Abstract
Background
In order to select for quantitative plant resistance to pathogens, high throughput approaches that can precisely quantify disease severity are needed. Automation and use of calibrated image analysis should provide more accurate, objective and faster analyses than visual assessments. In contrast to conventional visible imaging, chlorophyll fluorescence imaging is not sensitive to environmental light variations and provides single-channel images prone to a segmentation analysis by simple thresholding approaches. Among the various parameters used in chlorophyll fluorescence imaging, the maximum quantum yield of photosystem II photochemistry (Fv/Fm) is well adapted to phenotyping disease severity. Fv/Fm is an indicator of plant stress that displays a robust contrast between infected and healthy tissues. In the present paper, we aimed at the segmentation of Fv/Fm images to quantify disease severity.
Results
Based on the Fv/Fm values of each pixel of the image, a thresholding approach was developed to delimit diseased areas. A first step consisted in setting up thresholds to reproduce visual observations by trained raters of symptoms caused by Xanthomonas fuscans subsp. fuscans (Xff) CFBP4834-R on Phaseolus vulgaris cv. Flavert. In order to develop a thresholding approach valuable on any cultivars or species, a second step was based on modeling pixel-wise Fv/Fm-distributions as mixtures of Gaussian distributions. Such a modeling may discriminate various stages of the symptom development but over-weights artifacts that can occur on mock-inoculated samples. Therefore, we developed a thresholding approach based on the probability of misclassification of a healthy pixel. Then, a clustering step is performed on the diseased areas to discriminate between various stages of alteration of plant tissues. Notably, the use of chlorophyll fluorescence imaging could detect pre-symptomatic area. The interest of this image analysis procedure for assessing the levels of quantitative resistance is illustrated with the quantitation of disease severity on five commercial varieties of bean inoculated with Xff CFBP4834-R.
Conclusions
In this paper, we describe an image analysis procedure for quantifying the leaf area impacted by the pathogen. In a perspective of high throughput phenotyping, the procedure was automated with the software R downloadable at http://www.r-project.org/. The R script is available at http://lisa.univ-angers.fr/PHENOTIC/telechargements.html.
Background
Quantitative phenotyping is important in plant breeding to screen accessions for resistance to pathogens. Indeed, plant resistance to pathogens may either be qualitative or quantitative. Qualitative resistance is due to the presence of single major resistance genes that confer total resistance to pathogens carrying the cognate avirulence genes. However, these monogenic total resistances are often rapidly bypassed. In contrast, resistance conferred by multiple loci exhibit quantitative phenotype and is thought to contribute to durability [1-3]. Thus quantitative phenotyping methods are necessary to ensure a good evaluation of the disease severity and to make appropriate decisions in gauging cultivar resistance in plant breeding.
Visual assessments have often been used to quantify disease severity. They require rating scales to be finely tuned, and raters to be trained, which increases considerably the cost and the time devoted to phenotyping projects. Moreover, these methods highly depend on the subjectivity of the raters and thus often lack accuracy, reproducibility and traceability [4-7].
In contrast, automated image analysis-based phenotyping provides a powerful alternative to visual assessments. Indeed, automation eventually provides a calibrated image analysis, thereby eliminating any subjectivity of the raters and ensuring reproducibility [6]. Furthermore, automation allows high throughput phenotyping. Calibrated protocols and data storage may provide useful tools for traceability or inter-laboratory comparison of the results.
Image segmentation aims at partitioning the digital image into multiple sets of pixels to select the areas of interest. In plant pathology, imaging of the symptoms has been mainly achieved using conventional color imaging. Segmentation algorithms aimed at the automated quantification of the total amount of symptoms on leaves from conventional color images [8-12]. Some of them aim at the quantification of the chlorosis or the necrosis using the differential colors of diseased leaf tissues [10,12]. Indeed, yellow and brown coloration respectively characterize chlorotic and necrotic tissues. Such automated segmentation procedures rely on color-based thresholds to discriminate diseased from healthy tissues on leaves. However, heterogeneity during exposition may alter the contrast of visible images and interfere with any color-based thresholding. Batch segmentation of visible images using color-based thresholds may thus generate numerous artifacts if light conditions during exposure are not tightly controlled. Scanner imaging of detached leaves or adjusting the color balance using a color checker may help standardizing contrasts prior to the segmentation procedures [9]. As conventional color images are typically multichannel images (for instance RGB images are composed by red, green and blue channels), they need sophisticated image analysis methods [13]. As segmentation by simple thresholding can only be applied on single channel images, conventional color images should be transformed into grayscale images prior to the segmentation. Such a transformation may result in a loss of information. Moreover, conventional color image standards aim at reproducing human vision, and thus do not directly represent the physiology of plant leaves.
Among non-conventional imaging approaches, some approaches such as thermography or chlorophyll fluorescence depict the physiology of plant leaves through single channel images [14,15]. These single channel images may easily be segmented using automated thresholding procedures for the quantification of disease severity. Chlorophyll fluorescence analysis is a non-destructive technique that has been used for imaging plant pathogen interactions [16,17] and in particular to assess the resistance of plant to pathogen [18-23]. Indeed, symptoms result from the alteration of the tissues and many pathogens target the carbon metabolism and the photosynthetic apparatus [24-26]. Among all the chlorophyll fluorescence parameters that can be estimated, the maximum quantum yield of photosystem II (PSII) photochemistry (Fv/Fm = (Fm-F0)/Fm) [27] is interesting for phenotyping disease severity as it is an indicator of plant stress [17,28]. Fv/Fm is a parameter calculated from two measured fluorescence parameters, F0 (minimum fluorescence) and Fm (maximum fluorescence). Fv/Fm was reported to display a robust contrast between infected and healthy tissues [17,20,28,29]. Furthermore, healthy tissues were reported to yield Fv/Fm values around 0.84 for numerous plant species [30,31]. When tissues are altered by biotic or abiotic stress, Fv/Fm values decrease [32-34]. In many studies, mean Fv/Fm measurements were used to qualitatively discriminate between diseased and healthy leaves [23,33,35-37]. However, quantitative assessments of the total diseased area on leaves require each pixel to be classified as diseased or healthy. Thresholding based on Fv/Fm values may allow the segmentation of diseased areas on the imaged leaves.
Common Bacterial Blight (CBB) of bean is caused by Xanthomonas axonopodis pv. phaseoli and X. fuscans subsp. fuscans (Xff). These pathogens are listed by the European and Mediterranean Plant Protection Organization [38] as quarantine pathogens as CBB is the most destructive bacterial disease of the common bean Phaseolus vulgaris resulting in up to 60% yield losses in favorable conditions [39]. On leaves, visible symptoms start usually four to seven days after infection with pinpoint water soaked areas that enlarge and eventually form necrotic tissues surrounded by a chlorotic halo. Attack may also result in leaflet wilting and in severe cases to defoliation [38]. Furthermore, the agents of CBB may also accomplish their whole cycle in the absence of visible symptoms [40]. As for other plant bacterial diseases, no efficient chemical treatment is allowed in the European Union, and control of the disease mainly involves the use of resistant bean cultivars.
No total monogenic resistance to the agents of CBB is known in P. vulgaris. However, quantitative trait loci conditioning resistance to CBB have been identified and quantitative resistances may be bred into commercial cultivars of bean [41]. Therefore quantification of the total diseased area on bean leaflets is needed to monitor the resistance level of novel bean lines to the agents of CBB during the selection process. In the present study, we developed procedures for the automated segmentation of Fv/Fm images in order to quantify disease severity on plant leaflets in the pathosystem P. vulgaris/Xff CFBP4834-R. At first, we explored expert-defined thresholds after visual observations to discriminate in Fv/Fm images areas corresponding to necrotic, wilted, impacted tissues, and healthy tissues. Second, we tested a segmentation approach based on modeling pixel-wise Fv/Fm-distributions as mixtures of Gaussian distributions, each distribution representing a different stage of the alteration of plant tissues, from strongly altered to healthy tissues. Finally, we developed a thresholding approach based on the probability of misclassification of a healthy pixel into the class of diseased pixels. Then, the segmented diseased areas can be modeled as mixtures of Gaussian distributions to discriminate various stages of alteration of plant tissues, from strongly to weakly altered tissues.
Results
Datasets
Two datasets were used. The first one was used to setup the segmentation procedure for the quantification of symptoms by image analysis. It features images of leaflets of P. vulgaris cultivar (cv.) Flavert plants inoculated with the strain Xff CFBP4834-R or water. In this first dataset, the imaged leaflets were not detached from the plant to monitor the development of symptoms during 11 days after inoculation (dai). The same leaflets were imaged first at 1 dai then every day between 4 and 11 dai to monitor the development of the symptoms. For each leaflet, the symptomatic area was delimited either by: i) thresholding based on expert visual observations, ii), thresholding based on modeling pixel-wise Fv/Fm-distributions as mixtures of Gaussian distributions or iii) thresholding based on the probability of misclassification of a healthy pixel followed by a subsequent clustering of diseased pixels to describe the various stages of alteration of plant tissues.
