Abstract
The role of metabolic disturbance in polycystic ovary syndrome (PCOS) has been well established, with insulin resistance and the resulting compensatory hyperinsulinemia thought to promote hyperandrogenemia. Genome-wide association studies (GWAS) have established a large number of loci for metabolic conditions such as type 2 diabetes and obesity. A subset of these loci has been investigated for a role in PCOS; these studies generally have not revealed a confirmed role for these loci in PCOS risk. However, a large scale investigation of genes related to these pathways has not previously been performed. We conducted a two stage case control association study of 121,715 single nucleotide polymorphisms (SNPs) selected to represent susceptibility loci associated with traits such as type 2 diabetes, obesity measures, lipid levels and cardiovascular function using the Cardio-Metabochip in 847 PCOS cases and 845 controls. Several hypothesis-generating associations with PCOS were observed (top SNP rs2129107, P = 3.8 × 10−6). We did not find any loci definitively associated with PCOS after strict correction for multiple testing, suggesting that cardio-metabolic loci are not major risk factors underlying the susceptibility to PCOS.
Keywords: Candidate-wide association study, Polycystic ovary syndrome, Cardio-Metabochip, Single nucleotide polymorphism, Genetic association
1. Introduction
The genetic basis of polycystic ovary syndrome (PCOS) was established by familial aggregation and twin studies [1,2]. The literature of PCOS genetics is dominated by many studies each examining a single gene. Because few genes are widely accepted as PCOS risk factors, large-scale genome-wide association studies (GWAS) have been eagerly anticipated. To date, one GWAS has been published from China, implicating only three loci in PCOS pathogenesis [3]. Women with PCOS have an increased prevalence of type 2 diabetes, insulin resistance, central obesity, dyslipidemia, endothelial dysfunction, and subclinical vascular disease [4], possibly conferring an increased risk of ischemic cardiovascular disease [5]. Therefore, a potential approach to identifying PCOS susceptibility loci is the examination of validated genes for these traits. To date, only a subset of diabetes susceptibility loci have been tested for association with PCOS, with mixed results.
In the current study, we utilized the Cardio-MetaboChip (196,725 single nucleotide polymorphisms (SNPs)) to investigate the role of a broad set of genes selected based on GWAS results for diabetes-related traits (diabetes status, age at diabetes diagnosis, early onset of diabetes, fasting glucose and insulin, two hour glucose and hemoglobin A1c), obesity-related traits (body mass index (BMI), waist-hip ratio, percent fat mass and waist circumference), cardiovascular traits (myocardial infarction, coronary artery disease, systolic blood pressure, diastolic blood pressure and QT interval), lipid traits (total cholesterol, triglycerides, low density lipoprotein (LDL) cholesterol, high density lipoprotein (HDL) cholesterol), and several other measures such as height, platelet count, mean platelet volume and white blood count. Given the scope of this effort, we have termed this a “candidate-wide association study” (CWAS) [6]. Utilizing a discovery and a replication cohort approach, we found a number of loci associated with PCOS in both cohorts; however, none of these maintained significance after consideration of multiple testing.
2. Experimental
2.1. Subjects and phenotyping
All subjects gave written informed consent; each study was approved by the Institutional Review Boards of the recruiting centers and Cedars-Sinai Medical Center (CSMC). Clinical characteristics of the discovery and replication cohorts are presented in Table 1.
Table 1.
Clinical characteristics of PCOS and control subjects.
| Discovery cohort |
Replication cohort |
Combined |
||||
|---|---|---|---|---|---|---|
| Cases (n = 385) | Controls (n = 170) | Cases (n = 462) | Controls (n = 675) | Cases (n = 847) | Controls (n = 845) | |
| Age (year) | 26.7 (10.1)a | 33.5 (15.0) | 28.0 (8.0)a | 52.2 (18.7)b | 27.0 (9.0)a | 48.2 (21.6) |
| BMI (kg/m2) | 31.2 (14.7)a | 24.2 (6.8) | 34.5 (13.3)a,b | 26.7 (6.9)b | 33.4 (14.2)a | 26.2 (7.1) |
| Insulin (µIU/ml) | 14.0 (17.0)a | 7.1 (6.7) | 20.0 (17.2)a,b | 11.9 (8.0)b | 17.0 (17.0)a | 11.0 (8.3) |
| Glucose (mg/dl) | 85.0 (11.0) | 85.0 (11.0) | 87.5 (11.5)a,b | 92.0 (13.6)b | 86.5 (12.0)a | 91.2 (13.8) |
| HOMA-%B | 150.0 (100.8)a | 105.4 (56.2) | 190.8 (86.7)a,b | 122.8 (55.7)b | 176.3 (98.0)a | 120.6 (56.4) |
| HOMA-IR | 1.72 (1.96)a | 0.98 (0.88) | 2.47 (1.96)a,b | 1.57 (1.06)b | 2.18 (2.01)a | 1.48 (1.10) |
| Testosterone (ng/dl) | 69.0 (41.5)a | 40.0 (27.5) | 69.0 (33.0)a,c | 29.0 (14.0)b | 69.0 (36.4)a | 34.0 (27.0) |
| DHEAS (ng/ml) | 2203.0 (1802.5)a | 983.5 (739.5) | 2157.5 (1416.25)a | 1343.0 (670.8)b | 2183.0 (1629.8)a | 1070.0 (782.5) |
Data are median (interquartile range). In the replication control cohort, androgen measurements were not available for CAP subjects.
Abbreviations: BMI, body mass index; HOMA-IR, homeostasis model assessment of insulin resistance; HOMA-%B, homeostasis model assessment of beta-cell function (insulin secretion).
