Summary
Apoptosis is a conserved cellular pathway that results in the activation of cysteine-aspartyl proteases, or caspases. To dissect the non-redundant roles of the executioner caspases-3, -6 and -7 in orchestrating apoptosis, we have developed an orthogonal protease to selectively activate each isoform in human cells. Our approach uses a split-Tobacco Etch Virus (TEV) protease under small-molecule control, that we call the SNIPer, with caspase alleles containing genetically encoded TEV cleavage sites. These studies reveal that all three caspases are transiently activated but only activation of caspase-3 or -7 is sufficient to induce apoptosis. Proteomic analysis shown here and from others reveals that 20 of the 33 subunits of the 26S proteasome can be cut by caspases, and we demonstrate synergy between proteasome inhibition and dose-dependent caspase activation. We propose a model of proteolytic reciprocal negative regulation with mechanistic implications for the combined clinical use of proteasome inhibitors and proapoptotic drugs.
Introduction
Apoptosis, or programmed cell death, is a ubiquitous and conserved cellular pathway required for development, immune cell maturation and to prevent oncogenesis (Taylor et al., 2008). Apoptosis is triggered by signaling events involving a diverse array of protein networks, organelles, and macromolecular complexes that converge upon the activation of caspases (Fuentes-Prior and Salvesen, 2004). The executioner caspases are the final proteases to be activated in apoptosis. This leads to cleavage of upwards of 1000 proteins (Dix et al., 2008; Mahrus et al., 2008) to produce the characteristic apoptotic phenotypes of membrane blebbing, nuclear condensation, DNA fragmentation, and ultimately phagocytosis by immune cells. Executioner caspases have been recently linked to non-apoptotic phenotypes such as axonal pruning, stem cell differentiation, and red blood cell enucleation(Yi and Yuan, 2009). The cellular processes that stop caspases short of inducing apoptosis in these diverse biological settings remain a mystery.
There are 12 caspase isoforms in humans but their precise and non-redundant roles for mediating apoptosis, non-apoptotic phenotypes, and innate immunity are only beginning to be fully elucidated. Extensive biochemical and structural analyses indicate that the inflammatory and initiator caspases are recruited to larger complexes that induce caspase dimerization, auto-proteolysis, and protease activation (Shi, 2004). Upon activation, the apoptotic initiator caspases-8 and -9 cleave the executioner procaspases-3, -6 and -7. The executioner caspases are translated as inactive dimers; there are at least two sites of processing in the executioner caspases; and the specific role of each proteolytic event in regulating executioner caspase activity is still unclear, both in vitro and in cells, as is the role of the prodomain in executioner caspase function.
Gene knockout studies in murine embryonic fibroblasts demonstrate that cells deficient in both caspase-3 and -7 are more resistant to a variety of apoptotic stimuli as compared to single knockouts, suggesting functional redundancy in these two executioner isoforms (Lakhani et al., 2006). Furthermore, single-cell studies reveal that the induction of executioner caspase activity is highly stochastic (Albeck et al., 2008b; Rehm et al., 2002). To further probe the function of each caspase with sharp temporal resolution, we needed to activate an individual isoform selectively and synchronously. Recent progress was reported toward building small molecule activators of the procaspases (Wolan et al., 2009), but these compounds are not yet selective enough to trigger each one of the caspases individually.
To address the goal of activating a single executioner caspase isoform in a site-selective manner, we describe the optimization of a previously split NIa Tobacco Etch Virus (TEV) protease (Wehr et al., 2006). We demonstrate that an engineered variant of split-TEV can be robustly activated in cells to specifically cleave each of the executioner caspase isoforms. These studies reveal that activation of caspase-3 or -7, but not caspase-6, is sufficient to induce apoptosis. Remarkably, the activation is transient for each isoform. These data support a model of antagonistic proteolysis between the executioner caspases and the proteasome.
Results
The SNIPer: Design, Optimization, and Characterization in Mammalian Cells
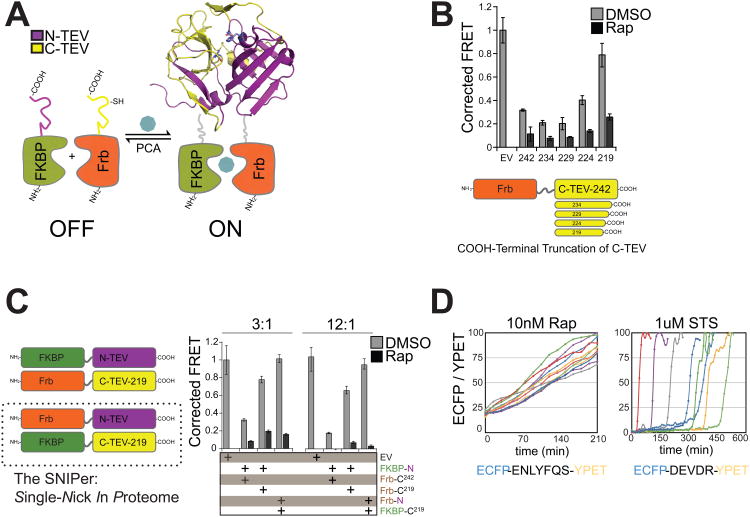
Our initial split-TEV construct was based on a previous design developed for protein-complementation assays (Wehr et al., 2006) (Figure 1A). The full-length TEV variant S219V was split into two component fragments: N-TEV (residues 1-118) and C-TEV (residues 119-242). This design inactivates TEV protease by separating the catalytic triad residues Asp44 and His81 from the Cys151 nucleophile. N-TEV was fused to the C-terminus of FKBP with a 10-amino acid (GGGGS)2× linker between the two domains, and C-TEV was fused to the C-terminus of Frb (the rapamycin-binding domain of mTOR kinase). Therefore, protease activity is conditionally restored by the addition of rapamycin, which promotes split-TEV fragment association and ligand-dependent protein folding.
Figure 1. Engineering the SNIPer for Conditional Proteolysis.
(A) Generalized scheme for a ligand-inducible orthogonal protease based on the protein complementation system. (B) Plasmids with deletions at the C-terminus of Frb-C-TEV or empty vector (EV) were co-transfected with FKBP-N-TEV and ECFP-TevS-YPET in 293T cells. 10nM Rap was added for 12h and FRET measurements were recorded using a fluorescent microplate reader. Data are presented as corrected averages of FRET/ECFP from an experiment performed in quadruplicate (Error Bars±SD). (C) N-TEV and C-TEV-219 were fused to either FKBP or Frb and assayed by FRET in quadruplicate as above. Transfections were performed with 3× or 12× molar excess of Tev constructs to reporter plasmid (Error Bars±SD). (D) The kinetics and cell-to-cell heterogeneity of SNIPer-mediated protease activity were compared to endogenous caspase activation by live-cell fluorescent imaging. HEK293 cells were transfected with the SNIPer and ECFP-TevS-YPET or ECFP-DEVDR-YPET, a reporter for caspase-3/-7 activity in cells, and treated with 10nM Rap or 1uM staurosporine. Live-cell FRET measurements were recorded every 15 minutes. See also Figure S1.
