Abstract
Sleep research in Drosophila is not only here to stay, but is making impressive strides towards helping us understand the biological basis for and the purpose of sleep—perhaps one of the most complex and enigmatic of behaviors. Thanks to over a decade of sleep-related studies in flies, more molecular methods are being applied than ever before towards understanding the genetic basis of sleep disorders. The advent of high-throughput technologies that can rapidly interrogate whole genomes, epigenomes and proteomes, has also revolutionized our ability to detect genetic variants that might be causal for a number of sleep disorders. In the coming years, mutational studies in model organisms such as Drosophila will need to be functionally connected to information being generated from these whole-genome approaches in humans. This will necessitate the development of appropriate methods for interpolating data and increased analytical power to synthesize useful network(s) of sleep regulatory pathways—including appropriate discriminatory and predictive capabilities. Ultimately, such networks will also need to be interpreted in the context of fundamental neurobiological substrates for sleep in any given species. In this review, we highlight some emerging approaches, such as network analysis and mathematical modeling of sleep distributions, which can be applied to contemporary sleep research as a first step to achieving these aims. These methodologies should favorably impact not only a mechanistic understanding of sleep, but also future pharmacological intervention strategies to manage and treat sleep disorders in humans.
Keywords: Drosophila, disease, distribution, genetics, modeling, networks, sleep
Sleep research in Drosophila melanogaster
All animals studied thus far display some form of sleep or a sleep/rest-like state. In many species including Drosophila, chronic sleep loss severely impacts normal physiological functions to the extent that prolonged sleep loss can lead to death.1,2 These simple observations, though not mechanistically insightful, do underscore the fundamental importance of sleep. Since sleep is also a state in which an individual is in a position of increased vulnerability to predators, it has been argued that sleep must perform a vital biological function(s) to have been so strongly conserved through evolution.3 However, the identity of this function continues to be debated and remains inconclusively demonstrated.
Typically, physiological and neurobiological correlates of sleep have been studied more readily in vertebrates, including humans. To some extent this has been due to a more robust functional definition for sleep in these animals that has aided initial observational studies. The advent of more sophisticated, non-invasive methods for measuring brain electrical activity has also led to consistently identifiable “electrical signatures” for sleep that now find routine use in the clinic. Consequently, regions in the vertebrate brain that are involved in the regulation of sleep have been identified and studied for some time.4-6 By contrast, the molecular basis for sleep regulation is relatively poorly understood. The highly complex and enigmatic nature of sleep and the absence of models that allow rapid multi-factorial genetic analysis have, in part, precluded a better molecular description of sleep. Given that a number of human sleep disorders are likely to have genetic components,7 an appreciation for molecular mechanisms underlying sleep is not only vital to a better understanding of these disorders, but also to understanding the fundamental nature of sleep itself.
Use of genetic methods to study sleep in the laboratory is a relatively modern phenomenon, though the significance of potential genetic determinants has been recognized for a long time. The deployment of models systems, such as Drosophila, that offer varied and powerful tools for genetic analysis, is an even more recent occurrence. Methodology and analytical tools of Drosophila sleep research derive heavily from a sizeable body of work on circadian rhythms. For example, the most widely used experimental setup to measure sleep in flies is the Drosophila Activity Monitor or DAM system.8 This technique was developed to overcome laborious recordings of circadian phenomena in flies such as eclosion rates (the rate at which adult flies emerge from their pupae). In this method, the locomotor activity of individual flies could be monitored over a period of days by measuring the number of times each fly walked through an infrared beam in a single glass tube (i.e., a beam break). Periods of increased locomotor activity, such as during the day, result in a larger number of beam breaks, while decreased activity at night leads to fewer beam breaks. Recordings can be averaged over suitable bin sizes (e.g., 1 or 5 min) and across a number of flies of the same age, sex and genotype, thereby increasing the power of this analysis. Periodicity of this rhythmic change in activity can be accurately measured using a number of statistical tools for the analysis of biological rhythm, and deviations from a circadian profile can be detected with ease. Thus, the DAM system allowed automation, scaling, reproducibility and the ability to monitor locomotor rhythms in single flies over circadian time. Experiments using this method have given rise to a vast amount of data that has contributed richly to a very sophisticated understanding of the molecular clock that regulates circadian rhythms in flies, as well as in other animals. More recently, this system has also been combined with natural variations in light and temperature to deduce the effect of seasonal variations on circadian rhythm.9,10
The first evidence that Drosophila could be used to study sleep came from two reports published in early 2000, both of which used the DAM system.11,12 Together, these studies showed that periods of rest in flies correspond to a sleep-like state. Several characteristics of mammalian sleep biology appeared to be conserved in flies. For instance, periods of rest in flies correspond to increased threshold for arousal by mechanical stimuli, and were responsive to stimulants and hypnotics, as well as to prior lack of rest/sleep. Thus, fly sleep showed a circadian pattern and was subject to strong homeostatic regulation. Once the correlates for sleep in Drosophila had been defined, the stage was set for the discovery of sleep-related genetic pathways, the elucidation of neural correlates for sleep in flies, and perhaps even the modeling of human sleep disorders in this model system. In the last decade, several Drosophila sleep mutants have been isolated through traditional forward genetic screens designed to isolate mutations with altered sleep patterns. Some of the genes implicated by these mutational studies lend additional support to known neurobiological pathways involved in sleep such as the dopaminergic system (e.g., fumin mutants are deficient in the Drosophila dopamine transporter13). Indeed, the dopaminergic system has been investigated extensively in the context of sleep in flies.13-17 Yet other mutations have led to the discovery of genes and pathways hitherto unstudied in the context of sleep. For example, sleepless/quiver mutants are deficient in a small GPI-anchored protein that has been shown to influence potassium currents carried by Shaker channels.18-20 Interestingly, previous mutational studies for short-sleeping mutants had already implicated Shaker channels in the regulation of sleep in Drosophila.21 Similarly, a recent study has reported the isolation of insomniac (inc) mutants that show disruptions in sleep consolidation. Transposon insertion induced inc mutants reduce expression of CG32810, a BTB-domain containing protein that is predicted to bind the E3 Ubiquitin ligase, Cullin-3.22 This study has implicated Ubiquitinating pathways as important regulators of sleep. Ubiquitination-dependent mechanisms have also been implicated in Drosophila sleep regulation by two other studies, both of which used reverse genetic analysis to model genes linked to human disorders – one to model Angelman’s syndrome23 and the other to test putative risk factors for Restless Legs Syndrome/Willis-Ekbom Disease in flies.24 Additionally, studies in Drosophila are also beginning to shed light on why sleep is so fundamentally required in all animals. Perhaps the best explored of these is the notion that normal sleep is necessary for proper cognitive function and neuronal plasticity.15,25-32 Overall, both forward and reverse genetic approaches are increasingly being combined in Drosophila to uncover molecular regulators of sleep.