P < 0.001 compared to control group in the same cohort.
P < 0.001 compared to the same group in the discovery cohort.
P = 0.04 compared to the same group in the discovery cohort.
2.2. Discovery cohort
We studied 443 White PCOS patients and 193 White control women recruited at two centers, the University of Alabama at Birmingham and CSMC. Cases were premenopausal, non-pregnant, on no hormonal therapy, including oral contraceptives, for at least three months, and met 1990 NIH criteria for PCOS [7]. Parameters for defining hirsutism, hyperandrogenemia, ovulatory dysfunction, and exclusion of related disorders were previously reported [8]. Controls were healthy women, with regular menstrual cycles and no evidence of hirsutism, acne, alopecia, or endocrine dysfunction and had not taken hormonal therapy (including oral contraceptives) for at least three months. Controls were recruited by word of mouth and advertisements calling for “healthy women.”
2.3. Replication cohort
We assembled a cohort of 462 White PCOS patients and 675 White control subjects from four sources: 356 PCOS subjects and 68 healthy controls previously recruited at Pennsylvania State University by Legro [1], 37 PCOS subjects and four healthy controls recruited at CSMC using the same criteria as those used in the discovery cohort, 69 PCOS women from the Pregnancy in PCOS (PPCOS) trial who consented to enter into the NIH Reproductive Medicine Research Network sample repository [9]; and 332 healthy White men and 271 White women derived from the Cholesterol and Pharmacogenetics (CAP) Study, a component of the Pharmacogenomics and Risk of Cardiovascular Disease (PARC) Study [10]. All PCOS subjects met the 1990 NIH criteria [7]. The CAP samples consist of general community controls.
2.4. Genotyping
All genotyping was performed at the Medical Genetics Institute at CSMC using Infinium II technology on the Cardio-Metabochip, following the manufacturer’s protocol (Illumina, San Diego, CA) [11,12]. The Cardio-Metabochip was designed to provide high throughput genotyping for replication of GWAS results and fine mapping purposes (see Supplementary Methods (Appendix A) for further details). Of the unique study samples genotyped (4.5% of which were whole genome amplified DNA) sample quality control measures removed 60 samples for low genotyping rate (<98%) or low p10GC (a sample statistic representing the tenth percentile of the distribution of genotype quality scores across all SNPs genotyped). Six subjects were removed for errors in gender estimates, which were calculated within Genome Studio (Illumina, San Diego, CA).
Case and control cohorts were investigated using Identity-By-Descent in PLINK (http://pngu.mgh.harvard.edu/purcell/plink) [13] in order to identify cryptic relatedness. Thirty three unknown duplicates and siblings (sister pairs in the replication cohort) were excluded. Following these quality control steps, 856 PCOS cases and 855 controls were included in the study. The genotyping rate in these subjects was 99.94%. Across the two projects (discovery and replication), 56 pairs of sample duplicates were run (representing 5% of the entire sample run as either within-plate, across plate and across project duplicates). One sample pair had an unacceptable reproducibility statistic (84.47%) and was removed. Of the remaining 55 sample pairs the average reproducibility was 99.99%. Across the two projects 10 Centre d’Etude du Polymorphisme Humain (CEPH) parent-child pairs were run with an average heritability estimate of 99.98%.
196,475 SNPs were available from Genome Studio for the analysis pipeline. SNPs were excluded if they met any of the following: a test of Hardy–Weinberg equilibrium gave a P < 0.0001 in the control subjects (163 SNPs); SNP failure rate >10%; non-polymorphic markers (74,760 SNPs). The final number of SNPs available for analysis was 121,715.
2.5. Statistical analysis
Population structure was detected using EIGENSTRAT implemented within Golden Helix (Bozeman, MT). In total, 25 principal components were calculated and plotted with Hapmap Caucasian (CEU, CEPH (Utah residents with ancestry from northern and western Europe)), African (YRI, Yoruba in Ibadan, Nigeria) and Asian (CHB/CHD, Han Chinese in Beijing China and Chinese in Metropolitan Denver, Colorado) reference populations for graphical representation of population substructure within the cohort (Fig. B.1). Subjects with significant estimated African or Asian admixture were excluded from the data set (21 subjects removed). Eigenvectors were computed within the case and control subjects only to test for association with the PCOS phenotype (yes/no) in order to detect possible confounding due to population structure (Golden Helix). No Eigenvalue was associated with the disease phenotype, so analyses were not adjusted for principal components. Association testing was then performed using a logistic regression model for association with disease status under an additive model in PLINK adjusting for age, BMI, recruitment site, and sex.
The Cardio-Metabochip entails a large number of SNPs and therefore a large number of statistical tests. Given the nature of this chip, we arbitrarily chose a suggestive P -value cut-off of 1 × 10−4 and sought to establish a fully definitive Bonferroni corrected cut-off based on the number of independent tests. Correcting for the total number of SNPs is not appropriate because, given the chip’s focus on fine-mapping, it contains many variants in linkage disequilibrium. A P-value cut-off to account for multiple testing correction was calculated by determining the number of independent (r2 < 0.8) SNPs in the set of 121,715 SNPs that was successfully genotyped, using a sliding window of 200 markers and a slide of 50 markers (indep-pairwise in PLINK). The Bonferroni multiple-testing corrected P-value cut-off was set as 0.05 divided by this number of independent SNPs.
