but not confirmed in two replication studies. CD36 is involved in microglial binding of fibrillar Aβ and consecutive activation of an innate immune response.[142,143]
CYP46A1
The product of the CYP46A1 gene, which is 43 kb long and is located at 14q32, is the cholesterol 24-hydroxylase, which converts cholesterol to 24-hydroxycholesterol. As described above, unlike cholesterol, 24-hydroxycholesterol can pass the blood brain barrier. Thus, conversion to 24-hydroxycholesterol is the major mechanism of cholesterol elimination from the CNS. Recently published data from mouse experiments support a role of CYP46A1 in AD, in which neuronal over-expression of CYP46A1 by means of an adeno-associated vector injection before or after the onset of amyloid plaques formation reduced Aβ pathology in two mouse models of AD.[144] Genetic association studies relating variants in the gene with AD were conflicting.[145,146,147,148]
DHCR24
The DHCR24 gene is 37.6 kb long and resides on chromosome 1p33-p31.1. It was first described as the selective AD indicator 1 (seladin 1) as it is downregulated in brain regions vulnerable to AD pathology in AD patients.[149,150] Its gene product is an oxidoreductase that catalyzes delta-24 double-bond reduction in sterol intermediates in the biosynthesis of cholesterol. It has a protective effect against oxidative stress and apoptosis induced e.g. by Aβ via reduction of caspase 3 activity. Moreover, it counteracts the β-secretase cleavage of AbetaPP and the formation of β.[150] In a study involving about 1000 individuals and four SNPs rs600491 and haplotypes involving this SNP were associated with AD.[150] However, this association has not been confirmed as yet.
HMGCR
The HMGCR gene is 24.8 kb long and lies on 5q13.3-q14. The HMG-CoA reductase is the rate-limiting enzyme for the biosynthesis of cholesterol and is the target of “statin” inhibitors of cholesterol synthesis. As described above, some clinical trials reported a protective effect of statin treatment on AD risk.[28,29,30,31,32] Association of the – 911 rs3761740 with AD was described in one study, [151] and a second study reported an interactive effect of HMGCR and ABCA1 on AD risk.[152] However, these findings were not replicated. Although statins have pleiotropic effects beyond HMG-CoA reductase inhibition all studies showing effects of statins on AD pathology may be quoted as functional support for a role of HMGCR in AD.
LIPC
The LIPC gene is 136 kb long and resides on the cytogenetic band 15q21–q23. LIPC encodes hepatic triglyceride lipase, which is expressed in liver. LIPC acts as a triglyceride hydrolase and as is involved in receptor-mediated lipoprotein uptake. There was a positive finding in one sub-sample of the initial study but all replication studies including one with fine mapping of the locus with 25 SNPs were negative.[153]
LPL
The LPL gene is 28kb long and is located at 8p22. LPL encodes lipoprotein lipase. The enzyme is predominantly expressed in heart, muscle, and adipose tissue. LPL forms a homodimer that has triglyceride hydrolase activity and contributes to receptor-mediated lipoprotein uptake. Mutation-caused LPL deficiency result in type I hyperlipoproteinemia and various disorders of lipoprotein metabolism are associated with LPL. Out of 8 genetic association studies that have been performed, four were positive.[110,154,155,156]
LRP1
The LRP1 gene is 84,9 kb long and is located at 12q13-q14. It encodes the low density lipoprotein-related protein 1, which is involved in cellular lipid homeostasis and contributes to the clearance of chylomicrons from the plasma. It serves as a receptor for ApoE. LRP1 has been extensively investigated for genetic association with AD. Most studies assessed only one or two variants in the gene, and yielded both positive and negative findings (www.alzgene.org). Moreover, a recent study with a haplotype approach which investigated the candidacy of 10 LRP1 SNPs in AD reported negative results.[157] The AlzGene meta-analysis for LRP1 (rs1799986), which comprises almost 15,000 individuals, is negative (www.alzgene.org). LRP1 mediates clearance of Aβ from the brain into the circulation at the blood brain barrier.[158] This process seems to be modulated by ApoE in an isoform-dependent manner. LRP1 is also involved in the hepatic clearance of Aβ. A soluble form of LRP1 is a major Aβ-binding protein in plasma and may influence the distribution of Aβ between brain and blood.[159] Moreover, LRP1 seems to regulate the transport of AbetaPP.[160] AbetaPP on its part influences ApoE levels and cholesterol metabolism in the brain by mechanisms involving LRP1.[161]
NPC2
NPC2 is 13,442 bases long and maps to 14q24.3. The gene product may be involved in the regulation of cholesterol transport through the late endosomal/lysosomal system. NPC2 mutations are associated with Niemann–Pick disease, type C2 and with frontal lobe atrophy. A SNP of NPC2 was positive in one small sub-sample of a study comprising samples from several European centres.[127] The association could not be confirmed in the other sub-samples in this study.
SOAT1
SOAT1 is 61.4 kb long and is located on chromosome 1q25. The gene product acyl-coenzyme A: cholesterol acyltransferase (ACAT1) is located in the endoplasmic reticulum and catalyses the formation of cholesterol esters. Two SNPs were found to be associated with AD risk, brain amyloid load, and cholesterol levels in CSF in two studies,[108,162] while 7 studies did not find an association. There is substantial experimental evidence, that ACAT1 deficiency reduces the generation of Aβ in vitro and in vivo and promotes the proteolytic cleavage of AbetaPP at a recently described Glu281 cleavage site.[163,164,165]
SREBF1
SREBF1 is 24.9 kb long and is located at 17p11.2. It encodes the sterol regulatory element binding transcription factor 1, which is a transcriptional activator with a role in lipid homeostasis. Amongst others it regulates the transcription of the LDL receptor gene and of genes involved in the cholesterol synthesis pathway. Association of one SREBF1 polymorphism with AD has been described in a medium size sample.[166] No replication studies have been performed.
VLDLR
The VLDLR gene comprises 42,693 bases on chromosome 9p24. The gene product, the very low density lipoprotein receptor binds lipoproteins including ApoE-containing particles. Unlike other ApoE receptors the very low density lipoprotein receptor binds ApoE independently of its lipidation status. Out of 14 genetic association studies performed, six were positive[167,168,169,170,171,172].
Support of an association between genes involved in lipid metabolism and AD comes in addition from a study that mined the two largest GWAS datasets to date for biologically meaningful information (ie. enrichment of significant genes in functional pathway categories) using ALIGATOR analysis. In this study, the two main themes that emerged were processes related to sterol and lipid metabolism and the immune response including as cholesterol transport, plasma lipoprotein remodeling, protein-lipid complex remodeling, cholesterol hemostasis and cholesterol metabolism.[173] Of note, some of the genes identified in this review are not expressed in the brain, including hepatic lipase (LIPC), apolipoprotein A1 (APOA1), and endothelial lipase (LIPG), but are important in the systemic control of sterol metabolism in the liver and blood. It is possible that they represent systemic biomarkers of disease progress.
DISCUSSION
The current evidence supports a possible involvement of lipid levels in the development of dementia and AD, suggesting dyslipidemia as one of the modifiable risk factors to be targeted by therapeutic interventions which are already widely available.
However, it has to be emphasized that the mechanisms involved are still unclear. First, while it is clear that dyslipidemia induces vascular disease, also alterations in brain cholesterol homeostasis have been linked to the main pathological features of AD in particular Aβ. Brain cholesterol and serum cholesterol, however, represent separate pools whose interaction has not been fully characterized. Second, dyslipidemia could be related to dementia risk through being a component of the metabolic syndrome (MetS). The MetS is a multifactorial disorder represented by the co-occurrence of several vascular conditions related to central obesity that also includes impaired glucose metabolism, dyslipidemia, and high BP. Each individual factor and MetS as a whole, have been repeatedly associated with cognitive decline and dementia.[174,175] [176,177,178] Third, the available data relating the various lipid types (LDL-C, HDL-C, triglycerides) with AD is still insufficient. The fact that only HDL-like lipoproteins are found in CSF and that HDL levels are lowest in APOE 4 carriers and/or persons with AD may indicate that this lipoprotein is particularly important. However, these notes need to be confirmed. Fourth, association and linkage studies exploring the role of cholesterol-metabolism-related genes in AD have clearly implicated APOE, CLU and SORL1, but remain inconsistent for the other choleseterol –related genes. While studying the association of genetic variation with a disease is invaluable as it avoids reverse causation by taking advantage of Mendelian Randomization, most studies that have been performed were underpowered and/or did not cover the complete genetic variation in the gene. Finally, it is clear that AD is a complex disease with multiple risk factors involved. In order to properly characterize the association of different risk factors with dementia/AD and to pinpoint the critical etiological pathways in which they are involved, early identification of the disease process (before its clinical expression) and long-term studies with multiple time points are needed. Monotherapy is not likely to be sufficiently effective in a complex disease and a more detailed risk profile can provide clues for a better multi-targeted interventional strategy.
Acknowledgments
Funding
This study was supported by grants AG07232 and AG07702 from the National Institute on Aging (Washington, DC), the Charles S. Robertson Memorial Gift for Research in Alzheimer’s disease, the Blanchette Hooker Rockefeller Foundation, and a Paul B. Beeson Career Development Award (K23AG034550).