We assayed for reconstituted protease activity in 293T cells by co-transfecting both N-and C-TEV halves and a FRET-based ECFP-YPET reporter that is cleaved by TEV. In the presence of rapamycin (Rap), the split-TEV construct extending to residue 242 cleaved the protease reporter as compared to an empty vector control (Figure 1B). However, significant processing also occurred in the absence of Rap, suggesting the two halves can associate and reconstitute activity in the absence of a small molecule dimerizer. Another group recently implemented the published N-TEV and C-TEV design in mammalian cells and similarly observed leaky TEV protease activity in the absence of Rap (Williams et al., 2009). They reduced Rap-independent activation by lowering the amount of split-TEV DNA that was transiently transfected or by targeting one fragment to the cell membrane via a genetically encoded myristoylation tag. However, our goal was to engineer cell death pathways using stable cell lines, which requires a split-TEV design with no background activity. We hypothesized that the C-terminus of the TEV was causing spontaneous refolding of the fragments in the absence of Rap (Figure S1A for a model). A systematic set of deletion mutants was created from the C-TEV119-242 fragment to generate four truncated C-TEV constructs. Co-expression of FKBP-N-TEV and Frb-C-TEV-219 yielded inducible proteolysis upon addition of Rap with reduced leaky proteolytic activity in mock-treated cells as confirmed by the FRET assay (Figure 1B) and immunoblot analysis (Figure S1B, top panel).
Further optimization was required due to detectable TEV activity for FKBP-N-TEV and Frb-C-TEV-219 in the absence of Rap (Figure 1C and Figure S2B, bottom panel). We tested the opposite FKBP and Frb protein fusions to N-TEV and to the enhanced C-TEV-219 fragment, and we observed that Frb-N-TEV(1-118) and FKBP-C-TEV(119-219) maintained robust TEV activity in the presence of Rap with little background. As an initial test of split-TEV stability, we found that proteolysis of the FRET reporter was not enhanced after treatment with Rap and 500nM PS-341 (Bortezomib), a clinically available proteasome inhibitor (Figure S1C), indicating that a proteasome inhibition was not sufficient to promote increased split-TEV activity in cells. Each split-TEV protein fragment was also detected by immunoblotting after blocking protein translation with cyclohexamide for 8hr, suggesting both were stable in cells (Figure S1D). Both removal of the C-terminal end of the C-TEV fragment and fusion pair optimization to FKBP and Frb were necessary to generate a rapamycin-inducible split-TEV protease for mammalian cells. We refer to this optimized construct pair as the SNIPer (Single Nick in Proteome) since there are no known natural TEV cleavage sites in the human proteome except for those we introduce.
We observed rapid activation of the SNIPer in individual cells (Figure 1D, left panel) using the TevS-FRET reporter in transiently transfected HEK293 cells. Cells were monitored over a 3 hr time frame over which a linear increase in TEV activity was measured by single-cell fluorescent microscopy. Approximately 50% of the ECFP-YPETENLYFQ|S substrate was cleaved by 100 ± 20 min. Importantly, activation of the SNIPer at the single-cell level was more synchronous than endogenous caspase pathways during experiments in which SNIPer-expressing cells were directly compared to HEK293 cells transfected with ECFP-YPETDEVD|R, a reporter for caspase-3 and -7 activity (Albeck et al., 2008a) (Figure 1D, right panel), and treated with 1uM staurosporine (STS). We conclude that SNIPer activation kinetics matches or exceeds endogenous DEVDase activation kinetics during apoptosis, which prompted its further use to probe executioner caspase signaling in cells.
Engineering Orthogonal Procaspase-3 and -7 Alleles for Selective Activation by the SNIPer
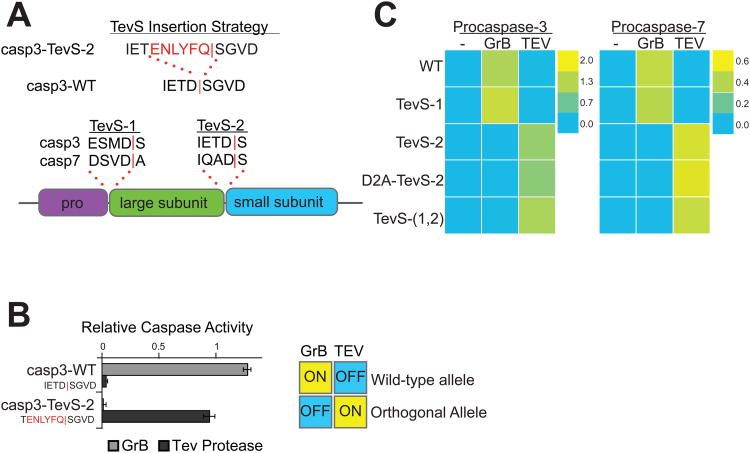
The executioner caspases are translated as inactive zymogens that are cleaved by initiator caspases, by other executioner caspases in trans, or by auto-activation. Two sites of processing exist in procaspase-3 and -7: one at the junction of the prodomain and large subunit (site-1) and the second between the large and small subunits in the intersubunit linker (site-2) (Earnshaw et al., 1999). To date, there are no reported crystal structures of procaspase-3 or procaspase-7 with defined electron density in the intersubunit linker or the prodomain. However, previous studies have shown that mutation of several residues within the “loop bundle” of the intersubunit linker of caspase-3 influences the activity of the protease (Feeney et al., 2006). Therefore, we inserted a TEV recognition sequence (TevS) into the P1 position at site-2 on procaspase-3, replacing the P1 Asp of the endogenous caspase site, P5-IETD-P1′ with the preferred P6 Glu for the Tev cleavage site (TevS), P7-ENLFYQ-P1′. This resulted in a chimeric substrate at site-2 (IETENLYFQ) that maintained the integrity of the caspase cleavage site from P4-P2 while eliminating the aspartic acid critical for cutting by the caspases themselves (Figure 2A, top panel). In principle, this approach would render the sites cleavable by the SNIPer only. Since caspases and TEV protease share similar preference for the small P1′ residues (Gly, Ser and Ala), the wild-type peptide sequence after the scissile bond was also maintained. Once the TevS is cleaved, a small peptide ‘scar’ is left behind with an ENLYFQ sequence on the C-terminus.
Figure 2. Design and In Vitro Characterization of Orthogonal Procaspase-3 and -7 Alleles.
(A) We employed the indicated TevS insertion strategy to convert caspase sites into SNIPer-cleavage sites as shown for caspase-3. The sequences (P4-P1′ with a red bar to denote the scissile bond) and location of site-1 and site-2 are shown in the context of executioner caspase domain structure below. (B) In vitro activation of wild-type procaspase-3 (Casp3-WT) or Casp3-TevS was assayed by expressing each allele in E. coli, treating with either rat Granzyme-B (20nM) or TEV Protease(1.1uM) for 1h, and measuring caspase activity with 10uM Ac-DEVD-AFC. Data are presented as the average rate of substrate cleavage (RFU s-1) from an experiment performed in triplicate (Error Bars±SD). (C) Caspase activity of the procaspase-3 and -7 matrix of orthogonal alleles was performed as above and is presented as a heat map. Abbreviations: WT, wild-type; TevS-1, TevS insertion at site-1; TevS-2, TevS insertion at site-2; D2A-TevS-2, TevS insertion at site-2 with a non-cleavable site-1; and TevS-(1,2), TevS insertions at site-1 and site-2. See also Figure S2.
With this insertion strategy we generated four alleles in both procaspase-3 and -7: one where TevS was inserted between the pro-domain and the large subunit (TevS-1), one where the TevS was inserted between the large and small domain (TevS-2), one where two insertions were made (TevS-1,2) and one that contained the TevS inserted between the large and small subunits but that also contained a mutation of Asp to Ala mutation at site-1 to prevent prodomain cleavage (D2A-TevS-2) (Figure 2A, bottom panel). To begin to test the impact of these mutations on processing and activity, a His-tagged version of the Casp3-D2A-TevS-2 protein was over-expressed in E. coli and purified by Ni-NTA affinity chromatography. Casp3-D2A-TevS-2 was co-incubated with recombinant TEV protease, and the processed samples were analyzed by SDS-PAGE to confirm that the engineered caspase construct was indeed cleaved at the expected single site (Figure S2A). Furthermore, the measured catalytic efficiency (kcat/KM) for the engineered caspase was in good agreement with the value for wild-type active caspase-3 produced in our lab (Figure S2B).
