Abstract
Background
Although gene overlapping is a common feature of prokaryote and mitochondria genomes, such genes have also been identified in many eukaryotes. The overlapping genes in eukaryotes are extensively rearranged even between closely related species. In this study, we investigated retention and rearrangement of positionally overlapping genes between the mosquitoes Aedes aegypti (dengue virus vector) and Anopheles gambiae (malaria vector). The overlapping gene pairs of A. aegypti were further compared with orthologs of other selected insects to conduct several hypothesis driven investigations relating to the evolution and rearrangement of overlapping genes.
Results
The results show that as much as ~10% of the predicted genes of A. aegypti and A. gambiae are localized in positional overlapping manner. Furthermore, the study shows that differential abundance of introns and simple sequence repeats have significant association with positional rearrangement of overlapping genes between the two species. Gene expression analysis further suggests that antisense transcripts generated from the oppositely oriented overlapping genes are differentially regulated and may have important regulatory functions in these mosquitoes. Our data further shows that synonymous and non-synonymous mutations have differential but non-significant effect on overlapping localization of orthologous genes in other insect genomes.
Conclusion
Gene overlapping in insects may be a species-specific evolutionary process as evident from non-dependency of gene overlapping with species phylogeny. Based on the results, our study suggests that overlapping genes may have played an important role in genome evolution of insects.
Keywords: Gene rearrangement, Culicidae, Genome evolution, Positionally overlapping genes, Negative selection
Background
Gene rearrangement is one of the necessary ingredients of genome evolution. Several well studied mechanisms such as chromosomal inversions, translocations, duplications and transpositions are known to have important roles in genomic rearrangement events [1-4]. Reshuffling of genomic DNA by gross chromosomal rearrangements generally involves a number of genes that undergo positional relocation in the genome. In addition to such large scale genomic rearrangements, genomic rearrangements at small scale levels facilitate relocation of genes which are otherwise positionally overlapping in a genome [5]. It has been suggested that transposition mechanisms may contribute to such gene arrangements [1,2,6,7], but the functional and evolutionary significance of such events is largely unknown.
Positional overlapping between genes is a common structural feature of prokaryote and mitochondria genomes [8-10]. However, overlapping genes have also been identified from whole genome sequences of several eukaryotes such as fruit fly, zebrafish, human, chimpanzee, orangutan, marmoset, rhesus, cow, dog, mouse, rat and chicken [11-13]. Studies show that overlapping genes in eukaryotes are extensively rearranged even between closely related species [5,12,14-16]. Bhutkar et al. 2007 [5] compared overlapping genes of Drosophila melanogaster and Anopheles gambiae with Apis mellifera (honey bee) and suggested that relocalization of overlapping genes may have played a significant role in genome evolution of these insects. Although several other insect genome sequences are now available, overlapping genes of most of these insects have not been studied.
The present study is an effort to investigate overlapping genes of Aedes aegypti, the primary global vector of dengue virus, in a comparative manner with those of A. gambiae, a major vector of malaria in subSaharan Africa. Understanding genome structure of these mosquitoes has become one of the major interests among insect vector biologists. At present, the draft genome sequences for three mosquito species have been completed [17-19]. These projects (http://www.vectorbase.org) have provided new insights on structure, function and evolution of mosquito genes, thus furthering our ability to study mosquito-parasite or mosquito-virus interactions at the molecular level [20-23].
We identified positional overlapping of genes at the whole genome level in A. aegypti and studied structural differences and evolutionary features by comparisons with orthologous genes of A. gambiae and other selected arthropod genomes. The primary aim was to test several common hypotheses relating to rearrangement of overlapping genes and determine factors that may have a role in relocalization of overlapping genes in insects. The results of our investigation show that positional overlapping among genes is a species specific evolutionary process as evident from non-dependency of gene overlapping with species phylogeny, and also show that specific factors, such as introns and repeat sequences, are significantly associated with retention/rearrangement of overlapping genes in mosquitoes. Based on these results, our study suggests that overlapping genes may have played an important role in genome evolution among insects.
Methods
Official gene sets and extraction of overlapping gene pairs
The overlapping gene pairs of A. aegypti and A. gambiae were identified in a genome-wide manner based on the coordinates of gene boundaries of official gene sets annotated from the genome assemblies. The other mosquito genome sequence for Culex quinquefasciatus was not used for this purpose because of differences in gene annotation of this species compared to A. aegypti or A. gambiae. That is, while nearly equally percentages (~60%) of the official gene sets of A. aegypti as well as A. gambiae have been annotated for gene boundaries that incorporated the 5’ and 3’ untranslated regions, only 15% of the C. quinquefasciatus genes have been annotated in this manner. Thus, incorporating C. quinquefasciatus could have produced biased results in the genome-wide comparison of overlapping gene pairs between A. aegypti and A. gambiae. However, we have used orthologs of A. aegypti overlapping gene pairs in C. quinquefasciatus and other selected insect species such as Drosophila melanogaster, Apis mellifera, Pediculus humanus, Bombyx mori and Acyrthosiphon pisum to determine if they are also localized in overlapping positions in the respective genomes. For genome-wide comparison of overlapping genes, the predicted gene sets of A. aegypti (AaegL1.1) and A. gambiae (AgamP3.4) along with coordinates of genes in the reference genome were downloaded from VectorBase (http://www.vectorbase.org/GetData/). The one-to-one orthologous genes (OrthoDB5; http://cegg.unige.ch/orthodb5) were compared to determine if they were also present in overlapping gene pairs across multiple genomes. To determine the relative position of the orthologous genes, the official gene lists along with their start and end positions in the genome sequences of the other six insects (C. quinquefasciatus: CpipJ1, D. melanogaster: BDGP 5, A. mellifera: Amel_2.0, P. humanus: PhumU1, B. mori: SilkDB V2.0 and A. pisum: Acyr2) were downloaded from either VectorBase (http://www.vectorbase.org/) or the SilkDB database (http://www.silkdb.org) or the ‘Ensembl Metazoa 10’ data sets at http://www.biomart.org.
Intron analysis
To determine if introns have an association with overlapping between genes, orthologous genes were categorized as intronless and intron-containing genes for overlapping and non-overlapping pairs in the A. aegypti and A. gambiae genomes. The exon structures predicted for A. aegypti and A. gambiae genes (obtained from Biomart.org) were used to classify genes into single exon genes (intronless) and multi exon genes (intron-containing). The number of introns in each gene was determined from the number of exons annotated in the genes. The 2x2 contingency analysis of counts of the intronless and intron-containing genes of both categories (overlapping/ non-overlapping) was performed using Yates Chi square tests to determine significance of association between introns and gene overlapping.