A second dataset featured images of leaflets of five bean cultivars (cvs. Flavert, Michelet, Pike, Caprice and Wonder) inoculated with Xff CFBP4834-R. The image analysis procedure previously calibrated on the first dataset was applied to images belonging to this second dataset for evaluating the resistance of these commercial bean cultivars to Xff CFBP4834-R. In this second dataset, leaflets were detached from inoculated plants just before imaging to ease and speed up the image acquisition.
Thresholding based on expert visual observations
Expert-based thresholding consisted in the comparison of conventional color images with Fv/Fm images of the same leaflet by trained raters to manually define the relevant thresholds to segment the Fv/Fm images. The segmented Fv/Fm images should visually reproduce the distribution of symptoms as visualized on conventional color images, i.e. the various segmented parts in Fv/Fm images should co-localize with the various stages of the symptom development as observed by the eye of trained raters. On bean leaflets of cv. Flavert harboring symptoms of Xff CFBP4834-R, we could discriminate between necrotic tissues, wilted, impacted and healthy tissues. Water soaked symptoms displayed Fv/Fm values similar to that of wilted tissues, therefore both are referred to as wilted tissues. Some tissues that did not harbor any visible symptoms displayed similar Fv/Fm values as chlorotic tissues, therefore both are referred to as impacted tissues. Subsequently, three Fv/Fm thresholds were determined to allow the automated segmentation of necrotic, wilted, impacted and healthy tissues (Figure 1).
Figure 1.
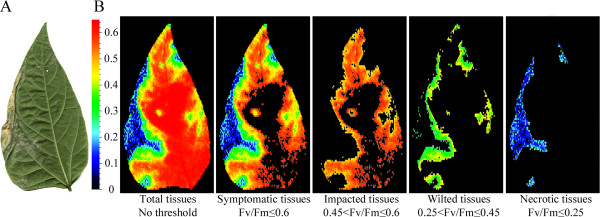
Expert-based thresholds allow the segmentation of various stages of the symptom development. Two weeks-old bean plants cv. Flavert were inoculated with either Xff CFBP4834-R (1.106 CFU ml_1) or mock. This leaflet inoculated with Xff CFBP4834-R was sampled on bean P.vulgaris cv. Flavert at 11 dai. A: visible image obtained by scanning. Necrosis is clearly visible on the left marge of the leaflet surrounded by wilted tissues. B: Fv/Fm image obtained by chlorophyll fluorescence imaging. The three stages of the symptom development, i.e. necrotic, wilted and impacted tissues, were segmented respectively with the thresholds 0.25 ≤ Fv/Fm, 0.25 < Fv/Fm ≤ 0.45 and 0.45 < Fv/Fm ≤ 0.6. Black areas represent non-selected pixels with the threshold. After the segmentation step, the proportion of pixels in each segment may be quantified.
A training subset of images was used by the trained raters to define thresholds. The comparison of visible and Fv/Fm images revealed that pixels displaying Fv/Fm values inferior to 0.6 co-localized with a diseased area. Fv/Fm values ranging from 0 to 0.25 co-localized with necrotic tissues, whereas Fv/Fm values ranging from 0.25 to 0.45 co-localized with wilted tissues. Fv/Fm values ranging from 0.45 to 0.6 corresponded to impacted tissues.
Expert-based thresholds were applied on all Fv/Fm images to quantify each stage of the symptom development. Significant symptom development began at 7 dai on leaflets inoculated with Xff CFBP4834-R (p-value < 0.01). From 9 dai on, the symptoms were predominantly composed by wilted tissues (Figure 2A).
Figure 2.
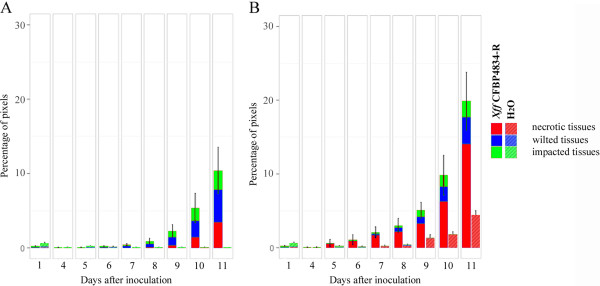
Evolution of the proportions of necrotic, wilted and impacted tissues on bean leaflets using expert-based thresholding. Two weeks-old bean plants cv. Flavert were inoculated with either Xff CFBP4834-R (1.106 CFU ml_1) or mock. Observations were made on bean leaflets sampled at 1 dai, and everyday after the fourth dai. Percentages of diseased tissues and standard error of the mean were calculated for 20 leaflets per sampling day. The percentages do not include (A) or include (B) the estimation of the shrinking of the leaflet. The shrinking of the leaflet is attributed to necrotic tissues.
Moreover, as in this first dataset we monitored the evolution of symptoms on the same leaflets over time, we could assess shrinking of leaflets induced by the pathogen. Shrinking corresponds to the difference between the maximum size of the leaflet and its current size at the observation timepoint. Shrinking of leaflets may represent up to 10% of the total leaflet area (Figure 2). It mostly corresponds to necrotic tissues. However, some shrinking may also originate from natural aging of leaflets as shrinking was also detected on mock-inoculated leaflets.
Thresholding based on modeling pixel-wise Fv/Fm distributions
Abiotic stresses during the experiment may affect the physiological status of plants, which will in turn impact the Fv/Fm values. In such cases, applying fixed thresholds may generate segmentation artifacts. To avoid such a drawback, mock-inoculated leaflets were used as internal daily controls to setup daily thresholds. Indeed, thresholds were defined daily to take into account the daily Fv/Fm variations.
From the Fv/Fm images of leaflets mock- or Xff CFBP4834-R-inoculated, we extracted the pixel-wise Fv/Fm-distributions. Analyzing these distributions may help defining appropriate non-overlapping clusters of Fv/Fm values that represent various stages of alteration of plant tissues. In statistics, Gaussian mixture model (i.e. weighted sum of Gaussian distribution) is often used for clustering analysis. A clustering method proposed by Hennig [42] was tested in the present study to identify clusters of tissues according to pixel-wise Fv/Fm-distributions. Gaussian mixture model can be used to formalize the underlying heterogeneous distribution of Fv/Fm values that are assumed to be composed of several clusters of pixels types, each cluster being modeled by a Gaussian distribution. From a practical point of view, Gaussian distributions are often too stiff to model true cluster shapes and Gaussian mixture modeling methods tend to select “larger optimal number of mixture components than what seems to be reasonable when looking to the data” [42]. Among the various methods proposed by Hennig [42] to solve this problem, we have tested the ridgeline unimodal method where clusters are merged if their fusion results in an unimodal Gaussian distribution [42,43].
In practice, using the R package MCLUST [44], the pixel-wise Fv/Fm-distributions were first fitted to Gaussian mixture models having from 1 to 4 components: one cluster should group pixels representing healthy tissues and three clusters should group pixels representing the various stages of the symptom development. The model best supported by the data is then determined using BIC criteria and lastly corrected by the ridgeline unimodal method [42,43]. For each pixel in the image, a probability of membership to each cluster is estimated. Clusters gathering less than 1% of the pixels were considered as not significant. Pixels initially attributed to these non-significant clusters were assigned according to their second better probability of membership.
On each image of mock-inoculated leaflets, pixel-wise Fv/Fm-distribution could be modeled as one single cluster per image. Predicted means of these clusters grouping pixels corresponding to healthy tissues may vary among images of mock-inoculated leaflets (e.g. from 0.71 to 0.85 at 4 dai). Hence, we calculated a confidence interval for predicted means of clusters grouping pixels corresponding to healthy tissues. Conversely, on diseased leaflets, Fv/Fm values were modeled by mixtures involving more than one cluster. Among these, one cluster grouped pixels corresponding to healthy tissues, while additional clusters contained pixels corresponding to diseased areas. To discriminate between healthy and diseased tissues, a threshold based on the lower limit of the confidence interval previously defined on mock-inoculated samples was used. Subsequently, the quantification of pixels corresponding to diseased area allows the calculation of the proportion of diseased tissues on each leaflet. As expected, the proportion of diseased area on inoculated leaflets increased from 7 dai to the end of the experiment (Table 1).
Table 1.