The Genetic Basis for Sleep Disorders in Humans
Sleep disorders and even specific sleep patterns tend to run in families, suggesting the presence of genetic factors that regulate sleep. In spite of this long-standing observation, genetic factors that have been systematically linked to sleep disorders are scarce. Narcolepsy with cataplexy is perhaps the best investigated among these. In general, genes identified from GWAS and genetic typing have confirmed the idea that there might be an autoimmune etiology underlying this sleep disorder. For instance, some HLA alleles have been shown to confer protection, while others to increase relative risk.33 Another candidate, the gene P2RY11 encoding a purinergic receptor that is coupled to both cAMP and IP3 signaling pathways and expressed highly in the brain and white blood cells, has been postulated to function as a sensor for extracellular ATP during immune regulation.34-36 Similarly, T-cell receptor α or TCRA, the major receptor for HLA-peptide presentation has been strongly associated with narcolepsy with cataplexy in Caucasians, Asians and African Americans.33,37 Finally, though not linked to immune function, an additional polymorphism between two genes CPT1B and CHKB has been associated with Japanese and Korean narcoleptics, but has not been replicated in European, African American or Chinese populations.38,39 Interestingly, this polymorphism has also been identified in essential hypersomnias.40 Restless Legs Syndrome (RLS, renamed Willis-Ekbom Disease, WED) is a disorder in which patients report an irresistible urge to move their limbs that worsens with rest and at night. RLS/WED patients display severe sleep fragmentation that can be managed clinically to some extent with dopaminergic agonists.41 Recent GWAS have identified a set of risk alleles that point to the genes BTBD9, MEIS1, MAP2K5/LBXCOR1, PTPRD and TOX3.42-46 While neuronal and/or developmental roles have been described for some of these genes, none of them immediately suggest molecular pathways that can be obviously connected to RLS/WED or sleep.47-49 However recent studies using model systems have begun to directly explore sleep-related functions for these genes in model systems24,50,51 (see next section for details).
Modeling Human Sleep Disorders in Drosophila
Drosophila has been used extensively as a powerful and rapid means to model human diseases.52-54 Similar modeling of sleep disorders in flies has been initiated by the twin developments of “sleep genetics” in flies and high throughput whole-genome searches for sleep disease genes in patient populations.7,55 Though still in its infancy, we highlight here a few instances where Drosophila has been used effectively to study disease genes in humans that also affect sleep.
Two single-gene neurological disorders that are known to precipitate intellectual disability and also lead to sleep disturbances are Angelman syndrome (loss of function of UBE3A) and Fragile X syndrome (loss of FMR1). Since Drosophila possess a single homolog of both these genes, sleep studies have been performed with loss-of-function mutations in the fly homolog of UBE3A (dUbe3a) and FMR1 (dFmr1).23,56 Loss of dUbe3a leads to strong defects in locomotion and circadian rhythm, though a specific defect in sleep has not been described. For dFmr1, mean sleep duration is bi-directionally influenced by a loss or gain of Fmr1, such that dFmr1 amorphs are long sleepers, while increased dFmr1 leads to short sleepers. Additionally, perturbations in dFmr1 also impair sleep homeostasis.56 Both these proteins are expressed in the mushroom bodies, anatomical regions of the fly brain that are involved in sleep as well as higher cognitive functions such as learning and memory formation. Consistent with a potential relationship between brain plasticity and sleep, Drosophila that express transgenic human DISC1 (a susceptibility factor in multiple mental illnesses including Schizophrenia) in the mushroom body also display significantly longer sleep bouts but normal bout frequency and circadian rhythms.57 These studies exemplify rational sleep disorder modeling in flies and also strengthen the connection between sleep and cognitive function.
Neurodegenerative disorders such as Parkinson disease (PD) and Huntington’s disease (HD) are often accompanied by sleep disturbances, though these are not classified as primary sleep disorders.58-63 Recent experiments in Drosophila have shown that this correlation is maintained in flies. For example in flies that express mutant forms of human huntingtin (mHtt) in the brain or in which the endogenous huntingtin protein (dhtt) is knocked down, displayed both sleep loss and sleep fragmentation.64 Similarly, expression of pre-fibrillar forms of human α-synuclein – a protein implicated in PD – in dopaminergic and serotonergic neurons precipitated non-motor symptoms such as abnormal sleep and circadian rhythm.65 Pan-neuronal expression of human α-synuclein also resulted in aberrant short-term memory formation when these flies were sleep deprived, once again highlighting the interconnectedness of sleep and cognitive function.25
While examples mentioned above demonstrate the utility of flies in exploring sleep phenotypes in a variety of neurological disorders, genetic regulators of human sleep are also beginning to be explored directly in flies. The gene ABCC9 – encoding the protein SUR2 a pore-forming subunit of an ATP-sensitive potassium channel66 – was identified through genome-wide association studies as a factor that might explain a fraction of the natural variation in sleep duration in human populations.67 Independently, this channel had also been identified to play a role in the etiology of cardiovascular disease and endocrine disorders that are associated with abnormalities in sleep durations.66 RNA interference mediated knock down of the Drosophila homolog, dSur, in the brain preferentially affected night-time sleep while leaving both day-time sleep and overall circadian rhythm intact. In another study, a gene BTBD9, first identified as a risk factor for Restless Legs Syndrome (Willis-Ekbom Disease, RLS/WED),42,43 was knocked out in flies. dBTBD9 mutants showed appreciable sleep fragmentation and increased motor activity, suggesting a physiological role for BTBD9 in the regulation of sleep architecture and RLS/WED pathophysiology.24 Taken together, these studies clearly demonstrate that the power and elegance of Drosophila genetics has the potential to shed light on conserved features of sleep and human sleep disorders. We predict an increasing application of Drosophila (and other genetic models) to the study of human sleep disorders, drug target discovery and the development of pharmacological screening platforms.
