Abstract
Background
Previous reports have suggested that malignant transformations originate from adult stem cells, and may thus express the stem-cell-associated markers. The purpose of this study is to investigate the differential expression and clinical significance of seven stem-cell-associated markers (Bmi1, CD133, CD44, Sox2, Nanog, OCT4 and Msi2) in lung cancer, providing new targets for the diagnosis and treatment of lung cancer.
Methods
In this study, we evaluated the differential expression of mRNA levels seven stem-cell-associated markers by semi-quantitative reverse transcription polymerase chain reaction (RT-PCR) from 112 human lung cancer and 18 non-cancer tissues obtained by bronchoscopy. We further verified the differential expression of these markers by immunohistochemistry in 50 lung cancer specimens, 30 benign inflammatory lesion tissues and 20 non-tumor adjacent lung tissues.
Results
With the exception of OCT4, other markers Bmi1, CD133, CD44, Sox2, Nanog and Msi2 mRNA and protein were abundantly expressed in lung cancer. Additionally, Nanog expression was highly upregulated in lung cancer tissues and rarely presented in non-cancerous lung tissues, the sensitivity and specificity of Nanog mRNA reached 63.4% and 66.7%, respectively. Nanog therefore possessed high diagnostic value, however, CD44, Bmi1 and CD133 showed poor diagnostic value in lung cancer.
Conclusion
Nanog may serve as a promising diagnostic marker of lung cancer and potential therapeutic target in lung cancer.
Keywords: Stem-cell-associated marker, Lung cancer, Therapeutic target, Diagnostic marker
Background
Lung cancer is one of the leading causes of cancer-related mortality in the world, and the incidence rates are increasing in many countries [1]. Although the prognosis is improving, the 5-year overall survival rate of lung cancer patients is still only approximately 16% [2]. In order to improve survival outcome, it is important to detect and surgically remove lung cancer at an early stage. Currently, the cancer stem cell (CSC) theory proposes that tumors contain a small subpopulation of CSC, which is responsible for tumor growth, invasion and metastasis [3]. CSC and normal tissue stem cells share important characteristics: self-renewal, multipotency and unlimited proliferation, and potentially overlapping molecular mechanisms [4,5]. In human adult tissues and tumors, several hundred stem-cell-associated markers have been identified. In lung cancer, the common stem-cell-associated markers include Bmi1, CD133, CD44, Sox2, OCT4 and so on [6,7]. Emerging evidences showed that these stem-cell-associated markers correlate with tumorigenesis, progression and metastasis, and may be as potential diagnostic markers for various human tumors [8-15].
Bmi1 is an oncogenic member of the polycomb group proteins involved in the self-renewal and differentiation of stem cells. The expression of Bmi1 mRNA has been shown to be a good marker to support the diagnosis of breast cancers in surgically resected specimens [8]. Likewise, CD133, a transmembrane glycoprotein which was first recognized in human hematopoietic stem cells, is considered the most representative marker to isolate CSC from lung cancer [9]. Recently, Moreira et al. [10] found CD133 is expressed in 58% of small cell lung cancers and 19% of lung adenocarcinomas, but not in normal lung tissue, suggesting that CD133 could be used as a potential diagnostic marker in lung cancer. Another surface marker, CD44, has also been used to isolate CSC from lung cancer [11]. A previous study using competitive RT-PCR to detect the expression of CD44 in urine for bladder cancer diagnosis was highly accurate and a potential non-invasive diagnostic marker for bladder cancer [12]. Transcription factors, Sox2, OCT4 and Nanog form a core regulatory network of self-renewal and differentiation in embryonic stem cells, which are essential in sustaining stem cell pluripotency [13]. Recent reports show that Sox2, OCT4 and Nanog are potential diagnostic markers for lung cancer [14-16]. Additionally, Musashi2 (Msi2), a RNA binding protein, play crucial roles in maintaining self-renewal and pluriopentency of embryonic stem cells. It have been demonstrated to participate in tumorigenesis and progression of multiple solid tumors [17,18], and are expressed in lung cancer [10]. However, these studies which are mainly based on surgical specimens to screen for new molecular markers have certain limitations in clinical application because most lung cancers are unresectable.
Bronchoscopy has become an essential method by which to analyze and diagnose lung cancer through technological advances and its widespread application. Common bronchoscopy techniques including forceps biopsy, brushing and washing can easily obtained adequate specimens for histological, cytological and molecular biological analysis [19]. The purpose of this study is to investigate the differential and clinical significance of these stem-cell-associated markers in bronchoscopy biopsy specimens.
In this study, we applied RT-PCR to examine the differential expression of Bmi1, CD133, CD44, Sox2, Nanog, OCT4 and Msi2 mRNA in bronchoscopic biopsy specimens from lung cancer and non-cancer patients. Furthermore immunohistochemistry was used to define the localization and expression patterns of these stem-cell-associated proteins in surgically resected lung cancer and non-malignant lung tissues. The diagnostic value of these seven stem-cell-associated markers was evaluated in lung cancer.