References
- 1.Brookmeyer R, Gray S, Kawas C. Projections of Alzheimer’s disease in the United States and the public health impact of delaying disease onset. Am J Public Health. 1998;88:1337–1342. doi: 10.2105/ajph.88.9.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.von Strauss E, Viitanen M, De Ronchi D, Winblad B, Fratiglioni L. Aging and the occurrence of dementia: findings from a population-based cohort with a large sample of nonagenarians. Arch Neurol. 1999;56:587–592. doi: 10.1001/archneur.56.5.587. [DOI] [PubMed] [Google Scholar]
- 3.Ferri CP, Prince M, Brayne C, Brodaty H, Fratiglioni L, Ganguli M, Hall K, Hasegawa K, Hendrie H, Huang Y, Jorm A, Mathers C, Menezes PR, Rimmer E, Scazufca M. Global prevalence of dementia: a Delphi consensus study. Lancet. 2005;366:2112–2117. doi: 10.1016/S0140-6736(05)67889-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayden KM, Zandi PP, Lyketsos CG, Khachaturian AS, Bastian LA, Charoonruk G, Tschanz JT, Norton MC, Pieper CF, Munger RG, Breitner JC, Welsh-Bohmer KA. Vascular risk factors for incident Alzheimer disease and vascular dementia: the Cache County study. Alzheimer Dis Assoc Disord. 2006;20:93–100. doi: 10.1097/01.wad.0000213814.43047.86. [DOI] [PubMed] [Google Scholar]
- 5.Kivipelto M, Helkala EL, Laakso MP, Hanninen T, Hallikainen M, Alhainen K, Soininen H, Tuomilehto J, Nissinen A. Midlife vascular risk factors and Alzheimer’s disease in later life: longitudinal, population based study. BMJ. 2001;322:1447–1451. doi: 10.1136/bmj.322.7300.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kivipelto M, Ngandu T, Fratiglioni L, Viitanen M, Kareholt I, Winblad B, Helkala EL, Tuomilehto J, Soininen H, Nissinen A. Obesity and vascular risk factors at midlife and the risk of dementia and Alzheimer disease. Arch Neurol. 2005;62:1556–1560. doi: 10.1001/archneur.62.10.1556. [DOI] [PubMed] [Google Scholar]
- 7.Li G, Shofer JB, Kukull WA, Peskind ER, Tsuang DW, Breitner JC, McCormick W, Bowen JD, Teri L, Schellenberg GD, Larson EB. Serum cholesterol and risk of Alzheimer disease: a community-based cohort study. Neurology. 2005;65:1045–1050. doi: 10.1212/01.wnl.0000178989.87072.11. [DOI] [PubMed] [Google Scholar]
- 8.Mainous AG, 3rd, Eschenbach SL, Wells BJ, Everett CJ, Gill JM. Cholesterol, transferrin saturation, and the development of dementia and Alzheimer’s disease: results from an 18-year population-based cohort. Fam Med. 2005;37:36–42. [PubMed] [Google Scholar]
- 9.Romas SN, Tang MX, Berglund L, Mayeux R. APOE genotype, plasma lipids, lipoproteins, and AD in community elderly. Neurology. 1999;53:517–521. doi: 10.1212/wnl.53.3.517. [DOI] [PubMed] [Google Scholar]
- 10.Tan ZS, Seshadri S, Beiser A, Wilson PW, Kiel DP, Tocco M, D’Agostino RB, Wolf PA. Plasma total cholesterol level as a risk factor for Alzheimer disease: the Framingham Study. Arch Intern Med. 2003;163:1053–1057. doi: 10.1001/archinte.163.9.1053. [DOI] [PubMed] [Google Scholar]
- 11.Notkola IL, Sulkava R, Pekkanen J, Erkinjuntti T, Ehnholm C, Kivinen P, Tuomilehto J, Nissinen A. Serum total cholesterol, apolipoprotein E epsilon 4 allele, and Alzheimer’s disease. Neuroepidemiology. 1998;17:14–20. doi: 10.1159/000026149. [DOI] [PubMed] [Google Scholar]
- 12.Whitmer RA, Sidney S, Selby J, Johnston SC, Yaffe K. Midlife cardiovascular risk factors and risk of dementia in late life. Neurology. 2005;64:277–281. doi: 10.1212/01.WNL.0000149519.47454.F2. [DOI] [PubMed] [Google Scholar]
- 13.Mielke MM, Zandi PP, Sjogren M, Gustafson D, Ostling S, Steen B, Skoog I. High total cholesterol levels in late life associated with a reduced risk of dementia. Neurology. 2005;64:1689–1695. doi: 10.1212/01.WNL.0000161870.78572.A5. [DOI] [PubMed] [Google Scholar]
- 14.Reitz C, Tang MX, Luchsinger J, Mayeux R. Relation of plasma lipids to Alzheimer disease and vascular dementia. Arch Neurol. 2004;61:705–714. doi: 10.1001/archneur.61.5.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuusisto J, Koivisto K, Mykkanen L, Helkala EL, Vanhanen M, Hanninen T, Kervinen K, Kesaniemi YA, Riekkinen PJ, Laakso M. Association between features of the insulin resistance syndrome and Alzheimer’s disease independently of apolipoprotein E4 phenotype: cross sectional population based study. BMJ. 1997;315:1045–1049. doi: 10.1136/bmj.315.7115.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stewart R, White LR, Xue QL, Launer LJ. Twenty-six-year change in total cholesterol levels and incident dementia: the Honolulu-Asia Aging Study. Arch Neurol. 2007;64:103–107. doi: 10.1001/archneur.64.1.103. [DOI] [PubMed] [Google Scholar]
- 17.Beydoun MA, Beason-Held LL, Kitner-Triolo MH, Beydoun HA, Ferrucci L, Resnick SM, Zonderman AB. Statins and serum cholesterol’s associations with incident dementia and mild cognitive impairment. J Epidemiol Community Health. 2010 doi: 10.1136/jech.2009.100826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trkanjec Z, Bene R, Martinic-Popovic I, Jurasic JM, Lisak M, Seric V, Demarin V. Serum HDL, LDL and total cholesterol in patients with late-life onset of Alzheimer’s disease versus vascular dementia. Acta Clin Croat. 2009;48:259–263. [PubMed] [Google Scholar]
- 19.Reitz C, Tang MX, Schupf N, Manly JJ, Mayeux R, Luchsinger JA. Association of higher levels of high-density lipoprotein cholesterol in elderly individuals and lower risk of late-onset Alzheimer disease. Arch Neurol. 2010;67:1491–1497. doi: 10.1001/archneurol.2010.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solfrizzi V, Scafato E, Capurso C, D’Introno A, Colacicco AM, Frisardi V, Vendemiale G, Baldereschi M, Crepaldi G, Di Carlo A, Galluzzo L, Gandin C, Inzitari D, Maggi S, Capurso A, Panza F. Metabolic syndrome and the risk of vascular dementia: the Italian Longitudinal Study on Ageing. J Neurol Neurosurg Psychiatry. 2010;81:433–440. doi: 10.1136/jnnp.2009.181743. [DOI] [PubMed] [Google Scholar]
- 21.Reynolds CA, Gatz M, Prince JA, Berg S, Pedersen NL. Serum lipid levels and cognitive change in late life. J Am Geriatr Soc. 2010;58:501–509. doi: 10.1111/j.1532-5415.2010.02739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalmijn S, Foley D, White L, Burchfiel CM, Curb JD, Petrovitch H, Ross GW, Havlik RJ, Launer LJ. Metabolic cardiovascular syndrome and risk of dementia in Japanese-American elderly men. The Honolulu-Asia aging study. Arterioscler Thromb Vasc Biol. 2000;20:2255–2260. doi: 10.1161/01.atv.20.10.2255. [DOI] [PubMed] [Google Scholar]
- 23.Hall K, Murrell J, Ogunniyi A, Deeg M, Baiyewu O, Gao S, Gureje O, Dickens J, Evans R, Smith-Gamble V, Unverzagt FW, Shen J, Hendrie H. Cholesterol, APOE genotype, and Alzheimer disease: an epidemiologic study of Nigerian Yoruba. Neurology. 2006;66:223–227. doi: 10.1212/01.wnl.0000194507.39504.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michikawa M. Cholesterol paradox: is high total or low HDL cholesterol level a risk for Alzheimer’s disease? J Neurosci Res. 2003;72:141–146. doi: 10.1002/jnr.10585. [DOI] [PubMed] [Google Scholar]
- 25.Yaffe K, Barrett-Connor E, Lin F, Grady D. Serum lipoprotein levels, statin use, and cognitive function in older women. Arch Neurol. 2002;59:378–384. doi: 10.1001/archneur.59.3.378. [DOI] [PubMed] [Google Scholar]
- 26.Moroney JT, Tang MX, Berglund L, Small S, Merchant C, Bell K, Stern Y, Mayeux R. Low-density lipoprotein cholesterol and the risk of dementia with stroke. JAMA. 1999;282:254–260. doi: 10.1001/jama.282.3.254. [DOI] [PubMed] [Google Scholar]
- 27.Furberg CD. Natural statins and stroke risk. Circulation. 1999;99:185–188. doi: 10.1161/01.cir.99.2.185. [DOI] [PubMed] [Google Scholar]