Multi-Factorial Analysis of Sleep: Future Challenges
Given the relentless wave of information in the post-genomic era, sleep research in Drosophila will have to overcome significant challenges if it is to continue to be a potent platform for interrogating the genetic basis for sleep and sleep disorders. Here, we explore two criteria that will help in discovering functional networks of genes that regulate sleep using Drosophila as a model system.
A. Analysis of sleep-regulatory networks
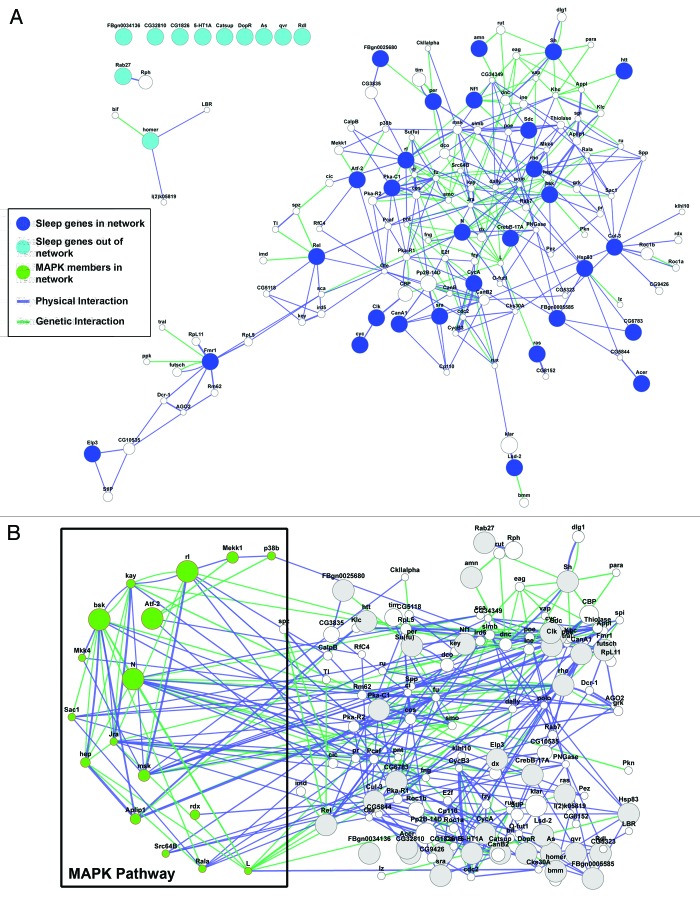
As more genes/proteins get implicated in the regulation of sleep, we will have to ask whether these genes belong to discrete signaling pathways and genetic networks that function in concert to control various aspects of sleep. Identification of these networks through computational approaches has become much more accessible and powerful in recent times. Algorithms can now draw from a number of databases that report genetic and physical interactions and collate this information with functional and structural information on proteins of interest.68 For instance, the program GeneMANIA, maintained by the University of Toronto and available either as a stand-alone web application or as a plugin in Cytoscape (an open source freeware for complex network analysis), is capable of generating gene networks based on user supplied criteria from a list of relevant genes.69-71 An example of this is shown in Figure 1, which depicts a network created from all genes that have identified sleep regulatory roles in flies. One can see immediately from Figure 1A that the majority of these genes fall within a network that is connected by either reported genetic or protein-protein interactions. Closer scrutiny of this interaction map reveals that MAP kinase pathways are over-represented (Fig. 1B, see figure legend for details). Therefore, such analysis, while confirming previous studies, also helps prioritize future experiments. In this specific instance, we would predict that other components of the MAP kinase cascade that have not been explicitly tested for sleep-related functions are likely to play a role in the regulation of sleep. Such predictions can be tested for parity with gene ontology enrichment using additional resources such as DEFOG, another web-based application that prioritizes GO analysis on top of GeneMANIA network outputs.72 With the discovery of additional genes, such networks are likely to become more enriched and lead to increased predictive power. Components from such networks will then have to be analyzed, most likely in a combinatorial fashion, using functional studies that are capable of identifying the unique ways in which these genes influence sleep architecture.
Figure 1.Network analysis of “sleep-related” genes in Drosophila. (A) A gene interaction network built based on gene ontology weighting using GeneMANIA from genes implicated in sleep regulation in Drosophila. Dark blue circles represent genes that are part of a network, while light blue circles are genes that are not part of a network based on current experimental evidence. Blue lines denote reported physical interactions between protein products, while green lines represent known genetic interactions from previous studies. (B) Gene Ontology (GO) analysis in GeneMANIA reveals a high degree of enrichment for signaling and MAP kinase pathway members. The network can be easily rearranged to highlight these components (green circles) and obtain an idea of the number and identity of these genes. Genes that are part of this network include the three MAP kinase members (known as basket (JNK), rolled (ERK) and p38b (p38 MAPK) in flies), upstream kinases such as Mekk1 and target transcription factors such as Fos (known as kayak in flies) and Jun (known as jra in flies). Such analysis suggests that future studies should focus on members of this network as potential sleep regulatory elements. The rest of the network shows the original sleep-related genes in gray and other members of the overall network in white.
B. High-resolution recordings of sleep
Since the first time Drosophila was shown to be a viable organism for examining sleep, the readily available DAM system has been crucial in providing a simple and scalable platform for rapid screening of molecular components that underlie the behavior. However, the traditional DAM detects fly locomotion by relying on the signal of a single infrared (IR) beam. Consequently, the technology has limited temporal resolution, almost no spatial resolution and in many cases vastly over-estimates sleep amount.73 In addition, the software tools that are currently available interpret the DAM data at a gross level, only yielding population-wide average quantities such as daily sleep and bout length. Powerful theoretical methods from time-series analysis and statistical theory, though applicable to fly activity recordings, have not seen wide applications in Drosophila sleep studies, thus leaving the information contained in the data relatively underutilized. Drawbacks of existing tools have meant that only severe variations in fly behavioral phenotypes, several standard deviations from the mean, can be reported with statistical confidence. Various subtle, yet physiologically interesting, changes in sleep pattern are likely being overlooked due to a lack of appropriate experimental and theoretical methods. Below, we examine a few improvements to methods presently employed in Drosophila sleep studies.