Materials and methods
Clinical samples from bronchoscope biopsy
This prospective study in 112 patients with histologically proven lung cancer and 18 non-cancer patients was performed at Guilin Medical University Hospital and Affiliated Nan Xi Shan Hospital in China from January, 2011 to January, 2012. These 112 lung cancer patients included 94 males and 18 females ranging from 29 to 80 years of age (median = 59.2). Fifty-six cases were squamous cell carcinomas (SCC), 17 cases adenocarcinomas (Ad), 28 cases small cell lung carcinomas (SCLC) and 11 cases of other types of lung cancer. Based on clinical and radiological findings, 100 cases had been evaluated for stages: 7 cases of stage I, 6 cases of stage II, 60 cases of stage III and 27 cases of stage IV of lung cancer. Among 18 cases of non-cancer, 7 cases were bronchitis, 7 cases tuberculosis, 3 cases pneumonia and 1 case brochiectasis. All patients had not received any anti-cancer therapy before receiving bronchoscopy. At least 5 biopsy specimens were obtained from one patient. One to two specimens were snap frozen and stored at -80°C for RT-PCR analysis under the condition of specimens were sufficient for routine diagnosis. The remaining specimens were fixed in buffered formalin for histopathological evaluation. This study was approved by the Guilin Medical University Review Board, and informed consent was obtained from all patients under the protocols prescribed by the Guilin University Ethics Committee.
Semi-quantitative RT-PCR
Total RNA was isolated from the biopsy tissue using Trizol reagent (TakaRa Bio Inc, Dalian, China) according to the manufacturer's instructions. One μg of the mRNA was reverse transcribed to cDNA using PrimeScript II 1st Strand cDNA Synthesis Kit (TakaRa). One μl of the cDNA was used in PCR for the amplification of β-actin and seven stem-cell-associated markers. The primers are presented in Table 1. The DNA thermal cycler conditions used were 94°C for 5 min (pre-denature), and 35 cycles of 94°C for 1 min, annealing for 30 s and extension at 72°C for 45 s, followed by a final extension of 72°C for 2 min. Six μl of each PCR-amplified product were separated on a 2% agarose gel, which was then visualized by ethidium bromide staining using a JS-780 Gel Image Analysis System (Peiqing Sci Tech, Ltd, Shanghai, China). The ratio of integrated density of target genes over corresponding β-actin was normalized as relative mRNA expression levels of stem-cell-associated markers.
Table 1.
The primers and primary antibody used in this study
|
Gene symbles |
Primers for RT-PCR |
|
Antibodies for IHC |
|
|
|
|---|---|---|---|---|---|---|
| Primer sequences | Annealing temperature (°C) | Antibody sources | Clone | Dilution | ||
| Bmi1 |
Reverse |
5’-ATT GTC TTT TCC GCC CGC TT-3’ |
58.2 |
ProMab Biotechnologies Inc |
3E3 |
1:800 |
| Forward |
5’-TGG CAT CAA TGA AGT ACC CTC-3’ |
|||||
| CD44 |
Reverse |
5’-TGC TAC TGA TTG TTT CAT TGC G-3’ |
56.2 |
ProMab Biotechnologies Inc |
8E2F3 |
1:30000 |
| Forward |
5’-GGA CCA GGC CCT ATT AAC CC-3’ |
|||||
| CD133 |
Reverse |
5’-AAA CAA TTC ACC AGC AAC GAG-3’ |
54.1 |
ProMab Biotechnologies Inc |
3 F10 |
1:400 |
| Forward |
5’-TAG TAC TTA GCC AGT TTT ACC G-3’ |
|||||
| Sox2 |
Reverse |
5’- GCT AGT CTC CAA GCG ACG AA-3’ |
56.2 |
ProMab Biotechnologies Inc |
10 F10 |
1:800 |
| Forward |
5’- TAC AGT CTA AAA CTT TTG CCC TT-3’ |
|||||
| Nanog |
Reverse |
5’-AGG CAA CTC ACT TTA TCC CAA-3’ |
54.1 |
Cell signaling technology |
D73G4 |
1:300 |
| Forward |
5’-GAT TCT TTA CAG TCG GAT GCT T-3’ |
|||||
| Oct-4 |
Reverse |
5’-TGC AGA AAG AAC TCG AGC AA-3’ |
56.2 |
Santa Cruz Biotechnology |
C-10 |
1:50 |
| Forward |
5’-CTC ACT CGG TTC TCG ATA CTG G-3’ |
|||||
| Msi2 |
Reverse |
5’-CAG ACC TCA CCA GAT AGC CTT-3’ |
56.2 |
ProMab Biotechnologies Inc |
2C11 |
1:1000 |
| Forward |
5’-TAC TGT GTT CGC AGA TAA CCC-3’ |
|||||
| β-actin (217 bp) |
Reverse |
5’GTG ACG TGG ACA TCC GCA AAG-3’ |
60.2 |
|
|
|
| Forward |
5’-ATC CAC ATC TGC TGG AAG GTG GAC-3’ |
|
|
|
||
| β-actin (417 bp) |
Reverse |
5’-ACA GAG CCT CGC CTT TGC CGA TC-3’ |
60.2 |
|
|
|
| Forward | 5’-TGG GTC ATC TTC TCG CGG TTG G-3’ | |||||
Immunohistochemistry (IHC)
A total of 50 cases of surgically resected lung cancer, 30 benign inflammatory lesion tissues and 20 normal or non-tumor adjacent lung tissues were used for IHC experiments. The lung cancer samples consisted of 17 adenocarcinomas (Ad), 3 bronchioloalveolar carcinomas (BAC), 23 squamous cell carcinomas (SCC) and 7 small cell lung carcinomas (SCLC). Thirty cases of benign inflammatory lesion samples included 12 cases of tuberculosis, 6 cases of pneumonia, 6 cases of inflammatory pseudotumor, 3 cases of brochiectasis, 2 cases of lung abscess and 1 case of benign fibroma of lung. In 50 non-cancer lung tissues 3 cases were squamous metaplasia including 2 cases of non-tumor adjacent lung tissues and 1 case of pneumonia. In all patients bronchoscopy and surgery were performed at Guilin Medical University Hospital from January, 2002 to December, 2011. None of the subjects received radiation therapy or chemotherapy before surgery.
