Abstract
Background
Secondary metabolite production, a hallmark of filamentous fungi, is an expanding area of research for the Aspergilli. These compounds are potent chemicals, ranging from deadly toxins to therapeutic antibiotics to potential anti-cancer drugs. The genome sequences for multiple Aspergilli have been determined, and provide a wealth of predictive information about secondary metabolite production. Sequence analysis and gene overexpression strategies have enabled the discovery of novel secondary metabolites and the genes involved in their biosynthesis. The Aspergillus Genome Database (AspGD) provides a central repository for gene annotation and protein information for Aspergillus species. These annotations include Gene Ontology (GO) terms, phenotype data, gene names and descriptions and they are crucial for interpreting both small- and large-scale data and for aiding in the design of new experiments that further Aspergillus research.
Results
We have manually curated Biological Process GO annotations for all genes in AspGD with recorded functions in secondary metabolite production, adding new GO terms that specifically describe each secondary metabolite. We then leveraged these new annotations to predict roles in secondary metabolism for genes lacking experimental characterization. As a starting point for manually annotating Aspergillus secondary metabolite gene clusters, we used antiSMASH (antibiotics and Secondary Metabolite Analysis SHell) and SMURF (Secondary Metabolite Unknown Regions Finder) algorithms to identify potential clusters in A. nidulans, A. fumigatus, A. niger and A. oryzae, which we subsequently refined through manual curation.
Conclusions
This set of 266 manually curated secondary metabolite gene clusters will facilitate the investigation of novel Aspergillus secondary metabolites.
Keywords: Aspergillus, Gene clusters, Gene Ontology, Genome annotation, Secondary metabolism, Sybil
Background
Secondary metabolites produced by fungi are a rich source of medically useful compounds because of their pharmaceutical and toxicological properties [1]. While secondary metabolites are not required for an organism’s growth or primary metabolism, they may provide important benefits in its environmental niche. For example, A. nidulans laeA mutants defective in the production of secondary metabolites are ingested more readily by the fungivorous arthropod, Folsomia candida, suggesting that secondary metabolite production can protect fungi from predation [2].
The Aspergilli are producers of a wide variety of secondary metabolites of considerable medical, industrial, agricultural and economic importance. For example, the antibiotic penicillin is produced by A. nidulans and the genes involved in the penicillin biosynthetic pathway have been extensively studied [3-5]. Sterigmatocystin (ST), an aflatoxin (AF) precursor, and many of the genes that are involved in its biosynthesis have also been extensively studied in A. nidulans[6-10]. AF is a secondary metabolite produced mainly by Aspergillus species growing in foodstuffs [11], and it is of both medical and economic importance as contaminated food sources are toxic to humans and animals when ingested. Gliotoxin is an extremely toxic secondary metabolite produced by several Aspergillus species during infection [12,13]. The ability of this toxin to modulate the host immune system and induce apoptosis in a variety of cell-types has been most studied in the ubiquitous fungal pathogen, A. fumigatus[14,15].
The availability of Aspergillus genomic sequences has greatly facilitated the identification of numerous genes involved in the production of other secondary metabolites. Based on the number of predicted secondary metabolite biosynthesis genes and the fact that the expression of many secondary metabolite gene clusters is cryptic [16], meaning that expression is not evident under standard experimental conditions, there appears to be the potential for production of many more secondary metabolites than currently known [17]. Secondary metabolite biosynthetic genes often occur in clusters that tend to be sub-telomerically located and are coordinately regulated under certain laboratory conditions [18-20]. Typically, a secondary metabolite biosynthetic gene cluster contains a gene encoding one of several key “backbone” enzymes of the secondary metabolite biosynthetic process: a polyketide synthase (PKS), a non-ribosomal peptide synthetase (NRPS), a polyketide synthase/non-ribosomal peptide synthetase hybrid (PKS-NRPS), a prenyltransferase known as dimethylallyl tryptophan synthase (DMATS) and/or a diterpene synthase (DTS).
Comparative sequence analysis based on known backbone enzymes has been used to identify potential secondary metabolite biosynthetic gene clusters for subsequent experimental verification. One approach for experimental verification is the deletion of genes with suspected roles in secondary metabolite biosynthesis followed by identification of the specific secondary metabolite profiles of the mutants by thin layer chromatography, NMR or other methods [7,8]. For example, the deletion of A. fumigatus encA, which encodes an ortholog of the A. nidulans non-reducing PKS (NR-PKS) mdpG, followed by analysis of culture extracts using high-performance liquid chromatography (HPLC) enabled the recent identification of endocrocin and its biosynthetic pathway intermediates [21]. Similarly, the deletion of the gene encoding the PKS, easB, enabled the identification of the emericellamide biosynthetic pathway of A. nidulans[22]. Another approach is the overexpression of predicted transcriptional regulators of secondary metabolism gene clusters with subsequent analysis of the gene expression and secondary metabolite profiles of the resulting strains, which has facilitated the identification of numerous secondary metabolites and the genes responsible for their synthesis [23,24]. For example, overexpression of laeA in A. nidulans, a global transcriptional regulator of secondary metabolism production, coupled with microarray analysis, facilitated the delineation of the cluster responsible for production of the anti-tumor compound, terrequinone A [18]. Thus, genome sequence analysis, coupled with targeted experimentation, has been a highly effective strategy for identifying novel secondary metabolites and the genes involved in their synthesis.
The Aspergillus Genome Database (AspGD; http://www.aspgd.org) is a web-based resource that provides centralized access to gene and protein sequences, analysis tools and manually curated information derived from the published scientific literature for A. nidulans, A. fumigatus, A. niger and A. oryzae[25,26]. AspGD curators read the published experimental literature to record information including gene names and synonyms, write free-text descriptions of each gene, record phenotypes and assign terms that describe functional information about genes and proteins using the Gene Ontology (GO; http://www.geneontology.org). These annotations are an important resource for the scientific research community, used both for reference on individual genes of interest as well as for analysis of results from microarray, proteomic experiments, or other screens that produce large lists of genes.
The GO is a structured vocabulary for describing the functions associated with genes products [27]. GO terms describe the activity of a gene product (Molecular Function; MF) within the cell, the biological process (Biological Process; BP) in which a gene product is involved and the location within the cell (Cellular Component; CC) where the gene product is observed [28]. Evidence codes are assigned to GO annotations based on the type of available experimental evidence.
At the start of this project most of the terms needed to describe secondary metabolite biosynthetic genes or regulators of secondary metabolism did not yet exist in the GO. Thus, in order to provide an improved annotation of secondary metabolite biosynthetic genes and their regulatory proteins, we developed new GO terms for secondary metabolite production in collaboration with the GO Consortium, and reannotated the entire set of genes associated with secondary metabolism in AspGD. We then performed a comprehensive analysis of the secondary metabolism biosynthetic genes and their orthologs across the genomes of A. nidulans, A. fumigatus, A. niger and A. oryzae and now provide a set of manually annotated secondary metabolite gene clusters. We anticipate that these new, more precise annotations will encourage the rapid and efficient experimental verification of novel secondary metabolite biosynthetic gene clusters in Aspergillus and the identification of the corresponding secondary metabolites.
Results
Identifying genes for reannotation
Many branches of the GO, such as apoptosis and cardiac development [29], have recently been expanded and revised to include new terms that are highly specific to these processes. The secondary metabolism literature has expanded over the last several years, allowing AspGD curators to make annotations to an increasing number of genes with roles in secondary metabolism. During routine curation, it became apparent that hundreds of Aspergillus genes that were candidates for annotation to the GO term ‘secondary metabolic process’ had the potential for more granular annotations, since, in many cases, the specific secondary metabolite produced by a gene product is known. At the inception of this project, only terms for ‘aflatoxin biosynthetic process, ’ ‘penicillin biosynthetic process’ and ‘sterigmatocystin biosynthetic process, ’ the 3 most well-studied secondary metabolites to date, were present in the GO (Additional file 1).
Candidate genes for reannotation were identified as those that had pre-existing GO annotations to ‘secondary metabolic process’ or curated mutant phenotypes that impact secondary metabolite production. For example, numerous genes in AspGD are annotated with mutant phenotypes affecting the production of secondary metabolites such as asperthecin [30], austinol and dehydroaustinol [31], emericellin [32], fumiquinazolines [33], orsellinic acid [34], pseurotin A [35], shamixanthones [32,36] and violaceol [37] among others. These genes were then analyzed and a list of new GO terms was generated to annotate these genes more specifically (Table 1 and Additional file 1).
Table 1.