To further probe the functional consequence of processing at site-1 and site-2 in vitro, we constructed a matrix of the three procaspase-3 and -7 alleles harboring TevS sites that replace one or both endogenous caspase processing. Granzyme-B (GrB), a serine protease secreted by cytotoxic T-lymphocytes, is capable of cleaving procaspase-3 at site-2 (Casciola-Rosen et al., 2007), but it does not process Casp3-TevS-2 since that cleavage site is mutated. Incubation of Casp3-WT and Casp3-TevS alleles with active GrB or Tev confirmed Casp3-TevS-2 is an orthogonal caspase; it is activated by TEV protease but not by GrB (Figure 2B).
Each procaspase-3 and -7 allele was expressed as a proenzyme in E. coli, treated with GrB or TevS for 1 hr, and relative caspase activity was measured with Ac-DEVD-AFC (Figure 2C). Immunoblots confirmed the predicted cleavage patterns for TEV protease or GrB for procaspase-3 allele series (Figure S2C). Cleavage by GrB at IETD175 at site-2 was sufficient to activate WT and TevS-1 procaspases variants, both of which are WT for site-2. TEV processing at TevS-2 was sufficient to activate both caspase-3 and -7-TevS-2, regardless if the prodomain is cleavable. TEV protease processing at TevS-1 was not sufficient to induce activity for either isoform. Together, these results confirm that processing at site-2 is both necessary and sufficient for activation in vitro by either TEV protease or GrB. Processing of the prodomain is neither sufficient for activation nor necessary for full levels of activation in conjunction with processing at the inter-subunit linker.
Selective Activation of Orthogonal Procaspase-3 and -7 Alleles in Stable Human Cell Lines
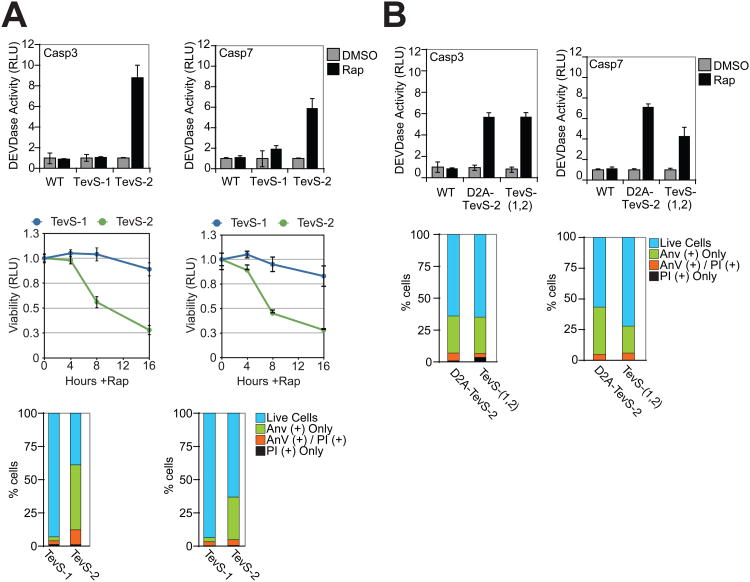
The matrix of caspase-TevS alleles was stably engineered as single-copy integrants into HEK293 cells as described in the Supplementary Experimental Procedures in order to minimize the over-expression phenotypes associated with transient transfection. HEK293 SNIPer cell lines expressing Casp3-WT, Casp3-TevS-1 or Casp3-TevS-2 were induced for 6 hr with 10nM Rap and assayed for DEVDase activity. Addition of Rap induced a 8-fold increase in caspase-3 activity in the Casp3-TevS-2 line as compared to the same cells treated with DMSO only (Figure 3A, left top and bottom panels). After 8 hr, the Casp3-TevS-2 cells showed robust GFP-Annexin-V staining (Ernst, 1998), a hallmark of apoptosis involved in the recruitment of macrophages for engulfment and clearance of apoptotic cells. By 16 hr, only 25% of the cells remained viable. In contrast, no increase in DEVDase activity, Annexin-V staining or cell death was observed with the Casp3-WT or Casp3-TevS-1 lines treated with Rap. Similar functional results were obtained for the Casp7-TevS-2 line, which showed a 6-fold induction of DEVDase activity by 6 hr, an increase Annexin-V staining by 8 hr, and a large decrease in cellular viability by 16 hr (Figure 3A). Overall, the relative phenotypic consequences of the WT, TevS-1 and TevS-2 lines were similar for procaspase-3 and procaspase-7, although total DEVDase activity and Annexin-V staining were less for activated Casp7-TevS-2 as compared to the corresponding procaspase-3 allele. These data establish that SNIPer-mediated cleavage and activation of either procaspase-3 or -7 at site-2 alone is sufficient to induce robust caspase activation and rapid apoptosis in human cells.
Figure 3. Phenotypic Analysis of SNIPer-Mediated Activation of Procaspase-3 and -7 in Cells.
(A) HEK293 cells stably expressing the SNIPer and indicated procaspase-3/-7 alleles were treated with DMSO or 10nM Rap over the course of 16 hr. The indicated cells lines were assayed for caspase activity using the Caspase-Glo-3/-7 assay at 6 hr (top row), for total cellular viability using the Cell-Titer-Glo assay the indicated times (line graph, bottom row), and for phosphatidylserine (PS) exposure/membrane integrity by staining with GFP-Annexin-V and propidium iodide (PI) and imaging by fluorescent microscopy at 8 hr. Each data point is presented as an average of triplicate experiments (Error Bars±SD). (B) HEK293 stably expressing procaspase-3/-7 D2A-TevS and TevS-(1,2) alleles were analyzed for caspase activity and for GFP-Annexin-V/PI staining exactly as above. See also Figure S3.
To further understand the functional consequences of prodomain cleavage in cells, we compared the D2A-TevS-2 and TevS-(1,2) alleles (Figure 3B). In parallel to the in vitro results, prodomain removal is neither necessary nor sufficient for caspase activation and apoptosis in cells. A comparison of the lines expressing TevS-2, D2A-TevS-2 and TevS-(1,2)-Casp7 suggested that prodomain removal results in modestly lower DEVDase activity at 6 hr (∼40%) and reduced Annexin-V staining.
It has been proposed that the executioner caspases may amplify apoptotic pathways via cleavage of Bid to generate tBid, which induces mitochondrial outer membrane permeability (MOMP), cytochrome-C release and activation of caspase-9 (Green, 2005) (Figure S3A). To examine if executioner caspase activation can ‘feedback’ and result in the activation of initiator caspases, we tested whether SNIPer-mediated activation of the Casp-7-TevS-2 allele was sufficient to promote MOMP. We observed several biochemical hallmarks of MOMP after 4 hr of Rap induction, including Bid cleavage and cleavage of procaspase-9 (Figure S3B). As expected, extensive PARP processing was also detected. These results further confirm the existence of positive feedback signaling between the executioner caspases and upstream apoptotic pathways.