Transcriptional analysis of overlapping genes
The expressed sequence tags (EST) of A. aegypti and A. gambiae mosquitoes used in this study were largely generated in conjunction with the individual genome sequencing projects (http://www.vectorbase.org). These ESTs were used to assist in the annotation of the official gene sets of the two mosquitoes. We used these ESTs to investigate expression patterns associated with the overlapping gene pairs. To further confirm correspondence of ESTs with overlapping gene pairs, we performed reciprocal BLAST analyses described as follows. The EST sequences were used to generate a local BLAST database and then searched by BLASTN with the sequences of overlapping genes. The EST ‘hits’ that had an e-value = 0 were used again as queries in another BLASTN search against all predicted gene sequences. If the reciprocal hits matched the same gene that was used as a query in the first BLAST, it was considered that the EST corresponded to that gene. Apart from analyzing the EST data, we also analyzed previously performed microarray expression data of A. aegypti[23] to determine expression patterns of the overlapping gene pairs. The A. gambiae microarray expression data was obtained from Baker et al. 2011 study [24]. The expression data of these studies [23,24] are publicly available with Gene Expression Omnibus (GEO) accession # GSE16563 and GSE21689 at http://www.ncbi.nlm.nih.gov/geo/. The Spearman’s rank correlation test was conducted to ascertain whether the overlapping gene pairs had significantly correlated expression levels throughout the genome.
Identification of microsatellites in overlapping genes
In order to determine if there is a significant association of microsatellites with retention or rearrangement of overlapping gene structures between A. aegypti and A. gambiae, we identified microsatellite sequences within the gene pairs in both genomes. SciRoKo, a simple sequence repeat (SSR) identification program [25], was used to detect both perfect and imperfect mono-, di-, tri-, tetra- and hexa-nucleotide repeats using the default parameters (mismatch, fixed penalty = 5). The repeats with more than 3 consecutive mismatch sites were excluded. The genes where one or more sites were ambiguous nucleotides (such as ‘N’s) were not used to report microsatellites. The length of orthologous genes may vary (primarily because of introns) that may contribute to varying amounts of microsatellite sequences in the orthologous gene copies. So, instead of comparing the absolute amounts of microsatellite sequences, their relative amounts were compared. The relative amounts were obtained from the total amount of microsatellites of genes normalized with the alignment length (common DNA sequences) of the orthologous genes between A. aegypti and A. gambiae.
Statistical and computational analyses
All statistical analyses were performed using the R statistical program. The p-value < 0.05 was considered statistical significance in all tests unless stated otherwise. Cluster analyses of gene pairs based on overlapping or non-overlapping structures across genomes were based on average correlation of city-block distance estimated using the Cluster3 program [26]. The phylogenetic analyses were performed by neighbor-joining method using MEGA4 [27]. The evolutionary distances were in the units of the number of base substitutions per site; and they were calculated using the Maximum Composite Likelihood method [28]. The Mantel procedure [29] was used to perform linear regression between matrices where the dependent matrix (representing 0 for non-overlapping and 1 for overlapping) was permutated 1000 times to test significance of the observed correlation with the independent matrix (that represented presence or absence of orthologs of overlapping gene pairs of A. aegypti) in the genomes used for comparison. The multi Mantel procedure was performed using an algorithm developed by Dr. Liam J. Revell (URL: http://anolis.oeb.harvard.edu/~liam/programs/). Maximum likelihood methods described elsewhere [30,31] were used to estimate the log likelihoods of models assuming either dependency or non-dependency of gene phylogeny with the discrete variation of gene traits (i.e. overlapping or non-overlapping localization in the respective genomes). The likelihood ratio tests were conducted to infer statistical significance of these two models. A binary logit model was developed to test marginal effects of the rates of synonymous (dS) and non-synonymous (dN) mutations in the orthologous gene pairs between A. aegypti and other select insect genomes (A. gambiae, C. quinquefasciatus, D. melanogaster and P. humanus). While each of the gene pairs (n =19) were localized in an overlapping manner in the A. aegypti genome, the orthologous genes showed variation in relative localization (overlapping = 1or non-overlapping = 0) in other species. The dN and dS values of orthologous genes were obtained from metazoan genes database at http://www.Biomart.org. A generalized linear model (described in detail in results section), fitting the dependent variable (0 or 1) and independent variables (dN and dS values for both genes), was used in R to estimate the logit coefficients.
Results and discussion
Identification of overlapping genes
A total of 761 and 565 overlapping gene pairs were identified in the assembled genomes of A. aegypti and A. gambiae, respectively (Additional file 1). They represent 8-10% of the annotated genes of the two mosquitoes. The frequencies of overlapping genes of A. aegypti and A. gambiae mosquitoes are within the range of overlapping gene frequencies reported in other eukaryotes [32,33]. More than two genes (overlapping gene clusters) were also found in overlapping locations in both genomes, with the majority of these overlapping gene clusters containing no more than three genes (21 clusters in A. aegypti and 19 in A. gambiae). These overlapping clustered genes constituted only a minor portion (less than 3%) of the total number of overlapping genes in either of the two genomes. Because of low frequency and also for simplicity of analysis, we have not included the gene clusters in our investigation. All the analyses performed in this study were based on overlapping gene pairs.
Orthology of overlapping genes between A. aegypti and A. gambiae
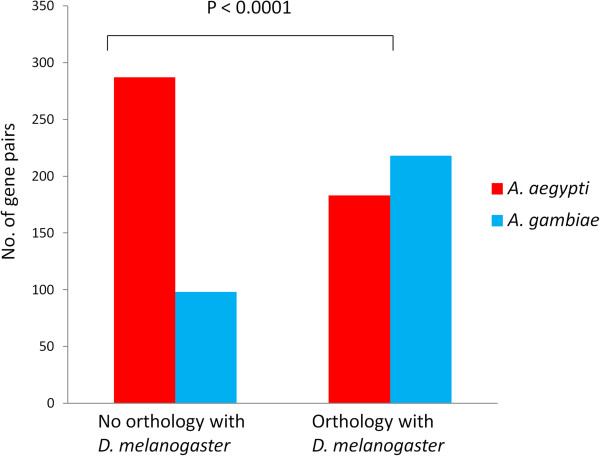
In A. aegypti as well as in A. gambiae, the overlapping gene pairs are localized either in nested form (one gene embedded within another gene, the embedded/host genes or E/H genes) or in partially overlapping form (henceforth abbreviated as P/O genes) as shown in Figure 1. Irrespective of whether gene pairs are in E/H or in P/O form, they are predominantly localized in opposite orientation to each other. Three possible patterns of evolution emerged from comparing the orthologs of overlapping genes between the two species: 1) the gene pairs are orthologous between the two species (‘old-old gene pairs’), 2) the gene pair is specific to the species (lack of orthology in the other species, ‘young-young gene pairs’), and 3) one of the genes is specific to the species and the other is common between the two species (‘young-old gene pairs’) (Figure 2). The number of young-young gene pairs (shown as circle A and C in Figure 2) and the old-old gene pairs (shown as circle B and D in Figure 2) vary between the two species. The 2x2 contingency tests based on the count statistics of old-old and young-young gene pairs between A. aegypti and A. gambiae (the A, C, B and D gene groups) shows that there is a significant bias in the distribution of these genes between the two species. It clearly shows that nearly the same number (~ 250) of orthologous gene pairs (old-old pairs) are present in overlapping manner in both the species, whereas a significantly larger number of A. aegypti specific genes (young-young pairs, circle A) are in overlapping position compared to A. gambiae (circle C). This shows that the young-young gene pairs show significant variation in overlapping patterns between these mosquitoes. When the A. aegypti and A. gambiae overlapping genes were compared with D. melanogaster orthologs, consistent patterns were observed (Figure 3). The data in Figure 3 shows that the old-old gene pairs (genes that have orthologous copies in D. melanogaster genome) are comparable in numbers between A. aegypti and A. gambiae whereas the young-young gene pairs (genes that lack orthologous copies in D. melanogaster genome) vary significantly between the two species. Consistent with the results shown in Figure 2, these results also suggest that young genes are major contributors to the positional overlapping of genes in these species.