Quantification of the diseased tissues using the expert-, the model- and the probability-based thresholding approaches
| 1 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Expert-based thresholding |
Necrotic tissues (%) |
0,02 |
0,00 |
0,00 |
0,02 |
0,03 |
0,08 |
0,39 |
1,46, |
3,44 |
| Wilted tissues (%) |
0,06 |
0,01 |
0,01 |
0,06 |
0,19 |
0,49 |
1,02 |
2,15 |
4,41 |
|
| Impacted tissues (%) |
0,16 |
0,05 |
0,06 |
0,13 |
0,20 |
0,37 |
0,90 |
1,74 |
2,56 |
|
| Total diseased tissues (%) |
0,24 |
0,06 |
0,08 |
0,22 |
0,42 |
0,93 |
2,30 |
5,35 |
10,41 |
|
| Model-based thresholding |
Total diseased tissues (%) |
0,00 |
5,26 |
5,26 |
23,87 |
42,96 |
5,64 |
35,62 |
49,01 |
30,39 |
| Probability-based thresholding |
Strong alteration (%) |
0,01 |
0,00 |
0,00 |
0,00 |
0,00 |
0,00 |
0,00 |
1,35 |
2,70 |
| Moderate alteration (%) |
0,08 |
0,01 |
0,03 |
0,11 |
0,28 |
0,70 |
1,85 |
2,52 |
6,67 |
|
| Weak alteration (%) |
0,00 |
0,95 |
0,04 |
0,14 |
0,45 |
0,66 |
1,74 |
2,96 |
7,27 |
|
| Total diseased tissues (%) | 0,09 | 0,95 | 0,07 | 0,25 | 0,73 | 1,36 | 3,59 | 6,83 | 16,64 |
Expert-based thresholding consists in defining Fv/Fm thresholds that enable the selection of areas on Fv/Fm images that match the various stages of the symptom development as observed by trained raters on conventional color images. Healthy, necrotic, wilted, or tissues impacted by the pathogen can be quantified.
Model-based thresholding consists in modeling the pixel-wise Fv/Fm-distributions extracted from each image by mixtures of Gaussian distributions. Such modeling results in the definition of clusters of pixels that correspond to various stages of the alteration of plant tissues. This step is based on the sole analysis of pixel-wise Fv/Fm-distributions and not on the visual observation of symptoms on conventional color images. Therefore, we use the terminology of strong, moderate and weak alteration, to emphasize that this classification is not based on visual observations, and does not necessarily correspond to the various stages of the symptom development as observed by trained raters. When applied directly without preliminary delimitation of the total diseased area, such a modeling over weights artifacts that can occur on healthy tissues, which results in a large overestimation of the proportion of diseased tissues.
Probability-based thresholds consists in the 500-quantile of the merged pixel-wise Fv/Fm-distributions of mock-inoculated samples. Each day of the experiment, the Fv/Fm probability-based threshold allows the splitting of pixels corresponding to healthy and diseased areas. Then whithin the diseased area only, pixel-wise Fv/Fm-distributions are modeled as mixtures of Gaussian distributions to quantify various stages of alteration of plant tissues.
Pixels were recolored according to the cluster they belong to (Figure 3). Such a model-based clustering allows the discrimination between various stages of alteration of the plant tissues. These various stages of alteration of plant tissues strictly depend on the structure of pixel-wise Fv/Fm-distributions and are thus independent of any a priori based on visual observation of symptoms (Figure 3).
Figure 3.
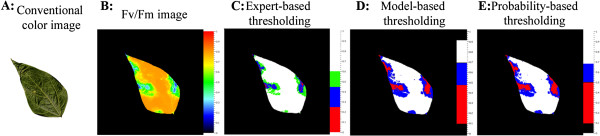
Mapping of the diseased areas segmented using the three thresholding approaches. Two weeks-old bean plants cv. Flavert were inoculated with either Xff CFBP4834-R (1.106 CFU ml_1) or mock. This leaflet inoculated with Xff CFBP4834-R was sampled on bean cv. Flavert at 11 dai. A: visible image obtained by scanning. B: Fv/Fm image obtained by chlorophyll fluorescence imaging. C-E: Segmentation of the Fv/Fm image for selection of the diseased area. C: Expert-based thresholds are defined after comparison with visual observations by trained raters. D: Model-based thresholds are defined by the clustering approach on the total surface af the leaflet. E: Probability-based thresholds are defined on the probability that a healthy pixel is misclassified with a specificity of 0.002. The healthy tissues are represented in white. The diseased tissues are colored in red, blue or green. Defioliation spots are represented in black.
Such an approach over-weights artifacts that can occur on non-symptomatic samples. Indeed, the rule for thresholding involves the mean of the predicted distribution, but not the own Fv/Fm value of each pixel. Therefore, on a mock-inoculated leaflet, if the mean of the single predicted distribution is slightly under the threshold, the whole leaflet area will be considered as diseased tissues. Such a caveat may also occur for the predicted distribution grouping pixels of healthy areas on Xff CFBP4834-R-inoculated leaflets, resulting in a strong overestimation of the diseased areas (Table 1).
Thresholding based on the probability of misclassification of a healthy pixel
To solve the caveat of the overestimation of the diseased area on inoculated leaflets, we decided to normalize on mock-inoculated samples by classifying each pixel based on the probability of misclassification of a healthy pixel. Probability-based thresholds were defined on the pixel-wise Fv/Fm-distributions of mock-inoculated leaflets. Thresholds were defined daily to take into account the daily Fv/Fm variations. Day by day, the pixel-wise Fv/Fm-distributions of all the mock-inoculated leaflets were merged. The resulting distribution thus represents all the possible values for a healthy pixel, including some abnormally low Fv/Fm values. The Fv/Fm values corresponding to the 100-quantile, 500-quantile or 1000-quantile, i.e. the Fv/Fm values splitting 1/100, 1/500 and 1/1000 of the pixels of the distribution of the mock-inoculated leaflets, were used as thresholds splitting infected and healthy tissues. For each Fv/Fm image, pixels were recolored according to these thresholds. The 1/1000 threshold was too stringent, as the segmented area does not contain the totality of the visible symptom. The 1/100 threshold was not stringent enough, as randomly distributed pixels were selected in addition to symptoms. Thus, we chose the 1/500 threshold as the totality of the symptom was segmented and no randomly distributed pixels were selected (data not shown). Using such a threshold the specificity of the approach is 0.002, i.e. there was a probability of 0.002 to misclassify a healthy pixel as diseased.
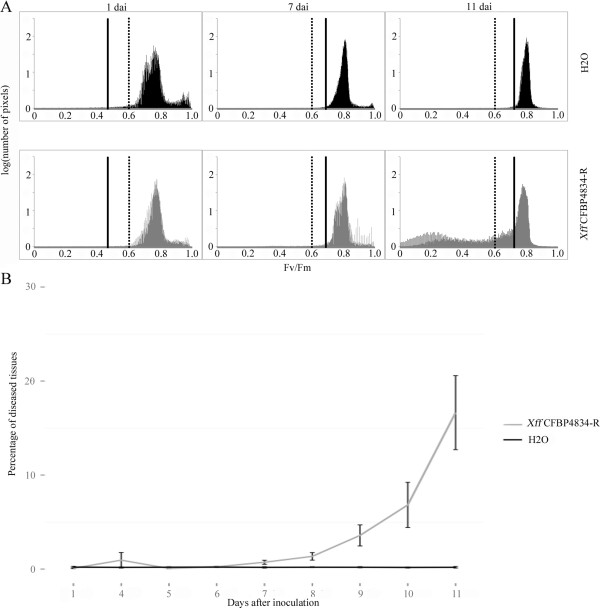
The probability-based thresholds are presented in Figure 4A by a vertical solid bar. Thresholds varied according to the day of the experiment (e.g. 0.467 at 1 dai, 0.689 at 7 dai and 0.722 at 11dai, Figure 4A) indicating that daily variations in the Fv/Fm status of plants occurred during the experiment. For each leaflet, pixels exhibiting Fv/Fm values lower than these probability-based thresholds were considered as diseased. Significant amounts of symptoms first arose at 7 dai (p-value < 0.01, Figure 4B, Table 1).
Figure 4.
Daily thresholds for calculation of the proportion of diseased tissues with the probability-based approach. Two weeks-old bean plants cv. Flavert were inoculated with either Xff CFBP4834-R (1.106 CFU ml_1) or mock. A: Pixel-wise Fv/Fm-distributions at 1, 7 and 11 dai of mock-inoculated leaflets (black) and Xff CFBP4834-R-inoculated leaflets (grey). The expert-based thresholds (dotted bars) are fixed over the whole experiment. Conversely, the probability-based thresholds (solid bars) may vary each day of the experiment, thereby taking into account daily physiological variations of plants. B: Evolution of the percentage of diseased areas on mock-inoculated (black curve) and Xff CFBP4834-R-inoculated (grey curve) leaflets calculated with probability-based thresholding approach.