- 28.Li G, Shofer JB, Rhew IC, Kukull WA, Peskind ER, McCormick W, Bowen JD, Schellenberg GD, Crane PK, Breitner JC, Larson EB. Age-varying association between statin use and incident Alzheimer’s disease. J Am Geriatr Soc. 2010;58:1311–1317. doi: 10.1111/j.1532-5415.2010.02906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cramer C, Haan MN, Galea S, Langa KM, Kalbfleisch JD. Use of statins and incidence of dementia and cognitive impairment without dementia in a cohort study. Neurology. 2008;71:344–350. doi: 10.1212/01.wnl.0000319647.15752.7b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dufouil C, Richard F, Fievet N, Dartigues JF, Ritchie K, Tzourio C, Amouyel P, Alperovitch A. APOE genotype, cholesterol level, lipid-lowering treatment, and dementia: the Three-City Study. Neurology. 2005;64:1531–1538. doi: 10.1212/01.WNL.0000160114.42643.31. [DOI] [PubMed] [Google Scholar]
- 31.Haag MD, Hofman A, Koudstaal PJ, Stricker BH, Breteler MM. Statins are associated with a reduced risk of Alzheimer disease regardless of lipophilicity. The Rotterdam Study. J Neurol Neurosurg Psychiatry. 2009;80:13–17. doi: 10.1136/jnnp.2008.150433. [DOI] [PubMed] [Google Scholar]
- 32.Horsdal HT, Olesen AV, Gasse C, Sorensen HT, Green RC, Johnsen SP. Use of statins and risk of hospitalization with dementia: a Danish population-based case-control study. Alzheimer Dis Assoc Disord. 2009;23:18–22. doi: 10.1097/WAD.0b013e318180f55b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones RW, Kivipelto M, Feldman H, Sparks L, Doody R, Waters DD, Hey-Hadavi J, Breazna A, Schindler RJ, Ramos H. The Atorvastatin/Donepezil in Alzheimer’s Disease Study (LEADe): design and baseline characteristics. Alzheimers Dement. 2008;4:145–153. doi: 10.1016/j.jalz.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 34.Feldman HH, Doody RS, Kivipelto M, Sparks DL, Waters DD, Jones RW, Schwam E, Schindler R, Hey-Hadavi J, DeMicco DA, Breazna A. Randomized controlled trial of atorvastatin in mild to moderate Alzheimer disease: LEADe. Neurology. 2010;74:956–964. doi: 10.1212/WNL.0b013e3181d6476a. [DOI] [PubMed] [Google Scholar]
- 35.Trompet S, van Vliet P, de Craen AJ, Jolles J, Buckley BM, Murphy MB, Ford I, Macfarlane PW, Sattar N, Packard CJ, Stott DJ, Shepherd J, Bollen EL, Blauw GJ, Jukema JW, Westendorp RG. Pravastatin and cognitive function in the elderly. Results of the PROSPER study. J Neurol. 2010;257:85–90. doi: 10.1007/s00415-009-5271-7. [DOI] [PubMed] [Google Scholar]
- 36.MRC/BHF Heart Protection Study of cholesterol lowering with simvastatin in 20,536 high-risk individuals: a randomised placebo-controlled trial. Lancet. 2002;360:7–22. doi: 10.1016/S0140-6736(02)09327-3. [DOI] [PubMed] [Google Scholar]
- 37.Sparks DL, Connor DJ, Sabbagh MN, Petersen RB, Lopez J, Browne P. Circulating cholesterol levels, apolipoprotein E genotype and dementia severity influence the benefit of atorvastatin treatment in Alzheimer’s disease: results of the Alzheimer’s Disease Cholesterol-Lowering Treatment (ADCLT) trial. Acta Neurol Scand Suppl. 2006;185:3–7. doi: 10.1111/j.1600-0404.2006.00690.x. [DOI] [PubMed] [Google Scholar]
- 38.Shepherd J, Blauw GJ, Murphy MB, Bollen EL, Buckley BM, Cobbe SM, Ford I, Gaw A, Hyland M, Jukema JW, Kamper AM, Macfarlane PW, Meinders AE, Norrie J, Packard CJ, Perry IJ, Stott DJ, Sweeney BJ, Twomey C, Westendorp RG. Pravastatin in elderly individuals at risk of vascular disease (PROSPER): a randomised controlled trial. Lancet. 2002;360:1623–1630. doi: 10.1016/s0140-6736(02)11600-x. [DOI] [PubMed] [Google Scholar]
- 39.Rockwood K, Kirkland S, Hogan DB, MacKnight C, Merry H, Verreault R, Wolfson C, McDowell I. Use of lipid-lowering agents, indication bias, and the risk of dementia in community-dwelling elderly people. Arch Neurol. 2002;59:223–227. doi: 10.1001/archneur.59.2.223. [DOI] [PubMed] [Google Scholar]
- 40.Grziwa B, Grimm MO, Masters CL, Beyreuther K, Hartmann T, Lichtenthaler SF. The transmembrane domain of the amyloid precursor protein in microsomal membranes is on both sides shorter than predicted. J Biol Chem. 2003;278:6803–6808. doi: 10.1074/jbc.M210047200. [DOI] [PubMed] [Google Scholar]
- 41.Pfrieger FW. Cholesterol homeostasis and function in neurons of the central nervous system. Cell Mol Life Sci. 2003;60:1158–1171. doi: 10.1007/s00018-003-3018-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arnaud C, Mach F. Pleiotropic effects of statins in atherosclerosis: role on endothelial function, inflammation and immunomodulation. Arch Mal Coeur Vaiss. 2005;98:661–666. [PubMed] [Google Scholar]
- 43.Jurevics H, Morell P. Cholesterol for synthesis of myelin is made locally, not imported into brain. J Neurochem. 1995;64:895–901. doi: 10.1046/j.1471-4159.1995.64020895.x. [DOI] [PubMed] [Google Scholar]
- 44.Fagan AM, Holtzman DM. Astrocyte lipoproteins, effects of apoE on neuronal function, and role of apoE in amyloid-beta deposition in vivo. Microsc Res Tech. 2000;50:297–304. doi: 10.1002/1097-0029(20000815)50:4<297::AID-JEMT9>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 45.Poirier J. Apolipoprotein E and cholesterol metabolism in the pathogenesis and treatment of Alzheimer’s disease. Trends Mol Med. 2003;9:94–101. doi: 10.1016/s1471-4914(03)00007-8. [DOI] [PubMed] [Google Scholar]
- 46.Heverin M, Bogdanovic N, Lutjohann D, Bayer T, Pikuleva I, Bretillon L, Diczfalusy U, Winblad B, Bjorkhem I. Changes in the levels of cerebral and extracerebral sterols in the brain of patients with Alzheimer’s disease. J Lipid Res. 2004;45:186–193. doi: 10.1194/jlr.M300320-JLR200. [DOI] [PubMed] [Google Scholar]
- 47.Papassotiropoulos A, Lutjohann D, Bagli M, Locatelli S, Jessen F, Buschfort R, Ptok U, Bjorkhem I, von Bergmann K, Heun R. 24S-hydroxycholesterol in cerebrospinal fluid is elevated in early stages of dementia. J Psychiatr Res. 2002;36:27–32. doi: 10.1016/s0022-3956(01)00050-4. [DOI] [PubMed] [Google Scholar]
- 48.Bogdanovic N, Bretillon L, Lund EG, Diczfalusy U, Lannfelt L, Winblad B, Russell DW, Bjorkhem I. On the turnover of brain cholesterol in patients with Alzheimer’s disease. Abnormal induction of the cholesterol-catabolic enzyme CYP46 in glial cells. Neurosci Lett. 2001;314:45–48. doi: 10.1016/s0304-3940(01)02277-7. [DOI] [PubMed] [Google Scholar]
- 49.Papassotiropoulos A, Lutjohann D, Bagli M, Locatelli S, Jessen F, Rao ML, Maier W, Bjorkhem I, von Bergmann K, Heun R. Plasma 24S-hydroxycholesterol: a peripheral indicator of neuronal degeneration and potential state marker for Alzheimer’s disease. Neuroreport. 2000;11:1959–1962. doi: 10.1097/00001756-200006260-00030. [DOI] [PubMed] [Google Scholar]
- 50.Refolo LM, Pappolla MA, LaFrancois J, Malester B, Schmidt SD, Thomas-Bryant T, Tint GS, Wang R, Mercken M, Petanceska SS, Duff KE. A cholesterol-lowering drug reduces beta-amyloid pathology in a transgenic mouse model of Alzheimer’s disease. Neurobiol Dis. 2001;8:890–899. doi: 10.1006/nbdi.2001.0422. [DOI] [PubMed] [Google Scholar]
- 51.Refolo LM, Malester B, LaFrancois J, Bryant-Thomas T, Wang R, Tint GS, Sambamurti K, Duff K, Pappolla MA. Hypercholesterolemia accelerates the Alzheimer’s amyloid pathology in a transgenic mouse model. Neurobiol Dis. 2000;7:321–331. doi: 10.1006/nbdi.2000.0304. [DOI] [PubMed] [Google Scholar]
- 52.Kojro E, Gimpl G, Lammich S, Marz W, Fahrenholz F. Low cholesterol stimulates the nonamyloidogenic pathway by its effect on the alpha -secretase ADAM 10. Proc Natl Acad Sci U S A. 2001;98:5815–5820. doi: 10.1073/pnas.081612998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simons M, Keller P, De Strooper B, Beyreuther K, Dotti CG, Simons K. Cholesterol depletion inhibits the generation of beta-amyloid in hippocampal neurons. Proc Natl Acad Sci U S A. 1998;95:6460–6464. doi: 10.1073/pnas.95.11.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wahrle S, Das P, Nyborg AC, McLendon C, Shoji M, Kawarabayashi T, Younkin LH, Younkin SG, Golde TE. Cholesterol-dependent gamma-secretase activity in buoyant cholesterol-rich membrane microdomains. Neurobiol Dis. 2002;9:11–23. doi: 10.1006/nbdi.2001.0470. [DOI] [PubMed] [Google Scholar]