Number of Aspergillus genes with manual and computational GO annotations to ‘secondary metabolic process’
| GO term ID | GO term name | Manual GO annotations | A. nidulans IEA annotations | A. fumigatus IEA annotations | A. niger IEA annotations | A. oryzae IEA annotations | Total IEA annotations* |
|---|---|---|---|---|---|---|---|
| GO:1900596 |
(+)-kotanin biosynthetic process |
1 |
1 |
1 |
0 |
1 |
3 |
| GO:1900581 |
(17Z)-protosta-17(20),24-dien-3beta-ol biosynthetic process |
1 |
0 |
0 |
0 |
0 |
0 |
| GO:0045122 |
aflatoxin biosynthetic process (PT) |
1 |
6 |
2 |
0 |
2 |
10 |
| GO:1900587 |
arugosin biosynthetic process |
1 |
0 |
3 |
2 |
1 |
6 |
| GO:1900554 |
asperfuranone biosynthetic process |
7 |
0 |
10 |
29 |
23 |
62 |
| GO:0036184 |
asperthecin biosynthetic process |
5 |
0 |
11 |
10 |
6 |
27 |
| GO:1900560 |
austinol biosynthetic process |
15 |
0 |
10 |
13 |
12 |
35 |
| GO:1900805 |
brevianamide F biosynthetic process |
1 |
2 |
0 |
4 |
4 |
10 |
| GO:1900566 |
chanoclavine-I biosynthetic process |
3 |
1 |
0 |
2 |
2 |
5 |
| GO:1900563 |
dehydroaustinol biosynthetic process |
16 |
0 |
11 |
14 |
13 |
38 |
| GO:1900599 |
demethylkotanin biosynthetic process |
1 |
1 |
1 |
0 |
1 |
2 |
| GO:1900617 |
emericellamide A biosynthetic process |
2 |
0 |
3 |
8 |
6 |
17 |
| GO:1900557 |
emericellamide biosynthetic process |
6 |
0 |
6 |
20 |
12 |
38 |
| GO:1900575 |
emodin biosynthetic process |
1 |
0 |
3 |
2 |
1 |
6 |
| GO:1900602 |
endocrocin biosynthetic process |
4 |
3 |
0 |
3 |
4 |
10 |
| GO:0035837 |
ergot alkaloid biosynthetic process (PT) |
5 |
6 |
0 |
4 |
7 |
17 |
| GO:1900611 |
F-9775A biosynthetic process |
3 |
0 |
0 |
0 |
0 |
0 |
| GO:1900614 |
F-9775B biosynthetic process |
3 |
0 |
0 |
0 |
0 |
0 |
| GO:0031171 |
ferricrocin biosynthetic process |
4 |
2 |
1 |
4 |
4 |
11 |
| GO:1900809 |
fumigaclavine C biosynthetic process |
2 |
2 |
0 |
1 |
1 |
4 |
| GO:1900778 |
fumiquinazoline A biosynthetic process |
2 |
2 |
0 |
2 |
2 |
6 |
| GO:2001310 |
gliotoxin biosynthetic process |
11 |
5 |
0 |
5 |
10 |
20 |
| GO:0006583 |
melanin biosynthetic process from tyrosine |
1 |
1 |
0 |
1 |
0 |
2 |
| GO:1900815 |
monodictyphenone biosynthetic process |
15 |
0 |
20 |
29 |
23 |
72 |
| GO:1900551 |
N',N'',N'''-triacetylfusarinine C biosynthetic process |
4 |
5 |
0 |
5 |
5 |
15 |
| GO:1900787 |
naphtho-gamma-pyrone biosynthetic process |
2 |
2 |
2 |
0 |
2 |
6 |
| GO:1900584 |
o-orsellinic acid biosynthetic process |
4 |
0 |
6 |
18 |
9 |
33 |
| GO:1900821 |
orlandin biosynthetic process |
1 |
1 |
1 |
0 |
1 |
3 |
| GO:0042318 |
penicillin biosynthetic process (PT) |
17 |
10 |
14 |
14 |
16 |
54 |
| GO:0030639 |
polyketide biosynthetic process (PT) |
18 |
0 |
0 |
1 |
0 |
1 |
| GO:1900793 |
shamixanthone biosynthetic process |
5 |
0 |
10 |
5 |
8 |
23 |
| GO:0019290 |
siderophore biosynthetic process (PT) |
5 |
3 |
0 |
4 |
3 |
10 |
| GO:0045461 |
sterigmatocystin biosynthetic process (PT) |
36 |
0 |
39 |
57 |
50 |
146 |
| GO:1900605 |
tensidol A biosynthetic process |
1 |
1 |
1 |
0 |
1 |
3 |
| GO:1900796 |
terrequinone A biosynthetic process |
1 |
0 |
0 |
0 |
1 |
1 |
| GO:1900590 |
violaceol I biosynthetic process |
2 |
0 |
1 |
2 |
4 |
7 |
| GO:1900593 | violaceol II biosynthetic process | 2 | 0 | 1 | 2 | 4 | 7 |
* Numbers are combined for A. nidulans, A. fumigatus, A. niger and A. oryzae.
We also used published SMURF (Secondary Metabolite Unknown Regions Finder) predictions [38] to annotate additional candidate gene cluster backbone enzymes (i.e., PKS, NRPS, DMATS). SMURF is highly accurate at predicting most of these cluster backbone enzymes; across the four species of Aspergillus analyzed it identified a total of 105 genes as encoding PKS or PKS-like enzymes, 65 genes encoding NRPS or NRPS-like enzymes, 8 genes encoding putative hybrid PKS-NRPS enzymes and 15 DMATS. Note that DTS genes are not predicted by the SMURF algorithm. The AspGD Locus Summary pages now indicate these annotations based on the cluster backbone predictions generated by SMURF and by direct experimental characterization from the secondary metabolism literature.
Expansion of the secondary metabolism branch of the GO
To improve the accuracy of the AspGD GO annotation in the area of secondary metabolite production, a branch of the GO in which terms were sparse, we worked in collaboration with the GO Consortium to add new, more specific terms to the BP aspect of the ontology, and then used many of these new GO terms to annotate the Aspergillus genes that had experimentally determined mutant phenotype data associated with one or more secondary metabolite. We focused on the BP annotations because the relevant processes are well-represented in the experimental literature, whereas experimental data to support CC annotations are relatively sparse in the secondary metabolism literature. Adequate MF terms exist for the PKS and NRPS enzymes, but annotations to them in AspGD are mostly based on computationally determined domain matches and Interpro2GO annotations, or by annotations with Reviewed Computational Analysis (RCA) as the evidence code, meaning that these functions are predicted, rather than directly characterized through experiments.
The new GO annotations that we have added now precisely specify the secondary metabolite produced. For example, mdpG is known to influence the production of arugosin, emodin, monodictyphenone, orsinellic acid, shamixanthones and sterigmatocystin in A. nidulans. The gene was formerly annotated to the fairly nonspecific parental term ‘secondary metabolic process’ (GO:0019748), but because the secondary metabolites produced by this protein are known and published, it is now annotated to the new and more informative child terms ‘arugosin biosynthetic process’ (GO:1900587), ‘emodin biosynthetic process’ (GO:1900575), ‘monodictyphenone biosynthetic process’ (GO:1900815), ‘o-orsellinic acid biosynthetic process’ (GO:1900584), ‘shamixanthone biosynthetic process’ (GO:1900793) and ‘sterigmatocystin biosynthetic process’ (GO:0045461).
In total, we added 290 new BP terms to the GO for 48 secondary metabolites produced by one or more Aspergillus species. There are over 400 Aspergillus genes in AspGD that have been manually or computationally annotated to more specific secondary metabolism BP terms, based on over 260 publications (Table 2). A complete list of the GO terms for secondary metabolic process annotations is available in Additional file 1. The addition of new terms is ongoing as new secondary metabolites and their biosynthetic genes are identified and described in the scientific literature. The process of adding new GO terms depends on the elucidation of the structure of the secondary metabolite as the structure is required for new ChEBI (Chemical Entities of Biological Interest; http://www.ebi.ac.uk/chebi/) terms to be assigned, and these chemical compound terms are a prerequisite for GO term assignments involving chemical compounds. These new and improved GO terms provide researchers with valuable clues to aid in the identification of proteins involved in the production of specific classes of Aspergillus secondary metabolites.
Table 2.
GO terms used for secondary metabolism annotations at AspGD
| A. nidulans | A. fumigatus | A. niger | A. oryzae | |
|---|---|---|---|---|
| Number of predicted protein-encoding genes |
10,287 |
9,793 |
13,870 |
11,896 |
| Number of genes with GO annotations to secondary metabolism |
248 |
171 |
228 |
195 |
| Number of genes with manual GO annotations to secondary metabolism* |
202 |
96 |
81 |
32 |
| Number of genes with computational GO annotations to secondary metabolism* | 58 | 98 | 170 | 166 |
* or to child terms of ‘secondary metabolic process’ (GO: 0019748).