Figure 1.
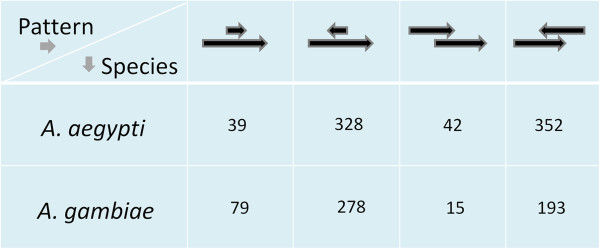
Patterns localization of E/H and P/O gene pairs in the genomes of three mosquito species. The gene pair counts and relative orientations are also shown.
Figure 2.
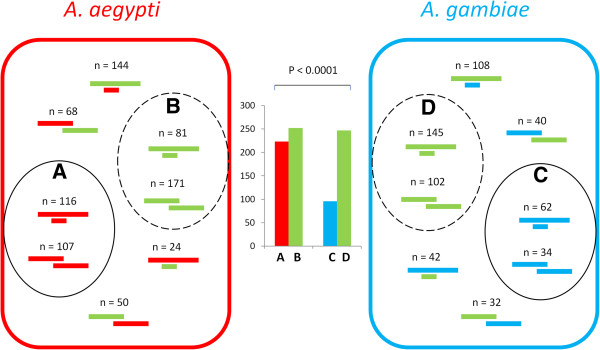
Orthology (one-to-one) of overlapping gene pairs between A. aegypti and A. gambiae. Genes have been color coded to represent orthologous relationships between the two species: red color represents A. aegypti and blue color represents A. gambiae. The one-to-one orthologous gene pairs that are localized in positionally overlapping manner in both the genomes are indicated by green color. The number of gene pairs is shown for each group. The circles A and C represent the young-young gene pairs, circles B and D represents the old-old gene pairs, and the gene pairs outside the circles represent young-old gene pairs in each species. The 2x2 contingency tests based on the count statistics of young-young and old-old genes between A. aegypti and A. gambiae (i.e. the A, B, C and D gene groups) shows that there is a significant bias in the distribution of these genes between the two species (shown as the column graph in the middle). In this graph, the y-axis shows the number of gene pairs corresponding to the four gene groups; and the gene groups are represented by the x-axis. The columns are color coded same manner as that of the gene groups (A, B, C and D).
Figure 3.
Significant variation in the number of overlapping gene pairs of A. aegypti and A. gambiae based on presence or absence of orthology in D. melanogaster genome. Y-axis shows number of gene pairs and x-axis shows whether the pairs have or don’t have orthology of A. aegypti and A. gambiae genes in the D. melanogaster genome. The p-value shows significance by Chi square test. It shows that the ancestral genes (where orthology is evident in D. melanogaster) are comparable in numbers between A. aegypti and A. gambiae whereas the young gene pairs vary significantly between the two species suggesting that the young genes are major contributors to the positional overlapping among genes.
Rearrangement of overlapping genes
One of the common hypotheses about positional overlapping of genes is that selection acts against the retention of gene overlap between genomes [14,33]. If the above hypothesis is correct, we expect that overlapping genes should be extensively rearranged between A. aegypti and A. gambiae. To test that expectation, the orthologous (one-to-one) copies of overlapping gene pairs were compared between the two species (Additional file 2). It was found that only 139 of the total 499 orthologous gene pairs are localized in overlapping manner in both the genomes. The other 360 gene pairs are localized in overlapping manner in one genome but in non-overlapping manner in the other (Table 1) suggesting that only a fraction of overlapping genes are retained across genomes. To determine if retention or rearrangement of overlapping localization of genes between A. aegypti and A. gambiae may be associated with loss or gain of terminal exons of genes, we investigated several gene pairs that contain multiple exons in the orthologous gene pairs (Additional file 3) and found no discrepancy in annotation of first and last exon of any gene pair between the two species.
Table 1.
Number of one-to-one orthologous gene pairs which are localized either in overlapping or non-overlapping manner relative to each other between the A. aegypti and A. gambiae genomes
| Localization pattern | Number of gene pairs |
|---|---|
| E/H in both A. aegypti and A. gambiae |
75 |
| P/O in both A. aegypti and A. gambiae |
54 |
| E/H in A. aegypti but P/O in A. gambiae |
3 |
| P/O in A. aegypti but E/H in A. gambiae |
7 |
| E/H in A. aegypti but non-overlapping in A. gambiae |
43 |
| P/O in A. aegypti but non-overlapping in A. gambiae |
140 |
| Non-overlapping in A. aegypti but E/H in A. gambiae |
103 |
| Non-overlapping in A. aegypti but P/O in A. gambiae | 74 |
The overlapping gene pairs where one gene is embedded in another are represented as E/H pairs. The gene pairs which are localized in partially overlapping manner with each other are represented as P/O gene pairs.