- 55.Zou K, Kim D, Kakio A, Byun K, Gong JS, Kim J, Kim M, Sawamura N, Nishimoto S, Matsuzaki K, Lee B, Yanagisawa K, Michikawa M. Amyloid beta-protein (Abeta)1–40 protects neurons from damage induced by Abeta1–42 in culture and in rat brain. J Neurochem. 2003;87:609–619. doi: 10.1046/j.1471-4159.2003.02018.x. [DOI] [PubMed] [Google Scholar]
- 56.McLaurin J, Darabie AA, Morrison MR. Cholesterol, a modulator of membrane-associated Abeta-fibrillogenesis. Pharmacopsychiatry. 2003;36(Suppl 2):S130–135. doi: 10.1055/s-2003-43054. [DOI] [PubMed] [Google Scholar]
- 57.Trousson A, Bernard S, Petit PX, Liere P, Pianos A, El Hadri K, Lobaccaro JM, Ghandour MS, Raymondjean M, Schumacher M, Massaad C. 25-hydroxycholesterol provokes oligodendrocyte cell line apoptosis and stimulates the secreted phospholipase A2 type IIA via LXR beta and PXR. J Neurochem. 2009;109:945–958. doi: 10.1111/j.1471-4159.2009.06009.x. [DOI] [PubMed] [Google Scholar]
- 58.Ma MT, Zhang J, Farooqui AA, Chen P, Ong WY. Effects of cholesterol oxidation products on exocytosis. Neurosci Lett. 2010;476:36–41. doi: 10.1016/j.neulet.2010.03.078. [DOI] [PubMed] [Google Scholar]
- 59.Pitas RE, Boyles JK, Lee SH, Hui D, Weisgraber KH. Lipoproteins and their receptors in the central nervous system. Characterization of the lipoproteins in cerebrospinal fluid and identification of apolipoprotein B, E(LDL) receptors in the brain. J Biol Chem. 1987;262:14352–14360. [PubMed] [Google Scholar]
- 60.Mauch DH, Nagler K, Schumacher S, Goritz C, Muller EC, Otto A, Pfrieger FW. CNS synaptogenesis promoted by glia-derived cholesterol. Science. 2001;294:1354–1357. doi: 10.1126/science.294.5545.1354. [DOI] [PubMed] [Google Scholar]
- 61.Eto M, Watanabe K, Chonan N, Ishii K. Familial hypercholesterolemia and apolipoprotein E4. Atherosclerosis. 1988;72:123–128. doi: 10.1016/0021-9150(88)90072-x. [DOI] [PubMed] [Google Scholar]
- 62.Murakami K, Shimizu M, Yamada N, Ishibashi S, Shimano H, Yazaki Y, Akanuma Y. Apolipoprotein E polymorphism is associated with plasma cholesterol response in a 7-day hospitalization study for metabolic and dietary control in NIDDM. Diabetes Care. 1993;16:564–569. doi: 10.2337/diacare.16.4.564. [DOI] [PubMed] [Google Scholar]
- 63.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 64.Graff-Radford NR, Green RC, Go RC, Hutton ML, Edeki T, Bachman D, Adamson JL, Griffith P, Willis FB, Williams M, Hipps Y, Haines JL, Cupples LA, Farrer LA. Association between apolipoprotein E genotype and Alzheimer disease in African American subjects. Arch Neurol. 2002;59:594–600. doi: 10.1001/archneur.59.4.594. [DOI] [PubMed] [Google Scholar]
- 65.Jarvik GP, Wijsman EM, Kukull WA, Schellenberg GD, Yu C, Larson EB. Interactions of apolipoprotein E genotype, total cholesterol level, age, and sex in prediction of Alzheimer’s disease: a case-control study. Neurology. 1995;45:1092–1096. doi: 10.1212/wnl.45.6.1092. [DOI] [PubMed] [Google Scholar]
- 66.Rubinsztein DC, Easton DF. Apolipoprotein E genetic variation and Alzheimer’s disease. a meta-analysis. Dement Geriatr Cogn Disord. 1999;10:199–209. doi: 10.1159/000017120. [DOI] [PubMed] [Google Scholar]
- 67.Craft S, Teri L, Edland SD, Kukull WA, Schellenberg G, McCormick WC, Bowen JD, Larson EB. Accelerated decline in apolipoprotein E-epsilon4 homozygotes with Alzheimer’s disease. Neurology. 1998;51:149–153. doi: 10.1212/wnl.51.1.149. [DOI] [PubMed] [Google Scholar]
- 68.Reiman EM, Chen K, Liu X, Bandy D, Yu M, Lee W, Ayutyanont N, Keppler J, Reeder SA, Langbaum JB, Alexander GE, Klunk WE, Mathis CA, Price JC, Aizenstein HJ, DeKosky ST, Caselli RJ. Fibrillar amyloid-beta burden in cognitively normal people at 3 levels of genetic risk for Alzheimer’s disease. Proc Natl Acad Sci U S A. 2009;106:6820–6825. doi: 10.1073/pnas.0900345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Greenberg SM, Rebeck GW, Vonsattel JP, Gomez-Isla T, Hyman BT. Apolipoprotein E epsilon 4 and cerebral hemorrhage associated with amyloid angiopathy. Ann Neurol. 1995;38:254–259. doi: 10.1002/ana.410380219. [DOI] [PubMed] [Google Scholar]
- 70.Christensen DZ, Schneider-Axmann T, Lucassen PJ, Bayer TA, Wirths O. Accumulation of intraneuronal Abeta correlates with ApoE4 genotype. Acta Neuropathol. 2010;119:555–566. doi: 10.1007/s00401-010-0666-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lambert JC, Pasquier F, Cottel D, Frigard B, Amouyel P, Chartier-Harlin MC. A new polymorphism in the APOE promoter associated with risk of developing Alzheimer’s disease. Hum Mol Genet. 1998;7:533–540. doi: 10.1093/hmg/7.3.533. [DOI] [PubMed] [Google Scholar]
- 72.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 74.Gelissen IC, Hochgrebe T, Wilson MR, Easterbrook-Smith SB, Jessup W, Dean RT, Brown AJ. Apolipoprotein J (clusterin) induces cholesterol export from macrophage-foam cells: a potential anti-atherogenic function? Biochem J. 1998;331 (Pt 1):231–237. doi: 10.1042/bj3310231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.DeMattos RB, Cirrito JR, Parsadanian M, May PC, O’Dell MA, Taylor JW, Harmony JA, Aronow BJ, Bales KR, Paul SM, Holtzman DM. ApoE and clusterin cooperatively suppress Abeta levels and deposition: evidence that ApoE regulates extracellular Abeta metabolism in vivo. Neuron. 2004;41:193–202. doi: 10.1016/s0896-6273(03)00850-x. [DOI] [PubMed] [Google Scholar]
- 76.Bell RD, Sagare AP, Friedman AE, Bedi GS, Holtzman DM, Deane R, Zlokovic BV. Transport pathways for clearance of human Alzheimer’s amyloid beta-peptide and apolipoproteins E and J in the mouse central nervous system. J Cereb Blood Flow Metab. 2007;27:909–918. doi: 10.1038/sj.jcbfm.9600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yerbury JJ, Poon S, Meehan S, Thompson B, Kumita JR, Dobson CM, Wilson MR. The extracellular chaperone clusterin influences amyloid formation and toxicity by interacting with prefibrillar structures. FASEB J. 2007;21:2312–2322. doi: 10.1096/fj.06-7986com. [DOI] [PubMed] [Google Scholar]
- 78.Matsubara E, Frangione B, Ghiso J. Characterization of apolipoprotein J-Alzheimer’s A beta interaction. J Biol Chem. 1995;270:7563–7567. doi: 10.1074/jbc.270.13.7563. [DOI] [PubMed] [Google Scholar]
- 79.Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, Katayama T, Baldwin CT, Cheng R, Hasegawa H, Chen F, Shibata N, Lunetta KL, Pardossi-Piquard R, Bohm C, Wakutani Y, Cupples LA, Cuenco KT, Green RC, Pinessi L, Rainero I, Sorbi S, Bruni A, Duara R, Friedland RP, Inzelberg R, Hampe W, Bujo H, Song YQ, Andersen OM, Willnow TE, Graff-Radford N, Petersen RC, Dickson D, Der SD, Fraser PE, Schmitt-Ulms G, Younkin S, Mayeux R, Farrer LA, St George-Hyslop P. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee JH, Cheng R, Honig LS, Vonsattel JP, Clark L, Mayeux R. Association between genetic variants in SORL1 and autopsy-confirmed Alzheimer disease. Neurology. 2008;70:887–889. doi: 10.1212/01.wnl.0000280581.39755.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee JH, Chulikavit M, Pang D, Zigman WB, Silverman W, Schupf N. Association between genetic variants in sortilin-related receptor 1 (SORL1) and Alzheimer’s disease in adults with Down syndrome. Neurosci Lett. 2007;425:105–109. doi: 10.1016/j.neulet.2007.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bettens K, Brouwers N, Engelborghs S, De Deyn PP, Van Broeckhoven C, Sleegers K. SORL1 is genetically associated with increased risk for late-onset Alzheimer disease in the Belgian population. Hum Mutat. 2008;29:769–770. doi: 10.1002/humu.20725. [DOI] [PubMed] [Google Scholar]