In order to discriminate between various stages of alteration of plant tissues, the pixel-wise Fv/Fm-distributions corresponding to the diseased tissues were extracted. Using MCLUST [44], these distributions were first fitted to Gaussian mixture models having up to 3 components to match the various stages of alteration of plant tissues (Table 1). Mapping these clusters on the recolored Fv/Fm images confirmed that they match various stages of the alteration of plant tissues, i.e. weak alteration, moderate alteration, strong alteration.
Evaluation of the resistance of commercial bean cultivars to Xff CFBP4834-R
Five bean cultivars (cvs. Flavert, Michelet, Pike, Wonder and Caprice) were inoculated with the strain Xff CFBP4834-R or water. Symptoms were quantified by our image analysis procedure. The Expert-based thresholding approach was not used as the thresholds were defined from a training set of images only on cv. Flavert but not on the other cultivars. Instead, the total amount of diseased areas was determined using probability-based thresholds. Then, using a clustering approach (that does not require training datasets), the various stages of the alteration of plant tissues were discriminated and quantified.
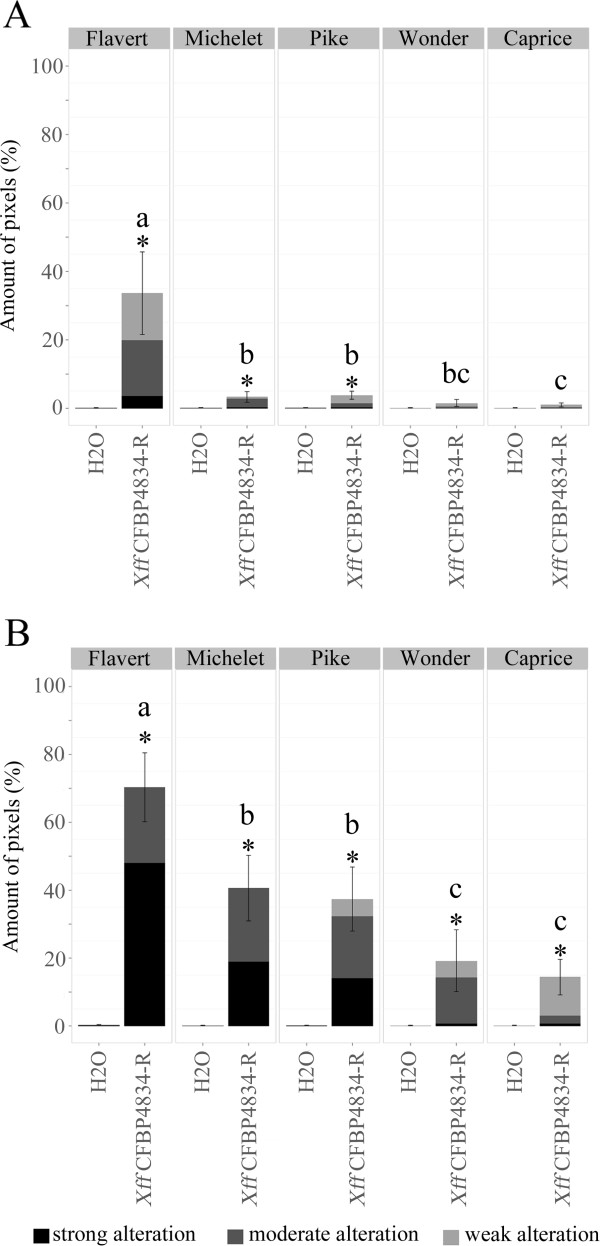
Fv/Fm images were taken on detached leaflets at 7 and 11 dai and the amounts of diseased tissues on leaflets were calculated (Figure 5). During the experiment, the amount of diseased tissues increased for all the cultivars tested. However, differential behaviors among the various cultivars tested could be observed. At both 7 and 11 dai, cv. Flavert exhibited a significantly higher amount of symptoms than the other bean cultivars (p-value < 0.05), indicating that cv. Flavert is the most sensitive cultivar to Xff CFBP4834-R. On the contrary, the amounts of symptoms detected on cvs. Wonder and Caprice significantly differed from the mock-inoculated samples only at 11 dai. At 11 dai, cvs. Caprice and Wonder displayed the weakest total amount of symptoms among all cultivars tested, indicating that these cultivars were the most tolerant to Xff CFBP4834-R in this study. At both 7 dai and 11 dai, cvs. Michelet and Pike exhibited amounts of symptoms significantly higher than mock-inoculated samples (p-value < 0.05). At 11 dai the total amounts of symptom detected on cvs. Michelet and Pike were significantly higher than that detected on cvs. Caprice and Wonder. Therefore, our study revealed three levels of tolerance to Xff CFBP4834-R at 11 dai: cv. Flavert was sensitive, cvs. Michelet and Pike were partially tolerant, and cvs. Caprice and Wonder were tolerant.
Figure 5.
Quantification of the proportion of diseased tissues caused by Xff CFBP4834-R on five cultivars of bean. Bean leaflets of cultivars Flavert, Michelet, Pike, Wonder and Caprice were inoculated with Xff CFBP4834-R (1.106 CFU ml_1) or mock. Observations were made at 7 (A), and 11 dai (B). On each leaflet diseased area was segmented using the probability-based thresholds. Means of percentages of diseased tissues and standard error of the mean were calculated from two repeats with 4 (7 dai) and 7 (11 dai) leaflets. Treatments denoted by different letters are significantly different (p-value < 0.01) based on the Mann–Whitney test. Asterisks mark significant differences between the mock-inoculated and CFBP4834-R-inoculated leaflets (p-value < 0.01) based on the Mann–Whitney test. Clusters representing the various stages of the alteration of plant tissues were determined by a clustering approach using MCLUST [44] on the diseased area only.
Then, the clustering approach was applied on the diseased areas to discriminate the various stages of the alteration of plant tissues i.e. weak alteration, moderate alteration and strong alteration. Therefore, an optimal number of three clusters was estimated. At 7 dai, symptoms on cv. Flavert, were essentially composed by moderately altered tissues. Some weakly altered tissues were also detected. At 11 dai, cv. Flavert exhibited a high amount of strongly altered tissues in comparison with the other cultivars. Symptoms segmented on cvs. Wonder and Caprice comprised weakly and moderately altered tissues. Cvs. Pike and Michelet presented an intermediary level of severity symptoms, i.e. moderately altered tissues and few strongly altered tissues.
Discussion
During the last decade, non-conventional imaging techniques such as chlorophyll fluorescence imaging were used for the study of the interactions between plant and pathogens [17]. Indeed, with chlorophyll fluorescence imaging, contrasts are enhanced compared to conventional color images, and depict more accurately the physiology of plant tissues [14,15]. Chlorophyll fluorescence imaging provides images that map on leaves the variations of single parameters associated to photosynthesis. Among the various fluorescence parameters, we monitored variations in the maximum quantum yield of photosystem II photochemistry (Fv/Fm). We used chlorophyll fluorescence imaging to map on bean (P. vulgaris) leaflets the areas altered by Xff CFBP4834-R. It must be pointed out that most stresses that decrease leaf health will affect photosynthesis. Therefore we first checked that visible symptoms of CBB on bean leaflets co-localized with decreased values of the Fv/Fm parameter. Other parameters such as Fv/F0 or F0/Fm were shown to yield a high contrast between healthy tissues and tissues affected with various pathogens [20,45,46]. However, we did not treat these parameters in this study as they do not have a clear physiological significance [20].
Images based on the single Fv/Fm parameter are easier to segment by thresholding approaches than conventional color images, thereby easing the image analysis process. In such a context, we developed an automated thresholding procedure to select pixels corresponding to symptoms. The respective amounts of pixels corresponding to diseased or healthy areas can then be quantified to assess the disease severity on inoculated plants. Even though the decrease in Fv/Fm values due to the pathogen attack is now well documented [32,33], only few studies developed approaches for the quantification of the diseased area on leaves. Most of the studies using the Fv/Fm parameter in plant pathology are based on the mean Fv/Fm value over the whole image [23,33,35-37,47]. The mean Fv/Fm value may qualitatively discriminate between healthy and diseased leaves, but does not quantify the amount of diseased tissues [23,33,35-37,47]. Only few studies attempted to analyze the pixel-wise Fv/Fm-distribution to discriminate between healthy and diseased organs using a threshold of Fv/Fm. For example, in the case of Fusarium culmorum, wheat ears were considered as infected when pixels displaying a Fv/Fm value lower than 0.3 could be observed in the image [18].