3. Results
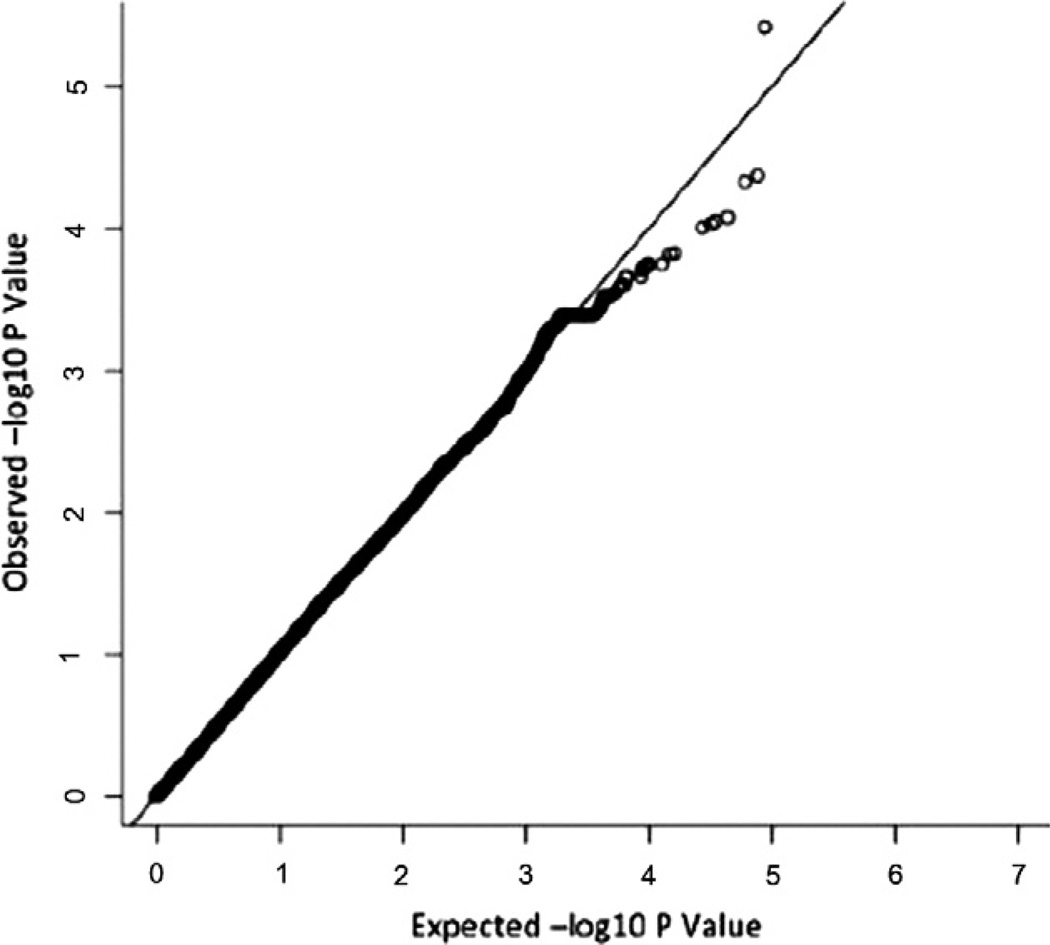
After exclusion of suboptimal DNA based on quality control criteria, data from 847 cases and 845 controls was available for statistical analyses with 121,715 autosomal SNPs. The overall results did not show a significant amount of genomic inflation, with minimal deviation from what was expected by chance (λGC = 1.066) (Fig. 1).
Fig. 1.
QQ plot displaying the expected and observed −log10 P-value for the combined (discovery plus replication) adjusted logistic regression for PCOS status.
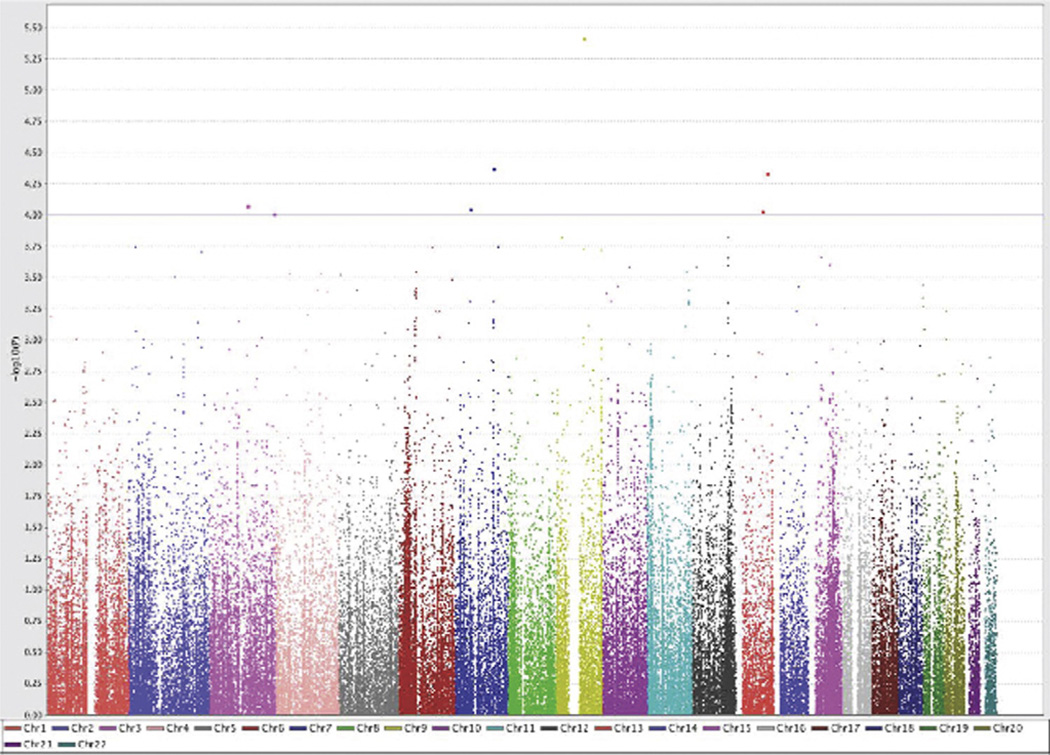
We identified seven distinct loci with suggestive evidence of association (P < 10−4) with PCOS in an adjusted analysis of the combined cohort, with P -values ranging from 9.81 × 10−5 to 3.81 × 10−6 (Table 2; Fig. 2; Fig. B.2). Three of these top seven loci nominally associated with PCOS susceptibility were mapped in or near genes included on the platform for diabetes related traits (rs2129107: TLE1: early onset type 2 diabetes; rs12428018: GPC6: fasting glucose; rs4603906: HES1: type 2 diabetes), supporting a role for metabolism loci in PCOS susceptibility (Table 2). Fig. B.2 displays regional association plots for the loci listed in Table 2. Several additional loci showed regional association, with two or more SNPs within 100 kb with P-value <0.001 (Table A.1). To account for multiple testing, we determined that the number of unlinked (r2 < 0.8) markers (representing the number of independent tests) represented by the SNPs successfully genotyped was 72,208, yielding a P value correction of 0.05/72,208 = 7 × 10−7. None of the SNP associations met this strict level of significance.