A natural extension of the single beam hardware is the multi-beam activity monitor (MAM), also from Trikinetics, Inc. which manufactures the commonly used DAM. In contrast to the DAM, the MAM is equipped with 17 IR sources per tube and can therefore detect fly movement approximately every 3 mm, an order-of-magnitude improvement in spatial resolution over current detection capability. The higher sensitivity of the MAM system should significantly reduce measurement errors inherent in the DAM.73 Also, its similarity to the previous single-beam technology implies it can be seamlessly integrated with existing equipment. The higher fidelity data, in conjunction with simple incorporation into ongoing experimental design, makes upgrading to the MAM system an attractive prospect.
Although a considerably different design from the DAM system, video-tracking of movement is perhaps the most accurate method to-date for studying Drosophila sleep. A basic system for video monitoring of flies can be set up with only an IR-sensitive digital camera, source of IR illumination, and a laptop computer. Flies can be monitored either in glass tubes that are standard with the DAM system or in a makeshift arena designed such that movement is confined to a fixed focal plane. Image acquisition and subsequent analysis can be performed with one of several freely available software packages such as pySolo,74 Tracker75 and Ctrx-Flytrax.76 Unlike the commercial DAM and MAM packages, setting up a video tracking system demands an initial investment of time for assembling and testing the individually procured components. In addition, video-tracking requires visual access to the flies at all times while the DAM and MAM systems can be operated with the individual monitors stacked on top of each other. In a large-scale study where the priority is to rapidly screen tens of thousands of flies, the demand on physical space can make video-tracking a less attractive option. However, the advantages gained from the new methodology far surpass the additional effort and space requirements. First, with just off-the-shelf components the researcher can achieve sub-millimeter and sub-second spatiotemporal resolution of locomotion. The higher precision measurement directly translates into a more accurate estimate of the standard fly sleep parameters. Second, the video images contain detailed information on a fly’s physical status and can be mined to investigate association between sleep and other behavioral attributes such as place preference, grooming, and food proximity. Third, it allows the researcher to design and execute experiments in arbitrary environments, with the only restriction being visibility to the camera. Finally, from an economic standpoint, a comparative study reported that the cost of tracking 30 flies using video-recording is at least one-fifth of that required for an equivalent IR detection system.74 Thus, the benefits of studying sleep through video-recording are many and should make the technique appealing to the fly community. Several Drosophila sleep laboratories have already adopted this new approach74,75,77 and we anticipate more will follow suit in the near future.
C. New computational methods to analyze sleep data
In addition to incorporating better measurement techniques, fly sleep studies can also benefit from taking advantage of more rigorous mathematical tools. Analysis of ion channel recordings with hidden Markov models,78 estimation of brain EEG parameters with time-series methods,79 and statistical modeling of animal movement patterns80 are examples where advanced mathematics has been successfully applied to address questions related to animal behavior. So far, most Drosophila sleep studies have relied on a handful of metrics that describe behavior averaged over many flies and several days. While these metrics have been useful in the initial characterization of novel mutants, they have limited ability to provide detailed insights into neuronal firing patterns modulating sleep or construct a systems-level link among the various regulators of the behavior. As a consequence of the former, the model of fly sleep still remains rudimentary while description of mammalian sleep is well-developed with the classification of several different stages of sleep and wake states. Access to real-time fly neuronal activity, such as EEG, would permit enhancement of the fly model but since such direct read-out remains technically challenging, an alternative approach may be to more carefully examine available locomotor data with the aim of establishing connections to brain activity.
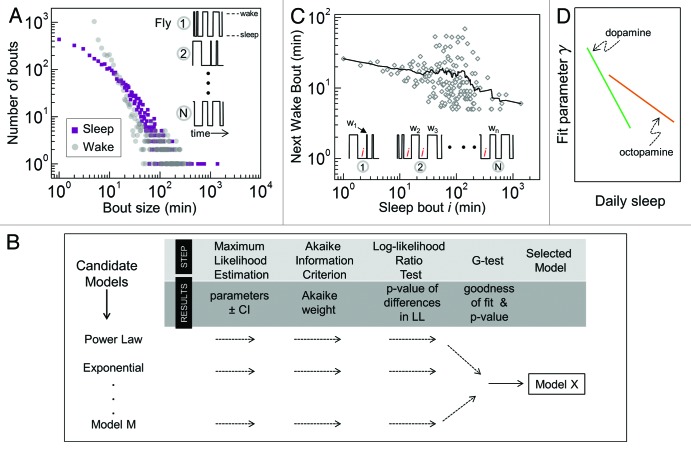
We sketch here a simple analytic strategy that may offer insights into the type of neuronal processes that underlie sleep-wake transitions. Instead of averaging over individual bouts, a running list of sleep bouts of all flies of one genotype can be pooled and the frequency of each distinct bout length can be determined to produce a distribution of the type shown in Figure 2A (purple squares). Similarly, an equivalent plot can be generated for the wake states (Fig. 2A, gray circles). Without any further quantification, data in Figure 2A makes a number of refinements to our current knowledge: (i) sleep durations are dominated by short bouts, < 50 min, underscoring the need for temporal resolution higher than 1 min; (ii) since the distribution is heavily skewed with the bouts spanning several orders of magnitude, quoting only the mean bout size is misleading; (iii) comparing overall shapes of sleep and wake distributions can provide clues as to whether similar or different processes regulate the two states. Well-established modeling and inference methods81 can be used for robust quantification of such data (Fig. 2B). For instance, if the data are best mimicked by a power-law model, it would suggest that sleep events are likely modulated by scale invariant neuronal dynamics. If instead the analysis favors an exponential or inverse Gaussian model, the evidence would point toward a process which has a characteristic timescale. These model dynamical processes have been investigated extensively in mammalian brains82 and therefore, establishing links between them and fly sleep-wake events could facilitate the gradual construction of a quantitative model of fly sleep (Syed and Kidd, in preparation).