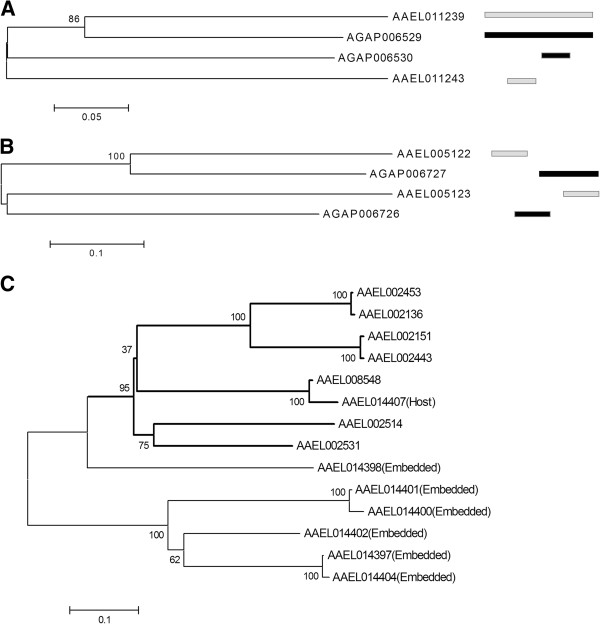
Additionally, we performed specific case studies of retention or rearrangement between the two species. A short-chain dehydrogenase gene (AAEL011239) acts as the host of another protein coding gene AAEL011243 (a paralog of AAEL011239) in A. aegypti. It was found that the corresponding orthologs in A. gambiae genome are also localized in E/H form (Figure 4A). In contrast to this, the gene (AAEL005122) of A. aegypti, that codes for a carboxylesterase, is localized in the genome in non-overlapping manner with one of its paralogs (AAEL005123) whereas the ortholog in A. gambiae (AGAP006727) is localized in P/O manner with the paralog AGAP006726 (Figure 4B). These genes associated with retention or rearrangement of positional localization in both the genomes are known to have significant changes in expression during blood feeding, growth and development of these mosquitoes [34-36]. In another case, we identified multiple genes that are embedded within intron sequences of a single host gene in A. aegypti. The gene AAEL014407 that putatively codes for the protein B-cell lymphoma/leukaemia (11A extra long form) harbors 6 paralogous genes within one intron along with several other paralogous genes that are localized in non-overlapping manner to AAEL014407 (Figure 4C). Importantly, these genes have been reported to be expressed in A. aegypti[36]. Moreover, the phylogenetic tree (Figure 4C) shows that the embedded genes tend to cluster together and are phylogenetically distinct from the genes located outside the host genes. This suggests a contrasting rate of evolution of embedded versus non-embedded paralogous copies of the gene AAEL014407 in A. aegypti.
Figure 4.
Phylogenetic (neighbor-joining trees) relationship among pairs of genes of A. aegypti (gene ID begins with AAEL; gene structural pattern shown as light color bars next to branches) and A. gambiae (gene ID begins with AGAP; gene structural pattern shown as dark color bars next to branches). The gene pairs in each case are paralogous to each other within species and at the same time they are orthologous to each other between the two species. They either retain positional overlapping structure in both the genomes (A) or show overlapping in one but non-overlapping in the other (B). The tree shown in (C) represents phylogenetic relationship of the host gene (AAEL014407) with paralogous copies in the A. aegypti genome which are either embedded (within AAEL014407) or non-embedded (located outside AAEL014407). The genes that are embedded are marked so within brackets. The scale for branch length is shown below each tree.
Gene overlapping is phylogeny independent
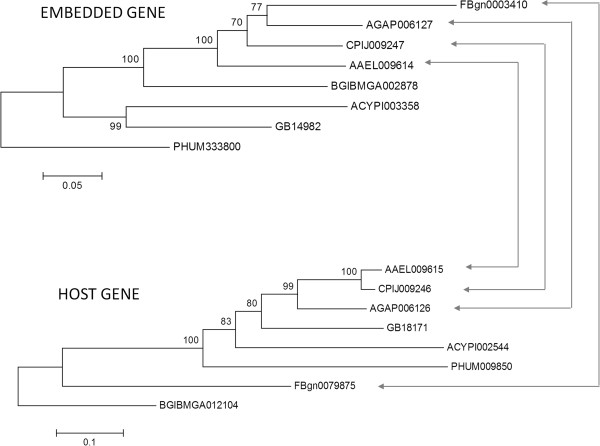
Having observed that orthologs of overlapping genes are extensively rearranged between species, we hypothesized that phylogenetic relationship has no correlation with the localization pattern (overlapping or non-overlapping) of orthologous genes across species. If this is true, one would expect that gene overlapping occurs as a trait which should be independent of species phylogenies. To test this hypothesis, we performed ‘discrete analysis’ of gene overlapping across phylogeny using maximum likelihood method [30] to determine if presence or absence of gene overlapping is correlated with phylogeny among species. Seven insect species (selected based on varying phylogenetic relationships with A. aegypti) were compared for orthology analysis with A. aegypti overlapping genes: A. gambiae (malaria mosquito), C. quinquefasciatus (southern house mosquito), D. melanogaster (fruit fly), A. mellifera (honey bee), P. humanus (body louse), B. mori (silkworm) and A. pisum (pea aphid). The results of this analysis showed that the likelihood estimates of the null model (independence of gene overlapping with phylogeny) consistently lack statistical significance when tested against the alternate model (dependency of gene overlapping with phylogeny) (Table 2). It clearly shows that there is no apparent relationship of positional overlapping with the phylogeny of the species. To illustrate this, a representation of phylogeny and gene overlapping pattern is shown in Figure 5 for orthologs of A. aegypti gene pair AAEL009614-AAEL009614 (E/H gene pair) among seven other insect species. It shows that retention or rearrangement of orthologous genes lacks correspondence with the phylogenetic relationships between species.
Table 2.
Positional overlapping/non-overlapping patterns of orthologs of A. aegypti gene pairs in 7 other insect species were compared with species phylogeny
| Orthologs of gene pairs | Model dependent | Model independent | Log likelihood difference | p-value |
|---|---|---|---|---|
| AAEL00241/ AAEL000233 |
−6.28154 |
−6.74766 |
0.466125 |
0.976714 |
| AAEL006054/ AAEL006056 |
−3.32599 |
−3.49681 |
0.170826 |
0.996554 |
| AAEL008942/ AAEL008940 |
−2.9433 |
−3.10494 |
0.161641 |
0.996905 |
| AAEL009614/ AAEL009615 | −1.81547 | −2.35038 | 0.534903 | 0.970015 |
The likelihoods of test and null models (dependence or independence models, respectively) and the p-values of log likelihood ratio tests are shown.
Figure 5.
Neighbor-joining phylogenetic trees of a representative gene pair among different insect species. The positional overlapping (one gene embedded in another), if present in a species, is indicated by arrowed lines connecting the corresponding genes between the two phylogenies. The alphabetic letters associated with gene IDs shown in the tree correspond to the species as follows: AAEL- A. aegypti, AGAP- A. gambiae, CPIJ- C. quinquefasciatus, FBgn- D. melanogaster, GB- A. mellifera, PHUM- P. humanus, BGIBMGA- B. mori and ACYP- A. pisum. The scale for branch length is shown below each tree.