Surgical specimens were fixed in 10% formaldehyde, and paraffin-embedded. After deparaffinization and rehydration, the 4 μm sections underwent antigen retrieval by boiling in 10 mM citrate buffer (pH 6.0) or EDTA (pH 8.0). The sections were immersed in H2O2 for 10 min and washed with PBS three times. Then the sections were incubated for 1 hr with the primary antibodies (Table 1) at 37°C. After a brief wash, the sections were incubated for 20 min with Polymer Helper (ZSGB-BIO, Beijing, China). Sections were washed three times with PBS and the antigen was visualized with polyperoxidase-anti-mouse/rabbit IgG (ZSGB-BIO) and DAB as substrate (ZSGB-BIO). The sections were counterstained with Mayer hematoxylin and mounted in Permount. Blank controls were obtained by replacing the primary antibodies with PBS.
The expression pattern criteria determined by IHC included: ‘diffuse’ when almost all cells expressed the antigen; ‘focal’ when isolated groups of positive cells were seen within a histological section; ‘isolated staining’ when single cells were positive for the marker. All slides were reviewed by a pathologist (Lu JY, Guilin, China) and a well-trained researcher in pathology (Li LD, Guilin, China) blinded to the patients' clinical information.
Statistical analysis
The Chi-Square test and the Mann–Whitney U test were applied to compare the expression of markers between lung cancer and non-cancer. The Chi-Square test was also performed to analyze the association between mRNA expression markers and lung cancer clinical factors. For evaluation of diagnostic value of seven stem-cell-associated markers mRNA, the following calculations were made: sensitivity (%) was calculated as true-positive/(true-positive +false-negative)×100, specificity (%) as true-negative/(true-negative + false-positive)×100 and diagnostic accuracy (%) as (true-positive + true-negative/(true-positive + false-negative + false-positive + true-negative)×100, where true-positive denotes specimens with stem-cell-associated marker positive expression and lung cancer diagnosis during follow up, true-negative denotes negative expression with no lung cancer, false-positive denotes positive expression with no lung cancer and false-negative denotes negative expression and diagnosed lung cancer. All statistical analyses were performed by SPSS 17.0 software package for Windows. P<0.05 was regarded statistically significant.
Results
The mRNA expression of seven stem-cell-associated markers in biopsy samples obtained through bronchoscopy
The expression of Bmi1, CD133, CD44, Sox2, Nanog, OCT4 and Msi2 mRNA in bronchoscopic biopsies of lung cancer and non-cancer patients are presented in Table 2 and Figure 1. Overall, the mRNA expression of seven markers was higher in the malignant group than in the benign group. However, the mRNA relative levels of Bmi1, CD133 and CD44 by RT-PCR were not significantly different between lung cancer and non-malignant lung tissues analyzed by Mann–Whitney U test, nor were the expression rates of CD44 and Msi2. We found that the Bmi1 positive expression rate was significantly correlated with histology types (P=0.007) and differentiation (P=0.027), while the positive rate of Nanog was negatively correlated with differentiation (0.032). However, the positive expression rates of CD133, CD44, Sox2, OCT4 and Msi2 did not correlate with age, gender, histological type, stage and differentiation of lung cancer (Table 3).
Table 2.
mRNA expression of stem cell makers in human lung cancer and non-cancer lung tissues
| |
Lung cancer |
Non-cancer |
P |
Lung cancer |
Non-cancer |
P |
|---|---|---|---|---|---|---|
| Positive rate, %(n) | Positive rate, %(n) | Expression, χ ± s | Expression, χ ± s | Value | ||
| Bmi1 |
88.4(99/112) |
66.7(12/18) |
0.026 |
0.60±0.73 |
0.32±0.29 |
0.118 |
| CD133 |
85.7(96/112) |
55.6(10/18) |
0.006 |
0.77±0.90 |
0.58±0.97 |
0.057 |
| CD44 |
98.2(110/112) |
88.9(16/18) |
0.092 |
1.67±1.77 |
1.44±1.33 |
0.606 |
| Sox2 |
98.2(110/112) |
83.3(15/18) |
0.019 |
2.06±2.15 |
0.99±1.53 |
0.001 |
| Nanog |
63.4(71/112) |
33.3(6/18) |
0.016 |
0.23±0.42 |
0.04±0.09 |
0.013 |
| OCT4 |
85.7(96/112) |
38.8(7/18) |
<0.0001 |
0.46±0.50 |
0.12±0.27 |
<0.0001 |
| Msi2 | 96.4(108/112) | 94.4(17/18) | 0.531 | 1.29±1.13 | 0.47±0.51 | <0.0001 |
Figure 1.