- 83.Cuenco KT, Lunetta KL, Baldwin CT, McKee AC, Guo J, Cupples LA, Green RC, St George-Hyslop PH, Chui H, DeCarli C, Farrer LA. Association of distinct variants in SORL1 with cerebrovascular and neurodegenerative changes related to Alzheimer disease. Arch Neurol. 2008;65:1640–1648. doi: 10.1001/archneur.65.12.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dodson SE, Gearing M, Lippa CF, Montine TJ, Levey AI, Lah JJ. LR11/SorLA expression is reduced in sporadic Alzheimer disease but not in familial Alzheimer disease. J Neuropathol Exp Neurol. 2006;65:866–872. doi: 10.1097/01.jnen.0000228205.19915.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Grear KE, Ling IF, Simpson JF, Furman JL, Simmons CR, Peterson SL, Schmitt FA, Markesbery WR, Liu Q, Crook JE, Younkin SG, Bu G, Estus S. Expression of SORL1 and a novel SORL1 splice variant in normal and Alzheimers disease brain. Mol Neurodegener. 2009;4:46. doi: 10.1186/1750-1326-4-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kimura R, Yamamoto M, Morihara T, Akatsu H, Kudo T, Kamino K, Takeda M. SORL1 is genetically associated with Alzheimer disease in a Japanese population. Neurosci Lett. 2009;461:177–180. doi: 10.1016/j.neulet.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 87.Kolsch H, Jessen F, Wiltfang J, Lewczuk P, Dichgans M, Kornhuber J, Frolich L, Heuser I, Peters O, Schulz JB, Schwab SG, Maier W. Influence of SORL1 gene variants: association with CSF amyloid-beta products in probable Alzheimer’s disease. Neurosci Lett. 2008;440:68–71. doi: 10.1016/j.neulet.2008.05.049. [DOI] [PubMed] [Google Scholar]
- 88.Kolsch H, Jessen F, Wiltfang J, Lewczuk P, Dichgans M, Teipel SJ, Kornhuber J, Frolich L, Heuser I, Peters O, Wiese B, Kaduszkiewicz H, van den Bussche H, Hull M, Kurz A, Ruther E, Henn FA, Maier W. Association of SORL1 gene variants with Alzheimer’s disease. Brain Res. 2009;1264:1–6. doi: 10.1016/j.brainres.2009.01.044. [DOI] [PubMed] [Google Scholar]
- 89.Lee JH, Barral S, Reitz C. The neuronal sortilin-related receptor gene SORL1 and late-onset Alzheimer’s disease. Curr Neurol Neurosci Rep. 2008;8:384–391. doi: 10.1007/s11910-008-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee JH, Shibata N, Cheng R, Mayeux R. Possible association between SORL1 and Alzheimer disease? Reanalysing the data of Shibata et al. Dement Geriatr Cogn Disord. 2008;26:482. doi: 10.1159/000167792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li Y, Rowland C, Catanese J, Morris J, Lovestone S, O’Donovan MC, Goate A, Owen M, Williams J, Grupe A. SORL1 variants and risk of late-onset Alzheimer’s disease. Neurobiol Dis. 2008;29:293–296. doi: 10.1016/j.nbd.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ma QL, Galasko DR, Ringman JM, Vinters HV, Edland SD, Pomakian J, Ubeda OJ, Rosario ER, Teter B, Frautschy SA, Cole GM. Reduction of SorLA/LR11, a sorting protein limiting beta-amyloid production, in Alzheimer disease cerebrospinal fluid. Arch Neurol. 2009;66:448–457. doi: 10.1001/archneurol.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Meng Y, Lee JH, Cheng R, St George-Hyslop P, Mayeux R, Farrer LA. Association between SORL1 and Alzheimer’s disease in a genome-wide study. Neuroreport. 2007;18:1761–1764. doi: 10.1097/WNR.0b013e3282f13e7a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reynolds CA, Hong MG, Eriksson UK, Blennow K, Johansson B, Malmberg B, Berg S, Gatz M, Pedersen NL, Bennet AM, Prince JA. Sequence variation in SORL1 and dementia risk in Swedes. Neurogenetics. 2009;11:139–142. doi: 10.1007/s10048-009-0210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shibata N, Ohnuma T, Baba H, Higashi S, Nishioka K, Arai H. Genetic association between SORL1 polymorphisms and Alzheimer’s disease in a Japanese population. Dement Geriatr Cogn Disord. 2008;26:161–164. doi: 10.1159/000149821. [DOI] [PubMed] [Google Scholar]
- 96.Tan EK, Lee J, Chen CP, Teo YY, Zhao Y, Lee WL. SORL1 haplotypes modulate risk of Alzheimer’s disease in Chinese. Neurobiol Aging. 2009;30:1048–1051. doi: 10.1016/j.neurobiolaging.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 97.Webster JA, Myers AJ, Pearson JV, Craig DW, Hu-Lince D, Coon KD, Zismann VL, Beach T, Leung D, Bryden L, Halperin RF, Marlowe L, Kaleem M, Huentelman MJ, Joshipura K, Walker D, Heward CB, Ravid R, Rogers J, Papassotiropoulos A, Hardy J, Reiman EM, Stephan DA. Sorl1 as an Alzheimer’s disease predisposition gene? Neurodegener Dis. 2008;5:60–64. doi: 10.1159/000110789. [DOI] [PubMed] [Google Scholar]
- 98.Reitz C, Cheng R, Rogaeva E, Lee JH, Tokuhiro S, Zou F, Bettens K, Sleegers K, Tan EK, Kimura R, Shibata N, Arai H, Kamboh MI, Prince JA, Maier W, Riemenschneider M, Owen M, Harold D, Hollingworth P, Cellini E, Sorbi S, Nacmias B, Takeda M, Pericak-Vance MA, Haines JL, Younkin S, Williams J, van Broeckhoven C, Farrer LA, St George-Hyslop PH, Mayeux R. Meta-analysis of the Association Between Variants in SORL1 and Alzheimer Disease. Arch Neurol. 2011;68:99–106. doi: 10.1001/archneurol.2010.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee JH, Cheng R, Graff-Radford N, Foroud T, Mayeux R. Analyses of the National Institute on Aging Late-Onset Alzheimer’s Disease Family Study: implication of additional loci. Arch Neurol. 2008;65:1518–1526. doi: 10.1001/archneur.65.11.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tang MX, Stern Y, Marder K, Bell K, Gurland B, Lantigua R, Andrews H, Feng L, Tycko B, Mayeux R. The APOE-epsilon4 allele and the risk of Alzheimer disease among African Americans, whites, and Hispanics. Jama. 1998;279:751–755. doi: 10.1001/jama.279.10.751. [DOI] [PubMed] [Google Scholar]
- 101.Seshadri S, DeStefano AL, Au R, Massaro JM, Beiser AS, Kelly-Hayes M, Kase CS, D’Agostino RB, Sr, Decarli C, Atwood LD, Wolf PA. Genetic correlates of brain aging on MRI and cognitive test measures: a genome-wide association and linkage analysis in the Framingham Study. BMC Med Genet. 2007;8(Suppl 1):S15. doi: 10.1186/1471-2350-8-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Reitz C, Tokuhiro S, Clark LN, Conrad C, Vonsattel JP, Hazrati LN, Palotas A, Lantigua R, Medrano M, IZJ-V, Vardarajan B, Simkin I, Haines JL, Pericak-Vance MA, Farrer LA, Lee JH, Rogaeva E, George-Hyslop PS, Mayeux R. SORCS1 alters amyloid precursor protein processing and variants may increase Alzheimer’s disease risk. Ann Neurol. 2011;69:47–64. doi: 10.1002/ana.22308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kim J, Castellano JM, Jiang H, Basak JM, Parsadanian M, Pham V, Mason SM, Paul SM, Holtzman DM. Overexpression of low-density lipoprotein receptor in the brain markedly inhibits amyloid deposition and increases extracellular A beta clearance. Neuron. 2009;64:632–644. doi: 10.1016/j.neuron.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Reiman EM, Webster JA, Myers AJ, Hardy J, Dunckley T, Zismann VL, Joshipura KD, Pearson JV, Hu-Lince D, Huentelman MJ, Craig DW, Coon KD, Liang WS, Herbert RH, Beach T, Rohrer KC, Zhao AS, Leung D, Bryden L, Marlowe L, Kaleem M, Mastroeni D, Grover A, Heward CB, Ravid R, Rogers J, Hutton ML, Melquist S, Petersen RC, Alexander GE, Caselli RJ, Kukull W, Papassotiropoulos A, Stephan DA. GAB2 alleles modify Alzheimer’s risk in APOE epsilon4 carriers. Neuron. 2007;54:713–720. doi: 10.1016/j.neuron.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cheng D, Huang R, Lanham IS, Cathcart HM, Howard M, Corder EH, Poduslo SE. Functional interaction between APOE4 and LDL receptor isoforms in Alzheimer’s disease. J Med Genet. 