By comparisons with predicted orthologous genes among sequenced arthropod genomes (OrthoDB5, http://cegg.unige.ch/orthodb5), we identified a total of 196 overlapping gene pairs of A. aegypti where at least one gene of each pair was also present among the other seven insect species. But only 19 of these gene pairs in A. aegypti had both the genes present as orthologs in all the other seven species (Additional file 4). To further confirm that overlapping or non-overlapping localization of genes has no correspondence with presence or absence of orthologs across genomes, we performed hierarchal cluster analysis among the above 19 orthologous gene pairs across the eight species (Additional file 5). The potential for correlation between gene orthology and gene positional overlapping was assessed for statistical significance by Mantel test (see Methods). The correlation was evaluated between binary data of orthologous genes in matrix forms (presence or absence of overlap) with the presence or absence of orthology of the gene pairs. The results showed non-significant correlation between the two (p > 0.8) suggesting that gene orthology has no relationship with overlapping localization of genes across species.
Role of selection on rearrangement of overlapping genes
Another hypothesis about overlapping genes is that mutations occurring within the shared region of overlapping gene pairs would mostly be negatively selected because such mutations may affect adaptation [37] and function of both genes [38,39]. If this is true, we expect to see a higher frequency of synonymous (dS) changes than non-synonymous (dN) changes between orthologous genes when they are present in overlapping manner. To test this expectation, the numbers of per site synonymous and non-synonymous changes in 19 orthologous gene pairs (Additional file 4) were determined among A. aegypti, C. quinquefasciatus, A. gambiae, D. melanogaster and P. humanus. The dS and dN values are the rate of synonymous and non-synonymous changes, respectively, between A. aegypti gene and the corresponding ortholog of other species mentioned above. As shown in Additional file 4, each of these gene pairs is localized in overlapping manner in A. aegypti. But, the orthologous genes in the other species are found either in overlapping or non-overlapping manner. A binary logit model was developed to fit the data of dS and dN values with the occurrence or non-occurrence of positional overlapping between genes among the species. The dependent variable assumed a value 1 when the genes were found in overlapping position but 0 when the gene pairs were in non-overlapping position in the genome. We performed generalized linear model fitting of the data that is represented by y = β0 + Xβ + e, where y = dependent variable (overlapping/ non-overlapping of genes), X = dS and dN values of both genes, β = coefficient of independent variable, β0 represents the value of y when the predictor is equal to zero, and e (error) is assumed to be independent of X and has a standard logistic distribution with mean zero. The dS and dN values of both genes (gene1 and gene2) of A. aegypti gene pairs, calculated by aligning the codon sequence to the orthologs of other insect species (Additional file 4), were used as the independent variables. The results of the regression analysis are shown in Table 3. It shows estimates of coefficients of each of the four independent (predictor variables) in fitting the model to explain the occurrence of the dependent (predicted) variable (i.e. y = 1 or genes are in overlapping position). The estimated regression coefficient shows variation (%) of the outcome with unit change in the predictor variable [because probability (p) of the outcome in the logit model is estimated as the logarithm of the odds {p/(1 – p)}]. The data in Table 3 shows that the coefficients of regression are positive for synonymous changes but negative for non-synonymous changes indicating differential effects of synonymous and non-synonymous mutations on overlapping localization between genes. However, the effects of dS or dN are statistically non-significant in each case (Table 3) indicating that, in these insect species (Additional file 4), the association of synonymous or non-synonymous mutations with overlapping localization of orthologous genes may be a random event. However, the lack of significance may also be due to differences in the reading frames of orthologous genes. Such differences are known to be associated with bias in codon phases of overlapping prokaryotic genes [40]. However, we have not determined from this study if there is a bias in codon phase distribution of overlapping genes that may influence the rate of synonymous and non-synonymous changes between orthologs.
Table 3.
Binary logit regression coefficients of rate of synonymous (dS) and non-synonymous (dN) mutations between A. aegypti overlapping genes and their 1-to-1 orthologs in other selected insects (A. gambiae, C. quinquefasciatus, D. melanogaster and P. humanus)
| Coefficient | Std. error | p-value | |
|---|---|---|---|
| Gene1_dS |
0.002 |
0.000975 |
0.84 |
| Gene1_dN |
−0.838 |
0.065533 |
0.61 |
| Gene2_dS |
0.009 |
0.000099 |
0.39 |
| Gene2_dN | −4.533 | 0.346861 | 0.05 |
The regression was performed in relation to presence or absence of positional overlapping of the orthologous gene pairs across species (see Additional file 4).
Association of microsatellites with gene overlapping
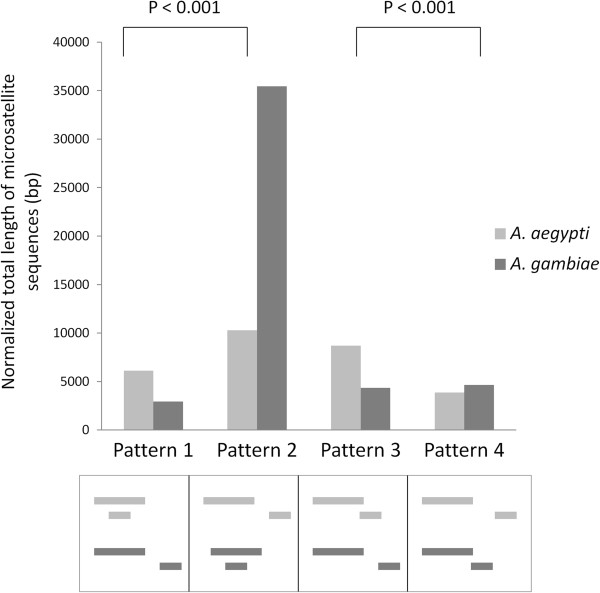
It is well recognized that transposition events contribute to positional rearrangement of genes in eukaryotes [5-7]. And as transposons are known to be intimately associated with simple sequence repeat elements (also known as microsatellites) [41-45], we hypothesized that microsatellites may have a role in positional overlapping of genes. Thus, one of our aims was to determine if there was a significant association between microsatellite contents with rearrangement of overlapping gene pairs between A. aegypti and A. gambiae. The amounts (total base pairs) of microsatellite sequences were normalized based on length of shared sequences between E/H gene pairs and their rearranged orthologous pairs in A. aegypti and A. gambiae. The results of the 2x2 contingency tests of these data show that positional rearrangement of E/H gene pairs is significantly associated with the amount of microsatellite sequences within the orthologous genes in the two mosquitoes (Figure 6). One scenario is that the repeat sequences, represented as common motifs between the two genes (Additional file 6), are involved in gene rearrangements possibly by facilitating cross over events associated with exchange of the flanking regions between microsatellites [46,47] that lead to positional rearrangements of genes.
Figure 6.
Association of microsatellites with retention or rearrangement of gene pairs between A. aegypti and A. gambiae. Y-axis shows the normalized amount of microsatellite sequences in the gene pairs and X-axis shows patterns (pattern 1 through 4) of overlapping or non-overlapping between the two species. Statistical significance of association of microsatellites between patterns 1 and 2 and between patterns 3 and 4 is shown.