Activated Caspase-3 and -7 are Proteasome Substrates and are Stabilized by a Proteasome Inhibitors
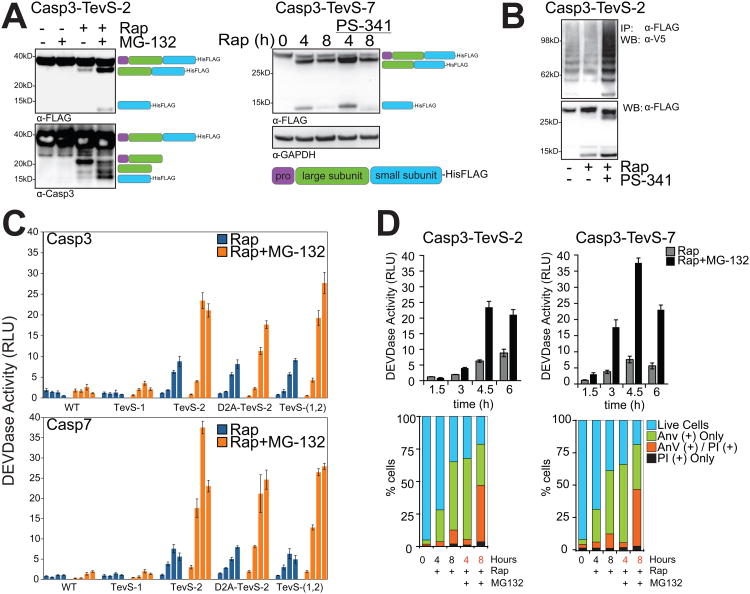
Processing of engineered procaspase alleles by the SNIPer was examined in cells by immunoblotting with an anti-FLAG antibody that detects the C-terminal His/FLAG epitope tag on both Casp3-TevS-2 and Casp7-TevS-2 constructs. The anti-FLAG antibody detects the full-length proenzyme (32 kDa), a processed form cleaved only at site-1 (29 kDa), and the form cleaved at site-2 to generate the small subunit (12 kDa). We also used a polyclonal caspase-3 antibody that recognizes both caspase-3 subunits. After the addition of Rap to the Casp3-TevS-2 cell line for 6hr, which correlates with the induction of DEVDase activity and apoptosis, the small subunit (∼12kDa) of cleaved caspase-3 could not be readily detected by immunoblot Similar results were observed for Casp7-Tevs-2 (Figure 4A).
Figure 4. Caspase-3 and -7 Regulation by the Proteasome.
(A) The Casp3-TevS-2 line was treated with DMSO or 10nM Rap ±5uM MG-132 for 6 hr and cell lysates were blotted with anti-FLAG M2 (left panel, top) or anti-caspase-3 (left panel, bottom) antibodies. The expected processing intermediates detected by each antibody are shown to the right of the immunoblots. The Casp7-TevS-2 line was treated with 10nM Rap for 4 or 8 hr ±500nM PS-341 and immunoblotted with anti-FLAG M2 (right panel) and anti-GAPDH. (B) The Casp3-TevS-2 line was transfected with pcDNA3.1-V5-Ubiquitin for 24 hr and treated with DMSO, 10nM Rap or 10nM Rap + 500nM PS-341 for 4 hr. Cell lysates were immunoprecipitated with anti-FLAG M2 agarose and immunoblotted for covalent ubiquitin with an anti-V5 antibody or with anti-flag to assess caspase-3 recovery. (C) The Casp3-TevS-2 and Casp7-TevS-2 lines were treated with 10nM Rap (blue bars) or 10nM Rap and 5uM MG-132 (orange bars). Caspase activity was measured every 1.5 hr in triplicate (Error Bars±SD). (D) Casp3-TevS-2 and Casp7-TevS-7 cell lines were treated with 10nM Rap ±5uM MG-132 and assayed for caspase activity as above (top) or by staining with GFP-Annexin-V and PI and imaging with fluorescent microscopy (bottom) for indicated time courses in triplicate (Error Bars±SD). See also Figure S4.
Recent studies have suggested that caspase-3 is ubiquitinated and degraded by the proteasome (Suzuki et al, 2001; Choi et al. 2009). To address the role of the Ubiquitin-Proteasome System (UPS) in mediating turnover of SNIPer-activated caspases, we added one of two different proteasome inhibitors, MG-132 or PS-341, in combination with Rap, which stabilized the cleaved caspase subunits for both caspase-3 and caspase-7, respectively (Figure 4A). Furthermore, Rap and PS-341 treatment promoted the accumulation of polyubiquitinated caspase-3 detected by immunoblotting as compared to Rap alone (Figure 4B). To evaluate the effects of the proteasome inhibition on the induction of caspase activity, a time course of Rap-induced processing of the procaspase-3 and -7 allele series was performed ±5uM MG-132. Addition of MG-132 resulted in a 6- to 8-fold enhancement in Rap-induced DEVDase activity for those caspase-3 and -7 alleles that were active in cells (TevS-2, D2A-TevS-2 and TevS-1,2 variants) (Figure 4C; Figure S4A).
An increase in caspase activity induced by MG-132 directly correlated to the severity and kinetics of the apoptotic phenotype (Figure 4D). For example, at 4h, MG-132 increased Annexin-V staining to 60% of cells compared to 25% with Rap alone. By 8 hr, almost 50% of the cells were double positive for Annexin-V and propidium iodide, a marker for the loss of cell membrane integrity. Thus, selective activation of either caspase-3 or -7 in combination with MG-132 robustly increased the level of DEVDase activity, accelerated apoptotic hallmarks such as Annexin-V staining and, by 8 hr, resulted in a large fraction of late-stage apoptotic cells. A minor population of cells were PI positive only, excluding necrotic cell death as the primary response to proteasome inhibitors. The addition of both Rap and MG-132 for as long as 8 hr to Casp3-WT or Casp7-WT control lines did not induce a increase in caspase activity (Figure 4C), which further established a direct relationship between the UPS and the activated forms of capase-3 and -7 generated by the SNIPer. We also tested the possibility that proteasome inhibitors stabilized the SNIPer itself. However, HEK293 cells stably expressing both SNIPer and the TevS-FRET reporter showed no detectable proteasome inhibitor-dependent enhancement of TEV activity by the FRET assay, which was further confirmed by immunoblotting to assess FRET reporter cleavage (Figure S1C).
We observed similar levels of Casp7-TevS-2 activation in experiments in which MG-132 or the more potent and selective PS-341 was directly compared (Figure S4B). To determine if proteasome inhibition promoted executioner caspases in a second cell line, we co-expressed the SNIPer in HeLa cells with retroviruses expressing the Casp3-WT and Casp3-TevS-2 alleles, and we successfully activated the SNIPer and Casp3-TevS-2 in a Rap-dependent fashion (Figure S4C). Moreover, the level of caspase-3 induction was potently enhanced by MG-132.
Activation of Caspase-6 is not Proapoptotic except in Conjunction with Proteasome Inhibition
Procaspase-6 activation was tested in vitro and in cells with the SNIPer. Although procaspase-6 is defined as an executioner caspase and is closely related to caspase-3 and -7 by sequence homology, several distinct features have been observed for this isoform. First, procaspase-6, unlike procaspase-3 and -7, has been reported to self-activate when over-expressed in 293T cells (Klaiman et al., 2009). There is an additional loop in the intersubunit linker that bisects a region known as the “safety catch” at the homologous site in procaspase-3. Second, procaspase-6 prefers Val or Thr at P4 unlike caspase-3/-7, which prefer acidic residues at this position (Thornberry et al., 1997). Procaspase-6, therefore, resembles the initiator caspases by a tendency to self-activate and by similar substrate preferences at P4.