2005;42:129–131. doi: 10.1136/jmg.2004.024968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zou F, Gopalraj RK, Lok J, Zhu H, Ling IF, Simpson JF, Tucker HM, Kelly JF, Younkin SG, Dickson DW, Petersen RC, Graff-Radford NR, Bennett DA, Crook JE, Estus S. Sex-dependent association of a common low-density lipoprotein receptor polymorphism with RNA splicing efficiency in the brain and Alzheimer’s disease. Hum Mol Genet. 2008;17:929–935. doi: 10.1093/hmg/ddm365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gopalraj RK, Zhu H, Kelly JF, Mendiondo M, Pulliam JF, Bennett DA, Estus S. Genetic association of low density lipoprotein receptor and Alzheimer’s disease. Neurobiol Aging. 2005;26:1–7. doi: 10.1016/j.neurobiolaging.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 108.Lamsa R, Helisalmi S, Herukka SK, Tapiola T, Pirttila T, Vepsalainen S, Hiltunen M, Soininen H. Genetic study evaluating LDLR polymorphisms and Alzheimer’s disease. Neurobiol Aging. 2008;29:848–855. doi: 10.1016/j.neurobiolaging.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 109.Papassotiropoulos A, Lambert JC, Wavrant-De Vrieze F, Wollmer MA, von der Kammer H, Streffer JR, Maddalena A, Huynh KD, Wolleb S, Lutjohann D, Schneider B, Thal DR, Grimaldi LM, Tsolaki M, Kapaki E, Ravid R, Konietzko U, Hegi T, Pasch T, Jung H, Braak H, Amouyel P, Rogaev EI, Hardy J, Hock C, Nitsch RM. Cholesterol 25-hydroxylase on chromosome 10q is a susceptibility gene for sporadic Alzheimer’s disease. Neurodegener Dis. 2005;2:233–241. doi: 10.1159/000090362. [DOI] [PubMed] [Google Scholar]
- 110.Papassotiropoulos A, Wollmer MA, Tsolaki M, Brunner F, Molyva D, Lutjohann D, Nitsch RM, Hock C. A cluster of cholesterol-related genes confers susceptibility for Alzheimer’s disease. J Clin Psychiatry. 2005;66:940–947. [PubMed] [Google Scholar]
- 111.Blankenberg S, Rupprecht HJ, Bickel C, Jiang XC, Poirier O, Lackner KJ, Meyer J, Cambien F, Tiret L. Common genetic variation of the cholesteryl ester transfer protein gene strongly predicts future cardiovascular death in patients with coronary artery disease. J Am Coll Cardiol. 2003;41:1983–1989. doi: 10.1016/s0735-1097(03)00408-x. [DOI] [PubMed] [Google Scholar]
- 112.Atzmon G, Rincon M, Rabizadeh P, Barzilai N. Biological evidence for inheritance of exceptional longevity. Mech Ageing Dev. 2005;126:341–345. doi: 10.1016/j.mad.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 113.Rodriguez E, Mateo I, Infante J, Llorca J, Berciano J, Combarros O. Cholesteryl ester transfer protein (CETP) polymorphism modifies the Alzheimer’s disease risk associated with APOE epsilon4 allele. J Neurol. 2006;253:181–185. doi: 10.1007/s00415-005-0945-2. [DOI] [PubMed] [Google Scholar]
- 114.Arias-Vasquez A, Isaacs A, Aulchenko YS, Hofman A, Oostra BA, Breteler M, van Duijn CM. The cholesteryl ester transfer protein (CETP) gene and the risk of Alzheimer’s disease. Neurogenetics. 2007;8:189–193. doi: 10.1007/s10048-007-0089-x. [DOI] [PubMed] [Google Scholar]
- 115.Koldamova R, Staufenbiel M, Lefterov I. Lack of ABCA1 considerably decreases brain ApoE level and increases amyloid deposition in APP23 mice. J Biol Chem. 2005;280:43224–43235. doi: 10.1074/jbc.M504513200. [DOI] [PubMed] [Google Scholar]
- 116.Wahrle SE, Jiang H, Parsadanian M, Hartman RE, Bales KR, Paul SM, Holtzman DM. Deletion of Abca1 increases Abeta deposition in the PDAPP transgenic mouse model of Alzheimer disease. J Biol Chem. 2005;280:43236–43242. doi: 10.1074/jbc.M508780200. [DOI] [PubMed] [Google Scholar]
- 117.Wahrle SE, Jiang H, Parsadanian M, Legleiter J, Han X, Fryer JD, Kowalewski T, Holtzman DM. ABCA1 is required for normal central nervous system ApoE levels and for lipidation of astrocyte-secreted apoE. J Biol Chem. 2004;279:40987–40993. doi: 10.1074/jbc.M407963200. [DOI] [PubMed] [Google Scholar]
- 118.Wollmer MA, Streffer JR, Lutjohann D, Tsolaki M, Iakovidou V, Hegi T, Pasch T, Jung HH, Bergmann K, Nitsch RM, Hock C, Papassotiropoulos A. ABCA1 modulates CSF cholesterol levels and influences the age at onset of Alzheimer’s disease. Neurobiol Aging. 2003;24:421–426. doi: 10.1016/s0197-4580(02)00094-5. [DOI] [PubMed] [Google Scholar]
- 119.Katzov H, Chalmers K, Palmgren J, Andreasen N, Johansson B, Cairns NJ, Gatz M, Wilcock GK, Love S, Pedersen NL, Brookes AJ, Blennow K, Kehoe PG, Prince JA. Genetic variants of ABCA1 modify Alzheimer disease risk and quantitative traits related to beta-amyloid metabolism. Hum Mutat. 2004;23:358–367. doi: 10.1002/humu.20012. [DOI] [PubMed] [Google Scholar]
- 120.Sundar PD, Feingold E, Minster RL, DeKosky ST, Kamboh MI. Gender-specific association of ATP-binding cassette transporter 1 (ABCA1) polymorphisms with the risk of late-onset Alzheimer’s disease. Neurobiol Aging. 2007;28:856–862. doi: 10.1016/j.neurobiolaging.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 121.Reynolds CA, Hong MG, Eriksson UK, Blennow K, Bennet AM, Johansson B, Malmberg B, Berg S, Wiklund F, Gatz M, Pedersen NL, Prince JA. A survey of ABCA1 sequence variation confirms association with dementia. Hum Mutat. 2009;30:1348–1354. doi: 10.1002/humu.21076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wavrant-De Vrieze F, Compton D, Womick M, Arepalli S, Adighibe O, Li L, Perez-Tur J, Hardy J. ABCA1 polymorphisms and Alzheimer’s disease. Neurosci Lett. 2007;416:180–183. doi: 10.1016/j.neulet.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shibata N, Kawarai T, Lee JH, Lee HS, Shibata E, Sato C, Liang Y, Duara R, Mayeux RP, St George-Hyslop PH, Rogaeva E. Association studies of cholesterol metabolism genes (CH25H, ABCA1 and CH24H) in Alzheimer’s disease. Neurosci Lett. 2006;391:142–146. doi: 10.1016/j.neulet.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 124.Mace S, Cousin E, Ricard S, Genin E, Spanakis E, Lafargue-Soubigou C, Genin B, Fournel R, Roche S, Haussy G, Massey F, Soubigou S, Brefort G, Benoit P, Brice A, Campion D, Hollis M, Pradier L, Benavides J, Deleuze JF. ABCA2 is a strong genetic risk factor for early-onset Alzheimer’s disease. Neurobiol Dis. 2005;18:119–125. doi: 10.1016/j.nbd.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 125.Wollmer MA, Kapaki E, Hersberger M, Muntwyler J, Brunner F, Tsolaki M, Akatsu H, Kosaka K, Michikawa M, Molyva D, Paraskevas GP, Lutjohann D, von Eckardstein A, Hock C, Nitsch RM, Papassotiropoulos A. Ethnicity-dependent genetic association of ABCA2 with sporadic Alzheimer’s disease. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:534–536. doi: 10.1002/ajmg.b.30345. [DOI] [PubMed] [Google Scholar]
- 126.Minster RL, DeKosky ST, Kamboh MI. No association of DAPK1 and ABCA2 SNPs on chromosome 9 with Alzheimer’s disease. Neurobiol Aging. 2009;30:1890–1891. doi: 10.1016/j.neurobiolaging.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wollmer MA, Sleegers K, Ingelsson M, Zekanowski C, Brouwers N, Maruszak A, Brunner F, Huynh KD, Kilander L, Brundin RM, Hedlund M, Giedraitis V, Glaser A, Engelborghs S, De Deyn PP, Kapaki E, Tsolaki M, Daniilidou M, Molyva D, Paraskevas GP, Thal DR, Barcikowska M, Kuznicki J, Lannfelt L, Van Broeckhoven C, Nitsch RM, Hock C, Papassotiropoulos A. Association study of cholesterol-related genes in Alzheimer’s disease. Neurogenetics. 2007;8:179–188. doi: 10.1007/s10048-007-0087-z. [DOI] [PubMed] [Google Scholar]
- 128.Tansley GH, Burgess BL, Bryan MT, Su Y, Hirsch-Reinshagen V, Pearce J, Chan JY, Wilkinson A, Evans J, Naus KE, McIsaac S, Bromley K, Song W, Yang HC, Wang N, DeMattos RB, Wellington CL. The cholesterol transporter ABCG1 modulates the subcellular distribution and proteolytic processing of beta-amyloid precursor protein. J Lipid Res. 2007;48:1022–1034. doi: 10.1194/jlr.M600542-JLR200. [DOI] [PubMed] [Google Scholar]