Role of introns in positional overlapping of genes
In A. aegypti and A. gambiae, most of the embedded genes (~ 87%) are localized within introns of host genes. Thus, intron loss/gain could contribute to gene relocalization. In that case, we expect that intron loss/gain between one-to-one orthologous genes should be significantly associated with rearrangement of overlapping gene pairs between the two mosquito species. To determine if introns have an association with retention/rearrangement of overlapping genes between the two species, the number of introns among orthologous gene pairs listed in Table 1 were quantified. Based on count statistics of introns between one-to-one orthologous gene pairs between A. aegypti and A. gambiae, we found that that loss/gain of introns is significantly (p < 0.0001) associated with retention or rearrangement of overlapping gene pairs between the two mosquitoes (Table 4). The rearranged genes show significant loss of introns compared to the orthologous gene pairs located in overlapping positions and vice versa suggesting that introns may have a role in gene overlapping and rearrangement. Although intron-mediated gene recombination [48] and references therein or other mechanisms such as intron-transposition [49-51] may be likely mechanisms for these processes, further investigations are needed to determine the exact role of introns in positional overlapping of genes.
Table 4.
Significant association of introns with rearrangement of overlapping gene pairs between A. aegypti (Aaeg) and A. gambiae (Agam)
| Gene pair structure | Aaeg Gene1 + Gene2 | Agam Gene1 + Gene2 | Yates Chi square | Two tailed p-value |
|---|---|---|---|---|
| Non-overlapping in Aaeg but E/H in Agam |
694 |
1056 |
31.29 |
p < 0.0001 |
| E/H in Aaeg but non-overlapping in Agam |
366 |
336 |
|
|
| P/O in Aaeg but non-overlapping in Agam |
1023 |
784 |
16.51 |
p < 0.0001 |
| Non-overlapping in Aaeg but P/O in Agam | 440 | 471 |
Expression of overlapping genes
Overlapping expression of more than one gene is well known in eukaryotes [33], [52]. We analyzed the expressed sequence tags (ESTs) datasets of A. aegypti and A. gambiae to determine if overlapping gene pairs may have overlapping transcripts. Using reciprocal BLASTN searches, we identified several ESTs of A. aegypti that represented the likely transcription product of overlapping gene pairs (Additional file 7). Although many of these gene pairs are oriented in opposite direction to each other, ESTs were also observed for gene pairs oriented in same direction. Whether these gene pairs are co-transcribed or co-regulated by common upstream/downstream sequences [52] are not known from this study. However, identification of ESTs of overlapping gene sequences clearly shows that these sequences are expressed. Moreover, we show that the annotation of overlapping genes is unaffected whether good evidence of expression (such as EST evidence) is available or not. The dataset of overlapping genes was also analyzed based on availability or non-availability of EST evidence. We found no significant difference in the number of genes that localized in positionally overlapping manner between the two groups (Additional file 8). In A. gambiae, the EST dataset didn’t reveal such transcripts except for a single gene pair. Although transcripts of overlapping genes were not available in the EST collections of A. gambiae, we found evidence of expression of these genes (Additional file 9) from published microarray data [24].
Generally, overlapping transcripts are processed by post-transcriptional events to produce individual transcripts of the genes [52]. To assess the expression level of individual gene transcripts of overlapping gene pairs, we examined the microarray expression data of A. aegypti[23]. Because overlapping genes are predominantly localized in opposite orientation to each other in the genome (Figure 1), we compared expression level of gene pairs (E/H genes) which are either oppositely oriented or oriented in same direction to each other. It was found that the expression levels of overlapping genes in opposite orientation lack significant correlation, whereas the overlapping genes which are oriented in the same direction to each other show statistically significant correlation (p < 0.01) (Additional file 10). Most of these genes code for known proteins and have been annotated with start and stop codons suggesting that these genes are not annotation artifacts, although a few genes were annotated as hypothetical proteins. Nevertheless, these results suggested that when the two genes are localized in overlapping manner and also oriented in the same direction, their expression may be co-regulated leading to similar transcription levels. On the other hand, when the two genes are localized in overlapping manner, but oriented in the opposite direction, their transcripts may have differential regulation. In fact, it is well documented that overlapping genes when transcribed in the opposite directions, give rise to sense-antisense transcript pairs which are differentially regulated to play a role in a variety of processes, including mRNA splicing and stability, RNA editing, genomic imprinting and control of translation [33 and references therein].
Conclusions
The results from this study provide insight into the common prevailing theories of origin and evolution of positionally overlapping genes. These are particularly important for better understanding of distribution and structure of overlapping genes in the genomes of A. aegypti and A. gambiae. The genome sequences of both A. gambiae and A. aegypti contain gaps that could affect our estimates of overlapping genes in the genome assemblies, but we find this unlikely based on our observation that the overlapping genes are distributed throughout the genome in each species without any bias to specific chromosomal region of A. gambiae or specific supercontigs of A. aegypti (data not shown). Furthermore, our estimated frequencies of overlapping genes in mosquitoes are within the range of overlapping gene frequencies reported in other eukaryotes [32,33]. Thus, it is unlikely that there may be large numbers of genes missing because of gaps in sequencing that are positionally overlapping. Nevertheless, the dynamic patterns of positional rearrangement of overlapping genes suggest that these genes may have important roles in genome evolution of vector mosquitoes. Importantly, the information from this investigation may help us in further studies pertaining to evolution and functional characterization of antisense transcripts among overlapping genes in mosquitoes.
Availability of supporting data
The manuscript is accompanied with the following listed Additional files in the form of supporting data for this study.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
Conceived and designed the experiments: SKB. Analyzed the data: SKB. Contributed reagents/materials/analysis tools: SKB, DWS. Wrote the paper: SKB, DWS. Both the authors read and approved the final manuscript.
Authors' information
SKB is a Research Assistant Professor in the Department of Biological Sciences and the Eck Institute for Global Health at the University of Notre Dame, Indiana. He has a broad interest in insect genomics and evolution with emphasis on disease transmitting vector species. DWS is a Professor of Biological Sciences and the Director of Eck Institute for Global Health at the University of Notre Dame, Indiana. His work focuses on genetic and genomic analysis of mosquito vector competence to various pathogens as well as on development and application of molecular tools to investigate population biology of mosquitoes.
Supplementary Material
List of E/H and P/O gene pairs of A. aegypti and A. gambiae.
Overlapping and non-overlapping patterns of orthologous genes between A. aegypti and A. gambiae.
List of gene pairs showing overlapping or non-overlapping localization in A. aegypti and A. gambiae genomes but have no changes in exon structure.
List of one-to-one orthologous genes of A. aegypti overlapping genes in other insect species and comparison of overlapping patterns across genomes.
Comparison of cluster patterns of retention or rearrangement of gene overlapping (left) with that of presence or absence of orthology of the corresponding gene pairs (right) among different insects. Red color indicates presence and black color indicates absence of overlapping/ orthology between genes.
List of common microsatellite motifs associated with the overlapping gene pairs that are rearranged between the two mosquitoes.