Figure 2. Analysis of sleep and wake bouts. (A) Frequency plot of wild-type Drosophila sleep and wake events show distributions with long tails. Schematic on the right indicates the data are pooled from N fly recordings. Here, n = 20 flies, each measured for 5 d in 1 min interval using the DAM system. (B) Workflow of a statistical approach to quantify sleep and wake distributions. Starting with arbitrary long-tailed distributions, maximum likelihood theory is first used to calculate parameters (CI, 95% confidence intervals) for each model, given the data. Next, the log-likelihood (LL) ratio test computes statistical evidence in favor of the model with the highest Akaike weight. If evidence is significant (according to p-value of LL test), the favored model subsequently undergoes a “goodness-of-fit” G- or Chisq-test, finally becoming the selected model with probability given by the test p-value. (C) Interrogating the data from (A) for correlation between sleep and wake bouts. All n sleep events of duration i minutes are located in each of the n = 20 recordings (see schematic below data) and average size of the subsequent wake bouts is computed, (w1+w2+…+wn)/n. The analysis shows on the whole a weak negative correlation between sleep bouts and average duration of the following wake bouts (solid line, smoothed data). (D) Parameters yielded by the types of analyses in (A-C) could be used to relate different genotypes. In a hypothetical scenario, fly strains with differentially modulated levels of arousal-promoting dopamine and octopomine may appear as two distinct “classes.” A future unknown strain can then be placed on such a graph to uncover its functional proximity to a known class of flies, thereby generating a more integrated view of various sleep models.
A different method of analysis can reveal time-dependent features of fly sleep-wake architecture by examining temporal correlations among bouts. For example, is a long sleep bout, on average, followed by a long wake bout? Such a question can be answered by first determining the distinct sleep epochs of a cohort and then computing the average wake event that immediately follows each distinct sleep bout. Any correlation among the opposing sleep-wake states can be deduced from the pooled data (Fig. 2C). The group of flies in Figure 2C does not exhibit a strong coupling between the duration of the two states, as evidenced by the line with an overall small negative slope, suggesting two partially independent mechanisms are likely generating sleep and wake events. In contrast, if strongly coupled processes underlay the two states, we would likely find significant positive or negative correlation in the data. Thus, this analytic strategy can potentially reveal temporal organization of the neuronal processes that signal sleep and wake events.
The two general analytic approaches sketched here introduce novel parameters that, since they quantify individual events rather than population-averaged behavior, should allow detection of previously elusive changes in sleep-wake patterns. Such subtle changes may be expected in response to novel stimuli such as new drugs and complex environmental variables like sinusoidal light modulation. These candidate effectors could, for instance, alter the temporal distribution of sleep bouts without changing the overall daily sleep amount. Moreover, the mathematical parameters could serve as diagnostic markers in the study of poorly understood sleep disorders in Drosophila. In the study of epilepsy, for example, rigorous analysis of EEG patterns has led to the development of powerful computational tools for human patient diagnosis.79 Additionally, the new strategies could help classify genetically distinct mutants into groups (Fig. 2D) or complement findings of virtual sleep gene networks such as those by GeneMANIA. Finally, the strategies seem capable of connecting behavioral output to certain aspects of activity of the fly sleep- and wake-promoting neurons. Ultimately, relevant findings from Drosophila studies will have to be explained in the context of human sleep and having a quantitative fly sleep model involving neuronal input and behavioral output will greatly facilitate such comparisons.
Summary
Although the application of Drosophila as a model system to the study of sleep has made serious inroads into this fascinating problem, future research must take into consideration the obvious complexity of biological systems. Specifically, we are now in a position to consider the role not just of individual genes, but rather interconnected gene networks functioning together as an ensemble. This comprehensive perspective will aid our understanding and highlight novel candidates for investigation. However, harnessing the full power of this model system will also require the integration of higher resolution observational tools and more sophisticated analytical approaches. With the application of these strategies, these tiny flies will continue to make vast contributions to our understanding of sleep and sleep disorders.
Acknowledgments
Drs. Freeman and Sanyal acknowledge grant support from the Sleep Research Society and the Restless Legs Syndrome/Willis-Ekbom Disease Foundation. Dr. Syed is grateful to Dr. Michael W. Young for support.