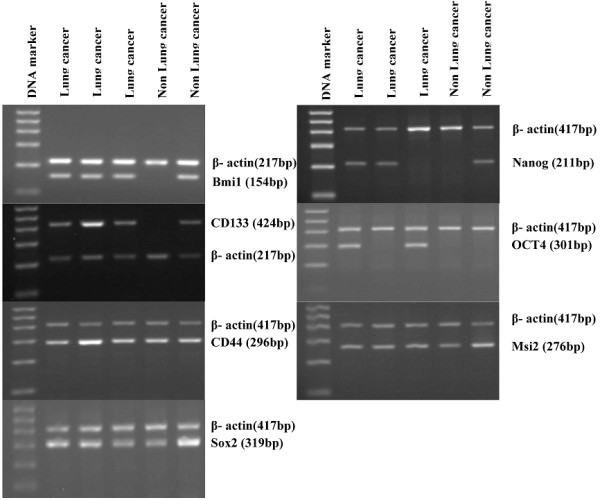
Example RT-PCR bands of human lung cancer and non-lung cancer biopsy tissues obtained from bronchoscopy. Total RNAs were isolated and reverse transcribed to cDNA from the biopsy tissues. RT-PCR Products of β-actin and stem-cell-associated markers were run on 2% agarose gels with ethidium bromide.
Table 3.
Correlation between stem cell mRNA expression of biopsy samples and lung cancer clinical features
| |
Analyzable |
Bmi1 expression |
P* |
CD133 expression |
P* |
CD44 expression |
P* |
Sox2 expression |
P* |
Nanog expression |
P* |
OCT4 expression |
P* |
MSi2 expression |
P* |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cases | Postive, n(%) | Postive, n(%) | Postive, n(%) | Postive, n(%) | Postive, n(%) | Postive, n(%) | Postive, n(%) | ||||||||
| Age |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| <60 |
57 |
51(89.5) |
0.716 |
48(84.2) |
0.643 |
56(98.2) |
1 |
55(96.5) |
0.496 |
36(63.2) |
0.958 |
47(82.5) |
0.448 |
54(94.7) |
0.618 |
| ≥60 |
55 |
48(87.3) |
|
48(87.3) |
|
54(98.2) |
|
55(100) |
|
35(63.6) |
|
49(89.1) |
|
54(98.2) |
|
| Gender |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Male |
94 |
84(89.4) |
0.436 |
80(85.1) |
1 |
92(97.9) |
1 |
92(97.9) |
1 |
57(60.6) |
0.167 |
79(84.0) |
0.462 |
90(95.7) |
1 |
| Female |
18 |
15(83.3) |
|
16(88.9) |
|
18(100) |
|
18(100) |
|
14(77.8) |
|
17(94.4) |
|
18(100) |
|
| Histology |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| SCLC |
28 |
27(96.4) |
0.007 |
27(96.4) |
0.066 |
26(92.9) |
0.171 |
27(96.4) |
1 |
22(78.6) |
0.068 |
26(92.9) |
0.601 |
27(96.4) |
1 |
| Ad |
17 |
11(64.7) |
|
16(94.1) |
|
17(100) |
|
17(100) |
|
13(76.5) |
|
14(82.4) |
|
16(94.1) |
|
| SCC |
56 |
52(92.9) |
|
43(76.8) |
|
56(100) |
|
55(98.2) |
|
31(55.4) |
|
46(82.1) |
|
54(96.4) |
|
| other |
11 |
9(81.8) |
|
10(90.9) |
|
11(100) |
|
11(100) |
|
5(45.5) |
|
10(90.9) |
|
11(100) |
|
| Stage |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| I~II |
13 |
13(100) |
0.601 |
11(84.6) |
1 |
13(100) |
1 |
13(100) |
1 |
7(53.8) |
0.369 |
10(76.9) |
0.407 |
13(100) |
1 |
| III~IV |
87 |
78(89.7) |
|
74(85.1) |
|
85(97.7) |
|
85(97.7) |
|
58(66.7) |
|
75(86.2) |
|
84(96.6) |
|
| Differentiation |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Well-Moderate |
28 |
21(75) |
0.027 |
21(75) |
0.216 |
28(100) |
1 |
27(96.4) |
0.337 |
12(42.9) |
0.032 |
22(78.6) |
0.537 |
26(92.9) |
0.262 |
| Poor | 55 | 52(94.5) | 48(87.3) | 55(100) | 55(100) | 37(67.3) | 47(85.5) | 54(96.4) |
* chi-square test . Ad = adenocarcinomas, SCC = squamous cell carcinomas, SCLC = small cell lung carcinomas and others = mucoepidermoid carcinoma, malignant mesothelioma or unclarified lung cancer.