List of gene pairs and overlapping ESTs in A. aegypti.
Number of gene pairs identified in positionally overlapping patterns with or without evidence of expressed sequence tags.
Expression level of gene pairs that are localized in positionally overlapping manner in A. gambiae genome.
Expression level of A. aegypti overlapping genes.
Contributor Information
Susanta K Behura, Email: sbehura@nd.edu.
David W Severson, Email: severson.1@nd.edu.
Acknowledgements
This work was supported, in part, by grants AI059342 and AI079125 from the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH).
References
- Lonnig WE, Saedler H. Chromosome rearrangements and transposable elements. Annu Rev Genet. 2002;36:389–410. doi: 10.1146/annurev.genet.36.040202.092802. [DOI] [PubMed] [Google Scholar]
- Lim JK, Simmons MJ. Gross chromosome rearrangements mediated by transposable elements in Drosophila melanogaster. Bioessays. 1994;16:269–275. doi: 10.1002/bies.950160410. [DOI] [PubMed] [Google Scholar]
- Vibranovski MD, Zhang Y, Long M. General gene movement off the X chromosome in the Drosophila genus. Genome Res. 2009;19:897–903. doi: 10.1101/gr.088609.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Bork P. Quantification of insect genome divergence. Trends Genet. 2007;23:16–20. doi: 10.1016/j.tig.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Bhutkar A, Russo SM, Smith TF, Gelbart WM. Genome-scale analysis of positionally relocated genes. Genome Res. 2007;17:1880–1887. doi: 10.1101/gr.7062307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan A, Wolfe KH. Fourfold faster rate of genome rearrangement in nematodes than in Drosophila. Genome Res. 2002;12:857–867. doi: 10.1101/gr.172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz JM, González J, Casals F, Ruiz A. Low occurrence of gene transposition events during the evolution of the genus Drosophila. Evolution. 2003;57:1325–1335. doi: 10.1111/j.0014-3820.2003.tb00340.x. [DOI] [PubMed] [Google Scholar]
- Normark S, Bergström S, Edlund T, Grundström T, Jaurin B, Lindberg FP, Olsson O. Overlapping genes. Annu Rev Genet. 1983;17:499–525. doi: 10.1146/annurev.ge.17.120183.002435. [DOI] [PubMed] [Google Scholar]
- Krakauer DC. Stability and evolution of overlapping genes. Evolution. 2000;54:731–739. doi: 10.1111/j.0014-3820.2000.tb00075.x. [DOI] [PubMed] [Google Scholar]
- Johnson ZI, Chisholm SW. Properties of overlapping genes are conserved across microbial genomes. Genome Res. 2004;14:2268–2272. doi: 10.1101/gr.2433104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S, Chen C, Gentles AJ, Cleary M. Associations between human disease genes and overlapping gene groups and multiple amino acid runs. Proc Natl Acad Sci USA. 2002;99:17008–17013. doi: 10.1073/pnas.262658799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeramachaneni V, Makałowski W, Galdzicki M, Sood R, Makałowska I. Mammalian overlapping genes: the comparative perspective. Genome Res. 2004;14:280–286. doi: 10.1101/gr.1590904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DS, Cho CY, Huh JW, Kim HS, Cho HG. EVOG: a database for evolutionary analysis of overlapping genes. Nucleic Acids Res. 2009;37:D698–D702. doi: 10.1093/nar/gkn813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldà G, Suyama M, Pelucchi P, Boi S, Guffanti A, Rizzi E, Bork P, Tenchini ML, Ciccarelli FD. Non-random retention of protein-coding overlapping genes in Metazoa. BMC Genomics. 2008;9:174. doi: 10.1186/1471-2164-9-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanna CR, Li WH, Zhang L. Overlapping genes in the human and mouse genomes. BMC Genomics. 2008;9:169. doi: 10.1186/1471-2164-9-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis R, Kondrashov AS, Koonin EV, Kondrashov FA. Nested genes and increasing organizational complexity of metazoan genomes. Trends Genet. 2008;24:475–478. doi: 10.1016/j.tig.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, Wides R, Salzberg SL, Loftus B, Yandell M, Majoros WH, Rusch DB, Lai Z, Kraft CL, Abril JF, Anthouard V, Arensburger P, Atkinson PW, Baden H, de Berardinis V, Baldwin D, Benes V, Biedler J, Blass C, Bolanos R, Boscus D, Barnstead M. et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M, Ren Q, Zdobnov EM, Lobo NF, Campbell KS, Brown SE, Bonaldo MF, Zhu J, Sinkins SP, Hogenkamp DG, Amedeo P, Arensburger P, Atkinson PW, Bidwell S, Biedler J, Birney E, Bruggner RV, Costas J, Coy MR, Crabtree J, Crawford M. et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arensburger P, Megy K, Waterhouse RM, Abrudan J, Amedeo P, Antelo B, Bartholomay L, Bidwell S, Caler E, Camara F, Campbell CL, Campbell KS, Casola C, Castro MT, Chandramouliswaran I, Chapman SB, Christley S, Costas J, Eisenstadt E, Feschotte C, Fraser-Liggett C, Guigo R, Haas B, Hammond M, Hansson BS, Hemingway J, Hill SR, Howarth C, Ignell R, Kennedy RC. et al. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science. 2010;330:86–88. doi: 10.1126/science.1191864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CA, Kafatos FC, Stansfield SK, Collins FH. Arthropod-borne diseases: vector control in the genomics era. Nat Rev Microbiol. 2005;3:262–268. doi: 10.1038/nrmicro1101. [DOI] [PubMed] [Google Scholar]
- Schneider DS, James AA. Bridging the gaps in vector biology. Workshop on the molecular and population biology of mosquitoes and other disease vectors. EMBO Re. 2006;7:259–262. doi: 10.1038/sj.embor.7400643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson DW, Behura SK. Mosquito genomics: progress and challenges. Annu Rev Entomol. 2012;57:143–166. doi: 10.1146/annurev-ento-120710-100651. [DOI] [PubMed] [Google Scholar]
- Behura SK, Gomez-Machorro C, Harker BW, deBruyn B, Lovin DD, Hemme RR, Mori A, Romero-Severson J, Severson DW. Global cross-talk of genes of the mosquito Aedes aegypti in response to dengue virus infection. PLoS Negl Trop Dis. 2011;5:e1385. doi: 10.1371/journal.pntd.0001385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DA, Nolan T, Fischer B, Pinder A, Crisanti A, Russell S. A comprehensive gene expression atlas of sex- and tissue-specificity in the malaria vector, Anopheles gambiae. BMC Genomics. 2011;12:296. doi: 10.1186/1471-2164-12-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Schlötterer C, Lelley T. SciRoKo: a new tool for whole genome microsatellite search and investigation. Bioinformatics. 2007;23:1683–1685. doi: 10.1093/bioinformatics/btm157. [DOI] [PubMed] [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, Miyano S. Open Source Clustering Software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- Pagel M. Detecting correlated evolution on phylogenies: a general method for the comparative analysis of discrete characters. Proc R Soc B. 1994;255:37–45. doi: 10.1098/rspb.1994.0006. [DOI] [Google Scholar]