Predictive annotation using orthology relationships in conjunction with experimentally-based GO term assignments
Manual curation of the genes of one species can be used to computationally annotate the uncharacterized genes in another species based on orthology relationships. The use of GO to describe gene products facilitates comparative analysis of functions of orthologous genes throughout the tree of life, including orthologous genes within the filamentous fungi. To augment the manual GO curation in AspGD, we leveraged orthology relationships to assign GO annotations to genes that lacked manual annotations of their own but which had an experimentally characterized ortholog in AspGD, the Saccharomyces Genome Database (SGD) (http://www.yeastgenome.org) or PomBase (http://www.pombase.org). A total of 492 GO annotations were made to secondary metabolism-related genes in A. nidulans, A. fumigatus, A. niger and A. oryzae based on their orthology relationships (Table 3). Files listing these orthology relationships are available for download at http://www.aspergillusgenome.org/download/homology/orthologs/ and the files describing all GO term annotations for each gene product in AspGD are available at http://www.aspergillusgenome.org/download/go/. A list of all genes annotated to the secondary metabolic process branch of the GO and their associated annotations can be obtained through the AspGD Advanced Search Tool (http://www.aspergillusgenome.org/cgi-bin/search/featureSearch).
Table 3.
Number of GO annotations for secondary metabolism that were transferred to and between Aspergillus species under curation at AspGD
| From: | To A. nidulans | To A. fumigatus | To A. niger | To A. oryzae |
|---|---|---|---|---|
|
S. cerevisiae |
3 |
1 |
0 |
4 |
|
S. pombe |
1 |
0 |
0 |
0 |
|
A. nidulans |
n/a |
96 |
138 |
131 |
|
A. fumigatus |
53 |
n/a |
47 |
55 |
|
A. niger |
2 |
1 |
n/a |
3 |
| A. oryzae | 4 | 3 | 5 | n/a |
Manual annotation of computationally predicted gene clusters
Algorithms such as SMURF [38] and antiSMASH (antibiotics and Secondary Metabolite Analysis SHell) [39] can be used to predict fungal secondary metabolite gene clusters. Both of these algorithms are based on the identification of backbone enzymes, usually one or more polyketide synthase (PKS), non-ribosomal peptide synthetase (NRPS), hybrid PKS-NRPS, NRPS-like enzyme or dimethylallyl tryptophan synthase (DMATS), and the use of a training set of experimentally characterized clusters. Adjacent genes are then scanned for the presence of common secondary metabolite gene domains and boundaries are predicted for each cluster. We used the pre-computed gene clusters for A. nidulans, A. fumigatus, A. niger and A. oryzae that were identified at the J. Craig Venter Institute (JCVI) with the SMURF algorithm [38]. We also used the antiSMASH algorithm [39] on these genomes to make gene cluster predictions and added 5 additional clusters for A. nidulans based on the presence of DTS/ent-kaurene synthase backbone enzymes.
Altogether, a total of 261 non-redundant clusters were predicted by SMURF and antiSMASH: 71 for A. nidulans, 39 for A. fumigatus, 81 for A. niger and 75 for A. oryzae (Tables 4, 5, 6, 7). Neither SMURF nor antiSMASH predict DTS-based clusters, so these clusters were manually identified based on their annotations. Because clusters with other types of non-PKS and non-NRPS backbone enzymes were included in the antiSMASH predictions and SMURF only analyzes PKS, NRPKS or DMATS-based clusters, antiSMASH identified more clusters than SMURF in every species except for A. niger (Table 8). For clusters identified by both algorithms, there were no cases where both the left and right boundary predictions were the same, although a small number of single boundary predictions did coincide with each other (Tables 4, 5, 6, 7). Both the experimentally and manually (see below) predicted clusters tend to be smaller than the SMURF and antiSMASH algorithms predict, as the algorithms are designed to err on the side of inclusivity while the manual boundaries are designed to provide increased precision of the cluster boundaries through the examination of inter- and intra-cluster genome synteny alignments across multiple Aspergillus species. SMURF was previously reported to overpredict boundaries by about 4 genes [38] and we found that antiSMASH performed similarly. Figure 1 shows an example of the disparity between these two prediction programs in cluster boundary determination and how intra- and inter-species cluster synteny data used in our analysis aids in the manual predictions of secondary metabolite gene cluster boundaries (see below).
Table 4.
A. nidulans secondary metabolite biosynthetic gene clusters determined by SMURF, antiSMASH and by manual annotation or experimental characterization
|
A. nidulans Cluster Summary | |||||||
|---|---|---|---|---|---|---|---|
| Backbone enzyme | SMURF | antiSMASH | Manual/experimental | Boundary 1 | Boundary 2 | Reference | |
| Asperfuranone (afo) cluster |
AN1036 |
AN1029 - AN11288 |
AN1026 - AN1040 |
AN1029 - AN1036 |
ED |
ED |
[40] |
| Asperthecin (apt) cluster |
AN6000 |
AN5989 - AN6007 |
AN5991 - AN6009 |
AN6000 - AN6002* |
ED, EP |
ED, EP |
[30] |
| Aspyridone (asp) cluster |
AN8412 |
AN8398 - AN8415 |
AN8404 - AN11609 |
AN8408 - AN8415* |
ED |
ED |
[41] |
| Austinol (aus) cluster 1 |
AN9244 |
AN9243 - AN9251 |
AN11817 - AN9254 |
AN9244 - AN9253 |
ED, FA |
ED |
[31] |
| Austinol (aus) cluster 2 |
AN8383 |
AN8382 - AN8395 |
AN8377 - AN8390 |
AN8379 - AN8384* |
ED, ECS |
ED, ECS |
[31] |
| Derivative of Benzaldehyde1 (dba) and F9775 hybrid cluster 1 |
AN7909 |
- |
AN7895 - AN7919 |
AN7896 - AN7903* |
ED |
ED, EP |
[34,42] |
| Derivative of Benzaldehyde1 (dba) and F9775 hybrid cluster 2 |
|
AN7905 - AN7923 |
AN7895 - AN7919 |
AN7907 - AN7916* |
ED |
ED |
[34,42] |
| Emericellamide (eas) cluster |
AN2547 |
AN2544 - AN2553 |
AN2538 - AN10326 |
AN2545 - AN2549* |
ED |
ED |
[22] |
| inp cluster |
AN3496 |
AN3491 - AN3506 |
AN3485 - AN3503 |
AN3490 - AN3496 |
EP |
EP |
[43] |
| ivo cluster |
AN10576 |
AN10576 - AN4649 |
AN4637 - AN4649 |
AN10573 - AN10576 |
EP |
EP |
This study |
| Microperfuranone (mic) cluster |
AN3396 |
AN3391 - AN3408 |
AN3391 - AN3404 |