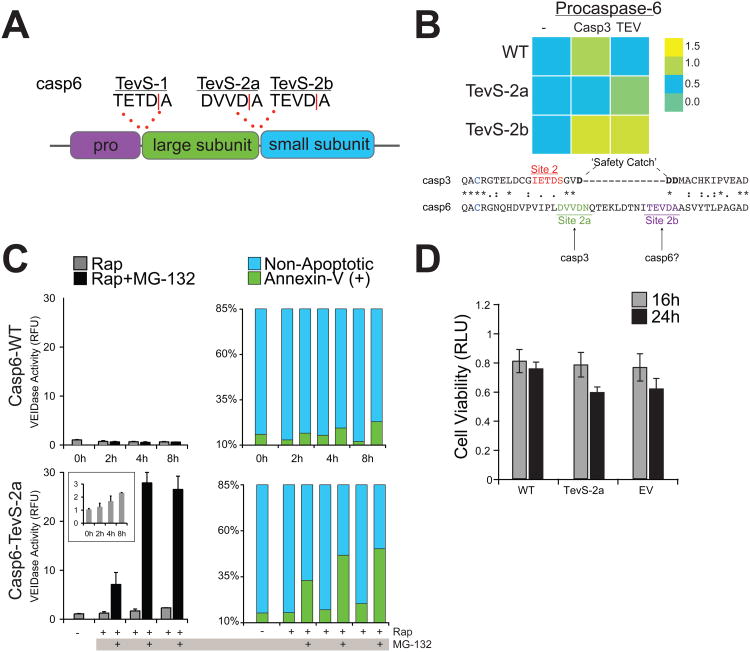
To probe caspase-6 signaling in apoptosis, we applied the TevS insertion strategy to engineer a series of orthogonal procaspase-6 alleles. In addition to site-1 at the junction of the prodomain and the large subunit, there are two reported cleavage sites in the intersubunit linker of procaspase-6 (Figure 5A): DVVD|N, which we refer to as site-2a, and TEVD|A, or site-2b. Although TEV protease can process sites with a P1′ Asn with reduced efficiency, an Ala was substituted at P1′ to enhance proteolysis at Site 2a. Thus, three TevS alleles were engineered by insertion at each of the three reported sites (site-1, site-2a with a P1′ Ala, and site 2b).
Figure 5. Proteasome Inhibition Converts Procaspase-6 to an Apoptotic Executioner Caspase Isoform.
(A) Sites for which the TevS insertion was applied for caspase-6. (B)In vitro caspase activity of the procaspase-6 matrix of orthogonal alleles is presented as a heat map (top). Caspase activity was measured by expressing each allele in E. coli, treating with active caspase-3 (2nM) or Tev Protease(1.1uM) for 1h with 30uM Ac-VEID-AFC in triplicate, correcting for caspase-3 VEIDase activity where appropriate. Abbreviations: WT, wild-type; TevS-2a, TevS insertion at site-2a with P1′ Ala; TevS-2b, TevS insertion at site-2b. Alignment of the protein sequences of procaspase-3 (CASP3_HUMAN) and procaspase-6 (CASP6_HUMAN) indicates an extra loop region in caspase-6 that harbors site-2a and -2b. (C) The Casp6-WT (top) line was treated with 10nM Rap ±5uM MG-132 over a course of 8h and assayed for caspase-6 activity with 50uM VEID-R110 (left) or by staining with GFP-Annexin-V imaging with fluorescent microscopy. Casp6-TevS-2 (bottom) was treated identically. The inset is the same assay in the range of 0-3 RFU. Data are presented as the average from an experiment performed in quadruplicate (Error Bars±SD). (D) The Casp3-TevS-2 line was treated with 10nM Rap for 16 or 24 hr and cell viability was measured with Cell Titer-Glo (Promega, WI). See also Figure S5.
Each construct was expressed in E. coli and processed in vitro with recombinant TEV protease or recombinant active caspase-3, which is capable of activating WT procaspase-6. A fluorogenic Ac-VEID-AFC peptide substrate, the preferred substrate for caspase-6, was used in these experiments (Figure 5B). TEV protease could process and activate the TevS-2 variants (2a and 2b), whereas recombinant caspase-3 activated Casp6-WT, and, unexpectedly, Casp6-TevS-2b. Because this allele is WT at site-2a, and caspase-3 was not able to activate the TevS variants at site-2a, we infer that site-2a is a functional caspase-3 processing site (Figure 5B). TEV protease did activate Casp6-TevS-2a with an Asn at P1′ (data not shown), but total VEIDase activity was less than that of the Ala mutant.
Casp6-TevS variants were transduced into HEK293 cells with the SNIPer, and cell-based assays were employed to measure caspase-6 activity in cells. Casp6-TevS-2a was nearly fully processed by the SNIPer by 6 hr (Figure S5A), but induced VEIDase activity was only 2.4-fold above the control (Figure 5C, left panels). Cellular viability was not significantly affected by caspase-6 activation at 16 or 24 hours post-Rap for the Casp6-TevS-2a line as compared to Casp6-WT or to an empty vector control (Figure 5D), and we did not observe a significant apoptotic response by Annexin-V staining (Figure 5C, right panels).
We hypothesized that the UPS may negatively regulate caspase-6 activity as well. Cells harboring the SNIPer and the Casp6-TevS-2a allele were treated with Rap±MG-132. Remarkably, VEIDase activity with Rap and MG-132 was elevated 14-fold at 4hr compared to Rap alone or to Casp6-WT or Casp6-TevS-1 control lines (Figure 5C, left panels). Amplification of VEIDase activity was accompanied by a dramatic increase (50%) in Annexin-V positive cells (Figure 5C, right panels). To further probe the mechanism by which MG-132 promotes caspase-6 activity and apoptosis, we immunoblotted for caspase-6 before and after SNIPer-mediated cleavage. Surprisingly, MG-132 stabilized the unprocessed Casp6-TevS-2a zymogen (Figure S5A, lanes 1 and 2). This result was independent of SNIPer-induced cleavage and in stark contrast to the mode of caspase-3 and -7 regulation by proteasome. When Casp6-TevS-2a cells were treated with Rap±MG132 (Figure S5A, lanes 3 and 4), the small subunit was also stabilized with MG-132. Similar results were obtained with PS-341. We next tested if caspase-6 activation induced MOMP, and after 8hr of sustained Casp6-TevS-2a activation with Rap only, we observed no evidence for MOMP. However, addition of PS-341 and Rap caused a large increase in Bid cleavage, procaspase-9 and PARP (Figure S5B). These results suggest that both procaspase-6 and activated caspase-6 are targets for negative regulation by the UPS, and caspase-6 is only proapoptotic in the presence of proteasome inhibitors. The severity of the apoptotic phenotype directly correlates with increased VEIDase activity in cells, stabilization of both the pro- and active forms (unlike casase-3 and -7), and recruitment of the endogenous apoptotic machinery via MOMP.