Glossary
Abbreviations:
- GWAS
genome wide association studies
- RLS
restless legs syndrome
- WED
Willis-Ekbom disease
- GO
gene ontology
- DAM
drosophila activity monitor
- MAM
multiple activity monitor
- IR
infrared
- GPI
glycophosphatidylinositol
- BTB
bric-a-brac, tramtrack broad complex
- cAMP
cyclic adenosine monophosphate
- IP3
inositol triphosphate
- HLA
human leukocyte antigen
- FMR1
Fragile X mental retardation 1
- DISC1
disrupted in schizophrenia 1
- PD
Parkinson's disease
- HD
Huntington's disease
- ABCC9
ATP-binding cassette, sub-family C member 9
- SUR2
sulfonylurea receptor 2
- DEFOG
discrete enrichment of functionally organized genes)
- EEG
electroencephalography
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/22733
References
- 1.Rechtschaffen A, Gilliland MA, Bergmann BM, Winter JB. Physiological correlates of prolonged sleep deprivation in rats. Science. 1983;221:182–4. doi: 10.1126/science.6857280. [DOI] [PubMed] [Google Scholar]
- 2.Reynolds AC, Banks S. Total sleep deprivation, chronic sleep restriction and sleep disruption. Prog Brain Res. 2010;185:91–103. doi: 10.1016/B978-0-444-53702-7.00006-3. [DOI] [PubMed] [Google Scholar]
- 3.Siegel JM. Sleep viewed as a state of adaptive inactivity. Nat Rev Neurosci. 2009;10:747–53. doi: 10.1038/nrn2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Landolt HP. Genetic determination of sleep EEG profiles in healthy humans. Prog Brain Res. 2011;193:51–61. doi: 10.1016/B978-0-444-53839-0.00004-1. [DOI] [PubMed] [Google Scholar]
- 5.Vyazovskiy VV, Cirelli C, Tononi G. Electrophysiological correlates of sleep homeostasis in freely behaving rats. Prog Brain Res. 2011;193:17–38. doi: 10.1016/B978-0-444-53839-0.00002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown EN, Lydic R, Schiff ND. General anesthesia, sleep, and coma. N Engl J Med. 2010;363:2638–50. doi: 10.1056/NEJMra0808281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sehgal A, Mignot E. Genetics of sleep and sleep disorders. Cell. 2011;146:194–207. doi: 10.1016/j.cell.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu JC, Low KH, Pike DH, Yildirim E, Edery I. Assaying locomotor activity to study circadian rhythms and sleep parameters in Drosophila. J Vis Exp. [DOI] [PMC free article] [PubMed]
- 9.Vanin S, Bhutani S, Montelli S, Menegazzi P, Green EW, Pegoraro M, et al. Unexpected features of Drosophila circadian behavioural rhythms under natural conditions. Nature. 2012;484:371–5. doi: 10.1038/nature10991. [DOI] [PubMed] [Google Scholar]
- 10.Bywalez W, Menegazzi P, Rieger D, Schmid B, Helfrich-Förster C, Yoshii T. The dual-oscillator system of Drosophila melanogaster under natural-like temperature cycles. Chronobiol Int. 2012;29:395–407. doi: 10.3109/07420528.2012.668505. [DOI] [PubMed] [Google Scholar]
- 11.Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–7. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 12.Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, et al. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–38. doi: 10.1016/S0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 13.Kume K, Kume S, Park SK, Hirsh J, Jackson FR. Dopamine is a regulator of arousal in the fruit fly. J Neurosci. 2005;25:7377–84. doi: 10.1523/JNEUROSCI.2048-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andretic R, van Swinderen B, Greenspan RJ. Dopaminergic modulation of arousal in Drosophila. Curr Biol. 2005;15:1165–75. doi: 10.1016/j.cub.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 15.Seugnet L, Suzuki Y, Vine L, Gottschalk L, Shaw PJ. D1 receptor activation in the mushroom bodies rescues sleep-loss-induced learning impairments in Drosophila. Curr Biol. 2008;18:1110–7. doi: 10.1016/j.cub.2008.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lebestky T, Chang JS, Dankert H, Zelnik L, Kim YC, Han KA, et al. Two different forms of arousal in Drosophila are oppositely regulated by the dopamine D1 receptor ortholog DopR via distinct neural circuits. Neuron. 2009;64:522–36. doi: 10.1016/j.neuron.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riemensperger T, Isabel G, Coulom H, Neuser K, Seugnet L, Kume K, et al. Behavioral consequences of dopamine deficiency in the Drosophila central nervous system. Proc Natl Acad Sci U S A. 2011;108:834–9. doi: 10.1073/pnas.1010930108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koh K, Joiner WJ, Wu MN, Yue Z, Smith CJ, Sehgal A. Identification of SLEEPLESS, a sleep-promoting factor. Science. 2008;321:372–6. doi: 10.1126/science.1155942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu MN, Joiner WJ, Dean T, Yue Z, Smith CJ, Chen D, et al. SLEEPLESS, a Ly-6/neurotoxin family member, regulates the levels, localization and activity of Shaker. Nat Neurosci. 2010;13:69–75. doi: 10.1038/nn.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dean T, Xu R, Joiner W, Sehgal A, Hoshi T. Drosophila QVR/SSS modulates the activation and C-type inactivation kinetics of Shaker K(+) channels. J Neurosci. 2011;31:11387–95. doi: 10.1523/JNEUROSCI.0502-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cirelli C, Bushey D, Hill S, Huber R, Kreber R, Ganetzky B, et al. Reduced sleep in Drosophila Shaker mutants. Nature. 2005;434:1087–92. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- 22.Stavropoulos N, Young MW. insomniac and Cullin-3 regulate sleep and wakefulness in Drosophila. Neuron. 2011;72:964–76. doi: 10.1016/j.neuron.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Y, Bolduc FV, Bell K, Tully T, Fang Y, Sehgal A, et al. A Drosophila model for Angelman syndrome. Proc Natl Acad Sci U S A. 2008;105:12399–404. doi: 10.1073/pnas.0805291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freeman A, Pranski E, Miller RD, Radmard S, Bernhard D, Jinnah HA, et al. Sleep fragmentation and motor restlessness in a Drosophila model of Restless Legs Syndrome. Curr Biol. 2012;22:1142–8. doi: 10.1016/j.cub.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seugnet L, Galvin JE, Suzuki Y, Gottschalk L, Shaw PJ. Persistent short-term memory defects following sleep deprivation in a drosophila model of Parkinson disease. Sleep. 