Abstract
The c-di-GMP riboswitch is a macromolecular target in the c-di-GMP second messenger signaling pathway. It regulates many genes related to c-di-GMP metabolism as well as genes involved in bacterial motility, virulence and biofilm formation. The riboswitch makes asymmetric contacts to the bases and phosphate backbone of this symmetric dinucleotide. The phylogenetics suggested and mutagenesis has confirmed that this is a flexible motif where variants can make alternative interactions with each of the guanine bases of c-di-GMP. A mutant riboswitch has been designed that can bind a related molecule, c-di-AMP, confirming the most important contacts made to the ligand. The binding kinetics reveal that this is a kinetically controlled riboswitch and mutations to the riboswitch lead to increases in the off-rate. This riboswitch is therefore flexible in sequence as well as kinetic properties.
Keywords: Riboswitch, c-di-GMP, signaling
c-di-GMP signaling and the c-di-GMP riboswitch
The dinucleotide bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP) functions as an important second messenger signaling molecule in bacteria. Many critical transitions are controlled via this pathway, including biofilm formation and expression of virulence genes [1]. While c-di-GMP has been studied for over 20 years [1,2], the macromolecular targets and the molecular details of these interactions have only recently begun to come to light. As expected, several proteins have been found to function in this pathway [1,3–14] and likely many more will be discovered. However, it was also realized that an RNA functions in this signaling pathway. In 2008, Breaker and coworkers reported the discovery of a riboswitch that binds c-di-GMP and modulates gene expression in response to this second messenger [15]. They recently identified a second class of c-di-GMP responsive riboswitches [16], further expanding the role of RNA in this signaling pathway.
Riboswitches are noncoding RNAs found primarily in the 5′ untranslated region (UTR) of mRNAs. The majority are in bacterial species although examples have been found in eukaryotic organisms as well [17]. Most riboswitches fold into complex tertiary structures that allow them to selectively bind small molecule metabolites. The domain that is responsible for recognition of the small molecule ligand is connected to a downstream element that controls gene expression either at the transcriptional or translational level. Most often this occurs through a conformational change in the ligand binding domain that leads to the stabilization of one of several mutually exclusive secondary structures in the downstream element [17,18].
The c-di-GMP responsive riboswitch (GEMM motif) is found upstream of genes that are involved in the metabolism of c-di-GMP and genes known to be involved in pathways controlled by this second messenger. Over 500 examples of this motif have been identified in many diverse bacterial species [15,19]. Examples of both transcriptional as well as translational regulation are found in this riboswitch class [15]. This RNA element binds c-di-GMP with an affinity of approximately 10 pM [20], making it the tightest binding c-di-GMP receptor as well as the highest affinity RNA-ligand interaction known.
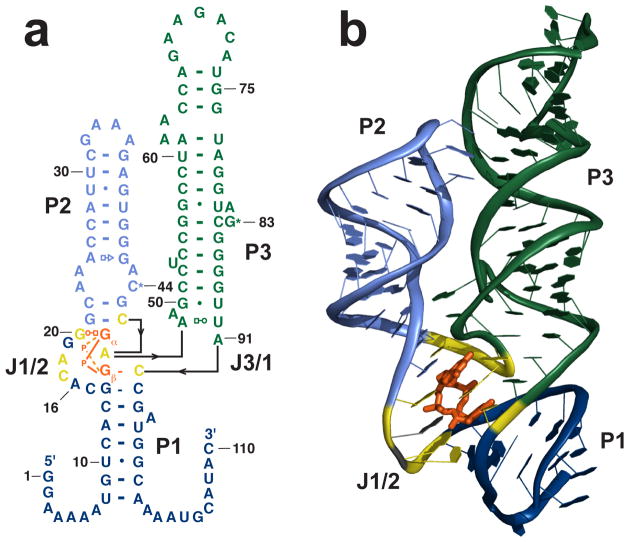
The secondary structure was predicted to consist of two stems that interacted via a tetraloop/tetraloop receptor (TL/TLR) interaction and undefined conformation in the 5′ and 3′ ends [15,19]. Biochemical evidence showed that these flanking regions became more structured upon c-di-GMP binding [15]. The crystal structure of a riboswitch from Vibrio cholerae was solved by both our group [20] and a second group in 2009 [21]. Both structures revealed that the two terminal ends of the RNA form a third helix (P1) when ligand is bound (Figure 1). We predict that the formation of this induced helix is the molecular trigger for this riboswitch class. The two predicted helices (P2 and P3) were observed as well as the TL/TLR motif. Additionally, there is a universally conserved CG (C44/G83) base pair that bridges the P2 and P3 helices. The overall architecture is y-shaped and c-di-GMP binds at the three helix junction. The binding pocket is formed by nucleotides from the P1 and P2 helices as well as the interhelical linker regions, J1/2 and J2/3.