AN3394 - AN3396 |
EP |
EP |
[16] |
| Monodictyphenone (mdp) cluster |
AN0150 |
AN0146 - AN0158 |
AN11846 - AN0156 |
AN10021 - AN10023* |
ED, EP |
ED, EP |
[16,44] |
| Penicillin cluster |
AN2621 |
AN2619 - AN2625 |
AN2616 - AN10329 |
AN2621 - AN2623* |
ED, EP |
ED, EP |
[16,45] |
| Nidulanin A (nptA) cluster |
AN11080 |
- |
AN8481 - AN8484 |
AN11082 - AN11080 |
IGD, FA |
ECS |
This study |
| pkb cluster |
AN6448 |
AN6436 - AN6462 |
AN6438 - AN6455 |
AN6444 - AN6451 |
EP |
EP |
[16] |
| pkdA cluster |
AN0523 |
AN0520 - AN0533 |
AN10104 - AN0534 |
AN0523 - AN0533 |
ECS |
ECS |
This study |
| pkf cluster |
AN3230 |
AN3217 - AN3237 |
AN3222 - AN3238 |
AN3225 -AN3230 |
EP |
ECS, EP |
This study |
| pkg cluster |
AN7071 |
AN7061 - AN7075 |
AN7064 - AN7078 |
AN10889 - AN7075 |
EP |
ECS, EP |
This study |
| pkh cluster |
AN2035 |
AN2032 - AN2043 |
AN2029 - AN2042 |
AN2030 - AN2038 |
ECS, IGD |
EP |
[16] |
| pki cluster |
AN3386 |
AN3380 - AN3386 |
AN3380 - AN3392 |
AN3379 - AN3386 |
ECS |
ECS |
This study |
| Sterigmatocystin (stc) cluster |
AN7815 |
AN7806 - AN7824 |
AN7815 - AN7832 |
AN7804 - AN7824* |
ED |
ED |
[46] |
| Terriquinone (tdi) cluster |
AN8514 |
AN8506 - AN8516 |
AN8508 - AN8522 |
AN8513 - AN8520* |
ED |
ED |
[47] |
| xptA-containing cluster |
AN6784 |
AN6770 - AN6791 |
AN6780 - AN6786 |
AN6784 - AN6791 |
FA, EP |
FA, EP |
FA, [16] |
| xptB-containing cluster |
AN12402 |
- |
AN7996 - AN12431 |
AN7999 - AN12431 |
ECS |
IGD |
This study |
| AN0016 cluster |
AN0016 |
AN0015 - AN0029 |
AN10009 - AN0025 |
AN0015 - AN0029 |
ECS, IGD |
FA |
This study |
| AN0607 (sidC) cluster |
AN0607 |
AN0592 - AN0607 |
AN0599 - AN0617 |
AN0607 - AN0609 |
ECS |
ECS, IGD |
This study |
| AN10289 cluster |
AN10289 |
AN10294 - AN2349 |
AN10294 - AN2347 |
AN10289 - AN2347 |
IGD |
FA, IGD |
This study |
| AN10297 cluster |
AN10297 |
AN2392 - AN10301 |
AN2395 - AN10298 |
AN2396 - AN11337 |
FA |
FA |
This study |
| AN10396 cluster |
AN10396 |
- |
AN3372 - AN3379 |
AN3375 - AN10401 |
ECS |
ECS, IGD |
This study |
| AN10430 cluster |
AN10430 |
AN3608 - AN3612 |
AN10429 - AN10448 |
AN3609 - AN3612 |
FA |
FA |
This study |
| AN10486 cluster |
AN10486 |
AN3911 - AN3917 |
AN10485 - AN3917 |
AN3911 - AN10486 |
ECS |
ECS |
This study |
| AN11065 cluster |
AN11065 |
- |
AN12204 - AN8263 |
- |
n/a |
n/a |
n/a |
| AN11191 cluster |
AN11191 |
AN9210 - AN11192 |
AN9212 - AN12146 |
AN11191 - AN9220 |
ECS, EP |
EP |
[16] |
| AN11194 cluster |
AN11194 |
AN9223 - AN9236 |
AN9220 - AN9236 |
AN8223 - AN9235 |
EP |
EP |
[16] |
| AN12331 cluster |
AN12331 |
AN7836 - AN7837 |
AN7831 - AN7842 |
AN7836 - AN7839 |
FA |
ECS |
This study |
| AN1242 cluster |
AN1242 |
AN1242 - AN1243 |
AN1236 - AN1250 |
AN1242 - AN1247 |
ECS, IGD |
ECS |
This study |
| AN1594 cluster |
AN1594 |
- |
- |
AN1592 - AN1599 |
EP |
EP |
[16] |
| AN1680 cluster |
AN1680 |
AN1677 - AN1691 |
AN1674 - AN1687 |
AN1678 - AN1681 |
ECS |
ECS |
This study |
| AN1784 cluster |
AN1784 |
AN1778 - AN1787 |
AN1776 - AN1790 |
AN1784 - AN1787 |
EP |
EP |
[16] |
| AN1793 cluster |
AN1793 |
- |
AN1790 - AN1802 |
AN1792 - AN1796 |
FA |
ECS |
ECS |
| AN2064 cluster |
AN2064 |
AN2061 - AN2064 |
AN2057 - AN2068 |
- |
n/a |
n/a |
n/a |
| AN2924 cluster |
AN2924 |
AN2921 - AN2925 |
AN2917 - AN2928 |
AN2921 - AN2924 |
EP |
EP |
[16] |
| AN3252 cluster |
AN3252 |
- |
- |
AN3252 - AN3257 |
EP |
EP |
[16] |
| AN3273 cluster |
AN3273 |
|
AN3267 - AN3285 |
AN10388 - AN3287 |
IGD |
IGD |
This study |
| AN3612 cluster |
AN3612 |
AN3608 - AN3612 |
AN10429 - AN10448 |
AN3605 - AN3612 |
FA |
ECS, FA |
This study |
| AN4827 cluster |
AN4827 |
AN4823 - AN4837 |
AN11232 - AN4836 |
AN10600 - AN11232 |
EP |
EP |
[16] |
| AN5318 cluster |
AN5318 |
AN5317 - AN5329 |
AN5312 - AN5325 |
AN5314 - AN5318 |
ECS |
ECS |
This study |
| AN5475 cluster |
AN5475 |
- |
- |
- |
n/a |
n/a |
n/a |
| AN5610 cluster |
AN5610 |
- |
AN5607 - AN5616 |
AN9473 - AN5610 |
ECS, IGD |
FA |
This study |
| AN6236 cluster |
AN6236 |
AN6236 - AN6238 |
AN6230 - AN11851 |
AN6234 - AN6236 |
ECS, EP |
ECS, EP |
ECS, [16] |
| AN6431 cluster |
AN6431 |
AN6429 - AN6434 |
AN10818 - AN6437 |
AN6431 - AN6437 |
ECS, EP |
EP |
[16] |
| AN6791 cluster |
AN6791 |
- |
AN11527 - AN10846 |
AN6787 - AN6791 |
IGD |
FA |
This study |
| AN7084 cluster |
AN7084 |
AN7081 - AN7089 |
AN7077 - AN7089 |
AN7080 - AN7086 |
EP |
EP |
[16] |
| AN7489 (mirC) cluster |
AN7489 |
AN7485 - AN7493 |
- |
AN7485 - AN7493 |
ECS |
ECS |
This study |
| AN7838 (AN12331) cluster |
AN7838 |
AN7836 - AN7839 |
- |
AN11024 - AN7839 |
FA |
FA |
This study |
| AN7884 cluster |
AN7884 |
AN7873 - AN7884 |
AN7879 - AN7892 |
AN7872 - AN7884 |
ECS, EP |
ECS, EP |
ECS, [16] |
| AN8105 cluster |
AN8105 |
AN8103 - AN8113 |
AN8098 - AN8113 |
AN8105 - AN8112 |
ECS, EP |
EP |
[16] |
| AN8142 cluster |
AN8142 |
- |
AN8133 - AN8147 |
AN12440 - AN8144 |
FA |
ECS |
This study |
| AN8209 (wA) cluster |
AN8209 |
- |
AN8202 - AN8214 |
AN12404 - AN8209 |
EP |
ECS, EP |
ECS, [16] |
| AN8249 cluster |
AN8249 |
- |
AN11063 - AN11069 |
- |
n/a |
n/a |
n/a |
| AN8504 cluster |
AN8504 |
- |
AN8495 - AN8508 |
AN8495 - AN8504 |
ECS |
FA |
This study |
| AN8910 cluster |
AN8910 |
AN8901 - AN8910 |
AN8903 - AN8915 |
AN8905 - AN8910 |
ECS |
ECS |
This study |
| AN9005 cluster |
AN9005 |
AN9002 - AN9013 |
AN9000 - AN9013 |
AN9002 - AN9007 |
EP |
EP |
[16] |
| AN9129 cluster |
AN9129 |
- |
- |
AN9129 - AN9130 |
ECS, EP |
ECS |
This study |
| AN9226 cluster |
AN9226 |
AN9223 - AN9236 |
AN9220 - AN9236 |
AN9223 - AN9234 |
EP |
EP |
[16] |
| AN9291 (AN11820) cluster |
AN11820 |
AN9280 - AN9307 |
- |
AN11820 - AN9294 |
EP |
ECS |
This study |
| AN9314 cluster |
AN9314 |
- |
- |
AN9313 - AN9314 |
EP |
EP |
[16] |
| No PKS/NRPS backbone 1 |
n/a |
- |
AN9184 - AN9194 |
AN9179 - AN9185 |
ECS |
ECS |
This study |
| No PKS/NRPS backbone 2 |
n/a |
- |
AN10325 - AN11348 |
- |
n/a |
n/a |
n/a |
| No PKS/NRPS backbone 3 |
n/a |
- |
AN0650 - AN0659 |
AN0653 - AN0660 |
ECS |
ECS |
This study |
| No PKS/NRPS backbone 4 | n/a | - | AN0039 - AN10006 | AN0042 - AN0044 | EP | EP | [16] |
* Experimentally characterized, Abbreviations: ECS, End of cluster synteny; IGD, Increase in intergenic distance; FA, Change in functional annotation; ED, Experimentally determined; EP, Expression Pattern.