Localization and expression patterns of stem-cell-associated markers protein in non-malignant lung tissues and lung cancer
Based on our RT-PCR results, most of the stem-cell-associated markers mRNA were expressed in non-malignant lung tissues although the expression levels were relative low. Therefore, we further examined the localization and expression patterns of stem cell markers in non-malignant lung tissues and lung cancer by IHC.
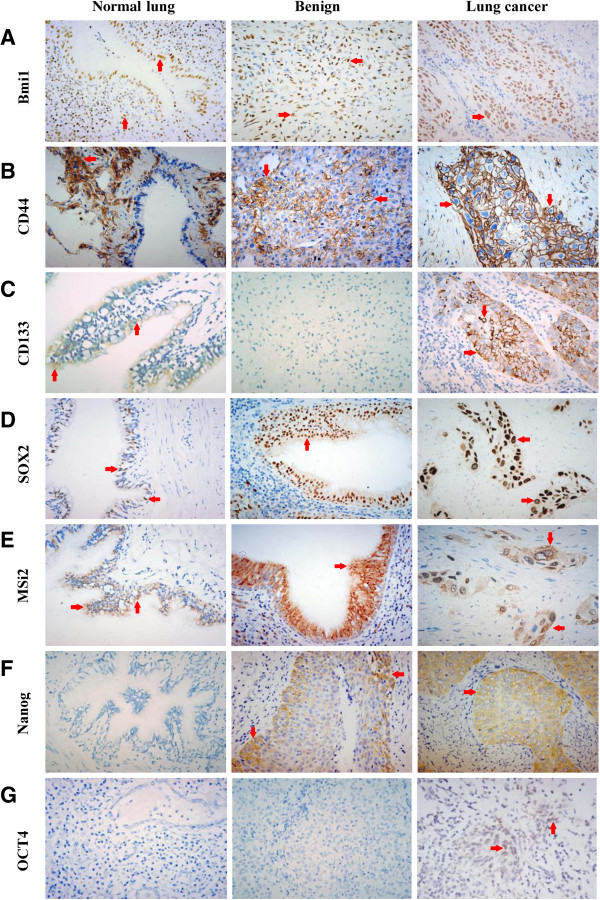
Bmi1 was diffusely expressed in bronchial epithelium cells, alveolar epithelium cells, lung interstitial cells and some inflammatory cells of all non-malignant lung tissues (Figure 2A), and was diffusely expressed in 47 cases of lung cancer and focally expressed in 1 case of Ad and 1 case of SCLC (Figure 2A). Similar to Bmi1, CD44 was abundantly expressed in alveolar epithelium cells, lung interstitial cells, macrophages, inflammatory cells and metaplastic squamous bronchial epithelium of non-malignant lung tissues (Figure 2B), but was absent in normal bronchial epithelium cells. 38 out of 50 lung cancer tissues were positive for CD44, of which 37 cases were diffusely positive and 1 case was focally positive expression (Figure 2B).
Figure 2.
Representative the expression of Bmi1, CD44, CD133, Sox2, Nanog, OCT4 and Msi2 in normal lung, benign lesion and lung cancer. (A) Nuclear staining of Bmi1 is expressed (red arrows) in normal lung, benign fibroma of lung and squamous cell carcinomas. (B) Membranous staining of CD44 in normal lung, benign fibroma of lung and squamous cell carcinomas. (C) Membranous and cytoplastic staining of CD133 in normal bronchial epithelium cells and squamous cell carcinomas, negative immunostaining signal of CD133 in benign fibroma of lung. (D) Nuclear staining of Sox2 in normal bronchial epithelium cells, squamous metaplasia and squamous cell carcinomas. (E) Cytoplastic and nuclear staining of Msi2 in normal bronchial epithelium cells, squamous metaplasia and squamous cell carcinomas. (F) Negative immunostaining signal of Nanog in normal lung, cytoplastic staining of Nanog in squamous metaplasia and squamous cell carcinomas. (G). Negative immunostaining signal of OCT4 in normal lung and tuberculosis, nuclear staining of OCT4 in small cell lung carcinomas. All images were taken at 400× magnification.
In non-malignant lung tissues, CD133 was exclusively expressed in some, but not all, bronchial epithelium cells and bronchial smooth muscle cells (Figure 2C). CD133+ bronchial epithelium cells were found in 74% of non-malignant lung tissues while CD133+ bronchial smooth muscle cells were 70%. In lung cancer tissues, about 56% of tumor samples were diffusely positive, 8% focally positive and 2% isolated positive for CD133 (Figure 2C).
In non-malignant lung tissues, all bronchial epithelium and squamous metaplasia showed positive expression of Sox2 (Figure 2D) and Msi2 (Figure 2E), the expression decreases in terminal bronchioles and was absent in alveolar epithelial. In lung cancer, the expression of Sox2 and Msi2 was 90% and 94% respectively, and more than 85% of tissues was diffusely positive for both of the markers (Figure 2D, E).