- 129.Kim WS, Rahmanto AS, Kamili A, Rye KA, Guillemin GJ, Gelissen IC, Jessup W, Hill AF, Garner B. Role of ABCG1 and ABCA1 in regulation of neuronal cholesterol efflux to apolipoprotein E discs and suppression of amyloid-beta peptide generation. J Biol Chem. 2007;282:2851–2861. doi: 10.1074/jbc.M607831200. [DOI] [PubMed] [Google Scholar]
- 130.Burgess BL, Parkinson PF, Racke MM, Hirsch-Reinshagen V, Fan J, Wong C, Stukas S, Theroux L, Chan JY, Donkin J, Wilkinson A, Balik D, Christie B, Poirier J, Lutjohann D, Demattos RB, Wellington CL. ABCG1 influences the brain cholesterol biosynthetic pathway but does not affect amyloid precursor protein or apolipoprotein E metabolism in vivo. J Lipid Res. 2008;49:1254–1267. doi: 10.1194/jlr.M700481-JLR200. [DOI] [PubMed] [Google Scholar]
- 131.Vollbach H, Heun R, Morris CM, Edwardson JA, McKeith IG, Jessen F, Schulz A, Maier W, Kolsch H. APOA1 polymorphism influences risk for early-onset nonfamiliar AD. Ann Neurol. 2005;58:436–441. doi: 10.1002/ana.20593. [DOI] [PubMed] [Google Scholar]
- 132.Koldamova RP, Lefterov IM, Lefterova MI, Lazo JS. Apolipoprotein A-I directly interacts with amyloid precursor protein and inhibits A beta aggregation and toxicity. Biochemistry. 2001;40:3553–3560. doi: 10.1021/bi002186k. [DOI] [PubMed] [Google Scholar]
- 133.Fagan AM, Christopher E, Taylor JW, Parsadanian M, Spinner M, Watson M, Fryer JD, Wahrle S, Bales KR, Paul SM, Holtzman DM. ApoAI deficiency results in marked reductions in plasma cholesterol but no alterations in amyloid-beta pathology in a mouse model of Alzheimer’s disease-like cerebral amyloidosis. Am J Pathol. 2004;165:1413–1422. doi: 10.1016/s0002-9440(10)63399-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Csaszar A, Kalman J, Szalai C, Janka Z, Romics L. Association of the apolipoprotein A-IV codon 360 mutation in patients with Alzheimer’s disease. Neurosci Lett. 1997;230:151–154. doi: 10.1016/s0304-3940(97)00500-4. [DOI] [PubMed] [Google Scholar]
- 135.Merched A, Xia Y, Papadopoulou A, Siest G, Visvikis S. Apolipoprotein AIV codon 360 mutation increases with human aging and is not associated with Alzheimer’s disease. Neurosci Lett. 1998;242:117–119. doi: 10.1016/s0304-3940(98)00025-1. [DOI] [PubMed] [Google Scholar]
- 136.Ji Y, Urakami K, Adachi Y, Nakashima K. No association between apolipoprotein A-IV codon 360 mutation and late-onset Alzheimer’s disease in the Japanese population. Dement Geriatr Cogn Disord. 1999;10:473–475. doi: 10.1159/000017192. [DOI] [PubMed] [Google Scholar]
- 137.Schellenberg GD, Deeb SS, Boehnke M, Bryant EM, Martin GM, Lampe TH, Bird TD. Association of an apolipoprotein CII allele with familial dementia of the Alzheimer type. J Neurogenet. 1987;4:97–108. [PubMed] [Google Scholar]
- 138.Zarif Yeganeh M, Mirabzadeh A, Khorram Khorshid HR, Kamali K, Heshmati Y, Gozalpour E, Veissy K, Olad Nabi M, Najmabadi H, Ohadi M. Novel extreme homozygote haplotypes at the human caveolin 1 gene upstream purine complex in sporadic Alzheimer’s disease. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:347–349. doi: 10.1002/ajmg.b.30985. [DOI] [PubMed] [Google Scholar]
- 139.Kang MJ, Chung YH, Hwang CI, Murata M, Fujimoto T, Mook-Jung IH, Cha CI, Park WY. Caveolin-1 upregulation in senescent neurons alters amyloid precursor protein processing. Exp Mol Med. 2006;38:126–133. doi: 10.1038/emm.2006.16. [DOI] [PubMed] [Google Scholar]
- 140.Gaudreault SB, Dea D, Poirier J. Increased caveolin-1 expression in Alzheimer’s disease brain. Neurobiol Aging. 2004;25:753–759. doi: 10.1016/j.neurobiolaging.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 141.Taguchi K, Yamagata HD, Zhong W, Kamino K, Akatsu H, Hata R, Yamamoto T, Kosaka K, Takeda M, Kondo I, Miki T. Identification of hippocampus-related candidate genes for Alzheimer’s disease. Ann Neurol. 2005;57:585–588. doi: 10.1002/ana.20433. [DOI] [PubMed] [Google Scholar]
- 142.Moore KJ, El Khoury J, Medeiros LA, Terada K, Geula C, Luster AD, Freeman MW. A CD36-initiated signaling cascade mediates inflammatory effects of beta-amyloid. J Biol Chem. 2002;277:47373–47379. doi: 10.1074/jbc.M208788200. [DOI] [PubMed] [Google Scholar]
- 143.Coraci IS, Husemann J, Berman JW, Hulette C, Dufour JH, Campanella GK, Luster AD, Silverstein SC, El-Khoury JB. CD36, a class B scavenger receptor, is expressed on microglia in Alzheimer’s disease brains and can mediate production of reactive oxygen species in response to beta-amyloid fibrils. Am J Pathol. 2002;160:101–112. doi: 10.1016/s0002-9440(10)64354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hudry E, Van Dam D, Kulik W, De Deyn PP, Stet FS, Ahouansou O, Benraiss A, Delacourte A, Bougneres P, Aubourg P, Cartier N. Adeno-associated virus gene therapy with cholesterol 24-hydroxylase reduces the amyloid pathology before or after the onset of amyloid plaques in mouse models of Alzheimer’s disease. Mol Ther. 2010;18:44–53. doi: 10.1038/mt.2009.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Borroni B, Archetti S, Agosti C, Akkawi N, Brambilla C, Caimi L, Caltagirone C, Di Luca M, Padovani A. Intronic CYP46 polymorphism along with ApoE genotype in sporadic Alzheimer Disease: from risk factors to disease modulators. Neurobiol Aging. 2004;25:747–751. doi: 10.1016/j.neurobiolaging.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 146.Chalmers KA, Culpan D, Kehoe PG, Wilcock GK, Hughes A, Love S. APOE promoter, ACE1 and CYP46 polymorphisms and beta-amyloid in Alzheimer’s disease. Neuroreport. 2004;15:95–98. doi: 10.1097/00001756-200401190-00019. [DOI] [PubMed] [Google Scholar]
- 147.Combarros O, Infante J, Llorca J, Berciano J. Genetic association of CYP46 and risk for Alzheimer’s disease. Dement Geriatr Cogn Disord. 2004;18:257–260. doi: 10.1159/000080025. [DOI] [PubMed] [Google Scholar]
- 148.Kolsch H, Lutjohann D, Ludwig M, Schulte A, Ptok U, Jessen F, von Bergmann K, Rao ML, Maier W, Heun R. Polymorphism in the cholesterol 24S-hydroxylase gene is associated with Alzheimer’s disease. Mol Psychiatry. 2002;7:899–902. doi: 10.1038/sj.mp.4001109. [DOI] [PubMed] [Google Scholar]
- 149.Greeve I, Hermans-Borgmeyer I, Brellinger C, Kasper D, Gomez-Isla T, Behl C, Levkau B, Nitsch RM. The human DIMINUTO/DWARF1 homolog seladin-1 confers resistance to Alzheimer’s disease-associated neurodegeneration and oxidative stress. J Neurosci. 2000;20:7345–7352. doi: 10.1523/JNEUROSCI.20-19-07345.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Crameri A, Biondi E, Kuehnle K, Lutjohann D, Thelen KM, Perga S, Dotti CG, Nitsch RM, Ledesma MD, Mohajeri MH. The role of seladin-1/DHCR24 in cholesterol biosynthesis, APP processing and Abeta generation in vivo. EMBO J. 2006;25:432–443. doi: 10.1038/sj.emboj.7600938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Porcellini E, Calabrese E, Guerini F, Govoni M, Chiappelli M, Tumini E, Morgan K, Chappell S, Kalsheker N, Franceschi M, Licastro F. The hydroxy-methyl-glutaryl CoA reductase promoter polymorphism is associated with Alzheimer’s risk and cognitive deterioration. Neurosci Lett. 2007;416:66–70. doi: 10.1016/j.neulet.2007.01.046. [DOI] [PubMed] [Google Scholar]