2009;32:984–92. doi: 10.1093/sleep/32.8.984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Yu F, Guo A. Sleep deprivation specifically impairs short-term olfactory memory in Drosophila. Sleep. 2009;32:1417–24. doi: 10.1093/sleep/32.11.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seugnet L, Suzuki Y, Donlea JM, Gottschalk L, Shaw PJ. Sleep deprivation during early-adult development results in long-lasting learning deficits in adult Drosophila. Sleep. 2011;34:137–46. doi: 10.1093/sleep/34.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerstner JR, Vanderheyden WM, Shaw PJ, Landry CF, Yin JC. Fatty-acid binding proteins modulate sleep and enhance long-term memory consolidation in Drosophila. PLoS One. 2011;6:e15890. doi: 10.1371/journal.pone.0015890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donlea JM, Thimgan MS, Suzuki Y, Gottschalk L, Shaw PJ. Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science. 2011;332:1571–6. doi: 10.1126/science.1202249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donlea JM, Ramanan N, Shaw PJ. Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science. 2009;324:105–8. doi: 10.1126/science.1166657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilestro GF, Tononi G, Cirelli C. Widespread changes in synaptic markers as a function of sleep and wakefulness in Drosophila. Science. 2009;324:109–12. doi: 10.1126/science.1166673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bushey D, Tononi G, Cirelli C. Sleep and synaptic homeostasis: structural evidence in Drosophila. Science. 2011;332:1576–81. doi: 10.1126/science.1202839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mignot E, Lin L, Rogers W, Honda Y, Qiu X, Lin X, et al. Complex HLA-DR and -DQ interactions confer risk of narcolepsy-cataplexy in three ethnic groups. Am J Hum Genet. 2001;68:686–99. doi: 10.1086/318799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore DJ, Chambers JK, Wahlin JP, Tan KB, Moore GB, Jenkins O, et al. Expression pattern of human P2Y receptor subtypes: a quantitative reverse transcription-polymerase chain reaction study. Biochim Biophys Acta. 2001;1521:107–19. doi: 10.1016/S0167-4781(01)00291-3. [DOI] [PubMed] [Google Scholar]
- 35.Communi D, Govaerts C, Parmentier M, Boeynaems JM. Cloning of a human purinergic P2Y receptor coupled to phospholipase C and adenylyl cyclase. J Biol Chem. 1997;272:31969–73. doi: 10.1074/jbc.272.51.31969. [DOI] [PubMed] [Google Scholar]
- 36.Bodin P, Burnstock G. Increased release of ATP from endothelial cells during acute inflammation. Inflamm Res. 1998;47:351–4. doi: 10.1007/s000110050341. [DOI] [PubMed] [Google Scholar]
- 37.Mignot E, Lin X, Arrigoni J, Macaubas C, Olive F, Hallmayer J, et al. DQB1*0602 and DQA1*0102 (DQ1) are better markers than DR2 for narcolepsy in Caucasian and black Americans. Sleep. 1994;17(Suppl):S60–7. doi: 10.1093/sleep/17.suppl_8.s60. [DOI] [PubMed] [Google Scholar]
- 38.Miyagawa T, Kawashima M, Nishida N, Ohashi J, Kimura R, Fujimoto A, et al. Variant between CPT1B and CHKB associated with susceptibility to narcolepsy. Nat Genet. 2008;40:1324–8. doi: 10.1038/ng.231. [DOI] [PubMed] [Google Scholar]
- 39.Han F, Lin L, Li J, Aran A, Dong SX, An P, et al. TCRA, P2RY11, and CPT1B/CHKB associations in Chinese narcolepsy. Sleep Med. 2012;13:269–72. doi: 10.1016/j.sleep.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hallmayer J, Faraco J, Lin L, Hesselson S, Winkelmann J, Kawashima M, et al. Narcolepsy is strongly associated with the T-cell receptor alpha locus. Nat Genet. 2009;41:708–11. doi: 10.1038/ng.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trotti LM, Rye DB. Restless legs syndrome. Handb Clin Neurol. 2011;100:661–73. doi: 10.1016/B978-0-444-52014-2.00047-1. [DOI] [PubMed] [Google Scholar]
- 42.Winkelmann J, Schormair B, Lichtner P, Ripke S, Xiong L, Jalilzadeh S, et al. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat Genet. 2007;39:1000–6. doi: 10.1038/ng2099. [DOI] [PubMed] [Google Scholar]
- 43.Stefansson H, Rye DB, Hicks A, Petursson H, Ingason A, Thorgeirsson TE, et al. A genetic risk factor for periodic limb movements in sleep. N Engl J Med. 2007;357:639–47. doi: 10.1056/NEJMoa072743. [DOI] [PubMed] [Google Scholar]
- 44.Kemlink D, Polo O, Frauscher B, Gschliesser V, Högl B, Poewe W, et al. Replication of restless legs syndrome loci in three European populations. J Med Genet. 2009;46:315–8. doi: 10.1136/jmg.2008.062992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Q, Li L, Yang R, Shen GQ, Chen Q, Foldvary-Schaefer N, et al. Family-based and population-based association studies validate PTPRD as a risk factor for restless legs syndrome. Mov Disord. 2011;26:516–9. doi: 10.1002/mds.23459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winkelmann J, Czamara D, Schormair B, Knauf F, Schulte EC, Trenkwalder C, et al. Genome-wide association study identifies novel restless legs syndrome susceptibility loci on 2p14 and 16q12.1. PLoS Genet. 2011;7:e1002171. doi: 10.1371/journal.pgen.1002171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mizuhara E, Nakatani T, Minaki Y, Sakamoto Y, Ono Y. Corl1, a novel neuronal lineage-specific transcriptional corepressor for the homeodomain transcription factor Lbx1. J Biol Chem. 2005;280:3645–55. doi: 10.1074/jbc.M411652200. [DOI] [PubMed] [Google Scholar]
- 48.Uetani N, Chagnon MJ, Kennedy TE, Iwakura Y, Tremblay ML. Mammalian motoneuron axon targeting requires receptor protein tyrosine phosphatases sigma and delta. J Neurosci. 2006;26:5872–80. doi: 10.1523/JNEUROSCI.0386-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan SH, Qiu Z, Ghosh A. TOX3 regulates calcium-dependent transcription in neurons. Proc Natl Acad Sci U S A. 2009;106:2909–14. doi: 10.1073/pnas.0805555106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Catoire H, Dion PA, Xiong L, Amari M, Gaudet R, Girard SL, et al. Restless legs syndrome-associated MEIS1 risk variant influences iron homeostasis. Ann Neurol. 2011;70:170–5. doi: 10.1002/ana.22435. [DOI] [PubMed] [Google Scholar]