- Pagel M. The maximum likelihood approach to reconstructing ancestral character states of discrete characters on phylogenies. Syst Biol. 1999;48:612–622. doi: 10.1080/106351599260184. [DOI] [Google Scholar]
- Boi S, Solda' G, Tenchini ML. Shedding light on the dark side of the genome: overlapping genes in higher eukaryotes. Curr Genom. 2004;5:509–524. doi: 10.2174/1389202043349020. [DOI] [Google Scholar]
- Makalowska I, Lin CF, Makalowski W. Overlapping genes in vertebrate genomes. Comput Biol Chem. 2005;29:1–12. doi: 10.1016/j.compbiolchem.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Marinotti O, Nguyen QK, Calvo E, James AA, Ribeiro JM. Microarray analysis of genes showing variable expression following a blood meal in Anopheles gambiae. Insect Mol Biol. 2005;14:365–373. doi: 10.1111/j.1365-2583.2005.00567.x. [DOI] [PubMed] [Google Scholar]
- Koutsos AC, Blass C, Meister S, Schmidt S, MacCallum RM, Soares MB, Collins FH, Benes V, Zdobnov E, Kafatos FC, Christophides GK. Life cycle transcriptome of the malaria mosquito Anopheles gambiae and comparison with the fruitfly Drosophila melanogaster. Proc Natl Acad Sci USA. 2007;104:11304–11309. doi: 10.1073/pnas.0703988104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissanayake SN, Ribeiro JM, Wang MH, Dunn WA, Yan G, James AA, Marinotti O. aeGEPUCI: a database of gene expression in the dengue vector mosquito, Aedes aegypti. BMC Res Notes. 2010;3:248. doi: 10.1186/1756-0500-3-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keese PK, Gibbs A. Origins of genes: "big bang" or continuous creation? Proc Natl Acad Sci USA. 1992;89:9489–9493. doi: 10.1073/pnas.89.20.9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott EM, Proudfoot NJ. Transcriptional collision between convergent genes in budding yeast. Proc Natl Acad Sci USA. 2002;99:8796–8801. doi: 10.1073/pnas.132270899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osato N, Suzuki Y, Ikeo K, Gojobori T. Transcriptional interferences in cis natural antisense transcripts of humans and mice. Genetics. 2007;176:1299–1306. doi: 10.1534/genetics.106.069484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Spiridonov AN, Sorokin AV, Wolf YI, Jordan IK, Tatusov RL, Koonin EV. Purifying and directional selection in overlapping prokaryotic genes. Trends Genet. 2002;18:228–232. doi: 10.1016/S0168-9525(02)02649-5. [DOI] [PubMed] [Google Scholar]
- von Sternberg RM, Novick GE, Gao GP, Herrera RJ. Genome canalization: the coevolution of transposable and interspersed repetitive elements with single copy DNA. Genetica. 1992;86:215–246. doi: 10.1007/BF00133722. [DOI] [PubMed] [Google Scholar]
- Jurka J, Pethiyagoda C. Simple repetitive DNA sequences from primates: compilation and analysis. J Mol Evol. 1995;40:120–126. doi: 10.1007/BF00167107. [DOI] [PubMed] [Google Scholar]
- Nadir E, Margalit H, Gallily T, Ben-Sasson SA. Microsatellite spreading in the human genome: evolutionary mechanisms and structural implications. Proc Natl Acad Sci USA. 1996;93:6470–6475. doi: 10.1073/pnas.93.13.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay L, Macaulay M, Cardle L, Morgante M, degli Ivanissevich S, Maestri E, Powell W, Waugh R. Intimate association of microsatellite repeats with retrotransposons and other dispersed repetitive elements in barley. Plant J. 1999;17:415–425. doi: 10.1046/j.1365-313X.1999.00392.x. [DOI] [PubMed] [Google Scholar]
- Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 2001;11:1441–1452. doi: 10.1101/gr.184001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meglecz E, Petenian F, Danchin E, D'Acier AC, Rasplus JY, Faure E. High similarity between flanking regions of different microsatellites detected within each of two species of Lepidoptera: Parnassius apollo and Euphydryas aurinia. Mol Ecol. 2004;13:1693–1700. doi: 10.1111/j.1365-294X.2004.02163.x. [DOI] [PubMed] [Google Scholar]
- Van’t Hof AE, Brakefield PM, Saccheri IJ, Zwaan BJ. Evolutionary dynamics of multilocus microsatellite arrangements in the genome of the butterfly Bicyclus anynana, with implications for other Lepidoptera. Heredity (Edinb) 2007;98:320–328. doi: 10.1038/sj.hdy.6800944. [DOI] [PubMed] [Google Scholar]
- Fedorova L, Fedorov A. Introns in gene evolution. Genetica. 2003;118:123–131. doi: 10.1023/A:1024145407467. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Selfish DNA and the origin of introns. Nature. 1985;315:283–284. doi: 10.1038/315283b0. [DOI] [PubMed] [Google Scholar]
- Logsdon JM Jr. Worm genomes hold the smoking guns of intron gain. Proc Natl Acad Sci USA. 2004;101:11195–11196. doi: 10.1073/pnas.0404148101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW. The origin of recent introns: transposons? Genome Biol. 2004;5:251. doi: 10.1186/gb-2004-5-12-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BA, Slamovits CH, Patron NJ, Fast NM, Keeling PJ. A high frequency of overlapping gene expression in compacted eukaryotic genomes. Proc Natl Acad Sci USA. 2005;102:10936–10941. doi: 10.1073/pnas.0501321102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of E/H and P/O gene pairs of A. aegypti and A. gambiae.
Overlapping and non-overlapping patterns of orthologous genes between A. aegypti and A. gambiae.
List of gene pairs showing overlapping or non-overlapping localization in A. aegypti and A. gambiae genomes but have no changes in exon structure.
List of one-to-one orthologous genes of A. aegypti overlapping genes in other insect species and comparison of overlapping patterns across genomes.
Comparison of cluster patterns of retention or rearrangement of gene overlapping (left) with that of presence or absence of orthology of the corresponding gene pairs (right) among different insects. Red color indicates presence and black color indicates absence of overlapping/ orthology between genes.
List of common microsatellite motifs associated with the overlapping gene pairs that are rearranged between the two mosquitoes.
List of gene pairs and overlapping ESTs in A. aegypti.
Number of gene pairs identified in positionally overlapping patterns with or without evidence of expressed sequence tags.
Expression level of gene pairs that are localized in positionally overlapping manner in A. gambiae genome.
Expression level of A. aegypti overlapping genes.