- 152.Rodriguez-Rodriguez E, Mateo I, Infante J, Llorca J, Garcia-Gorostiaga I, Vazquez-Higuera JL, Sanchez-Juan P, Berciano J, Combarros O. Interaction between HMGCR and ABCA1 cholesterol-related genes modulates Alzheimer’s disease risk. Brain Res. 2009;1280:166–171. doi: 10.1016/j.brainres.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 153.Zhu H, Taylor JW, Bennett DA, Younkin SG, Estus S. Lack of association of hepatic lipase polymorphisms with late-onset Alzheimer’s disease. Neurobiol Aging. 2008;29:793–794. doi: 10.1016/j.neurobiolaging.2006.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Baum L, Wiebusch H, Pang CP. Roles for lipoprotein lipase in Alzheimer’s disease: an association study. Microsc Res Tech. 2000;50:291–296. doi: 10.1002/1097-0029(20000815)50:4<291::AID-JEMT8>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 155.Blain JF, Aumont N, Theroux L, Dea D, Poirier J. A polymorphism in lipoprotein lipase affects the severity of Alzheimer’s disease pathophysiology. Eur J Neurosci. 2006;24:1245–1251. doi: 10.1111/j.1460-9568.2006.05007.x. [DOI] [PubMed] [Google Scholar]
- 156.Scacchi R, Gambina G, Broggio E, Moretto G, Ruggeri M, Corbo RM. The H+ allele of the lipoprotein lipase (LPL) HindIII intronic polymorphism and the risk for sporadic late-onset Alzheimer’s disease. Neurosci Lett. 2004;367:177–180. doi: 10.1016/j.neulet.2004.05.111. [DOI] [PubMed] [Google Scholar]
- 157.Chalmers KA, Barker R, Passmore PA, Panza F, Seripa D, Solfrizzi V, Love S, Prince JA, Kehoe PG. LRP-1 variation is not associated with risk of Alzheimer’s disease. Int J Mol Epidemiol Genet. 2010;1:104–113. [PMC free article] [PubMed] [Google Scholar]
- 158.Deane R, Bell RD, Sagare A, Zlokovic BV. Clearance of amyloid-beta peptide across the blood-brain barrier: implication for therapies in Alzheimer’s disease. CNS Neurol Disord Drug Targets. 2009;8:16–30. doi: 10.2174/187152709787601867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sagare A, Deane R, Bell RD, Johnson B, Hamm K, Pendu R, Marky A, Lenting PJ, Wu Z, Zarcone T, Goate A, Mayo K, Perlmutter D, Coma M, Zhong Z, Zlokovic BV. Clearance of amyloid-beta by circulating lipoprotein receptors. Nat Med. 2007;13:1029–1031. doi: 10.1038/nm1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Waldron E, Heilig C, Schweitzer A, Nadella N, Jaeger S, Martin AM, Weggen S, Brix K, Pietrzik CU. LRP1 modulates APP trafficking along early compartments of the secretory pathway. Neurobiol Dis. 2008;31:188–197. doi: 10.1016/j.nbd.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 161.Liu Q, Zerbinatti CV, Zhang J, Hoe HS, Wang B, Cole SL, Herz J, Muglia L, Bu G. Amyloid precursor protein regulates brain apolipoprotein E and cholesterol metabolism through lipoprotein receptor LRP1. Neuron. 2007;56:66–78. doi: 10.1016/j.neuron.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wollmer MA, Streffer JR, Tsolaki M, Grimaldi LM, Lutjohann D, Thal D, von Bergmann K, Nitsch RM, Hock C, Papassotiropoulos A. Genetic association of acyl-coenzyme A: cholesterol acyltransferase with cerebrospinal fluid cholesterol levels, brain amyloid load, and risk for Alzheimer’s disease. Mol Psychiatry. 2003;8:635–638. doi: 10.1038/sj.mp.4001296. [DOI] [PubMed] [Google Scholar]
- 163.Puglielli L, Konopka G, Pack-Chung E, Ingano LA, Berezovska O, Hyman BT, Chang TY, Tanzi RE, Kovacs DM. Acyl-coenzyme A: cholesterol acyltransferase modulates the generation of the amyloid beta-peptide. Nat Cell Biol. 2001;3:905–912. doi: 10.1038/ncb1001-905. [DOI] [PubMed] [Google Scholar]
- 164.Huttunen HJ, Greco C, Kovacs DM. Knockdown of ACAT-1 reduces amyloidogenic processing of APP. FEBS Lett. 2007;581:1688–1692. doi: 10.1016/j.febslet.2007.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bryleva EY, Rogers MA, Chang CC, Buen F, Harris BT, Rousselet E, Seidah NG, Oddo S, LaFerla FM, Spencer TA, Hickey WF, Chang TY. ACAT1 gene ablation increases 24(S)-hydroxycholesterol content in the brain and ameliorates amyloid pathology in mice with AD. Proc Natl Acad Sci U S A. 2010;107:3081–3086. doi: 10.1073/pnas.0913828107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Spell C, Kolsch H, Lutjohann D, Kerksiek A, Hentschel F, Damian M, von Bergmann K, Rao ML, Maier W, Heun R. SREBP-1a polymorphism influences the risk of Alzheimer’s disease in carriers of the ApoE4 allele. Dement Geriatr Cogn Disord. 2004;18:245–249. doi: 10.1159/000080023. [DOI] [PubMed] [Google Scholar]
- 167.Brookes AJ, Howell WM, Woodburn K, Johnstone EC, Carothers A. Presenilin-I, presenilin-II, and VLDL-R associations in early onset Alzheimer’s disease. Lancet. 1997;350:336–337. doi: 10.1016/S0140-6736(05)63387-9. [DOI] [PubMed] [Google Scholar]
- 168.Helbecque N, Richard F, Cottel D, Neuman E, Guez D, Amouyel P. The very low density lipoprotein (VLDL) receptor is a genetic susceptibility factor for Alzheimer disease in a European Caucasian population. Alzheimer Dis Assoc Disord. 1998;12:368–371. doi: 10.1097/00002093-199812000-00019. [DOI] [PubMed] [Google Scholar]
- 169.McIlroy SP, Vahidassr MD, Savage DA, Patterson CC, Lawson JT, Passmore AP. Risk of Alzheimer’s disease is associated with a very low-density lipoprotein receptor genotype in Northern Ireland. Am J Med Genet. 1999;88:140–144. doi: 10.1002/(sici)1096-8628(19990416)88:2<140::aid-ajmg9>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 170.Okuizumi K, Onodera O, Namba Y, Ikeda K, Yamamoto T, Seki K, Ueki A, Nanko S, Tanaka H, Takahashi H, Oyanagi K, Mizusawa H, Kanazawa I, Tsuji S. Genetic association of the very low density lipoprotein (VLDL) receptor gene with sporadic Alzheimer’s disease. Nat Genet. 1995;11:207–209. doi: 10.1038/ng1095-207. [DOI] [PubMed] [Google Scholar]
- 171.Yamanaka H, Kamimura K, Tanahashi H, Takahashi K, Asada T, Tabira T. Genetic risk factors in Japanese Alzheimer’s disease patients: alpha1-ACT, VLDLR, and ApoE. Neurobiol Aging. 1998;19:S43–46. doi: 10.1016/s0197-4580(98)00035-9. [DOI] [PubMed] [Google Scholar]
- 172.Perry RT, Collins JS, Harrell LE, Acton RT, Go RC. Investigation of association of 13 polymorphisms in eight genes in southeastern African American Alzheimer disease patients as compared to age-matched controls. Am J Med Genet. 2001;105:332–342. doi: 10.1002/ajmg.1371. [DOI] [PubMed] [Google Scholar]
- 173.Jones L, Holmans PA, Hamshere ML, Harold D, Moskvina V, Ivanov D, Pocklington A, Abraham R, Hollingworth P, Sims R, Gerrish A, Pahwa JS, Jones N, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, van den Bussche H, Heuser I, Peters O, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Ruther E, Carrasquillo MM, Pankratz VS, Younkin SG, Hardy J, O’Donovan MC, Owen MJ, Williams J. Genetic evidence implicates the immune system and cholesterol metabolism in the aetiology of Alzheimer’s disease. PLoS One. 2010;5:e13950. doi: 10.1371/journal.pone.0013950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Luchsinger JA. Diabetes, related conditions, and dementia. J Neurol Sci. 2010;299:35–38. doi: 10.1016/j.jns.2010.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Frisardi V, Solfrizzi V, Seripa D, Capurso C, Santamato A, Sancarlo D, Vendemiale G, Pilotto A, Panza F. Metabolic-cognitive syndrome: a cross-talk between metabolic syndrome and Alzheimer’s disease. Ageing Res Rev. 2010;9:399–417. doi: 10.1016/j.arr.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 176.Panza F, Frisardi V, Seripa D, Imbimbo BP, Sancarlo D, D’Onofrio G, Addante F, Paris F, Pilotto A, Solfrizzi V. Metabolic Syndrome, Mild Cognitive Impairment, and Dementia. Curr Alzheimer Res. 2011 doi: 10.2174/156720511796391818. [DOI] [PubMed] [Google Scholar]
- 177.Reitz C, Tang MX, Schupf N, Manly JJ, Mayeux R, Luchsinger JA. A summary risk score for the prediction of Alzheimer disease in elderly persons. Arch Neurol. 2010;67:835–841. doi: 10.1001/archneurol.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Li J, Wang YJ, Zhang M, Xu ZQ, Gao CY, Fang CQ, Yan JC, Zhou HD. Vascular risk factors promote conversion from mild cognitive impairment to Alzheimer disease. Neurology. 2011;76:1485–1491. doi: 10.1212/WNL.0b013e318217e7a4. [DOI] [PubMed] [Google Scholar]