Table 5.
A. fumigatus secondary metabolite biosynthetic gene clusters determined by SMURF, antiSMASH and by manual annotation or experimental characterization
|
A. fumigatus cluster summary | |||||||
|---|---|---|---|---|---|---|---|
| Backbone enzyme | SMURF | antiSMASH | Manual/experimental | Boundary 1 | Boundary 2 | Reference | |
| Fumigaclavine C (fga) cluster* |
Afu2g18040 |
Afu2g17930 - Afu2g18070 |
Afu2g17950 - Afu2g18070 |
Afu2g17960 - Afu2g18060 |
ED |
ED |
[48] |
| Fumitremorgin B (ftm) cluster* |
Afu8g00210 |
Afu8g00280 - Afu8g00100 |
Afu8g00100 - Afu8g00110 |
Afu8g00260 - Afu8g00170 |
ED |
ED |
[49] |
| Gliotoxin (gli) cluster* |
Afu6g09650 |
Afu6g09580 - Afu6g09740 |
Afu6g09520 - Afu6g09745 |
Afu6g09630 - Afu6g09740 |
ED, IGD |
ED |
[50] |
| Pseurotin A cluster* |
Afu8g00540 |
- |
Afu8g00342 - Afu8g00595 |
Afu8g005340 - Afu8g00570 |
ED |
ED |
[35] |
| Siderophore (sid) cluster* |
Afu3g03420 |
Afu3g03280 - Afu3g03580 |
Afu3g03270 - Afu3g03490 |
Afu3g03300 - Afu3g03460 |
ED |
ED |
[48] |
| Afu1g01010 cluster |
Afu1g01010 |
Afu1g00970 - Afu1g01010 |
Afu1g00970 - Afu1g01030 |
Afu1g00980 - Afu1g01010 |
FA |
IGD |
This study |
| Afu1g10380 (nrps1) cluster |
Afu1g10380 |
Afu1g10310 - Afu1g10380 |
Afu1g10310 - Afu1g10420 |
Afu1g10270 - Afu1g10380 |
ECS |
IGD |
This study |
| Afu1g17200 (sidC) cluster |
Afu1g17200 |
Afu1g17080 - Afu1g17240 |
Afu1g17090 - Afu1g17270 |
Afu1g17200 - Afu1g17240 |
ECS |
ECS |
This study |
| Afu1g17740 cluster |
Afu1g17740 |
Afu1g17710 - Afu1g17740 |
Afu1g17720 - Afu1g17750 |
Afu1g17710 - Afu1g17740 |
ECS |
ECS |
This study |
| Afu2g01290 cluster |
Afu2g01290 |
Afu2g01170 - Afu2g01400 |
Afu2g01210 - Afu2g01390 |
Afu2g01280 - Afu2g01330 |
ECS |
ECS |
This study |
| Afu2g05760 cluster |
Afu2g05760 |
Afu2g05730 - Afu2g05840 |
- |
Afu2g05740 - Afu2g05830 |
IGD |
IGD |
This study |
| Afu2g17600 cluster |
Afu2g17600 |
Afu2g17511 - Afu2g17600 |
Afu2g17490 - Afu2g17690 |
Afu2g17480 - Afu2g17600 |
IGD |
IGD |
This study |
| Afu3g01410 cluster |
Afu3g01410 |
Afu3g01400 - Afu3g01560 |
Afu3g01360 - Afu3g01560 |
Afu3g01400 - Afu3g01480 |
IGD |
IGD |
This study |
| Afu3g02530 cluster |
Afu3g02530 |
Afu3g02450 - Afu3g02540 |
Afu3g02450 - Afu3g02650 |
Afu3g02520 - Afu3g02650 |
FA |
IGD |
This study |
| Afu3g02570 cluster |
Afu3g02570 |
Afu3g02570 - Afu3g02650 |
Afu3g02450 - Afu3g02650 |
Afu3g02520 - Afu3g02650 |
FA |
IGD |
This study |
| Afu3g02670 cluster |
Afu3g02670 |
Afu3g02670 - Afu3g02760 |
Afu3g02670 - Afu3g02760 |
Afu3g02670 - Afu3g02720 |
ECS, IGD |
ECS, IGD |
This study |
| Afu3g12920 cluster |
Afu3g12920 |
Afu3g12960 - Afu3g12750 |
Afu3g13020 - Afu3g12820 |
Afu3g12960 - Afu3g12890 |
ECS |
ECS |
This study |
| Afu3g12930 cluster |
Afu3g12930 |
Afu3g13000 - Afu3g12750 |
Afu3g13020 - Afu3g12820 |
Afu3g12960 - Afu3g12890 |
ECS |
ECS |
This study |
| Afu3g13730 cluster |
Afu3g13730 |
Afu3g13730 - Afu3g13600 |
Afu3g13790 - Afu3g13600 |
Afu3g13750 - Afu3g13670 |
IGD |
FA |
This study |
| Afu3g14700 cluster |
Afu3g14700 |
Afu3g14880 - Afu3g14690 |
Afu3g14820 - Afu3g14620 |
Afu3g14730 - Afu3g14690 |
ECS |
FA |
This study |
| Afu3g15270 cluster |
Afu3g15270 |
Afu3g15290 - Afu3g15250 |
Afu3g15350 - Afu3g15190 |
Afu3g15290 - Afu3g15240 |
ECS, IGD |
ECS |
This study |
| Afu4g00210 cluster |
Afu4g00210 |
Afu4g00260 - Afu4g00210 |
Afu4g00290 - Afu4g00150 |
Afu4g00260 - Afu4g00200 |
IGD |
ECS |
This study |
| Afu4g14560 cluster |
Afu4g14560 |
Afu4g14730 - Afu4g14420 |
Afu4g14660 - Afu4g14440 |
Afu4g14610 - Afu4g14450 |
ECS |
ECS |
This study |
| Afu5g10120 cluster |
Afu5g10120 |
Afu5g10250 - Afu5g09970 |
Afu5g10240 - Afu5g10010 |
Afu5g10130 - Afu5g10040 |
ECS |
FA |
This study |
| Afu5g12730 cluster |
Afu5g12730 |
Afu5g12840 - Afu5g12720 |
Afu5g12830 - Afu5g12680 |
Afu5g12770 - Afu5g12730 |
FA |
ECS |
This study |
| Afu6g03480 cluster |
Afu6g03480 |
Afu6g03620 - Afu6g03430 |
Afu6g03550 - Afu6g03400 |
Afu6g03490 - Afu6g03430 |
ECS |
ECS, IGD |
This study |
| Afu6g08560 cluster |
Afu6g08560 |
Afu6g08540 - Afu6g08560 |
Afu6g08520 - Afu6g08640 |
Afu6g08550 - Afu6g08560 |
ECS |
ECS |
ECS, IGD |
| Afu6g12080 cluster |
Afu6g12080 |
Afu6g12040 - Afu6g12160 |
Afu6g11980 - Afu6g12145 |
Afu6g12040 - Afu6g12080 |
ECS |
ECS |
This study |
| Afu6g13930 cluster |
Afu6g13930 |
Afu6g13830 - Afu6g14050 |
Afu6g13820 - Afu6g14030 |
Afu6g13920 - Afu6g14000 |
ECS |
ECS |
This study |
| Afu7g00170 cluster |
Afu7g00170 |
Afu7g00200 - Afu7g00120 |
Afu7g00220 - Afu7g00100 |
Afu7g00190 - Afu7g00120 |
ECS |
ECS, IGD |
This study |
| Afu8g00540 cluster |
Afu8g00540 |
Afu8g00370 - Afu8g00370 |
Afu8g00490 - Afu8g00310 |
- |
n/a |
n/a |
This study |
| Afu8g00620 cluster |
Afu8g00620 |
Afu8g00640 - Afu8g00470 |
Afu8g00720 - Afu8g00390 |
- |
n/a |
n/a |
This study |
| Afu8g01640 cluster |
Afu8g01640 |
Afu8g01640 - Afu8g01640 |
Afu8g01600 - Afu8g01730 |
Afu8g01630 - Afu8g01640 |
ECS, IGD |
ECS, IGD |
This study |
| Afu8g02350 cluster |
Afu8g02350 |
Afu8g02460 - Afu8g02350 |
Afu8g02490 - Afu8g02280 |
Afu8g02430 - Afu8g02350 |
ECS, IGD |
ECS, IGD |
This study |
| No PKS or NRPS backbone 1 |
n/a |
- |
Afu4g11170 - Afu4g11300 |
- |
n/a |
n/a |
|
| No PKS or NRPS backbone 2 |
n/a |
- |
Afu4g11980 - Afu4g12070 |
- |
n/a |
n/a |
|
| No PKS or NRPS backbone 3 |
n/a |
- |
Afu5g00100 - Afu5g04130 |
- |
n/a |
n/a |
|
| No PKS or NRPS backbone 4 |
n/a |
- |
Afu7g00230 -Afu7g00350 |
Afu5g00100 - Afu5g00135 |
ECS, IGD |
ECS, IGD |
This study |
| No PKS or NRPS backbone 5 | n/a | - | Afu7g01180 - Afu7g01270 | Afu7g00260 - Afu7g00270 | ECS, FA | ECS | This study |
* Experimentally characterized; Abbreviations: ECS, End of cluster synteny; IGD, Increase in intergenic distance; FA, Change in functional annotation; ED, Experimentally determined (1).