Figure 1.
c-di-GMP riboswitch from V. cholerae. a. Secondary structure of the riboswitch upstream of the tfoX-like gene in V. cholerae. The P1 helix is shown in dark blue, P2 in light blue, P3 in green. The asterisks next to C44 and G83 indicate that these residues are base paired. Nucleotides that directly contact the bases of c-di-GMP are show in yellow. c-di-GMP is shown in orange. b. Crystal structure. The U1A protein used for cocrystallization has been removed for clarity. Coloring is the same as in part a.
Recognition of c-di-GMP by GEMM
These structural studies revealed the details of molecular recognition of c-di-GMP by the riboswitch [20,21]. The two guanine bases, Ga and Gβ, are recognized asymmetrically by the RNA. Both are contacted by multiple riboswitch nucleotides but the number and types of interactions are quite different. Gα is primarily recognized on its Hoogsteen face by the conserved nucleotide G20. Additional contacts are provided by backbone atoms of C46 and A48, as well as a water molecule present in the binding pocket (Figure 2a). Gβ is more extensively recognized by the RNA. The highly conserved cytidine, C92, forms a standard Watson-Crick base pair with Gβ. N2 and N3 are contacted by nucleotides from the J1/2 linker region, C17 and A18. Finally, the 2′-OH of A47 forms a hydrogen bond with N7 (Figure 2b). This base pair effectively becomes the first pair in the P1 helix.
Figure 2.
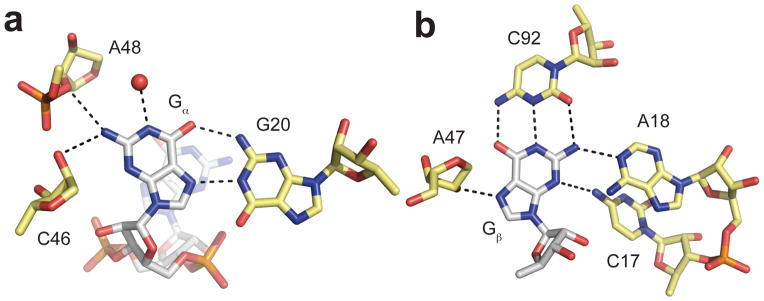
Riboswitch interactions with c-di-GMP. a. Contacts made to Gα. c-di-GMP is colored by atom with nitrogen colored blue, oxygen colored red, phosphorus colored orange, and carbon colored white. Riboswitch nucleotides are colored by atom with nitrogen colored blue, oxygen colored red, phosphorus colored orange, and carbon colored yellow. A water molecule is shown as a red sphere. Hydrogen bonds are denoted by black dashed lines. b. Contacts made to Gβ, Coloring is the same as in part a.
In addition to direct hydrogen bonding contacts to the bases of c-di-GMP, the riboswitch also provides stacking interactions [20,21]. A universally conserved nucleotide, A47, stacks between the two guanine bases. GC base pairs both above and below c-di-GMP continue the stacking network into the P2 and P1 helices, respectively. These stacking interactions are predicted to contribute to the exceptionally tight binding affinity observed in this system [20].
The riboswitch makes contacts to the phosphodiester backbone of c-di-GMP as well. Both sugars interact with RNA atoms as well as solvent molecules [20]. Similarly to the guanine bases, there is asymmetric recognition of the two phosphates. The phosphate 5′ of Gα (PGα) is more heavily contacted than the phosphate 5′ of Gβ (PGβ). PGα forms hydrogen bonds with the exocyclic amines of A47 and A18. Additionally, it is coordinated by a fully hydrated magnesium ion. PGβ primarily interacts with water molecules [20].
RNA requirements for second messenger binding
Despite the many specific interactions that the riboswitch makes with c-di-GMP, very few nucleotides in this RNA motif are universally conserved [15,20]. Surprisingly, G20 or C92, the nucleotides that most directly interact with the ligand, are not among the most conserved nucleotides. Instead, nucleotides in the J2/3 region, including A47, the nucleotide that stacks between the two bases, as well as nucleotides involved in tertiary interactions, are the majority of the universally conserved bases. This observation led us to hypothesize that alternative interactions could be made with c-di-GMP and to define the RNA requirements for ligand binding through a comprehensive mutagenesis study [22].