Table 2.
SNPs with suggestive evidence of association with PCOS (P < 1.0 × 10−4) in the combined analysis of cases and controls.
| Chr | SNP | Selection category |
Combined |
Discovery |
Replication |
Locia | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| MAF | OR (95% CI) | P | OR OR (95% CI) | P | OR (95% CI) | P | ||||
| 9 | rs2129107 | Early onset T2D | 0.21 | 0.55 (0.41–0.74) | 3.81 × 10−6 | 0.60 (0.32–0.70) | 0.002 | 0.50 (0.44–1.13) | 5.95 × 10−4 | TLE1 |
| 7 | rs42476 | QT | 0.09 | 0.45 (0.30–0.67) | 4.23 × 10−5 | 0.46 (0.33–0.98) | 0.002 | 0.43 (0.17–0.57) | 0.008 | CAV1, MET |
| 13 | rs12428018 | Fasting glucose | 0.44 | 1.72 (0.47–0.78) | 4.68 × 10−5 | 1.69 (0.47–0.90) | 0.002 | 1.77 (0.35–0.79) | 0.008 | GPC6 |
| 3 | rs7643975 | BMI | 0.10 | 0.55 (0.30–0.66) | 8.34 × 10−5 | 0.47 (0.28–0.75) | 1.58 × 10−4 | 0.71 (0.23–0.80) | 0.145 | ZBTB20 |
| 7 | rs2119050 | QT | 0.36 | 0.60 (043–0.71) | 8.91 × 10−5 | 0.65 (0.43–0.83) | 0.009 | 0.52 (0.34–0.75) | 0.002 | CCM2, TBRG4, RAMP3 |
| 13 | rs9545133 | LDL-C | 0.04 | 0.31 (0.17–0.56) | 9.29 × 10−5 | 0.24(0.11–0.50) | 1.56 × 10−4 | 0.52 (0.19–1.45) | 0.213 | NDFIP2 |
| 3 | rs4603906 | T2D | 0.42 | 0.45 (1.32–2.23) | 9.81 × 10−5 | 0.57 (0.34–0.98) | 0.041 | 0.32 (1.16–2.69) | 1.57 × 10−4 | HES1 |
Abbreviations: BMI, body mass index; Chr, chromosme; LDL-C, low density lipoprotein cholesterol; MAF, minor allele frequency; OR, odds ratio; P, P-value; QT, QT Interval; SNP, single nucleotide polymorphism; T2D, type 2 diabetes.
Loci indicating genes within a 200 kb window around the SNP. Genes shown in bold contain the marker within the gene.
Fig. 2.
Manhattan plot displaying −log10 P-values for all SNPs (121,175 markers across 22 autosomes) using an adjusted logistic model in the combined data set. Chr, chromosome.
4. Discussion
Despite extensive efforts, the genetic basis of PCOS is not fully elucidated. Insulin resistance and hyperinsulinemia are critical contributors to the phenotype of PCOS. Insulin stimulates androgen production via luteinizing hormone (LH) and insulin-like growth factor 1 (IGF-1) [14–16], promotes luteinization of premature follicles by increasing follicle-stimulating hormone (FSH)-induced granulosa cell differentiation [17], and elevates serum free testosterone levels by decreasing hepatic sex hormone-binding globulin production [18]. With the hypothesis that inherited dysfunction in glucose homeostasis or cardiovascular function may contribute to the pathophysiology of PCOS, we used a CWAS approach to simultaneously analyse over 100,000 SNPs implicated in metabolic and cardiovascular traits for a potential role in PCOS susceptibility. The Cardio-Metabochip was the ideal platform for this effort, especially considering that PCOS predisposes to diabetes and is characterized by cardiovascular risk factors such as dyslipidemia, central obesity, and inflammation [4], all of which are represented on the Cardio-Metabochip.