Table 6.
A. niger secondary metabolite biosynthetic gene clusters determined by SMURF, antiSMASH and by manual annotation or experimental characterization
|
A. niger cluster summary | ||||||
|---|---|---|---|---|---|---|
| Backbone enzyme | SMURF | antiSMASH | Manual | Boundary 1 | Boundary 2 | |
| An01g01130 cluster |
An01g01130 |
An01g01080 - An01g01150 |
An01g01050 - An01g01220 |
An01g01110 - An01g01230 |
FA |
ECS |
| An01g06930/ An01g06950 cluster |
An01g06930 |
An01g06730 - An01g06950 |
An01g06770 - An01g07020 |
An01g06810 - An01g06970 |
ECS |
ECS. FA |
| An01g11770 cluster |
An01g11770 |
An01g11740 - An01g11900 |
An01g11690 - An01g11880 |
An01g11760 - An01g11830 |
IGD, ECS |
ECS, IGD |
| An02g00210 cluster |
An02g00210 |
An02g00150 - An02g00290 |
An02g00140 - An02g00320 |
An02g00210 - An02g00260 |
ECS |
ECS |
| An02g00450 cluster |
An02g00450 |
An02g00430 - An02g00580 |
An02g00340 - An02g00540 |
- |
n/a |
n/a |
| An02g00840 cluster |
An02g00840 |
An02g00710 - An02g00860 |
An02g00740 - An02g00950 |
An02g00700 - An02g00840 |
ECS, IGD |
ECS. FA |
| An02g05070 cluster |
An02g05070 |
An02g05050 - An02g05190 |
An02g04970 - An02g05170 |
An02g05050 - An02g05170 |
ECS |
ECS |
| An02g08290 cluster |
An02g08290 |
An02g08130 - An02g08370 |
An02g08210 - An02g08380 |
An02g08290 - An02g08310 |
ECS |
ECS |
| An02g09430 cluster |
An02g09430 |
An02g09420 - An02g09450 |
An02g09310 - An02g09530 |
An02g09390 - An02g09430 |
ECS |
ECS |
| An02g10140 cluster |
An02g10140 |
An02g10110 - An02g10170 |
An02g10080 - An02g10200 |
An02g10140 - An02g10200 |
ECS |
FA |
| An02g14220 cluster |
An02g14220 |
An02g14190 - An02g14240 |
- |
An02g14170 - An02g14240 |
ECS |
ECS |
| An03g00650 cluster |
An03g00650 |
An03g00580 - An03g00790 |
An03g00530 - An03g00760 |
- |
n/a |
n/a |
| An03g01820 cluster |
An03g01820 |
An03g01750 - An03g01850 |
An03g01730 - An03g01930 |
An03g01790 - An03g01820 |
ECS |
ECS |
| An03g03520 cluster |
An03g03520 |
An03g03380 - An03g03620 |
An03g03450 - An03g03600 |
An03g03490 - An03g03620 |
ECS |
ECS, IGD |
| An03g05140 cluster |
An03g05140 |
An03g05050 - An03g05270 |
An03g05040 - An03g05230 |
An03g05140 - An03g05170 |
ECS |
ECS |
| An03g05440 cluster |
An03g05440 |
An03g05300 - An03g05500 |
An03g05330 - An03g05540 |
An03g05430 - An03g05500 |
ECS |
ECS |
| An03g05680 cluster |
An03g05680 |
An03g05660 - An03g05800 |
An03g05600 - An03g05760 |
An03g05660 - An03g05710 |
ECS |
ECS |
| An03g06010 cluster |
An03g06010 |
An03g05810 - An03g06020 |
An03g05910 - An03g06100 |
An03g05880 - An03g06020 |
ECS |
ECS, IGD |
| An03g06380 cluster |
An03g06380 |
An03g06370 - An03g06520 |
An03g06310 - An03g06470 |
An03g06310 - An03g06440 |
ECS, IGD |
ECS |
| An04g01150 cluster |
An04g01150 |
An04g01120 - An04g01170 |
An04g01070 - An04g01230 |
An04g01080 - An04g01180 |
ECS |
ECS |
| An04g04340 cluster |
An04g04340 |
An04g04220 - An04g04520 |
An04g04280 - An04g04500 |
An04g04330 - An04g04400 |
ECS |
ECS |
| An04g04380 cluster |
An04g04380 |
An04g04340 - An04g04520 |
- |
An04g06240 - An04g06320 |
ECS |
ECS |
| An04g06260 cluster |
An04g06260 |
An04g06230 - An04g06290 |
An04g06180 - An04g06370 |
An04g06240 - An04g06320 |
ECS |
ECS |
| An04g09530 cluster |
An04g09530 |
An04g09500 - An04g09650 |
An04g09420 - An04g09600 |
An04g09510 - An04g09570 |
ECS |
ECS |
| An04g10030 cluster |
An04g10030 |
An04g09890 - An04g10090 |
An04g09900 - An04g10100 |
- |
n/a |
n/a |
| An05g01060 cluster |
An05g01060 |
An05g01020 - An05g01120 |
An05g00930 - An05g01140 |
An05g01060 - An05g01120 |
ECS |
ECS |
| An06g01300 cluster |
An06g01300 |
An06g01290 - An06g01410 |
An06g01200 - An06g01390 |
An06g01290 - An06g01320 |
ECS |
ECS |
| An07g01030 cluster |
An07g01030 |
An07g00900 - An07g01100 |
An07g00900 - An07g01130 |
- |
n/a |
n/a |
| An07g02560 cluster |
An07g02560 |
An07g02540 - An07g02650 |
An07g02520 - An07g02620 |
An07g02510 - An07g02560 |
ECS |
ECS |
| An08g02310 cluster |
An08g02310 |
An08g02180 - An08g02330 |
An08g02210 - An08g02360 |
An08g02170 - An08g02310 |
ECS |
ECS |
| An08g03790 cluster |
An08g03790 |
An08g03610 - An08g03870 |
An08g03700 - An08g03880 |
An08g03730 - An08g03790 |
ECS |
ECS |
| An08g04820 cluster |
An08g04820 |
An08g04710 - An08g04940 |
An08g04710 - An08g04900 |
An08g04790 - An08g04830 |
ECS |
ECS |
| An08g09220 cluster |
An08g09220 |
An08g09150 - An08g09400 |
An08g09140 - An08g09360 |
- |
n/a |
n/a |
| An08g10930 cluster |
An08g10930 |
An08g10880 - An08g11000 |
- |
An08g10860 - An08g10930 |
ECS |
ECS |
| An09g00450 cluster |
An09g00450 |
An09g00400 - An09g00610 |
An09g00350 - An09g00610 |
An09g00390 - An09g00450 |
ECS |
ECS |
| An09g00520 cluster |
An09g00520 |
An09g00390 - An09g00620 |
- |
An09g00520 - An09g00620 |
ECS |
ECS |
| An09g01290 cluster |
An09g01290 |
An09g01140 - An09g01460 |
An09g01200 - An09g01390 |
An09g01260 - An09g01340 |
ECS |
ECS |
| An09g01690 cluster |
An09g01690 |
An09g01650 - An09g01860 |
An09g01560 - An09g01800 |
An09g01630 - An09g01860 |
ECS |
ECS |
| An09g01860/An09g01930 cluster |
An09g01860 |
An09g01690 - An09g02020 |