In non-malignant lung tissues, only 2 cases of squamous metaplasia in non-tumor adjacent lung tissues were positive for Nanog (Figure 2F), whereas, Nanog staining was detected in 36 of 50 (72%) cases of lung cancer, in which 29 cases were diffusely positive, 6 cases were focally positive and 1 case was isolated positive (Figure 2F).
In all non-malignant lung tissues, no positivity for OCT4 was observed (Figure 2G). In lung cancer group, only one case of SCC and one case of SCLC were focally positive for OCT4 (Figure 2G).
Potential value of the expression of stem-cell-associated markers as diagnostic markers
Table 4 describes the specificity, accuracy and sensitivity of seven stem-cell-associated markers mRNA in bronchoscopic biopsies of lung cancer and non-cancer patients. The stem-cell-associated markers with the highest sensitivity for malignancy were CD44 (98.2%), Sox2 (98.2%) and Msi2 (96.4%), but their specificity were too low to be considered of no clinical significance. Nanog exhibited the highest specificity which was 66.7%, and its sensitivity was 63.4%.
Table 4.
The specificity, accuracy and sensitivity of seven stem-cell-associated markers mRNA in biopsy samples obtained from bronchoscopy
| Specificity, % | Accuracy, % | Sensitivity, % | |
|---|---|---|---|
| Bmi1 |
33.3 |
80.8 |
88.4 |
| CD133 |
44.4 |
80 |
85.7 |
| CD44 |
11.1 |
86.2 |
98.2 |
| Sox2 |
16.7 |
86.9 |
98.2 |
| Nanog |
66.7 |
63.8 |
63.4 |
| OCT4 |
61.2 |
82.3 |
85.7 |
| Msi2 | 5.6 | 83.8 | 96.4 |
Discussion
In the present study, we evaluated the differential expression of seven stem-cell-associated markers in bronchoscopic biopsies of lung cancer and non-cancer patients by RT-PCR, and assessed their diagnostic value potential. Our data found Nanog mRNA had the highest specificity in lung cancer. We further confirmed the high diagnostic value of Nanog protein levels by IHC, Nanog was overexpressed in lung cancer tissues, but rarely expressed in non-malignant lung tissue. Taken together, these results demonstrate that Nanog mRNA is a potential diagnostic marker for lung cancer.
Nanog is a transcription factor that plays an important role in maintaining self-renewal of embryonic stem cells. Current studies have reported that the expression of Nanog was higher in multiple cancerous tissues than in their normal counterparts, including breast cancer [20], gastric adenocarcinomas [21], colorectal cancer [22], gliomas [23] and ovarian serous cystadenocarcinomas [24]. In this study, we found the expression of Nanog mRNA in bronchoscopic biopsies of lung cancer patients was significantly higher compared to that in non-cancer patients. Although Nirasawa et al. [16] have also reported that the expression of Nanog mRNA was higher in surgically resected lung cancer tissues than in non-cancerous tissues, it is not known what cells express Nanog in non-cancerous lung tissues. Using IHC, we found Nanog was only expressed in metaplastic squamous bronchial epithelium cells in 2 out of 50 non-malignant lung tissues, and was negative in normal airway epithelia. Therefore, Nanog may be a good diagnostic marker for lung cancer.
In this study, our results showed that the mRNA levels of Bmi1, CD44 and CD133 were not significantly different between lung cancer and non-malignant lung tissues. Further analyzed by IHC, we observed that Bmi1, CD44 and CD133 were not only expressed in lung cancers, Bmi1 and CD44 were also abundantly expressed in lung interstitial cells, inflammatory cells and bronchial epithelium cells, and CD133 was diffusely expressed in some normal bronchial epithelium cells and bronchial smooth muscle cells, consistent with previous studies [11,25,26]. Hence, Bmi1, CD44 and CD133 are poor diagnostic markers for lung cancer.
Likewise, although the expression levels of Sox2 and Msi2 mRNA in lung cancer tissues were significantly higher as compared with non-malignant tissues, we found more than 80% of bronchoscopic biopsy specimens of non-cancer patients were positive for Sox2 and Msi2 mRNA, and all non-malignant tissues were positive for Sox2 and Msi2 protein expression, consistent with previous findings [10,27,28]. Therefore, Sox2 and Msi2 have poor diagnostic specificity in lung cancer.
It is still controversial whether lung cancer cells express OCT4. Some researchers believe that OCT4 is involved in tumorigenesis and metastasis of lung cancer, and therefore is a potential diagnostic marker and useful therapeutic target of lung cancer [9,15,29], while others did not detect OCT4 expression in lung cancer [10,30]. Although we observed OCT4 mRNA expression in 85.7% of lung cancer and 38.8% of non-cancer bronchoscopic biopsy specimens, but OCT4 protein was nearly absent in 50 cases of lung cancer tissues. The reason for this discrepancy is unclear, but may be due to complex mechanism of post-transcriptional regulation, or potential presence of unknown OCT4 pseudogenes which cause false positive detection by RT-PCR. Therefore, the diagnostic value of OCT4 mRNA in bronchoscopic biopsy specimens requires further investigation.