Caspases Attack the 26S Proteasome, and Caspase Activation Synergizes with Proteasome Inhibition
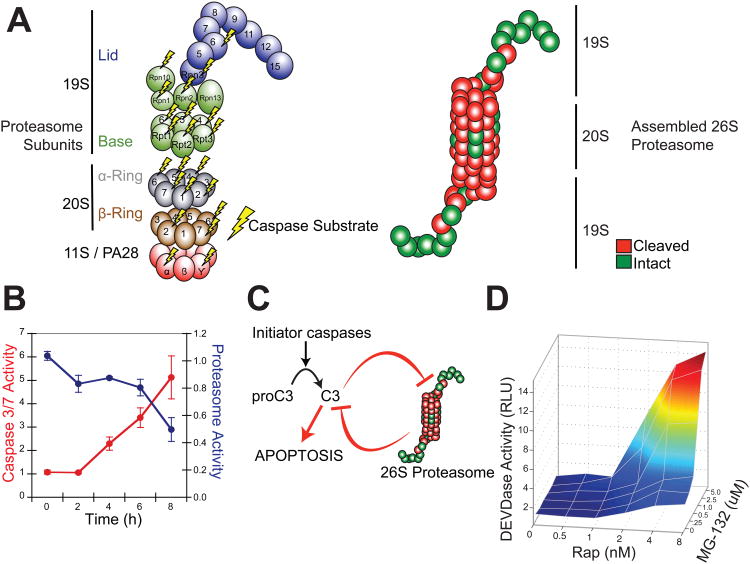
Recently gel-based proteomic data from several other laboratories have identified 10 proteins from the 26S proteasome, mostly in the 19S cap, that are cleaved during apoptosis (Sun et al., 2004; Adrain et al., 2004; Van Damme et al., 2005; Thiede et al., 2005 Jang et al., 2007). We have described a new proteomics strategy for the global identification of protease substrates in mammalian cells (Mahrus et al., 2008). This method utilizes an engineered protein ligase called subtiligase to biotin-tag and enrich for nascent N-termini produced during proteolysis, which are then sequenced by LC/MS/MS. A distinct advantage to this technology is that the exact site of proteolysis is identified. Our group has identified >1000 caspase substrates in several cell lines induced by a variety of apoptotic drug stimuli (S. Mahrus, H. Nguyen, and J. Wells, manuscript in preparation). Within this data set, we found 12 caspase substrates in the 26S proteasome and defined their cleavage sites from MS/MS data (Table S1). Ten of these are novel. Surprisingly, the substrate list included two additional α-subunit sites and three β-subunit sites of the 20S catalytic core particle. One of the three β-subunit cleavage sites identified by our degradomics experiments, PSB7_HUMAN or β2, is a bona fide proteasome catalytic subunit, providing trypsin-like hydrolase activity to the complex. The site of caspase proteolysis in the β2 subunit is only 10 residues from the catalytic N-terminal nucleophile and was likely missed in previous gel-based studies. These mass spectrometry data do not address the extent of cutting for each of these subunits by caspases, nor their functional impact. Nonetheless, the aggregate of our data plus previous publications show that 20 of 33 of the subunits in the 26S proteasome can be cleaved in a cellular context by caspases (Figure 6A). Further studies will be required to assess the functional impact and timing of these individual cuts on proteasome function.
Figure 6. Reciprocal Negative Regulation between the 26S Proteasome and Executioner Caspases.
(A) Novel and published proteasome subunits identified as caspase substrates are mapped to an inventory of 20S core, 19S lid/base and 11S activator subunits (left). Cleaved subunits (red) are also mapped onto a model of the fully assembled, multimeric 26S proteasome (right). (B) The Casp3-TevS-2 line was treated with 10nM Rap over a course of 8 hr and assayed for caspase activity with Caspase-Glo-3/-7 and protease activity with Proteasome-Glo (Chymotrypsin-Like). Data are presented as the average from an experiment performed in triplicate (Error Bars±SD; note different scales). (C) A model for reciprocal negative regulation between activated executioner caspases and the 26S proteasome-ubiquitin system predicts synergy between caspase activation and proteasome inhibition. (D) The Casp3-TevS-2 line was co-treated with the indicated concentrations of Rap (0-8nM) and MG-132 (0-5uM) for 4 hr and caspase activation was measured with Caspase-Glo-3/-7 in triplicate. See also Table S1.
We then assayed for proteasome function during apoptosis in our system. As caspase activity increased upon activation of the Casp3-TevS-2 allele by the SNIPer, proteasome activity decreased coincidently (Figure 6B). These data lead us to hypothesize that there is a negative feedback loop between the executioner caspases and the proteasome (Figure 6C). This model predicts that caspase activation should synergize with proteasome inhibition. To test this prediction, we modulated the activation of Casp3-TevS-2 by the SNIPer by titrating Rap (EC50 of ∼4 nM for SNIPer activation of the caspases) in combination with titration of MG-132 to achieve a range of proteasome inhibition (IC50 of 0.8 uM). We observed dramatic synergy between caspase-3 activation and inhibition of the proteasome (Figure 6D). For example, at the EC50 for SNIPer activation by Rap (∼2 nM), the addition of saturating MG-132 (5uM) yielded a 7.7-fold induction of caspase activity as compared to 1.6-fold induction with Rap only. Similarly, at the EC50 for MG-132 (0.8 uM), addition of saturating amounts of Rap (8nM) induced a 8.9-fold increase in caspase activity as compared to 2.8-fold with Rap alone.
Discussion
Design Considerations for SNIPer
We envisioned an inducible orthogonal protease and substrate system with the following design principles: (1) the orthogonal protease should have no reported substrates in the human or mouse proteome (2) expression and activation must be well tolerated in mammalian cell lines that stably express the protease (3) the time-course of induced protease activity should match or exceed the expected time-course of apoptosis (4) the cognate protease substrate sequence must be tolerated within the context of the target protein and (5) there can be no ‘leaky’ protease activity when the orthogonal protease and an engineered caspase isoform are co-expressed in living cells.
The remarkable specificity of TEV protease is attributed to the 7-mer cleavage sequence ENLYFQ/S (Parks et al., 1994). Although TEV is most often used as recombinant protein in vitro, the Naysmith group demonstrated that TEV protease is well tolerated when expressed in S. cervisiae or D. melanogaster (Pauli et al., 2008; Uhlmann et al., 2000), and that ectopic TEV protease expression was capable of cleaving specific protein targets with genetically encoded TevS sequences. Wehr and co-workers elegantly expanded upon this approach by engineering a split-TEV construct based on protein complementation assay technology, and they demonstrated regulated TEV protease activity in several diverse cell-culture systems (Wehr et al., 2006). However, when we implemented this design as fusions to the ligand-binding domains FKBP and Frb, there was only modest control of activity upon addition of Rap, especially at higher expression levels, and Williams and colleagues recently observed a similar phenomenon (Williams et al., 2009).
We made two essential changes to the previous split-TEV construct: (1) removal of the C-terminal peptide from the C-domain in order to prevent the spontaneous association between N-and C-TEV fragments in the absence of Rap and (2) optimization of the fusion domain partners such that Frb is fused to N-TEV and FKBP to the truncated C-TEV variant. We detect little to no background caspase activity for TevS-caspase alleles in the absence of Rap, and stable cell lines harboring these alleles and the SNIPer were readily produced and showed no impairment of growth (data not shown). Both fragments of the SNIPer exhibited reasonable post-translational stability in cells, and Rap-induced activation was rapid and synchronous as compared endogenous caspase pathways.
Orthogonal Activation of the Executioner Caspases Reveals Regulation by the UPS
The SNIPer can be used to synchronize populations of cells expressing caspase alleles that mimic discrete states of caspase activation, and this approach has revealed the specific processing requirements for executioner caspase activation and maturation, which are difficult to deconvolute in cells (Pop and Salvesen, 2009). Targeted and synchronous caspase activation may facilitate additional functional studies to understand how cell-to-cell variability, pathway redundancy and multiple levels of positive and negative feedback regulate cell death processes.
The constructs presented here should also serve as important tools for further studying the role of the UPS in restraining caspase activity. A recent study demonstrated that prodomain release was necessary for cIAP-1 binding to caspase-7, which is then polyubiquitinated in vitro upon binding cIAP-1 (Choi et al., 2009). Our TevS insertion strategy maintains the native P1′ residue, and cIAP-1 binding to cleaved caspase-7 explicitly depends the presence of the N-terminal Ala of the large subunit. We have conducted preliminary studies showing that RNAi-mediated knockdown of cIAP-1, cIAP-2 or XIAP has a modest effect (1.5-2 fold) on enhancing caspase-3 or -7 activity induced by the SNIPer (data not shown). Nevertheless, depletion of individual IAPs did not phenocopy pharmacological inhibition of the proteasome. Several factors could explain this result, including redundancy between the IAPs, different cellular compartment(s) where the SNIPer is activating caspases as compared to DISC or the apoptosome, the stoichiometry of orthogonal caspase to IAP expression, and the kinetics of the SNIPer relative to endogenous initiator caspases. Alternatively, additional E3-ligases may mediate caspase degradation.