- 51.DeAndrade MP, Johnson RL, Jr., Unger EL, Zhang L, van Groen T, Gamble KL, et al. Motor restlessness, sleep disturbances, thermal sensory alterations and elevated serum iron levels in Btbd9 mutant mice. Hum Mol Genet. 2012;21:3984–92. doi: 10.1093/hmg/dds221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bilen J, Bonini NM. Drosophila as a model for human neurodegenerative disease. Annu Rev Genet. 2005;39:153–71. doi: 10.1146/annurev.genet.39.110304.095804. [DOI] [PubMed] [Google Scholar]
- 53.Bonini NM, Fortini ME. Human neurodegenerative disease modeling using Drosophila. Annu Rev Neurosci. 2003;26:627–56. doi: 10.1146/annurev.neuro.26.041002.131425. [DOI] [PubMed] [Google Scholar]
- 54.Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–20. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 55.van Alphen B, van Swinderen B. Drosophila strategies to study psychiatric disorders. Brain Res Bull. [DOI] [PubMed]
- 56.Bushey D, Tononi G, Cirelli C. The Drosophila fragile X mental retardation gene regulates sleep need. J Neurosci. 2009;29:1948–61. doi: 10.1523/JNEUROSCI.4830-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sawamura N, Ando T, Maruyama Y, Fujimuro M, Mochizuki H, Honjo K, et al. Nuclear DISC1 regulates CRE-mediated gene transcription and sleep homeostasis in the fruit fly. Mol Psychiatry 2008; 13:1138-48, 069. [DOI] [PMC free article] [PubMed]
- 58.Louter M, van der Marck MA, Pevernagie DA, Munneke M, Bloem BR, Overeem S. Sleep matters in Parkinson's disease: use of a priority list to assess the presence of sleep disturbances. Eur J Neurol. [DOI] [PubMed]
- 59.Suzuki K, Miyamoto M, Miyamoto T, Iwanami M, Hirata K. Sleep disturbances associated with Parkinson’s disease. Parkinsons Dis. 2011;2011:219056. doi: 10.4061/2011/219056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schulte EC, Winkelmann J. When Parkinson’s disease patients go to sleep: specific sleep disturbances related to Parkinson’s disease. J Neurol. 2011;258(Suppl 2):S328–35. doi: 10.1007/s00415-011-5933-0. [DOI] [PubMed] [Google Scholar]
- 61.Menza M, Dobkin RD, Marin H, Bienfait K. Sleep disturbances in Parkinson’s disease. Mov Disord. 2010;25(Suppl 1):S117–22. doi: 10.1002/mds.22788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goodman AO, Morton AJ, Barker RA. Identifying sleep disturbances in Huntington’s disease using a simple disease-focused questionnaire. PLoS Curr. 2010;2:RRN1189. doi: 10.1371/currents.RRN1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gagnon JF, Petit D, Latreille V, Montplaisir J. Neurobiology of sleep disturbances in neurodegenerative disorders. Curr Pharm Des. 2008;14:3430–45. doi: 10.2174/138161208786549353. [DOI] [PubMed] [Google Scholar]
- 64.Gonzales E, Yin J. Drosophila Models of Huntington's Disease exhibit sleep abnormalities. PLoS Curr; 2. [DOI] [PMC free article] [PubMed]
- 65.Gajula Balija MB, Griesinger C, Herzig A, Zweckstetter M, Jäckle H. Pre-fibrillar α-synuclein mutants cause Parkinson’s disease-like non-motor symptoms in Drosophila. PLoS One. 2011;6:e24701. doi: 10.1371/journal.pone.0024701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Akrouh A, Halcomb SE, Nichols CG, Sala-Rabanal M. Molecular biology of K(ATP) channels and implications for health and disease. IUBMB Life. 2009;61:971–8. doi: 10.1002/iub.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Allebrandt KV, Amin N, Muller-Myhsok B, Esko T, Teder-Laving M, Azevedo RV, et al. A K(ATP) channel gene effect on sleep duration: from genome-wide association studies to function in Drosophila. Mol Psychiatry. [DOI] [PubMed]
- 68.Guruharsha KG, Rual JF, Zhai B, Mintseris J, Vaidya P, Vaidya N, et al. A protein complex network of Drosophila melanogaster. Cell. 2011;147:690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(Suppl 1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(Web Server issue):W214-20. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Montojo J, Zuberi K, Rodriguez H, Kazi F, Wright G, Donaldson SL, et al. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics. 2010;26:2927–8. doi: 10.1093/bioinformatics/btq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wittkop T, Berman AE, Fleisch KM, Mooney SD. DEFOG: discrete enrichment of functionally organized genes. Integr Biol (Camb); 4:795-804. [DOI] [PMC free article] [PubMed]
- 73.Zimmerman JE, Raizen DM, Maycock MH, Maislin G, Pack AI. A video method to study Drosophila sleep. Sleep. 2008;31:1587–98. doi: 10.1093/sleep/31.11.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gilestro GF. Video tracking and analysis of sleep in Drosophila melanogaster. Nat Protoc. 2012;7:995–1007. doi: 10.1038/nprot.2012.041. [DOI] [PubMed] [Google Scholar]
- 75.Donelson N, Kim EZ, Slawson JB, Vecsey CG, Huber R, Griffith LC. High-resolution positional tracking for long-term analysis of Drosophila sleep and locomotion using the “tracker” program. PLoS One. 2012;7:e37250. doi: 10.1371/journal.pone.0037250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Simon JC, Dickinson MH. A new chamber for studying the behavior of Drosophila. PLoS One. 2010;5:e8793. doi: 10.1371/journal.pone.0008793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ueno T, Tomita J, Kume S, Kume K. Dopamine modulates metabolic rate and temperature sensitivity in Drosophila melanogaster. PLoS One. 2012;7:e31513. doi: 10.1371/journal.pone.0031513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Becker JD, Honerkamp J, Hirsch J, Fröbe U, Schlatter E, Greger R. Analysing ion channels with hidden Markov models. Pflugers Arch. 1994;426:328–32. doi: 10.1007/BF00374789. [DOI] [PubMed] [Google Scholar]
- 79.Prusseit J, Lehnertz K. Stochastic qualifiers of epileptic brain dynamics. Phys Rev Lett. 2007;98:138103. doi: 10.1103/PhysRevLett.98.138103. [DOI] [PubMed] [Google Scholar]
- 80.Humphries NE, Queiroz N, Dyer JR, Pade NG, Musyl MK, Schaefer KM, et al. Environmental context explains Lévy and Brownian movement patterns of marine predators. Nature. 2010;465:1066–9. doi: 10.1038/nature09116. [DOI] [PubMed] [Google Scholar]
- 81.Hilborn R, Mangel M. The ecological detective: confronting models with data. Princeton, N.J.: Princeton University Press, 1997. [Google Scholar]
- 82.Laing C, Lord GJ. Stochastic methods in neuroscience. Oxford: Oxford University Press.