While GWAS studies have reproducibly identified loci for many common traits, such as type 2 diabetes [19], obesity [20], myocardial infarction [21], and lipid levels [22], the role of these genes in PCOS has not previously been assessed in a large scale approach. A handful of type 2 diabetes and obesity loci have been examined in PCOS. Individual studies of the Pro12Ala variant in PPARG (peroxisome proliferator-activated receptor gamma) were largely negative; however, a meta-analysis suggested that the Ala allele, a well-established protective factor in type 2 diabetes, may also confer protection against PCOS [23]. Several studies of the diabetes-associated variants in TCF7L2 (transcription factor 7 like 2), the gene with the largest effect size in type 2 diabetes, found no effect on PCOS risk [24–29]; however, one study found that single nucleotide polymorphisms (SNPs) at the other end of the gene were associated with PCOS [25]. Regarding FTO (fat mass and obesity associated) genetic variants that affect diabetes risk via body mass index (BMI), the balance of the evidence in PCOS found association with BMI, fat mass or fat distribution and other metabolic traits and rarely reproductive hormones, with some studies also describing association with PCOS; some but not all of these associations lost significance after BMI adjustment [28,30–35]. Other diabetes loci found not to have any association with PCOS or its component traits include KCNJ11 (potassium inwardly rectifying channel, subfamily J, member 11) [26,36], CDKAL1 (CDK5 regulatory subunit-associated protein 1 like 1) [27], and SLC30A8 (solute carrier 30 (zinc transporter) member 8) [37]. The diabetogenic variant in the HHEX/IDE (hematopoietically expressed homeobox/insulin-degrading enzyme) region was not associated with PCOS [29]; however, a different SNP in IDE was associated with PCOS and insulin levels [38].
All of the above studies examined one or two diabetes/obesity genes, except Tan et al., wherein four genes were studied [28]. We more broadly approached obesity genes in PCOS, genotyping 15 SNPs from 9 obesity genes, and found SNPs in FTO and MC4R (melanocortin 4 receptor) to be associated with BMI [32] but not PCOS itself. Our subsequent study of 18 variants in 10 diabetes genes found no association with PCOS [39]. Of note, the latter studies of multiple genes focused on the actual variants previously associated with obesity and diabetes by GWAS. As observed for TCF7L2 and IDE [25,38], variants elsewhere in such gene regions may affect the odds of PCOS, necessitating more comprehensive coverage across the entire genes. This need inspired our study of a large number of metabolic loci in our PCOS case control cohort. The Cardio-Metabochip was specifically designed to densely fine map these loci.
Given the lack of significance after multiple testing correction, the loci and genes identified by our analysis must be considered hypothesis generating and not definitively established. Nevertheless, some of the genes identified are of particular interest. TLE1 (transducin-like enhancer of split 1) codes for a transcriptional co-repressor that participates in repression of androgen receptor signaling [40]; however, it may also promote estrogen receptor (ER) binding to certain ER binding sites, enhancing ER activity [41]. The potential effects of TLE1 on sex steroid signaling are clearly relevant to PCOS. Another locus, CAV1 (caveolin-1) is possibly relevant to PCOS given that genetic variation in this gene has been associated with insulin resistance and hypertension [42].
The loci identified in this CWAS for PCOS susceptibility in a case control cohort of ethnically matched Caucasian subjects did not attain SNP-wide significance, likely for several reasons. Examining a large number of markers requires correction for multiple testing in order to minimize false positive results. We used a modified Bonferroni approach, where the number of independent tests was calculated in order to appropriately adjust our results, yielding a P-value significance level of 7 × 10−7. No SNPs in our adjusted logistic model reached this significance level. Despite studying a relatively large sample size compared to prior genetic epidemiologic studies of PCOS, our power was affected by the large number of tests. Using an alpha of 7 × 10−7 in power calculations, the sample size of 847 cases and 845 controls has good power (≥80%) to detect association of risk alleles of frequency ≥0.2 with PCOS at odds ratio ≥2.0 and fair-to-good power (37–87%) to detect association at odds ratio 1.75. Detailed power calculations given in Table A.2 reveal lower power to detect association of rare risk alleles (frequency ≤ 0.1) with PCOS at odds ratios less than 1.75. Thus, it is possible that SNPs on the Cardio-Metabochip with modest effect sizes may be associated with PCOS, but were not detected in the current study.
Clinical heterogeneity may have reduced our ability to detect significant loci, as subjects were recruited at multiple centers. However, our cohort was homogeneous for diagnosis (all met 1990 NIH criteria) and race (White, with subjects of mixed ancestry removed from analysis). Another factor affecting power in our study was the inclusion of a large number of community controls from the CAP study, wherein subjects did not undergo an evaluation for PCOS. On the other hand, use of community controls allows significant expansion of sample size, offsetting the loss of power from undiagnosed cases; this approach to control groups has been applied with much success in recent GWAS efforts in common disease [43,44].
The CWAS approach lacks a key advantage of the GWAS approach. The flood of new genes and pathways that have been discovered in many common diseases as a result of GWAS can in large part be attributed to the hypothesis-free approach of performing an unbiased genome scan. By including complete coverage of the genome, GWAS have been able to identify loci from previously unknown and unexpected pathways relative to the diseases and traits studied. On the other hand, the Cardio-Metabochip was designed primarily for extensive fine mapping coverage of known metabolic loci. The largely negative findings of our study suggest that the role of metabolic genes in PCOS susceptibility may be modest to minimal, highlighting the need for unbiased GWAS to discover novel loci in PCOS.