Orthogonal caspase activation by the SNIPer revealed synergy between caspase activation and pharmacological inhibition of the proteasome, and these data are consistent with a model in which the UPS potently restricts executioner caspase activity while the caspases disable the proteasome via subunit proteolysis during apoptosis. Our new proteomics data reveals that caspase proteolysis of the proteasome subunits is more extensive than previously reported. Strikingly, caspase-6 can induce apoptosis only when activated in combination with proteasome inhibitors, and in this context the UPS appears to control the propagation of positive feedback from SNIPer-induced caspase activation to MOMP.
This is the first example of reciprocal negative regulation between proteases that we are aware of. Mutual antagonism between signaling molecules is metabolically ‘expensive,’ but it is a common regulatory motif that allows for rapid pulses of activity (Rosenfeld et al., 2002), and we believe that proteasome may also constrain homeostatic (non-apoptotic) caspase activity in this fashion. Our data also provide an additional mechanism for bistability in apoptotic signaling. There are additional feedback loops between the executioner caspases and their substrates in apoptosis pathways, including caspase proteolysis of substrates yielding neo-epitopes that mimic SMAC/Diablo, an IAP inhibitor (Hell et al., 2003).
The caspase-6 zymogen is essentially converted to an apoptotic effector caspase by MG-132. Recently, a targeted knockout of PSMC1 (a 19S proteasome subunit) in the mouse neuron results in the depletion the 26S proteasome and yielded a neurodegenerative-like phenotype including the presence of cleaved caspases (Bedford et al., 2008). Similarly, genetically encoded proteasome reporters are potently induced in the presence of pathogenic Huntington's Disease (HD) alleles (Wang et al., 2008). Caspase-6 has already been implicated in the pathogenesis of the disease, but in the context of the present data, a second proteasome-caspase axis of mutual antagonism could exist in which HD patients could have impaired neuronal UPS function, stabilizing and potentially activating procaspase-6, which has been implicated in cleaving Htt (Graham et al., 2006), a crucial event in the etiology of the disease.
The proteasome inhibitor Bortezemib (PS-341) is approved as first-line therapy for treating multiple myeloma (MM), a severe hematopoetic malignancy caused the clonal proliferation of plasma cells (Raab et al., 2009). Proteasome inhibitors induce apoptosis in these cell types (Lee et al., 2003), which can be blocked by caspase inhibitors. Much research has focused on Bortezemib's therapeutic effect on NF-κB signaling, although recent evidence has also pointed to down-regulation of MAPK signaling, p53 activation and stabilization of p27, a cell-cycle inhibitor. Our model implies an additional mechanism: stabilization of the executioner caspases. As a single agent in MM, Bortezemib-mediated caspase stabilization may reduce the apoptotic threshold in these cells. Our data further suggest that combinations of proapoptotic agents to activate the caspases and proteasome inhibitors may synergize to induce apoptosis in additional cancer types.
Experimental Procedures
Cell Culture and Transfections
The 293T (ATCC#CRL-11268) and HEK293 (ATCC#CRL-1573) cell lines were obtained from the UCSF Cell Culture Facility, and the HeLa (ATCC#CCL-2) cell line was obtained from ATCC. Cells were passaged in MEM with Earle's Balanced Salt Solution supplemented 10% FBS (Invitrogen), 1mM Sodium Pyruvate, 2mM L-Glutamine, 1× Non-Essential Amino Acids and 100ug/mL Normocin (Invivogen).
Transient transfection of plasmids was performed at the 12- or 6-well plates scale. Wells were coated with poly-d-lysine prior to plating target cells. Cells were plated to reach ∼90% confluence the following day, and transfection mixes were prepared according to the standard Lipofectamine 2000 protocol. For FRET studies, N- and C-TEV fragments were co-transfected over a range of 3:1 to 12:1 molar excess to the TEV-FRET reporter. Rap (Calbiochem) and MG-132 (Sigma) were dissolved in DMSO and were stored as aliquots at -20°C. Please see the Supplemental Experimental Procedures for the details of plasmid construction and Table S2 for all DNA oligonucleotide sequences used in this paper.
Phenotypic Analysis of Caspase Activation in Cells
Caspase-Glo-3/-7, Proteasome-Glo (Chymotrypsin-like activity) and CellTiter-Glo Assays (Promega) were performed according to the manufacture's protocol. Replicate wells in a 96-well plate were mock-treated or with 10nM Rap. At defined endpoints, equal volume of either reagent was added to cells. Cellular lysis proceeded at room temperature for 30 minutes, and luciferase activity was recorded in the SpectraMax M5 plate reader with an integration time of 500 ms. To measure caspase-6 activity, a VEID-R110 (rhodamine 110-based caspase-6 substrate) was used. VEID-R110 was added to a final concentration of 50uM. To assay for PS exposure and membrane integrity, 1uL GFP-Annexin-V, a gift of Dr. Joel Ernst (New York University), and 5ug/mL propidium iodide were added to cells with Annexin-V assay buffer (1× final concentration 10uM HEPES, pH 7.4; 140 uM NaCl; and 2.5 mM CaCl2). After 10 minutes at 37°C, fluorescent imaging was performed in the Incell Analyzer 1000 (GE Healthcare).
Supplementary Material
Acknowledgments
We are grateful to members of the Wells lab for useful discussions and to Emily Crawford for curation of the laboratory caspase substrate database. We also thank the UCSF NIKON Imaging Center and Dr. Kurt Thorn for assistance with live cell imaging. We also thank Drs. Al Burlingame, David Maltby, and Jon Trinidad for use of the Mass Spectrometry Faculity at the Mission Bay Campus of UCSF. This work was supported by NIH R01 GM081051 and CA136779 (JAW), The Hartwell Foundation (JAW) and the University of California Systemwide Biotechnology Research & Education Program GREAT Training Grant 2008-23 (DG).