In addition, we examined the correlation of seven stem cell markers expression in bronchoscopic biopsy specimens of lung cancer with patient clinical features. As we know, poorly differentiated cancers show stronger aggressive and metastatic ability [21]. We found the positive expression rates of Nanog and Bmi1 mRNA was inversely correlated to differentiation of lung cancer, indicating these two markers may be useful to predict tumor progression and poor prognosis in lung cancer. Chiou et al. [29] reported that Nanog expression in surgically resected lung cancer tissues is an independent prognostic factors of poor prognosis for patients. Vrzalikova and colleagues [31] also believed that the expression of Bmi1 in surgically resected lung cancer tissues is a prognostic marker in lung cancer. However, surgical resection is not an option for all lung cancer patients, and therefore the use of these markers in bronchoscopic biopsies to predict prognosis would be a great clinical advantage.
Conclusions
In conclusion, the expression of Nanog mRNA in bronchoscopic biopsy specimens is useful diagnostic marker for lung cancer. Further investigation of the diagnostic potential of Nanog in early stages of lung cancer may have a profound clinical impact.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LDL and HNY collected data and specimens, carried out the RT-PCR and immunochemistry staining, analyzed the results and drafted the manuscript. XYW conceived and designed the experiments, drafted and revised the manuscript critically and gave final approval of the version to be published. JRZ and DYL helped to collected bronchoscopic biopsy specimens. JYL helped to carry out the immunochemistry staining and assessed the slides. CMW, JYW, JHW and MJ participated in study coordination and statistical analysis. BWM conceived and designed of the study, performed the interpretation of data, literature search, writing and revising. All authors read and approved the final manuscript.
Contributor Information
Laodong Li, Email: lilaodong@qq.com.
Huina Yu, Email: 946419304@qq.com.
Xiaoyang Wang, Email: xiaoyangwang@gmail.com.
Jinrong Zeng, Email: zjr@glmc.edu.cn.
Dangyu Li, Email: ldy6171@163.com.
Jingyan Lu, Email: 53459838@qq.com.
Changming Wang, Email: wcm@glmc.edu.cn.
Jiying Wang, Email: jonewang0773@yahoo.com.cn.
Jianghong Wei, Email: weijianghong4321@163.com.
Ming Jiang, Email: gljmjmjm@163.com.
Biwen Mo, Email: mobiwen@yahoo.com.
Acknowledgements
This work was supported by the Key Research Project Grant of Guangxi Health Department (#2012003). We thank NIH Fellows Editorial Board for editing the manuscript.
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours:accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- Hassan KA, Chen G, Kalemkerian GP, Wicha MS, Beer DG. An embryonic stem cell-like signature identifies poorly differentiated lung adenocarcinoma but not squamous cell carcinoma. Clin Cancer Res. 2009;15:6386–6390. doi: 10.1158/1078-0432.CCR-09-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen GH, Murph MM, Chang JY. Cancer stem cell radioresistance and enrichment: where frontline radiation therapy May fail in lung and esophageal cancers. Cancers. 2011;3:1232–1252. doi: 10.3390/cancers3011232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pine SR, Marshall B, Varticovski L. Lung cancer stem cells. Dis Markers. 2008;24:257–266. doi: 10.1155/2008/396281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki M, Kobayashi D, Tsuji N, Kuribayashi K, Watanabe N. Diagnostic importance of overexpression of Bmi-1 mRNA in early breast cancers. Int J Oncol. 2009;35:511–515. doi: 10.3892/ijo_00000362. [DOI] [PubMed] [Google Scholar]
- Chen YC, Hsu HS, Chen YW, Tsai TH, How CK, Wang CY. et al. Oct-4 expression maintained cancer stem-like properties in lung cancer-derived CD133-positive cells. PLoS One. 2008;3:e2637. doi: 10.1371/journal.pone.0002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira AL, Gonen M, Rekhtman N, Downey RJ. Progenitor stem cell marker expression by pulmonary carcinomas. Mod Pathol. 2010;23:889–895. doi: 10.1038/modpathol.2010.68. [DOI] [PubMed] [Google Scholar]
- Leung EL, Fiscus RR, Tung JW, Tin VP, Cheng LC, Sihoe AD. et al. Non-small cell lung cancer cells expressing CD44 are enriched for stem cell-like properties. PLoS One. 2010;5:e14062. doi: 10.1371/journal.pone.0014062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake H, Hara I, Gohji K, Yamanaka K, Arakawa S, Kamidono S. Urinary cytology and competitive reverse transcriptase-polymerase chain reaction analysis of a specific CD44 variant to detect and monitor bladder cancer. J Urol. 1998;160:2004–2008. doi: 10.1016/S0022-5347(01)62225-0. [DOI] [PubMed] [Google Scholar]