The final potential contributor to our lack of definitive loci is the content of the Cardio-Metabochip itself. Designed to interrogate known GWAS loci, many of which resulted from meta-analyses of tens of thousands of subjects, many of the loci on the platform have small effect sizes or very low frequency. The sample sizes required to detect significant findings with such markers could be significantly greater than that available to us in this study. The recent Chinese PCOS GWAS [3] utilized a total sample size of 4082 PCOS and 6687 subjects (one discovery cohort and two replication cohorts); the susceptibility locus with the largest effect size in that study had an odds ratio of 1.51 (rs10818854 in DENND1A: P = 9.4 × 10−18 in a meta analysis of all three Chinese cohorts), suggesting very large sample sizes are needed for studies of this nature. One of the loci associated with PCOS in the Chinese GWAS is THADA, a known diabetes gene. The reported odds ratios for this locus (2p21) ranged from 0.67 to 0.72, which we were not well powered to detect at an alpha of 7 × 10−7.
As a complex and common disease, the mechanism underlying PCOS has been difficult to elucidate. Candidate gene studies have yielded some clues to the role of diabetes loci; however, concordance within the field has been infrequent. Using a CWAS approach in a case control cohort we simultaneously screened 127,715 SNPs from known diabetes, glucose homeostasis, obesity, cardiovascular and lipid loci. We did not identify any significant associations between these loci and PCOS susceptibility that withstood correction for multiple testing, suggesting these loci are not major risk factors for PCOS. The susceptibility genes for PCOS may be very unique, requiring GWAS for their discovery, or may include genes whose effect size will require substantially greater sample sizes than have been available for PCOS genetic studies to date.
Supplementary Material
Acknowledgements
This study was supported by National Institutes of Health Grants U54-HD034449 to RSL, RR10732 and C06-RR016499 to Pennsylvania State University General Clinical Research Center (GCRC), R01-HD029364 to RA, R01-HL069757 to RMK, R01-DK079888 to MOG, CTSI Grant UL1RR033176, DERC Grant P30-DK063491, Grants to the Reproductive Medicine Network (U01-HD55925, U10-HD27049, U01-HD38997, U10-HD39005, U10-HD27011, U10-HD33172, U10-HD38988, U10-HD38992, U10-HD38998, U10-HD38999), and the Winnick Clinical Scholars Award to MOG.
Abbreviations
- BMI
body mass index
- CEPH
Centre d’Etude du Polymorphisme Humain
- CWAS
candidate-wide association study
- GWAS
genome-wide association study
- HDL-C
high density lipoprotein cholesterol
- LDL-C
low density lipoprotein cholesterol
- NIH
National Institutes of Health
- PCOS
polycystic ovary syndrome
- SNP
single nucleotide polymorphism
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.steroids.2011.12.005.
Contributor Information
Michelle R. Jones, Email: michelle.jones@cshs.org.
Angela K. Chua, Email: angela.chua@cshs.org.
Emebet A. Mengesha, Email: emebet.mengesha@cshs.org.
Kent D. Taylor, Email: kent.taylor@cshs.org.
Yii-Der I. Chen, Email: ida.chen@cshs.org.
Xiaohui Li, Email: xiaohui.li@cshs.org.
Ronald M. Krauss, Email: RKrauss@chori.org.
Jerome I. Rotter, Email: jerome.rotter@cshs.org.
Richard S. Legro, Email: rsl1@psu.edu.
Ricardo Azziz, Email: razziz@georgiahealth.edu.
Mark O. Goodarzi, Email: mark.goodarzi@cshs.org.
References
- 1.Legro RS, Driscoll D, Strauss JF, Fox J, Dunaif A. Evidence for a genetic basis for hyperandrogenemia in polycystic ovary syndrome. Proc Natl Acad Sci USA. 1998;95:14956–14960. doi: 10.1073/pnas.95.25.14956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vink JM, Sadrzadeh S, Lambalk CB, Boomsma DI. Heritability of polycystic ovary syndrome in a Dutch twin-family study. J Clin Endocrinol Metab. 2006;91:2100–2104. doi: 10.1210/jc.2005-1494. [DOI] [PubMed] [Google Scholar]
- 3.Chen ZJ, Zhao H, He L, Shi Y, Qin Y, Shi Y, et al. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat Genet. 2011;43:55–59. doi: 10.1038/ng.732. [DOI] [PubMed] [Google Scholar]
- 4.Goodarzi MO, Dumesic DA, Chazenbalk G, Azziz R. Polycystic ovary syndrome: etiology, pathogenesis and diagnosis. Nat Rev Endocrinol. 2011;7:219–231. doi: 10.1038/nrendo.2010.217. [DOI] [PubMed] [Google Scholar]
- 5.Shaw LJ, Bairey Merz CN, Azziz R, Stanczyk FZ, Sopko G, Braunstein GD, et al. Postmenopausal women with a history of irregular menses and elevated androgen measurements at high risk for worsening cardiovascular event-free survival: results from the National Institutes of Health–National Heart, Lung, and Blood Institute sponsored Women’s Ischemia Syndrome Evaluation. J Clin Endocrinol Metab. 2008;93:1276–1284. doi: 10.1210/jc.2007-0425. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Rich SS, Norris JM, Rotter JI. Genes associated with risk of type 2 diabetes identified by a candidate-wide association scan: as a trickle becomes a flood. Diabetes. 2008;57:2915–2917. doi: 10.2337/db08-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zawadzki J, Dunaif A. Diagnostic criteria for polycystic ovary syndrome: towards a rational approach. In: Dunaif A, Givens JR, Haseltine FP, Merriam GR, editors. Polycystic ovary syndrome. Boston: Blackwell Scientific; 1992. pp. 377–384. [Google Scholar]