Several mutations can be accommodated in the binding pocket without eliminating c-di-GMP binding [22]. The least detrimental alterations retained a purine at position 20 and a pyrimidine at position 92. Crystal structures of both the G20A and C92U mutants confirmed their ability to interact with c-di-GMP by contacting the Hoogsteen face and formation of a GU wobble pair, respectively. These mutations are, unsurprisingly, the most common variants at each of these positions, suggesting that these riboswitches are functional in vivo. The flexibility at positions 20 and 92 was not observed at position 47. All mutations to this nucleotide resulted in substantial loss of affinity for c-di-GMP, consistent with is absolute conservation [22].
Other binding pocket nucleotides, specifically those in the J1/2 segment, are tolerant of mutation except in cases in which the sequence becomes complementary to other regions in the RNA and promotes misfolding of the riboswitch. The clearest example is the C15G mutant, which can extend the P1 helix into the binding pocket by base pairing with C92 [22].
The tertiary elements close to the binding pocket are essential for ligand binding while those farther away appear to be less critical. The conserved C44/G83 base pair cannot be mutated without substantial loss in c-di-GMP binding while the TL/TLR can be altered with minimal effect [22].
From structural and biochemical data it is known that the P1 helix is formed when c-di-GMP is bound [15,20,21]. Only the first nucleotide of this helix is required for nanomolar binding of c-di-GMP [22]. Once the RNA polymerase has cleared this nucleotide, the riboswitch is now binding competent.
In summary, the requirements for c-di-GMP binding appear to be a purine at position 20, a pyrimidine at position 92, A47, the C44/G83 element, and the first nucleotide of the P1 helix. If these elements are present, the riboswitch is able to bind its ligand with nanomolar affinity [22]. The cellular concentration of c-di-GMP is predicted to be between nanomolar and low micromolar [1], making riboswitches with binding affinities in this range or tighter responsive in vivo.
Recognition of an alternative ligand
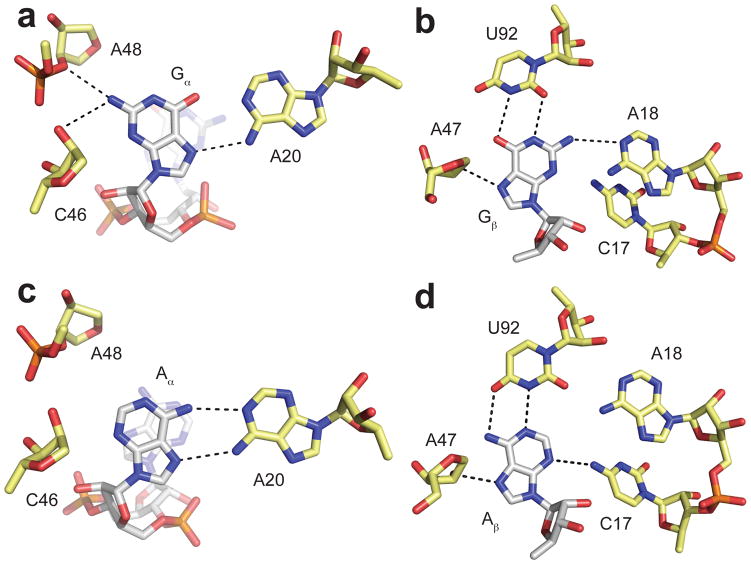
In order to more fully define the interactions made by the riboswitch with its ligand, a double mutant riboswitch, G20A/C92U, was designed that recognizes both c-di-GMP and a related molecule, c-di-AMP. We obtained a 4-fold preference for c-di-AMP over c-di-GMP from this mutant [20]. Structural characterization confirms that both molecules are bound in the same orientation, but that interactions with the bases are unique [22].
Recognition of each of the bases of c-di-GMP is identical to the single G20A and C92U mutants [22] (Figure 3a,b). A hydrogen bond is lost at both Gα and Gβ relative to the wild-type interactions. In the c-di-AMP structure, two hydrogen bonds are made with A20 in an AA Hoogsteen interaction, but hydrogen bonds with C46 and A48 are lost due to the absence of an N2 in adenosine (Figure 3c). Aβ forms a Watson-Crick base pair with U92 but again loses a hydrogen bond because of its lack of an exocyclic amine at position 2 (Figure 3d).
Figure 3.
Interactions between a mutant riboswitch and c-di-GMP and c-di-AMP. a. Interactions made by the G20A/C92U mutant riboswitch and Gα. Coloring is the same as in Figure 2. b. Interactions made by the G20A/C92U mutant riboswitch and Gβ. Coloring is the same as in Figure 2. c. Interactions made by the G20A/C92U mutant riboswitch and Aα. c-di-AMP is colored by atom with nitrogen colored blue, oxygen colored red, phosphorus colored orange, and carbon colored white. Riboswitch nucleotides are colored by atom with nitrogen colored blue, oxygen colored red, phosphorus colored orange, and carbon colored yellow. Hydrogen bonds are denoted by black dashed lines. d. Interactions made by the G20A/C92U mutant riboswitch and Aβ. Coloring is the same as in part c.