In the present study, we investigated thresholding approaches for the quantification of the diseased area on infected leaves. At first, trained raters compared a subset of visible images and Fv/Fm images of bean leaves of cv. Flavert inoculated with Xff CFBP4834-R. We could associate non-overlapping clusters of Fv/Fm values to each stage of the symptom development (necrotic, wilted, impacted and healthy tissues) caused by Xff CFBP4834-R on P. vulgaris cv. Flavert. Subsequently Fv/Fm thresholds could be defined to discriminate between the various stages of the symptom development. Counting the pixels associated to each Fv/Fm cluster enables the quantification of the leaf area corresponding to each stage of the symptom development on cv. Flavert. Defining non-overlapping clusters of Fv/Fm values to segment symptomatic areas can also be performed on a broad range of plant species to quantify areas affected by biotic or abiotic stresses. For example, on grapevine affected by lime-induced iron chlorosis, chlorotic areas displayed lower Fv/Fm values than healthy tissues [48].
Interestingly, lower Fv/Fm values may be observed on tissues located in the margin of symptomatic areas, but that do not display any visible symptoms. These areas evolve into symptoms over time. Therefore, as previously observed on Arabidopsis thaliana or Nicotiana benthamiana inoculated with Pseudomonas syringae, pre-symptomatic areas may also be phenotyped using Fv/Fm[33,36]. Fv/Fm values in these tissues may not differ from those observed in chlorotic tissue and both chlorotic and pre-symptomatic tissues were grouped into impacted tissues in the present study. The decrease of the Fv/Fm values in pre-symptomatic areas is not fully understood. Indeed, neither these areas are yet colonized by bacteria, nor can be observed increased levels of ammonia or a restricting water movement [36].
However, non-overlapping clusters matching the various stages of the symptom development should be defined by trained raters in each pathosystem studied. Indeed, Fv/Fm clusters defined on Flavert do not match the visual observation on other cultivars. For example, the cluster corresponding to visually chlorotic tissues on cv. Michelet overlaps with that corresponding to necrosis on cv. Flavert (Additional file 1: Figure S1). Therefore fixed thresholds defined on a correspondence with visual observations by trained raters are valuable only within a single cultivar, and cannot be extrapolated to other cultivars. Using an expert-based thresholding approach on other cultivars needs a calibration step on a training set of pictures. Such a need is a limitation for this thresholding approach in the perspective of high throughput phenotyping, as visual assessment is time consuming.
Therefore, we aimed at defining Fv/Fm thresholds that could be extrapolated to any cultivar or plant species. As a decrease of Fv/Fm depicts the alteration of PSII, Fv/Fm threshold can be defined independently of visual observations. Clusters of Fv/Fm would depict objective stages of alteration of plant tissues. Moreover, the Fv/Fm parameter may be impacted by the physiological status of plants [49,50] or abiotic stresses [16,34,51]. Fixed thresholds may therefore bias the quantification of the diseased area on leaves. Defining the thresholds on control plants for each experimental round helps avoid such a bias.
To avoid the use of fixed thresholds, we normalized our segmentation on mock-inoculated plants. We defined the threshold as the Fv/Fm value under which a healthy pixel only has a probability of 0.002 to be misclassified. Such a thresholding does not allow the discrimination of various stages of the symptom development. Therefore, within the diseased area, the pixel-wise Fv/Fm-distribution was modeled as a mixture of predicted Gaussian distributions. Such a modeling is largely used for image analysis in medical sciences [52,53]. As well, in plant sciences such a modeling was recently applied to the automated recognition of individual Arabidopsis rosettes, in order to monitor independently the growth of each plant in the image [54]. In the present study, the clustering of pixels according to Gaussian distributions aims at describing the various stages of alteration of plant tissues without any a priori based on visual observations. For each Fv/Fm image, a mixture of Gaussian is fitted independently, and such an approach does not need any calibration set of images. Based on these Gaussian mixture distributions, we could define non-overlapping clusters of pixels displaying similar Fv/Fm values, corresponding to the various stages of alteration of plant tissues.
Pathogen attack may also result in dwarfing or shrinking of leaves. Such a phenotype is rarely quantified [9,12], but the use of non-destructive image analysis approaches may help solve such a caveat. In the present study, we monitored the size of leaflets over time. We considered the maximum size of each leaflet as a reference and the size decrease compared to this reference was considered as shrinking. Such a phenotyping is difficult to assess by visual observation only. A similar approach was used to analyze the leaf area impacted by herbivory [9]. Other approaches were proposed to evaluate the leaf deformation, for example using a sphericity index [12,55]. However, using such an index does not allow the quantification of the leaf area impacted by the shrinking.
To test the applicability of our segmentation approach for the evaluation of plant resistance, we quantified the symptoms caused by Xff CFBP4834-R on five commercial cultivars of bean (cvs. Flavert, Michelet, Pike, Wonder and Caprice). When looking at the total amounts of symptoms, the cv. Flavert appeared to be the most sensitive to Xff CFBP4834-R. The cvs. Caprice and Wonder exhibit few symptoms and can be considered as tolerant. The cvs. Michelet and Pike are impacted to an intermediary extent. On top of the amount of symptomatic tissues, selection for resistance may also focus on the stage of development of the symptom. Indeed, it may be of interest for breeders to notice that cvs. Caprice and Wonder exhibit different symptoms topologies, even though they displayed similar total amounts of symptomatic tissues.
Finally, in order to select for quantitative plant resistance to pathogens, high throughput procedures aiming at precisely quantifying disease severity need to be developed. Robotic imaging procedures can increase the number of images taken [56] but few automatic analysis of chlorophyll fluorescence images procedures are available. The procedure presented in this study was automated under R and the R script is available at http://lisa.univ-angers.fr/PHENOTIC/telechargements.html. Running our procedure on the 1080 images of our dataset, two minutes only are needed for the Expert- and Probability-based thresholding analyses. The use of MCLUST [44] to discriminate various stages of alteration of plant tissues increases up to one hour the calculation time, which remains much faster than rating disease severity by visual observations.
Conclusions
In this paper, we described new procedures to quantify the impact of a pathogen on a plant, easy to automate, objective and accurate.
The expert-based thresholding approach aims at reproducing the visual observations. Such an approach allows the accurate quantification of the various stages of the symptoms development but needs to be calibrated by trained raters on each pathosystem. In contrast, a probability-based thresholding approach may accurately discriminate between healthy and diseased tissues. Within the diseased area, a clustering approach may accurately describe the various stages of alteration of plant tissues. This latter segmentation approach is expert-independent and is normalized on mock-inoculated plants at each day of the experiment. Moreover, the probability-based thresholding approach may allow the phenotyping of pre-symptomatic areas, which cannot be achieved by calibrating thresholds on visual observations by trained raters. A clustering approach applied on the diseased areas allows the quantification of each stage of the alteration of plant tissues. The segmentation approach developed in this study was automated using R, and the script is available at http://lisa.univ-angers.fr/PHENOTIC/telechargements.html.
Such a development of automated segmentation approach speeds up the assessment of disease severity on plants. It may reveal a significant improvement for high throughput testing of the plant resistance to pathogens during breeding.
Methods
Biological material
The bacterial strain Xff CFBP4834-R used in this study was obtained from the French Collection of Bacteria associated to Plants (CFBP, IRHS, Angers, France, http://www.angers.inra.fr/cfbp/, accession n°4885). The strain was grown at 28°C in 10% TSA medium (tryptone at 1.7 g/L, soybean peptone at 0.3 g/L, glucose at 0.25 g/L, NaCl at 0.5 g/L, K2HPO4 at 0.5 g/L, agar at 15 g/L, pH 7.2).
Beans were individually seeded in plastic pots (7 × 7 × 8 cm) containing prewetted compost (NEUHAUS HUMINSUBSTRAT *N4*, NFU 44–551). Plants were grown in a controlled climatic room at 23°C/20°C (day/night) with a photoperiod of 16 h. Plants were watered three times per week and supplemented with N-P-K (18:14:18) at 0.3 g/liter once a week.
Two sets of plants were used in this study. The first set was used to setup approaches for quantification of symptoms by image analysis and was composed by forty plants of bean Flavert. This experiment was performed three times. A second set was used to evaluate the resistance of commercial bean cultivars to Xff CFBP4834-R and was composed by twenty-eight plants of five cultivars of bean obtained from Vilmorin (La Ménitré, France): Flavert, Caprice, Michelet, Pike and Wonder. Twenty-eight plants of each cultivar were used. This experimentation was repeated twice.