- 8.Azziz R, Woods KS, Reyna R, Key TJ, Knochenhauer ES, Yildiz BO. The prevalence and features of the polycystic ovary syndrome in an unselected population. J Clin Endocrinol Metab. 2004;89:2745–2749. doi: 10.1210/jc.2003-032046. [DOI] [PubMed] [Google Scholar]
- 9.Legro RS, Myers ER, Barnhart HX, Carson SA, Diamond MP, Carr BR, et al. The Pregnancy in Polycystic Ovary Syndrome study: baseline characteristics of the randomized cohort including racial effects. Fertil Steril. 2006;86:914–933. doi: 10.1016/j.fertnstert.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 10.Simon JA, Lin F, Hulley SB, Blanche PJ, Waters D, Shiboski S, et al. Phenotypic predictors of response to simvastatin therapy among African-Americans and Caucasians: the Cholesterol and Pharmacogenetics (CAP) Study. Am J Cardiol. 2006;97:843–850. doi: 10.1016/j.amjcard.2005.09.134. [DOI] [PubMed] [Google Scholar]
- 11.Gunderson KL, Steemers FJ, Lee G, Mendoza LG, Chee MS. A genome-wide scalable SNP genotyping assay using microarray technology. Nat Genet. 2005;37:549–554. doi: 10.1038/ng1547. [DOI] [PubMed] [Google Scholar]
- 12.Gunderson KL, Steemers FJ, Ren H, Ng P, Zhou L, Tsan C, et al. Whole-genome genotyping. Methods Enzymol. 2006;410:359–376. doi: 10.1016/S0076-6879(06)10017-8. [DOI] [PubMed] [Google Scholar]
- 13.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nestler J, Jakubowicz DJ, Falcon de Vargas A, Brik C, Quintero N, Medina F. Insulin stimulates testosterone biosynthesis by human thecal cells from women with polycystic ovary syndrome by activating its own receptor and using inositoglycan mediators as the signal transduction system. J Clin Endocrinol Metab. 1998;83:2001–2005. doi: 10.1210/jcem.83.6.4886. [DOI] [PubMed] [Google Scholar]
- 15.Willis D, Franks S. Insulin action in human granulosa cells from normal and polycystic ovaries is mediated by the insulin receptor and not the the type-I insulin-like growth factor receptor. J Endocrinol Metabo. 1995;80:3788–3790. doi: 10.1210/jcem.80.12.8530637. [DOI] [PubMed] [Google Scholar]
- 16.Bergh C, Carlsson B, Olsson JH, Selleskog U, Hillensjo T. Regulation of androgen production in cultured human thecal cells by insulin-like growth factor I and insulin. Fertil Steril. 1993;59:323–331. doi: 10.1016/s0015-0282(16)55675-1. [DOI] [PubMed] [Google Scholar]
- 17.Franks S, Gilling-Smith C, Watson H, Willis D. Insulin action in the normal and polycystic ovary. Endocrinol Metab Clin North Am. 1999;28:361–378. doi: 10.1016/s0889-8529(05)70074-8. [DOI] [PubMed] [Google Scholar]
- 18.Sharp PS, Kiddy DS, Reed MJ, Anyaoku V, Johnston DG, Franks S. Correlation of plasma insulin and insulin-like growth factor-I with indices of androgen transport and metabolism in women with polycystic ovary syndrome. Clin Endocrinol (Oxf) 1991;35:253–257. doi: 10.1111/j.1365-2265.1991.tb03531.x. [DOI] [PubMed] [Google Scholar]
- 19.McCarthy MI. Genomics, type 2 diabetes, and obesity. N Engl J Med. 2010;363:2339–2350. doi: 10.1056/NEJMra0906948. [DOI] [PubMed] [Google Scholar]
- 20.Hebebrand J, Volckmar AL, Knoll N, Hinney A. Chipping away the ‘missing heritability’: GIANT steps forward in the molecular elucidation of obesity - but still lots to go. Obes Facts. 2010;3:294–303. doi: 10.1159/000321537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Demirkan A, Amin N, Isaacs A, Jarvelin MR, Whitfield JB, Wichmann HE, et al. Genetic architecture of circulating lipid levels. Eur J Hum Genet. 2011;19:813–819. doi: 10.1038/ejhg.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.San-Millan JL, Escobar-Morreale HF. The role of genetic variation in peroxisome proliferator-activated receptors in the polycystic ovary syndrome (PCOS): an original case-control study followed by systematic review and meta-analysis of existing evidence. Clin Endocrinol (Oxf) 2010;72:383–392. doi: 10.1111/j.1365-2265.2009.03679.x. [DOI] [PubMed] [Google Scholar]
- 24.Barber TM, Bennett AJ, Groves CJ, Sovio U, Ruokonen A, Martikainen H, et al. Disparate genetic influences on polycystic ovary syndrome (PCOS) and type 2 diabetes revealed by a lack of association between common variants within the TCF7L2 gene and PCOS. Diabetologia. 2007;50:2318–2322. doi: 10.1007/s00125-007-0804-z. [DOI] [PubMed] [Google Scholar]
- 25.Biyasheva A, Legro RS, Dunaif A, Urbanek M. Evidence for association between polycystic ovary syndrome (PCOS) and TCF7L2 and glucose intolerance in women with PCOS and TCF7L2. J Clin Endocrinol Metab. 2009;94:2617–2625. doi: 10.1210/jc.2008-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christopoulos P, Mastorakos G, Gazouli M, Panidis D, Deligeoroglou E, Katsikis I, et al. Genetic variants in TCF7L2 and KCNJ11 genes in a Greek population with polycystic ovary syndrome. Gynecol Endocrinol. 2008;24:486–490. doi: 10.1080/09513590802196379. [DOI] [PubMed] [Google Scholar]