While a G20A/C92U riboswitch has not been found in nature, this structure reveals that a c-di-AMP riboswitch could exist, evolving from a c-di-GMP-binding RNA. A few subtle modifications to the binding pocket could greatly improve the affinity for c-di-AMP by shifting residues so that they could interact with other heteroatoms of the adenosine moiety.
Kinetic control and genetic regulation
The exceptionally high affinity of this RNA-ligand interaction reflects the multitude of contacts made between the riboswitch and c-di-GMP. It also is a product of the kinetics of ligand binding. Both kon and koff are quite slow, 1 × 106 M−1 min−1 and 1 ×10−5 min−1, respectively [20]. This off-rate translates into a half-life for the complex of approximately 1 month. Once c-di-GMP is bound by the riboswitch, it will stay bound until the genetic decision is made. Consequently, the majority of the riboswitch response is dependent on the bimolecular on-rate, making this an example of a kinetically controlled riboswitch [23, 24]. The in vivo response will thus be highly sensitive to the cellular concentration of c-di-GMP.
Many of the mutants investigated for ligand binding have also been assayed for binding kinetics. The majority of mutations result in a faster off-rate, leaving the on-rate unchanged [22]. This implies that not only does this riboswitch exhibit sequence flexibility, but naturally occurring variants may exhibit a range of kinetic properties that could be tailored to specific, localized concentrations of c-di-GMP.
Acknowledgments
We thank Sarah Lipchock, Alison Livingston, and Carly Shanahan for many of the experiments described here. We also thank Elaine Lee, Tyler Ames, Ron Breaker and other members of the Breaker Laboratory for collaboration and many helpful conversations.
Funding
This work as well as ongoing riboswitch research in the Strobel laboratory is supported by the National Institutes of Health grant GM022778.
Abbreviations
- c-di-GMP
bis-(3′–5′)-cyclic dimeric guanosine monophosphate
- TL
tetraloop
- TLR
tetraloop receptor
References
- 1.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 2.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 3.Schirmer T, Jenal U. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol. 2009;7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 4.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell. 2010;38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, Kaever V, Sourjik V, Roth V, Jenal U. Second messenger-mediated adjustment of bacterial swimming velocity. Cell. 2010;141:107–116. doi: 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 6.Krasteva PV, Fong JCN, Shikuma NJ, Beyhan S, Navarro MVAS, Yildiz FH, Sondermann H. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leduc JL, Roberts GP. Cyclic di-GMP allosterically inhibits the CRP-like protein (Clp) of Xanthomonas axonopodis pv. citri. J Bacteriol. 2009;191:7121–7122. doi: 10.1128/JB.00845-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chin K-H, Lee Y-C, Tu Z-L, Chen C-H, Tseng Y-H, Yang J-M, Ryan RP, McCarthy Y, Dow JM, Wang AH-J, Chou S-H. The c-AMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris. J Mol Biol. 2010;396:646–662. doi: 10.1016/j.jmb.2009.11.076. [DOI] [PubMed] [Google Scholar]
- 9.Tao F, He Y-W, Wu D-H, Swarup S, Zhang L-H. The cNMP domain of Xanthomonas campestris global regulator Clp defines a new class of c-di-GMP effectors. J Bacteriol. 2009;192:1020–1029. doi: 10.1128/JB.01253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol. 2008;69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 2009;23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newell PD, Monds RD, O’Toole GA. LapD is a bis-(3′,5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc Natl Acad Sci USA. 2009;106:3461–3466. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol. 2007;65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 15.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker R. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010;329:845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dambach MD, Winkler WC. Expanding roles for metabolite-sensing regulatory RNAs. Curr Opin Microbiol. 2009;12:161–169. doi: 10.1016/j.mib.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, Neph S, Tompa M, Ruzzo WL, Breaker RR. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007;35:4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith K, Lipchock S, Ames T, Wang J, Breaker R, Strobel S. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol. 2009;16:1218–1223. doi: 10.1038/nsmb.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulshina N, Baird N, Ferré-D’Amaré A. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat Struct Mol Biol. 2009;16:1212–1217. doi: 10.1038/nsmb.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith KD, Lipchock SV, Livingston AL, Shanahan CA, Strobel SA. Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch. Biochemistry. 2010;49:7351–7359. doi: 10.1021/bi100671e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 24.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]