- Müller FJ, Laurent LC, Kostka D, Ulitsky I, Williams R, Lu C. et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman CJ, Thorpe AJ, Murray A, Parsy-Kowalska CB, Allen J, Stafford KM. et al. Immunobiomarkers in small cell lung cancer: potential early cancer signals. Clin Cancer Res. 2011;17:1474–1480. doi: 10.1158/1078-0432.CCR-10-1363. [DOI] [PubMed] [Google Scholar]
- Karoubi G, Cortes-Dericks L, Gugger M, Galetta D, Spaggiari L, Schmid RA. Atypical expression and distribution of embryonic stem cell marker, OCT4, in human lung adenocarcinoma. J Surg Oncol. 2010;102:689–698. doi: 10.1002/jso.21665. [DOI] [PubMed] [Google Scholar]
- Nirasawa S, Kobayashi D, Tsuji N, Kuribayashi K, Watanabe N. Diagnostic relevance of overexpressed nanog gene in early lung cancers. Oncol Rep. 2009;22:587–591. doi: 10.3892/or_00000476. [DOI] [PubMed] [Google Scholar]
- Sakakibara S, Nakamura Y, Satoh H, Okano H. Rna-binding protein Musashi2: developmentally regulated expression in neural precursor cells and subpopulations of neurons in mammalian CNS. J Neurosci. 2001;21:8091–8107. doi: 10.1523/JNEUROSCI.21-20-08091.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharas MG, Lengner CJ, Al-Shahrour F, Bullinger L, Ball B, Zaidi S. et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat Med. 2010;16:903–908. doi: 10.1038/nm.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Bayoumi E, Silvestri GA. Bronchoscopy for the diagnosis and staging of lung cancer. Semin Respir Crit Care Med. 2008;29:261–270. doi: 10.1055/s-2008-1076746. [DOI] [PubMed] [Google Scholar]
- Ezeh UI, Turek PJ, Reijo RA, Clark AT. Human embryonic stem cell genes OCT4, NANOG, STELLAR, and GDF3 are expressed in both seminoma and breast carcinoma. Cancer. 2005;104:2255–2265. doi: 10.1002/cncr.21432. [DOI] [PubMed] [Google Scholar]
- Lin T, Ding YQ, Li JM. Overexpression of nanog protein is associated with poor prognosis in gastric adenocarcinoma. Med Oncol. 2012;29:878–885. doi: 10.1007/s12032-011-9860-9. [DOI] [PubMed] [Google Scholar]
- Meng HM, Zheng P, Wang XY, Liu C, Sui HM, Wu SJ. et al. Overexpression of nanog predicts tumor progression and poor prognosis in colorectal cancer. Cancer Biol Ther. 2010;9:295–302. doi: 10.4161/cbt.9.4.10666. [DOI] [PubMed] [Google Scholar]
- Niu CS, Li DX, Liu YH, Fu XM, Tang SF, Li J. Expression of NANOG in human gliomas and its relationship with undifferentiated glioma cells. Oncol Rep. 2011;26:593–601. doi: 10.3892/or.2011.1308. [DOI] [PubMed] [Google Scholar]
- Pan Y, Jiao J, Zhou C, Cheng Q, Hu Y, Chen H. Nanog is highly expressed in ovarian serous cystadenocarcinoma and correlated with clinical stage and pathological grade. Pathobiology. 2010;77:283–288. doi: 10.1159/000320866. [DOI] [PubMed] [Google Scholar]
- Kikuchi J, Kinoshita I, Shimizu Y, Kikuchi E, Konishi J, Oizumi S. et al. Distinctive expression of the polycomb group proteins Bmi1 polycomb ring finger oncogene and enhancer of zeste homolog 2 in nonsmall cell lung cancers and their clinical and clinicopathologic significance. Cancer. 2010;116:3015–3024. doi: 10.1002/cncr.25128. [DOI] [PubMed] [Google Scholar]
- Woo T, Okudela K, Mitsui H, Yazawa T, Ogawa N, Tajiri M. et al. Prognostic value of CD133 expression in stage I lung adenocarcinomas. Int J Clin Exp Pathol. 2010;4:32–42. [PMC free article] [PubMed] [Google Scholar]
- Sholl LM, Long KB, Hornick JL. Sox2 Expression in pulmonary non-small cell and neuroendocrine carcinomas. Appl Immunohistochem Mol Morphol. 2010;18:55–61. doi: 10.1097/PAI.0b013e3181b16b88. [DOI] [PubMed] [Google Scholar]
- Lu Y, Futtner C, Rock JR, Xu X, Whitworth W, Hogan BL. et al. Evidence that SOX2 overexpression is oncogenic in the lung. PLoS One. 2010;5:e11022. doi: 10.1371/journal.pone.0011022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou SH, Wang ML, Chou YT, Chen CJ, Hong CF, Hsieh WJ. et al. Coexpression of oct4 and nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer Res. 2010;70:10433–10444. doi: 10.1158/0008-5472.CAN-10-2638. [DOI] [PubMed] [Google Scholar]
- Cantz T, Key G, Bleidissel M, Gentile L, Han DW, Brenne A. et al. Absence of OCT4 expression in somatic tumor cell lines. Stem Cells. 2008;26:692–697. doi: 10.1634/stemcells.2007-0657. [DOI] [PubMed] [Google Scholar]
- Vrzalikova K, Skarda J, Ehrmann J, Murray PG, Fridman E, Kopolovic J. et al. Prognostic value of Bmi-1 oncoprotein expression in NSCLC patients: a tissue microarray study. J Cancer Res Clin Oncol. 2008;134:1037–1042. doi: 10.1007/s00432-008-0361-y. [DOI] [PubMed] [Google Scholar]