An09g01740 - An09g02020 |
An09g01790 - An09g01950 |
ECS, FA |
ECS, FA |
| An09g02100 cluster |
An09g02100 |
An09g01980 - An09g02230 |
An09g01980 - An09g02220 |
- |
n/a |
n/a |
| An09g05110 cluster |
An09g05110 |
An09g04960 - An09g05130 |
An09g05010 - An09g05180 |
An09g05060 - An09g05150 |
ECS |
ECS |
| An09g05340 cluster |
An09g05340 |
An09g05310 - An09g05360 |
An09g05270 - An09g05400 |
An09g05300 - An09g05350 |
ECS |
ECS |
| An09g05730 cluster |
An09g05730 |
An09g05710 - An09g05750 |
An09g05655 - An09g05810 |
An09g05625 - An09g05730 |
ECS |
ECS |
| An10g00140 cluster |
An10g00140 |
An10g00010 - An10g00230 |
An10g00050 - An10g00240 |
An10g00100 - An10g00210 |
ECS |
ECS |
| An10g00630 cluster |
An10g00630 |
An10g00540 - An10g00700 |
- |
An10g00620 - An10g00700 |
ECS |
ECS |
| An11g00050 cluster |
An11g00050 |
An11g00030 - An11g00080 |
An11g00010 - An11g00100 |
An11g00040 - An11g00080 |
ECS |
ECS |
| An11g00250 cluster |
An11g00250 |
An11g00130 - An11g00380 |
An11g00170 - An11g00350 |
An11g00250 - An11g00300 |
ECS |
ECS |
| An11g03920 cluster |
An11g03920 |
An11g03750 - An11g04040 |
An11g03820 - An11g04010 |
An11g03870 - An11g03940 |
ECS |
ECS |
| An11g04280 cluster |
An11g04280 |
An11g04140 - An11g04310 |
An11g04180 - An11g04350 |
An11g04250 - An11g04320 |
ECS |
ECS |
| An11g05500 cluster |
An11g05500 |
An11g05470 - An11g05570 |
An11g05420 - An11g05560 |
An11g05480 - An11g05530 |
ECS |
ECS, IGD |
| An11g05570 cluster |
An11g05570 |
An11g05440 - An11g05590 |
An11g05510 - An11g05660 |
- |
n/a |
n/a |
| An11g05940 cluster |
An11g05940 |
An11g05820 - An11g05960 |
An11g05860 - An11g06060 |
An11g05940 - An11g05960 |
ECS |
ECS |
| An11g05960 cluster |
An11g05960 |
An11g05790 - An11g05980 |
An11g05860 - An11g06060 |
An11g05940 - An11g05960 |
ECS |
ECS |
| An11g06460 cluster |
An11g06460 |
An11g06420 - An11g06490 |
An11g06350 - An11g06570 |
An11g06420 - An11g06490 |
ECS |
ECS |
| An11g07310 cluster |
An11g07310 |
An11g07280 - An11g07350 |
An11g07210 - An11g07460 |
An11g07270 - An11g07350 |
ECS |
ECS |
| An11g09720 cluster |
An11g09720 |
An11g09700 - An11g09740 |
An11g09620 - An11g09790 |
- |
n/a |
n/a |
| An12g02670 cluster |
An12g02670 |
An12g02650 - An12g02830 |
An12g02560 - An12g02920 |
An12g02620 - An12g02750 |
ECS |
ECS |
| An12g02730 cluster |
An12g02730 |
An12g02670 - An12g02900 |
- |
- |
n/a |
n/a |
| An12g02840 cluster |
An12g02840 |
An12g02680 - An12g02880 |
- |
- |
n/a |
n/a |
| An12g07070 cluster |
An12g07070 |
An12g07050 - An12g07110 |
An12g06930 - An12g07140 |
An12g07060 - An12g07110 |
ECS |
ECS |
| An12g07230 cluster |
An12g07230 |
An12g07120 - An12g07250 |
An12g07150 - An12g07330 |
An12g07220 - An12g07280 |
ECS |
ECS |
| An12g10090 cluster |
An12g10090 |
An12g09930 - An12g10220 |
An12g09980 - An12g10190 |
An12g10000 - An12g10160 |
ECS |
ECS |
| An12g10860 cluster |
An12g10860 |
An12g10840 - An12g10870 |
An12g10770 - An12g10960 |
An12g10790 - An12g10860 |
ECS |
ECS |
| An13g01840 cluster |
An13g01840 |
An13g01820 - An13g01860 |
An13g01790 - An13g01880 |
An13g01810 - An13g01860 |
ECS |
ECS |
| An13g02430 cluster |
An13g02430 |
An13g02290 - An13g02540 |
An13g02350 - An13g02600 |
An13g02390 - An13g02470 |
ECS |
ECS |
| An13g02460 cluster |
An13g02460 |
An13g02400 - An13g02540 |
- |
An13g02390 - An13g02480 |
ECS |
ECS |
| An13g02960 cluster |
An13g02960 |
An13g02940 - An13g03090 |
An13g02830 - An13g03120 |
- |
n/a |
n/a |
| An13g03040 cluster |
An13g03040 |
An13g02940 - An13g03110 |
An13g02830 - An13g03120 |
- |
n/a |
n/a |
| An14g01910 cluster |
An14g01910 |
An14g01850 - An14g01970 |
An14g01820 - An14g02010 |
An14g01910 - An14g01960 |
ECS |
ECS |
| An14g04850 cluster |
An14g04850 |
An14g04830 - An14g04870 |
An14g04750 - An14g04940 |
An14g04830 - An14g04890 |
ECS |
ECS |
| An15g02130 cluster |
An15g02130 |
An15g02070 - An15g02190 |
An15g02040 - An15g02230 |
An15g02130 - An15g02200 |
ECS |
ECS |
| An15g04140 cluster |
An15g04140 |
An15g04120 - An15g04220 |
An15g04050 - An15g04250 |
An15g04130 - An15g04150 |
ECS |
ECS |
| An15g05090 cluster |
An15g05090 |
An15g05030 - An15g05150 |
An15g05000 - An15g05180 |
An15g05090 - An15g05150 |
ECS |
ECS |
| An15g07530 cluster |
An15g07530 |
An15g07510 - An15g07600 |
An15g07400 - An15g07640 |
An15g07530 - An15g07480 |
ECS |
ECS |
| An15g07910 cluster |
An15g07910 |
An15g07830 - An15g07930 |
An15g07810 - An15g07930 |
An15g07890 - An15g07920 |
ECS |
ECS |
| An15g07920 cluster |
An15g07920 |
An15g07830 - An15g07930 |
- |
- |
n/a |
n/a |
| An16g00600 cluster |
An16g00600 |
An16g00520 - An16g00740 |
An16g00460 - An16g00730 |
An16g00520 - An16g00600 |
ECS |
ECS |
| An16g06720 cluster |
An16g06720 |
An16g06570 - An16g06780 |
An16g06650 - An16g06790 |
An16g06720 - An16g06750 |
ECS |
ECS |
| An18g00520 cluster | An18g00520 | An18g00350 - An18g00550 | An18g00440 - An18g05820 | An18g00460 - An18g00530 | ECS | ECS |
* Experimentally characterized; Abbreviations: ECS, End of cluster synteny; IGD, Increase in intergenic distance; FA, Change in functional annotation; ED, Experimentally determined.
Table 7.