Footnotes
Supplemental Data: Supplemental Data includes Supplemental Experimental Procedures, Supplemental References, two tables (ST1 and ST2) and five figures (S1-S5).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adrain C, Creagh EM, Cullen SP, Martin SJ. Caspase-dependent inactivation of proteasome function during programmed cell death in Drosophila and man. J Biol Chem. 2004;279:36923–36930. doi: 10.1074/jbc.M402638200. [DOI] [PubMed] [Google Scholar]
- Albeck JG, Burke JM, Aldridge BB, Zhang M, Lauffenburger DA, Sorger PK. Quantitative analysis of pathways controlling extrinsic apoptosis in single cells. Mol Cell. 2008a;30:11–25. doi: 10.1016/j.molcel.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albeck JG, Burke JM, Spencer SL, Lauffenburger DA, Sorger PK. Modeling a snap-action, variable-delay switch controlling extrinsic cell death. Plos Biol. 2008b;6:2831–2852. doi: 10.1371/journal.pbio.0060299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford L, Hay D, Devoy A, Paine S, Powe DG, Seth R, Gray T, Topham I, Fone K, Rezvani N, et al. Depletion of 26S proteasomes in mouse brain neurons causes neurodegeneration and Lewy-like inclusions resembling human pale bodies. J Neurosci. 2008;28:8189–8198. doi: 10.1523/JNEUROSCI.2218-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casciola-Rosen L, Garcia-Calvo M, Bull H, Becker J, Hines T, Thornberry N, Rosen A. Mouse and Human Granzyme B Have Distinct Tetrapeptide Specificities and Abilities to Recruit the Bid Pathway. Journal of Biological Chemistry. 2007;282:4545. doi: 10.1074/jbc.M606564200. [DOI] [PubMed] [Google Scholar]
- Choi YE, Butterworth M, Malladi S, Duckett CS, Cohen GM, Bratton SB. The E3 Ubiquitin Ligase cIAP1 Binds and Ubiquitinates Caspase-3 and -7 via Unique Mechanisms at Distinct Steps in Their Processing. J Biol Chem. 2009;284:12772–12782. doi: 10.1074/jbc.M807550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clem R, Sheu TT, Richter B, He WW, Thornberry N, Duckett C, Hardwick J. c-IAP1 Is Cleaved by Caspases to Produce a Proapoptotic C-terminal Fragment. Journal of Biological Chemistry. 2001;276:7602. doi: 10.1074/jbc.M010259200. [DOI] [PubMed] [Google Scholar]
- Dix MM, Simon GM, Cravatt BF. Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell. 2008;134:679–691. doi: 10.1016/j.cell.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Martins LM, Kaufmann SH. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu Rev Biochem. 1999;68:383–424. doi: 10.1146/annurev.biochem.68.1.383. [DOI] [PubMed] [Google Scholar]
- Ernst J. Preparation and Characterization of an Endogenously Fluorescent Annexin for Detection of Apoptotic Cells. Anal Biochem. 1998;260:18–23. doi: 10.1006/abio.1998.2677. [DOI] [PubMed] [Google Scholar]
- Feeney B, Pop C, Swartz P, Mattos C, Clark AC. Role of loop bundle hydrogen bonds in the maturation and activity of (Pro)caspase-3. Biochemistry. 2006;45:13249–13263. doi: 10.1021/bi0611964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes-Prior P, Salvesen GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J. 2004;384:201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, Pearson J, Shehadeh J, Bertram L, Murphy Z, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Green DR. Apoptotic pathways: ten minutes to dead. Cell. 2005;121:671–674. doi: 10.1016/j.cell.2005.05.019. [DOI] [PubMed] [Google Scholar]
- Hell K, Saleh M, Crescenzo GD, O'Connor-McCourt MD, Nicholson DW. Substrate cleavage by caspases generates protein fragments with Smac/Diablo-like activities. Cell Death Differ. 2003;10:1234–1239. doi: 10.1038/sj.cdd.4401298. [DOI] [PubMed] [Google Scholar]
- Jang M, Park BC, Lee AY, Na KS, Kang S, Bae KH, Myung PK, Chung BC, Cho S, Lee DH, Park SG. Caspase-7 mediated cleavage of proteasome subunits during apoptosis. Biochem Biophys Res Commun. 2007;363:388–394. doi: 10.1016/j.bbrc.2007.08.183. [DOI] [PubMed] [Google Scholar]
- Klaiman G, Champagne N, LeBlanc AC. Self-activation of Caspase-6 in vitro and in vivo: Caspase-6 activation does not induce cell death in HEK293T cells. Biochim Biophys Acta. 2009;1793:592–601. doi: 10.1016/j.bbamcr.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Lakhani SA, Masud A, Kuida K, Porter GA, Booth CJ, Mehal WZ, Inayat I, Flavell RA. Caspases 3 and 7: key mediators of mitochondrial events of apoptosis. Science. 2006;311:847–851. doi: 10.1126/science.1115035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc Natl Acad Sci U S A. 2003;100:9946–9951. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 2008;134:866–876. doi: 10.1016/j.cell.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks TD, Leuther KK, Howard ED, Johnston SA, Dougherty WG. Release of proteins and peptides from fusion proteins using a recombinant plant virus proteinase. Anal Biochem. 1994;216:413–417. doi: 10.1006/abio.1994.1060. [DOI] [PubMed] [Google Scholar]
- Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, Dickson BJ, Nasmyth K. Cell-Type-Specific TEV Protease Cleavage Reveals Cohesin Functions in Drosophila Neurons. Dev Cell. 2008;14:239–251. doi: 10.1016/j.devcel.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop C, Salvesen GS. Human caspases: activation, specificity, and regulation. J Biol Chem. 2009;284:21777–21781. doi: 10.1074/jbc.R800084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab MS, Podar K, Breitkreutz I, Richardson PG, Anderson KC. Multiple myeloma. Lancet. 2009;374:324–339. doi: 10.1016/S0140-6736(09)60221-X. [DOI] [PubMed] [Google Scholar]
- Rehm M, Dussmann H, Janicke RU, Tavare JM, Kogel D, Prehn JHM. Single-cell fluorescence resonance energy transfer analysis demonstrates that caspase activation during apoptosis is a rapid process. Role of caspase-3. J Biol Chem. 2002;277:24506–24514. doi: 10.1074/jbc.M110789200. [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Elowitz MB, Alon U. Negative autoregulation speeds the response times of transcription networks. Journal of Molecular Biology. 2002;323:785–793. doi: 10.1016/s0022-2836(02)00994-4. [DOI] [PubMed] [Google Scholar]
- Shi Y. Caspase activation, inhibition, and reactivation: a mechanistic view. Protein Sci. 2004;13:1979–1987. doi: 10.1110/ps.04789804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XM, Butterworth M, MacFarlane M, Dubiel W, Ciechanover A, Cohen GM. Caspase activation inhibits proteasome function during apoptosis. Mol Cell. 2004;14:81–93. doi: 10.1016/s1097-2765(04)00156-x. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Nakabayashi Y, Takahashi R. Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death. Proc Natl Acad Sci USA. 2001;98:8662–8667. doi: 10.1073/pnas.161506698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol. 2008;9:231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- Thiede B, Treumann A, Kretschmer A, Sohlke J, Rudel T. Shotgun proteome analysis of protein cleavage in apoptotic cells. Proteomics. 2005;5:2123–2130. doi: 10.1002/pmic.200401110. [DOI] [PubMed] [Google Scholar]
- Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, Houtzager VM, Nordstrom PA, Roy S, Vaillancourt JP, et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- Van Damme P, Martens L, Van Damme J, Hugelier K, Staes A, Vandekerckhove J, Gevaert K. Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat Methods. 2005;2:771–777. doi: 10.1038/nmeth792. [DOI] [PubMed] [Google Scholar]
- Vaux DL, Silke J. IAPs, RINGs and ubiquitylation. Nat Rev Mol Cell Biol. 2005;6:287–297. doi: 10.1038/nrm1621. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang CE, Orr A, Tydlacka S, Li SH, Li XJ. Impaired ubiquitin-proteasome system activity in the synapses of Huntington's disease mice. J Cell Biol. 2008;180:1177–1189. doi: 10.1083/jcb.200709080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehr MC, Laage R, Bolz U, Fischer TM, Grünewald S, Scheek S, Bach A, Nave KA, Rossner MJ. Monitoring regulated protein-protein interactions using split TEV. Nat Methods. 2006;3:985–993. doi: 10.1038/nmeth967. [DOI] [PubMed] [Google Scholar]
- Williams DJ, Puhl HL, Ikeda SR. Rapid modification of proteins using a rapamycin-inducible tobacco etch virus protease system. PLoS ONE. 2009;4:e7474. doi: 10.1371/journal.pone.0007474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolan DW, Zorn JA, Gray DC, Wells JA. Small-molecule activators of a proenzyme. Science. 2009;326:853–858. doi: 10.1126/science.1177585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi CH, Yuan J. The Jekyll and Hyde functions of caspases. Dev Cell. 2009;16:21–34. doi: 10.1016/j.devcel.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.