Pathogenicity assay
Bacterial suspensions calibrated at 1.108 CFU.ml-1 were made by harvesting bacterial cells from agar plates and suspending them in sterile distilled water. The inoculations were made at the trifoliate step by deeping half of batch of plants during 30 seconds in the diluted bacterial suspension to 1.106 CFU.ml-1. The other half served as control plants and was deeped in water. The first set of plants was incubated at 28°C/25°C (day/night) with a photoperiod of 16 h during 11 days and under high (70%) relative humidity. The second set of plant was incubated at 28°C/25°C (day/night) with a photoperiod of 16 h during 11 days and under high (95%) relative humidity. Plant inoculations were carried out under quarantine at UMR1345 IRHS, Centre INRA, Beaucouzé, France.
Technical setup and image acquisition
The PSI Open FluorCam FC 800-O (PSI, Brno, Czech Republic) was used to capture chlorophyll fluorescence images and to estimate the maximum quantum yield of PSII (Fv/Fm) of inoculated and control leaflets. The system sensor is a CCD camera with a pixel resolution of 512 by 512 and a 12-bit dynamic. The system includes 4 LED panels divided to 2 pairs. One pair provides an orange actinic light with a wavelength of around 618 nm, with an intensity that can vary from 200 to 400 μmol/m2/s. It provides a 2s pulse that allows the measurement of the initial fluorescent state (F0). The other pair provides a saturating pulse during 1s in blue wavelength, typically 455 nm, with an intensity of up to 3000 μmol/m2/s. The saturating pulse allows collecting of the maximum fluorescence (Fm). Fluorescence chlorophyll imaging was used in a dark adapted mode after a dark period of 45 min [28] to produce maps with the fluorescent quantum efficiency Fv/Fm = (Fm - F0)/Fm. For all image acquisitions, the observed leaflet is maintained horizontally for this sub-section. A kinetic analysis was performed: image acquisitions were done at 1 dai, and everyday after the fourth dai. The experiment was repeated three times, and in total 1080 images were collected. Another analysis consisted in collecting images of detached leaflets at 1, 7 and 11 dai, in total 280 images were collected. False color images representing Fv/Fm values of the pixels and pixel-wise Fv/Fm-distributions were extracted for each leaflet. For a subset of leaflets, visible images were also taken with a digital camera to compare visual symptoms and Fv/Fm values.
Shrinking of the leaflets
The shrinking of the leaflets was calculated when a kinetic analysis was performed on each leaflet. The shrinking corresponded to the difference between the maximum size of the leaflet during the experiment and the current size of the leaflet. The shrinking of the leaflet was added to the class presenting the lower Fv/Fm values. The amounts of the other stages of the symptom development were calculated using the maximum size of the leaflet.
Thresholding based on expert visual observations
The expert-based thresholding approach consisted in the determination of values characterizing the diseased tissues. Three stages of the symptom development were discriminated: necrotic, wilted and impacted tissues. Conventional color images and Fv/Fm images were manually compared by trained raters to determine clusters of Fv/Fm values matching with each stage of the symptom development. On the conventional color image, the various stages of the symptom development were manually delimited and the delimitations were superimposed on the Fv/Fm image. The maximum and minimum Fv/Fm values of each area that co-localized with each stage of the symptom development were determined. These values corresponded to the expert-based thresholds. The amounts of pixels contained in each subgroup were calculated.
Thresholding based on modeling pixel-wise Fv/Fm distributions
The R package MCLUST [44] was run to select the number of Gaussian distributions that compose the Gaussian mixture model best supported by the data and to estimate the mean, variance and weight in the mixture distribution of each cluster. For each pixel, a probability of membership to each cluster is also returned. Gaussian mixture models including from 1 to 4 clusters with unequal variance were fitted to each leaflet pixel-wise Fv/Fm-distribution. The clusters representing less than 1% of the pixel-wise Fv/Fm-distribution were suppressed as they were considered as artifactual clusters. Pixels initially attributed to these non-significant clusters were assigned according to their second better probability of membership. The clusters of the Gaussian mixture model selected were then merged if their fusion was unimodal according to the ridgeline unimodal method implemented in the R package fpc [42,43]. A threshold was calculated for each dai to discriminate between distributions characterizing the healthy tissues and distributions characterizing the diseased tissues. The means of the clusters found on mock-inoculated samples were averaged. Then, the confidence interval was calculated. A cluster displaying a mean inferior to the threshold was considered as characterizing diseased tissues.
Thresholding based on the probability of misclassification of a healthy pixel
The probability based-thresholds were built based a pixel-wise Fv/Fm-distribution resulting from the merging of the pixel-wise Fv/Fm-distributions of all the mock-inoculated leaflets. The thresholds corresponded to the 100-quantile, 500-quantile or 1000-quantile, i.e. the Fv/Fm values splitting 1/100, 1/500 and 1/1000 of the pixels of the distribution. The thresholds were determined for each dai and each cultivar to take into consideration the possible daily variations and the differences between cultivars. A pixel displaying a Fv/Fm value inferior to the threshold was considered as diseased. The total diseased proportion was then calculated for each leaflet. Then, a clustering method using the R package MCLUST [44] was performed on the diseased area detected by the Probability-based thresholds to segment the diseased area according to three stages of alteration of plant tissues. Gaussian mixture models including from 1 to 3 clusters with unequal variance were fitted to the pixel-wise Fv/Fm-distribution of each leaflet. In order to classify the clusters into the various stages of alteration of plant tissues, a second clustering procedure with an optimal number of clusters between 1 and 3 was performed on the means of all the clusters. The clusters were then classified according to their membership to the clusters representing the stages of alteration of plant tissues.
Visualization of the diseased tissues and statistical test
For the three thresholding steps, the stages of the symptom development were colored using the R package EBImage [57]. The pixels were colored according to the various thresholds.
Mann–Whitney test [58] was performed to compare amounts of diseased tissues between mock-inoculated and Xff CFBP4834-R-inoculated leaflets and between the various cultivars of bean.
Abbreviations
PSII: Photosystem II; Fm: Maximum fluorescence; Fv: Minimum fluorescence; Dai: Day(s) after inoculation; Xff: Xanthomonas fuscans subsp. fuscans.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CR conceived and designed the experiments, carried out acquisition of chlorophyll fluorescence data, conceived and carried out the R-analysis, interpreted the data, wrote and revised the manuscript. EBe helped setting up the acquisition of chlorophyll fluorescence data, and revised the manuscript. EBo helped setting up the acquisition of chlorophyll fluorescence data and carried out acquisition of chlorophyll fluorescence data. DR helped setting up the acquisition of chlorophyll fluorescence data, and revised the manuscript. FF designed the statistical analysis of the data and revised the manuscript. RB designed the statistical analysis of the data and revised the manuscript. JG helped in the acquisition of the chlorophyll fluorescence data. CM revised the manuscript. MAJ conceived and designed the experiments, and revised the manuscript. TB conceived and designed the experiments, carried out the acquisition of the chlorophyll fluorescence data, conceived the statistical analysis, interpreted the data, and wrote and revised the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Expert-based thresholds needs to be calibrated on each cultivar. Symptoms of Xff CFBP4834-R on leaflets of cv. Flavert (A) and Michelet (B). Beans were inoculated at 1.106 CFU ml_1 and leaflets were sampled at 11 dai. Expert-based thresholds are defined after comparison by trained raters of Fv/Fm images and visual observations only on P. vulgaris cv. Flavert harboring symptoms of Xff CFBP4834-R. Using expert-based thresholds defined on cv. Flavert, chlorotic tissues on cv. Michelet are misclassified and considered as necrotic. A: visible image of a leaflet of cv. Flavert obtained by conventional color imaging and by chlorophyll fluorescence imaging. The various stages of the symptom development segmented using expert-based thresholds co-localize with visual observations. B: visible image of a leaflet of cv. Michelet obtained by conventional color imaging and by chlorophyll fluorescence imaging. The major part of the diseased tissues is composed by chlorotic tissues and misclassify as necrotic tissues with expert-based thresholds calibrated on cv. Flavert.
Contributor Information
Céline Rousseau, Email: celine.rousseau@angers.inra.fr.
Etienne Belin, Email: etienne.belin@univ-angers.fr.
Edouard Bove, Email: edouard.bove@gmail.com.
David Rousseau, Email: david.rousseau@univ-angers.fr.
Frédéric Fabre, Email: frederic.fabre@avignon.inra.fr.
Romain Berruyer, Email: romain.berruyer@univ-angers.fr.
Jacky Guillaumès, Email: jacky.guillaumes@angers.inra.fr.
Charles Manceau, Email: charles.manceau@anses.fr.
Marie-Agnès Jacques, Email: marie-agnes.jacques@angers.inra.fr.
Tristan Boureau, Email: tristan.boureau@univ-angers.fr.