- 27.Liu X, Li L, Chen ZJ, Lu Z, Shi Y, Zhao Y. Genetic variants of cyclin-dependent kinase 5 regulatory subunit associated protein 1-like 1 and transcription factor 7-like 2 are not associated with polycystic ovary syndrome in Chinese women. Gynecol Endocrinol. 2010;26:129–134. doi: 10.3109/09513590903215490. [DOI] [PubMed] [Google Scholar]
- 28.Tan S, Scherag A, Janssen OE, Hahn S, Lahner H, Dietz T, et al. Large effects on body mass index, insulin resistance of fat mass, obesity associated gene (FTO) variants in patients with polycystic ovary syndrome (PCOS) BMC Med Genet. 2010;11:12. doi: 10.1186/1471-2350-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu P, Che Y, Cao Y, Wu X, Sun H, Liang F, et al. Polymorphisms of TCF7L2 and HHEX genes in Chinese women with polycystic ovary syndrome. J Assist Reprod Genet. 2010;27:23–28. doi: 10.1007/s10815-009-9377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Attaoua R, Ait El Mkadem S, Radian S, Fica S, Hanzu F, Albu A, et al. FTO gene associates to metabolic syndrome in women with polycystic ovary syndrome. Biochem Biophys Res Commun. 2008;373:230–234. doi: 10.1016/j.bbrc.2008.06.039. [DOI] [PubMed] [Google Scholar]
- 31.Barber TM, Bennett AJ, Groves CJ, Sovio U, Ruokonen A, Martikainen H, et al. Association of variants in the fat mass and obesity associated (FTO) gene with polycystic ovary syndrome. Diabetologia. 2008;51:1153–1158. doi: 10.1007/s00125-008-1028-6. [DOI] [PubMed] [Google Scholar]
- 32.Ewens KG, Jones MR, Ankener W, Stewart DR, Urbanek M, Dunaif A, TO F, et al. MC4R gene variants are associated with obesity in polycystic ovary syndrome. PLoS One. 2011;6:e16390. doi: 10.1371/journal.pone.0016390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kowalska I, Malecki MT, Straczkowski M, Skupien J, Karczewska-Kupczewska M, Nikolajuk A, et al. The FTO gene modifies weight, fat mass and insulin sensitivity in women with polycystic ovary syndrome, where its role may be larger than in other phenotypes. Diabetes Metab. 2009;35:328–331. doi: 10.1016/j.diabet.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 34.Wehr E, Schweighofer N, Moller R, Giuliani A, Pieber TR, Obermayer-Pietsch B. Association of FTO gene with hyperandrogenemia and metabolic parameters in women with polycystic ovary syndrome. Metabolism. 2010;59:575–580. doi: 10.1016/j.metabol.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 35.Yan Q, Hong J, Gu W, Zhang Y, Liu Q, Su Y, et al. Association of the common rs9939609 variant of FTO gene with polycystic ovary syndrome in Chinese women. Endocrine. 2009;36:377–382. doi: 10.1007/s12020-009-9257-0. [DOI] [PubMed] [Google Scholar]
- 36.Barber TM, Bennett AJ, Gloyn AL, Groves CJ, Sovio U, Ruokonen A, et al. Relationship between E23K (an established type II diabetes-susceptibility variant within KCNJ11), polycystic ovary syndrome and androgen levels. Eur J Hum Genet. 2007;15:679–684. doi: 10.1038/sj.ejhg.5201802. [DOI] [PubMed] [Google Scholar]
- 37.Li L, Zhao Y, Wang L, Yan J, Chen ZJ. Genetic variations of solute carrier family 30 (zinc transporter) member 8 (SLC30A8) are not associated with polycystic ovary syndrome. Fertil Steril. 2009;91:1598–1601. doi: 10.1016/j.fertnstert.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 38.Wang K, You L, Shi Y, Wang L, Zhang M, Chen ZJ. Association of genetic variants of insulin degrading enzyme with metabolic features in women with polycystic ovary syndrome. Fertil Steril. 2008;90:378–384. doi: 10.1016/j.fertnstert.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 39.Ewens KG, Jones MR, Ankener W, Stewart DR, Urbanek M, Dunaif A, et al. Type 2 diabetes susceptibility single-nucleotide polymorphisms are not associated with polycystic ovary syndrome. Fertil Steril. 2011;95:2538–2541. doi: 10.1016/j.fertnstert.2011.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi L, Ko S, Kim S, Echchgadda I, Oh TS, Song CS, et al. Loss of androgen receptor in aging and oxidative stress through Myb protooncoprotein-regulated reciprocal chromatin dynamics of p53 and poly(ADP-ribose) polymerase PARP-1. J Biol Chem. 2008;283:36474–36485. doi: 10.1074/jbc.M805980200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holmes KA, Hurtado A, Brown GD, Launchbury R, Ross-Innes CS, Hadfield J, et al. Transducin-like enhancer protein 1 mediates estrogen receptor binding and transcriptional activity in breast cancer cells. Proc Natl Acad Sci USA. 2011 doi: 10.1073/pnas.1018863108. [Epub May 2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pojoga LH, Underwood PC, Goodarzi MO, Williams JS, Adler GK, Jeunemaitre X, et al. Variants of the caveolin-1 gene: a translational investigation linking insulin resistance and hypertension. J Clin Endocrinol Metab. 2011;96:E1288–E1292. doi: 10.1210/jc.2010-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.