Acknowledgements
This work was supported by Angers Loire Métropole, Conseil Général Département de Maine-et-Loire and Région Pays de la Loire (Phenotic) and by Institut National de la Recherche Agronomique and Région Pays de la Loire. We are grateful to Valérie Grimault at Station Nationale Essais de Semences, Valérie Caffier, Carolyne Dürr and Philippe Simoneau (IRHS, Angers, France) and Laurent Noël (LIPM, Toulouse, France) for helpful discussions. We thank Marion Le Saux, Perrine Portier, Martial Briand and Géraldine Taghouti at French Collection of Plant-associated Bacteria (CFBP) for providing the bacterial strain. We wish to thank Elise Vandoeuvre at Vilmorin for providing the seeds. We thank Installations Expérimentales Mutualisées (INEM) for growing the plants.
References
- Brun H, Chèvre A-M, Fitt BDL, Powers S, Besnard A-L, Ermel M, Huteau V, Marquer B, Eber F, Renard M, Andrivon D. Quantitative resistance increases the durability of qualitative resistance to Leptosphaeria maculans in Brassica napus. New Phytol. 2010;185:285–299. doi: 10.1111/j.1469-8137.2009.03049.x. [DOI] [PubMed] [Google Scholar]
- Palloix A, Ayme V, Moury B. Durability of plant major resistance genes to pathogens depends on the genetic background, experimental evidence and consequences for breeding strategies. New Phytol. 2009;183:190–199. doi: 10.1111/j.1469-8137.2009.02827.x. [DOI] [PubMed] [Google Scholar]
- Quenouille J, Montarry J, Palloix A, Moury B. Farther, slower, stronger: how the plant genetic background protects a major resistance gene from breakdown. Mol Plant Pathol. 2013;14:109–118. doi: 10.1111/j.1364-3703.2012.00834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland JA, Nelson RJ. In the eye of the beholder: the effect of rater variability and different rating scales on QTL mapping. Phytopathology. 2011;101:290–298. doi: 10.1094/PHYTO-03-10-0087. [DOI] [PubMed] [Google Scholar]
- Bock CH, Parker PE, Cook AZ, Gottwald TR. Visual rating and the Use of image analysis for assessing different symptoms of citrus canker on grapefruit leaves. Plant Disease. 2001;92:412–424. doi: 10.1094/PDIS-92-4-0530. [DOI] [PubMed] [Google Scholar]
- Bock CH, Poole GH, Parker PE, Gottwald TR. Plant disease severity estimated visually, by digital photography and image analysis, and by hyperspectral imaging. Crit Rev Plant Sci. 2010;29:59–107. doi: 10.1080/07352681003617285. [DOI] [Google Scholar]
- Sherwood RT, Berg CC, Hoover MR, Zeiders KE. Illusions in visual assessment of Stagonospora leaf spot of Orchardgrass. Phytopathology. 1983;73:173–177. doi: 10.1094/Phyto-73-173. [DOI] [Google Scholar]
- Camargo A, Smith JS. An image-processing based algorithm to automatically identify plant disease visual symptoms. Biosystems Engineer. 2009;102:9–21. doi: 10.1016/j.biosystemseng.2008.09.030. [DOI] [Google Scholar]
- Green JM, Appel H, Rehrig EM, Harnsomburana J, Chang J-F, Balint-Kurti P, Shyu C-R. PhenoPhyte: a flexible affordable method to quantify 2D phenotypes from imagery. Plant Methods. 2012;8:45. doi: 10.1186/1746-4811-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Yu K, Pauls KP, Navabi A. Application of image analysis in studies of quantitative disease resistance, exemplified using common bacterial blight-common bean pathosystem. Phytopathology. 2012;102:434–442. doi: 10.1094/PHYTO-06-11-0175. [DOI] [PubMed] [Google Scholar]
- Wijekoon CP, Goodwin PH, Hsiang T. Quantifying fungal infection of plant leaves by digital image analysis using Scion Image software. J Microbiol Methods. 2008;74:94–101. doi: 10.1016/j.mimet.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Contreras-Medina LM, Osornio-Rios RA, Torres-Pacheco I, Romero-Troncoso RDJ, Guevara-González RG, Millan-Almaraz JR. Smart sensor for real-time quantification of common symptoms present in unhealthy plants. Sensors (Basel, Switzerland) 2012;12:784–805. doi: 10.3390/s120100784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thilagamani S, Shanthi N. A survey on image segmentation through clustering. Int J Res Rev Info Sci. 2011;1:16–19. [Google Scholar]
- Chaerle L, Van Der Straeten D. Seeing is believing: imaging techniques to monitor plant health. Biochim Biophys Acta. 2001;1519:153–166. doi: 10.1016/S0167-4781(01)00238-X. [DOI] [PubMed] [Google Scholar]
- Sankaran S, Mishra A, Ehsani R, Davis C. A review of advanced techniques for detecting plant diseases. Computers Electro Agri. 2010;72:1–13. doi: 10.1016/j.compag.2010.02.007. [DOI] [Google Scholar]
- Gorbe E, Calatayud A. Applications of chlorophyll fluorescence imaging technique in horticultural research: A review. Sci Hortic. 2012;138:24–35. [Google Scholar]
- Rolfe SA, Scholes JD. Chlorophyll fluorescence imaging of plant–pathogen interactions. Protoplasma. 2010;247:163–175. doi: 10.1007/s00709-010-0203-z. [DOI] [PubMed] [Google Scholar]
- Bauriegel E, Giebel A, Herppich WB. Hyperspectral and chlorophyll fluorescence imaging to analyse the impact of Fusarium culmorum on the photosynthetic integrity of infected wheat ears. Sensors (Basel, Switzerland) 2011;11:3765–3779. doi: 10.3390/s110403765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin É, Rousseau D, Boureau T, Caffier V. Thermography versus chlorophyll fluorescence imaging for detection and quantification of apple scab. Computers Electron Agri. 2013;90:159–163. [Google Scholar]
- Berger S, Benediktyová Z, Matouš K, Bonfig KB, Mueller MJ, Nedbal L, Roitsch T. Visualization of dynamics of plant-pathogen interaction by novel combination of chlorophyll fluorescence imaging and statistical analysis: differential effects of virulent and avirulent strains of P. syringae and of oxylipins on A. thaliana. J Exp Botany. 2007;58:797–806. doi: 10.1093/jxb/erl208. [DOI] [PubMed] [Google Scholar]
- Chaerle L, Hagenbeek D, De Bruyne E, Valcke R, Van Der Straeten D. Thermal and chlorophyll-fluorescence imaging distinguish plant-pathogen interactions at an early stage. Plant Cell Physiol. 2004;45:887–896. doi: 10.1093/pcp/pch097. [DOI] [PubMed] [Google Scholar]
- Pérez-Bueno ML, Ciscato M, VandeVen M, García-Luque I, Valcke R, Barón M. Imaging viral infection: studies on Nicotiana benthamiana plants infected with the pepper mild mottle tobamovirus. Photosynthesis Res. 2006;90:111–123. doi: 10.1007/s11120-006-9098-0. [DOI] [PubMed] [Google Scholar]
- Pineda M, Olejníčková J, Cséfalvay L, Barón M. Tracking viral movement in plants by means of chlorophyll fluorescence imaging. J Plant Physiol. 2011;168:2035–2040. doi: 10.1016/j.jplph.2011.06.013. [DOI] [PubMed] [Google Scholar]
- Bilgin DD, Zavala JA, Zhu J, Clough SJ, Ort DR, De Lucia EH. Biotic stress globally downregulates photosynthesis genes. Plant Cell Environ. 2010;33:1597–1613. doi: 10.1111/j.1365-3040.2010.02167.x. [DOI] [PubMed] [Google Scholar]
- Jelenska J, Yao N, Vinatzer BA, Wright CM, Brodsky JL, Greenberg JT. A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. CB. 2007;17:499–508. doi: 10.1016/j.cub.2007.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gürlebeck D, Jahn S, Gürlebeck N, Szczesny R, Szurek B, Hahn S, Hause G, Bonas U. Visualization of novel virulence activities of the Xanthomonas type. Mol Plant Pathol. 2009;10:175–188. doi: 10.1111/j.1364-3703.2008.00519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler WL. Energy distribution in the photochemical apparatus of photosynthesis. Ann Rev Plant Physiol. 1978;29:345–378. doi: 10.1146/annurev.pp.29.060178.002021. [DOI] [Google Scholar]
- Baker NR. Chlorophyll fluorescence: a probe of photosynthesis in vivo. Ann Rev Plant Biol. 2008;59:89–113. doi: 10.1146/annurev.arplant.59.032607.092759. [DOI] [PubMed] [Google Scholar]
- Matouš K, Benediktyova Z, Berger S, Roitsch T, Nedbal L. Case study of combinatorial imaging: what protocol and what chlorophyll fluorescence image to use when visualizing infection of Arabidopsis thaliana by pseudomonas syringae? Photosynthesis Res. 2006;243:243–253. doi: 10.1007/s11120-006-9120-6. [DOI] [PubMed] [Google Scholar]
- Björkman O, Demmig B. Planta characteristics at 77 K among vascular plants of diverse origins. Planta. 1987;170:489–504. doi: 10.1007/BF00402983. [DOI] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN. Chlorophyll fluorescence - a practical guide. J Exp Bot. 2000;51:659–668. doi: 10.1093/jexbot/51.345.659. [DOI] [PubMed] [Google Scholar]
- Kuckenberg J, Tartachnyk I, Noga G. Temporal and spatial changes of chlorophyll fluorescence as a basis for early and precise detection of leaf rust and powdery mildew infections in wheat leaves. Precision Agri. 2008;10:34–44. [Google Scholar]
- Bonfig KB, Schreiber U, Gabler A, Roitsch T, Berger S. Infection with virulent and avirulent P. syringae strains differentially affects photosynthesis and sink metabolism in Arabidopsis leaves. Planta. 2006;225:1–12. doi: 10.1007/s00425-006-0303-3. [DOI] [PubMed] [Google Scholar]
- Woo NS, Badger MR, Pogson BJ. A rapid, non-invasive procedure for quantitative assessment of drought survival using chlorophyll fluorescence. Plant Methods. 2008;4:27. doi: 10.1186/1746-4811-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuchelian K, Porta NLA, Bertamini M, Nedunchezhian N. Cypress canker induced inhibition of photosynthesis in field grown cypress (Cupressus sempervirens L.) needles. Physiol Mol Plant Pathol. 2005;67:33–39. doi: 10.1016/j.pmpp.2005.08.007. [DOI] [Google Scholar]
- Iqbal MJ, Goodwin PH, Leonardos ED, Grodzinski B. Spatial and temporal changes in chlorophyll fluorescence images of Nicotiana benthamiana leaves following inoculation with Pseudomonas syringae pv. tabaci. Plant Pathol. 2012;61:1052–1062. doi: 10.1111/j.1365-3059.2012.02592.x. [DOI] [Google Scholar]
- Zou J, Rodriguez-Zas S, Aldea M, Li M, Zhu J, Gonzalez DO, Vodkin LO, DeLucia E, Clough SJ. Expression profiling soybean response to Pseudomonas syringae reveals new defense-related genes and rapid HR-specific downregulation of photosynthesis. MPMI. 2005;18:1161–1174. doi: 10.1094/MPMI-18-1161. [DOI] [PubMed] [Google Scholar]
- European and Mediterranean Plant Protection Organization. http://www.eppo.int/
- Broughton WJ, Hern G, Blair M, Beebe S, Gepts P, Vanderleyden J. Beans (Phaseolus spp.) – model food legumes. Plant Soil. 2003;252:55–128. [Google Scholar]
- Darrasse A, Bureau C, Samson R, Morris CE, Jacques M-A. Contamination of bean seeds by Xanthomonas axonopodis pv. phaseoli associated with low bacterial densities in the phyllosphere under field and greenhouse conditions. Eur J Plant Pathol. 2007;119:203–215. doi: 10.1007/s10658-007-9164-2. [DOI] [Google Scholar]
- Duncan RW, Singh SP, Gilbertson RL. Interaction of common bacterial blight bacteria with disease resistance quantitative trait Loci in common bean. Phytopathology. 2011;101:425–435. doi: 10.1094/PHYTO-03-10-0095. [DOI] [PubMed] [Google Scholar]
- Hennig C. Methods for merging Gaussian mixture components. Adv Data Anal Classification. 2010;4:3–34. doi: 10.1007/s11634-010-0058-3. [DOI] [Google Scholar]
- Ray S, Lindsay BG. The topography of multivariate normal mixtures. Ann Stat. 2005;33:2042–2065. doi: 10.1214/009053605000000417. [DOI] [Google Scholar]
- Fraley C, Raftery AE. MCLUST version 3 for R: normal mixture modeling and technical report No. 504. Department. 2006. pp. 1–57. http://www.stat.washington.edu/www/research/reports/2006/tr504.pdf.
- Nedbal L, Soukupová J, Whitmarsh J, Trtilek M. Postharvest imaging of chlorophyll fluorescence from lemons can be used to predict fruit quality. Photosynthetica. 2000;38:571–579. doi: 10.1023/A:1012413524395. [DOI] [Google Scholar]
- Soukupova J, Smatanova S, Nedbal L, Jegorov A. Plant response to destruxins visualized by imaging of chlorophyll fluorescence. Physiol Plant. 2003;118:399–405. doi: 10.1034/j.1399-3054.2003.00119.x. [DOI] [Google Scholar]
- Mandal K, Saravanan R, Maiti S, Kothari IL, Plants A, Vidyanagar V. Effect of downy mildew disease on photosynthesis and chlorophyll fluorescence in Plantago. J Plant Dis Protect. 2009;116:164–168. [Google Scholar]
- Bavaresco L, Bertamini M, Iacono F. Lime-induced chlorosis and physio- logical responses in grapevine (Vitis vinifera L . cv . Pinot blanc) leaves. Vitis. 2006;45:45–46. [Google Scholar]
- Aranda I, Castro L, Alía R, Pardos JA, Gil L. Low temperature during winter elicits differential responses among populations of the Mediterranean evergreen cork oak (Quercus suber) Tree Physiol. 2005;25:1085–1090. doi: 10.1093/treephys/25.8.1085. [DOI] [PubMed] [Google Scholar]
- Ogaya R, Peñuelas J. Comparative field study of Quercus ilex and Phillyrea latifolia: photosynthetic response to experimental drought conditions. Environment Exp Bot. 2003;50:137–148. doi: 10.1016/S0098-8472(03)00019-4. [DOI] [Google Scholar]
- Jonaviciene K, Studer B, Asp T, Jensen LB, Paplauskienơ V, Lazauskas S. Identification of genes involved in a water stress response in timothy and mapping of orthologous loci in perennial ryegrass. Biologia Plantarum. 2012;56:473–483. doi: 10.1007/s10535-012-0110-6. [DOI] [Google Scholar]
- Wehrens R, Simonetti AW, Buydens LMC. Mixture modelling of medical magnetic resonance data. J Chem. 2002;16:274–282. doi: 10.1002/cem.721. [DOI] [Google Scholar]
- Li Q, Fraley C, Bumgarner RE, Yeung KY, Raftery AE. Donuts, scratches and blanks: robust model-based segmentation of microarray images. Bioinformatics (Oxford, England) 2005;21:2875–2882. doi: 10.1093/bioinformatics/bti447. [DOI] [PubMed] [Google Scholar]
- De Vylder J, Vandenbussche F, Hu Y, Philips W, Van Der Straeten D. Rosette tracker: an open source image analysis tool for automatic quantification of genotype effects. Plant Physiol. 2012;160:1149–1159. doi: 10.1104/pp.112.202762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt WK. Digital Image Processing: PIKS Inside. Third. New York, NY, USA: John Wiley; 2001. pp. 623–650. 5. [Google Scholar]
- Harbinson J, Prinzenberg AE, Kruijer W, Aarts MGM. High throughput screening with chlorophyll fluorescence imaging and its use in crop improvement. Curr Opin Biotechnol. 2012;23:221–226. doi: 10.1016/j.copbio.2011.10.006. [DOI] [PubMed] [Google Scholar]
- Pau G, Fuchs F, Sklyar O, Boutros M, Huber W. EBImage - an R package for image processing with applications to cellular phenotypes. Bioinformatics (Oxford, England) 2010;26:979–981. doi: 10.1093/bioinformatics/btq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry: the principles and practice of statistics in biological research. 4. New York: W. H. Freeman and Co; 2012. p. 937. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expert-based thresholds needs to be calibrated on each cultivar. Symptoms of Xff CFBP4834-R on leaflets of cv. Flavert (A) and Michelet (B). Beans were inoculated at 1.106 CFU ml_1 and leaflets were sampled at 11 dai. Expert-based thresholds are defined after comparison by trained raters of Fv/Fm images and visual observations only on P. vulgaris cv. Flavert harboring symptoms of Xff CFBP4834-R. Using expert-based thresholds defined on cv. Flavert, chlorotic tissues on cv. Michelet are misclassified and considered as necrotic. A: visible image of a leaflet of cv. Flavert obtained by conventional color imaging and by chlorophyll fluorescence imaging. The various stages of the symptom development segmented using expert-based thresholds co-localize with visual observations. B: visible image of a leaflet of cv. Michelet obtained by conventional color imaging and by chlorophyll fluorescence imaging. The major part of the diseased tissues is composed by chlorotic tissues and misclassify as necrotic tissues with expert-based thresholds calibrated on cv. Flavert.