A. oryzae secondary metabolite biosynthetic gene clusters determined by SMURF, antiSMASH and by manual annotation or experimental characterization
|
A. oryzaecluster summary | ||||||
|---|---|---|---|---|---|---|
| Backbone enzyme | SMURF | antiSMASH | Manual | Boundary 1 | Boundary 2 | |
| Csypyrone B1 cluster |
AO090701000566 |
- |
AO090701000569 - AO090701000568 |
AO090701000561 - AO090701000568 |
ED |
ED |
| Dipeptidyl Peptidase IV 2 Inhibitor (WYK-1) cluster |
AO090001000009 |
AO090001000031 - AO090001000009 |
AO090001000019 - AO090001000001 |
AO090001000019 - AO090001000009 |
ED |
ED |
| AO090001000043 cluster |
AO090001000043 |
AO090001000051 - AO090001000021 |
AO090001000055 - AO090001000021 |
AO090001000051 - AO090001000028 |
ECS, FA |
FA |
| AO090001000262 cluster |
AO090001000262 |
- |
AO090001000253 - AO090001000268 |
AO090001000260 - AO090001000264 |
ECS |
FA, ECS |
| AO090001000277 cluster |
AO090001000277 |
AO090001000293 - AO090001000256 |
AO090001000289 - AO090001000267 |
AO090001000292 - AO090001000277 |
ECS |
ECS |
| AO090001000293 cluster |
AO090001000293 |
- |
AO090001000757 - AO090001000295 |
- |
n/a |
n/a |
| AO090001000402 cluster |
AO090001000402 |
AO090001000404 - AO090001000390 |
AO090001000408 - AO090001000390 |
AO090001000408 - AO090001000390 |
ECS |
ECS |
| AO090001000506 cluster |
AO090001000506 |
AO090001000506 - AO090001000505 |
AO090001000512 - AO090001000495 |
AO090001000506 - AO090001000505 |
FA |
ECS |
| AO090001000516 cluster |
AO090001000516 |
AO090001000516 - AO090001000515 |
AO090001000523 - AO090001000512 |
AO090001000516 - AO090001000515 |
ECS |
ECS |
| AO090001000768 cluster |
AO090001000768 |
AO090001000704 - AO090001000692 |
- |
AO090001000768 - AO090001000687 |
ECS |
ECS |
| AO090003000945 cluster |
AO090003000945 |
AO090003000954 - AO090003000945 |
AO090003000954 - AO090003000936 |
AO090003000946 - AO090003000941 |
ECS |
ECS |
| AO090003001097 cluster |
AO090003001097 |
- |
AO090003001090 - AO090003001106 |
AO090003001094 - AO090003001099 |
ECS |
ECS |
| AO090003001545 cluster |
AO090003001545 |
AO090003001556 - AO090003001541 |
AO090003001552 - AO090003001535 |
AO090003001556 - AO090003001537 |
ECS |
ECS |
| AO090005000688 cluster |
AO090005000688 |
AO090005000688 - AO090005000693 |
AO090005000681 - AO090005000696 |
AO090005000687 - AO090005000695 |
FA |
ECS |
| AO090005000798 cluster |
AO090005000798 |
- |
AO090005000791 - AO090005000805 |
AO090005000796 - AO090005000805 |
ECS |
ECS |
| AO090005000952 cluster |
AO090005000952 |
AO090005000952 - AO090005000955 |
AO090005000942 - AO090005000959 |
AO090005000952 - AO090005000956 |
ECS |
ECS |
| AO090005000961 cluster |
AO090005000961 |
AO090005000961 - AO090005000968 |
AO090005000956 - AO090005000970 |
AO090005000961 - AO090005000968 |
ECS |
ECS |
| AO090005000993 cluster |
AO090005000993 |
AO090005000990 - AO090005001001 |
AO090005000986 - AO090005001002 |
AO090005000990 - AO090005000993 |
ECS |
ECS |
| AO090005001079 cluster |
AO090005001079 |
AO090005001078 - AO090005001087 |
AO090005001075 - AO090005001081 |
- |
n/a |
n/a |
| AO090005001551 cluster |
AO090005001551 |
- |
AO090005001668 - AO090005001555 |
- |
n/a |
n/a |
| AO090009000052 cluster |
AO090009000052 |
- |
AO090009000046 - AO090009000717 |
AO090009000051 - AO090009000717 |
IGD |
IGD |
| AO090009000071 cluster |
AO090009000071 |
AO090009000071 - AO090009000068 |
AO090009000079 - AO090009000065 |
AO090009000071 - AO090009000066 |
ECS, IGD |
ECS |
| AO090009000131 cluster |
AO090009000131 |
AO090009000143 - AO090009000131 |
AO090009000141 - AO090009000122 |
AO090009000140 - AO090009000131 |
FA |
ECS |
| AO090010000048 (aoi) cluster* |
AO090010000048 |
AO090010000035 - AO090010000054 |
AO090010000040 - AO090010000056 |
AO090010000040 - AO090010000056* |
ED |
ED |
| AO090010000082 cluster |
AO090010000082 |
AO090010000070 - AO090010000082 |
- |
AO090010000074 - AO090010000083 |
ECS |
ECS |
| AO090010000114 cluster |
AO090010000114 |
AO090010000097 - AO090010000114 |
AO090010000108 - AO090010000122 |
AO090010000104 - AO090010000115 |
ECS |
ECS |
| AO090010000204 cluster |
AO090010000204 |
- |
AO090010000202 - AO090010000207 |
AO090010000202 - AO090010000204 |
ECS |
ECS |
| AO090010000349 cluster |
AO090010000349 |
AO090010000349 - AO090010000350 |
AO090010000340 - AO090010000353 |
AO090010000348 - AO090010000349 |
ECS |
ECS |
| AO090010000404 cluster |
AO090010000404 |
AO090010000390 - AO090010000407 |
AO090010000388 - AO090010000414 |
AO090010000390 - AO090010000407 |
ECS, FA |
ECS |
| AO090010000426 cluster |
AO090010000426 |
AO090010000424 - AO090010000426 |
AO090010000417 - AO090010000435 |
AO090010000422 - AO090010000427 |
IGD |
IGD, ECS |
| AO090010000498 cluster |
AO090010000498 |
AO090010000488 - AO090010000500 |
AO090010000490 - AO090010000507 |
- |
n/a |
n/a |
| AO090011000015 cluster |
AO090011000015 |
AO090011000023 - AO090011000004 |
AO090011000022 - AO090011000009 |
AO090011000957 - AO090011000009 |
ECS |
ECS |
| AO090011000043 cluster |
AO090011000043 |
AO090011000043 - AO090011000031 |
AO090011000049 - AO090011000037 |
AO090011000043 - AO090011000040 |
ECS |
ECS |
| AO090011000103 cluster |
AO090011000103 |
- |
AO090011000098 - AO090011000107 |
AO090011000099 - AO090011000105 |
ECS |
ECS |
| AO090011000328 cluster |
AO090011000328 |
AO090011000328 - AO090011000961 |
AO090011000333 - AO090011000320 |
AO090011000329 - AO090011000326 |
ECS, IGD |
ECS, IGD |
| AO090011000408 cluster |
AO090011000408 |
- |
AO090011000403 - AO090011000413 |
- |
n/a |
n/a |
| AO090011000738 cluster |
AO090011000738 |
AO090011000744 - AO090011000729 |
AO090011000743 - AO090011000733 |
AO090011000740 - AO090011000738 |
ECS, FA |
ECS, FA |
| AO090012001034 cluster |
AO090012001034 |
AO090012001034 - AO090012000510 |
AO090012000526 - AO090012000510 |
AO090012000521 - AO090012001034 |
IGD, ECS |
ECS, IGD |
| AO090020000159 cluster |
AO090020000159 |
- |
AO090020000154 - AO090020000163 |
AO090020000156 - AO090020000162 |
ECS |
ECS |
| AO090020000186 cluster |
AO090020000186 |
AO090020000177 - AO090020000192 |
AO090020000172 - AO090020000191 |
AO090020000186 - AO090020000188 |
ECS |
ECS |
| AO090020000194 cluster |
AO090020000194 |
- |
AO090020000191 - AO090020000200 |
AO090020000194 - AO090020000202 |
ECS |
FA, ECS |
| AO090020000240 cluster |
AO090020000240 |
AO090020000236 - AO090020000241 |
AO090020000232 - AO090020000247 |
AO090020000234 - AO090020000241 |
ECS, IGD |
ECS, IGD |
| AO090020000380 cluster |
AO090020000380 |
AO090020000380 - AO090020000386 |
AO090020000369 - AO090020000388 |
AO090020000366 - AO090020000387 |
ECS, FA |
IGD, FA |
| AO090020000527 cluster |
AO090020000527 |
AO090020000515 - AO090020000535 |
AO090020000521 - AO090020000531 |
- |
n/a |
n/a |
| AO090023000082 cluster |
AO090023000082 |
AO090023000097 - AO090023000072 |
AO090023000096 - AO090023000074 |
AO090023000082 - AO090023000077 |
ECS |
FA |
| AO090023000444 cluster |
AO090023000444 |
AO090023000450 - AO090023000444 |
AO090023000450 - AO090023000436 |
AO090023000447 - AO090023000443 |
FA |
ECS |
| AO090023000528 cluster |
AO090023000528 |
AO090023000528 - AO090023000523 |
AO090023000536 - AO090023000521 |
AO090023000529 - AO090023000523 |
ECS, IGD |
ECS |
| AO090023000877 cluster |
AO090023000877 |
AO090023000881 - AO090023000875 |
AO090023000884 - AO090023000869 |
- |
n/a |
n/a |
| AO090026000009 (afl) cluster* |
AO090026000009 |
AO090026000004 - AO090026000029 |
AO090026000001 - AO090026000015 |
AO090026000009 - AO090026000021 |
ED |
ED |
| AO090026000149 cluster |
AO090026000157 |
AO090026000149 - AO090026000157 |
AO090026000141 - AO090026000157 |
AO090026000147 - AO090026000157 |
ECS, IGD |
ECS, IGD |
| AO090026000378 cluster |
AO090026000378 |
AO090026000378 - AO090026000388 |
AO090026000369 - AO090026000390 |
AO090026000378 - AO090026000384 |
ECS |
ECS, FA |
| AO090026000585 cluster |
AO090026000585 |
AO090026000575 - AO090026000585 |
AO090026000575 - AO090026000591 |
AO090026000575 - AO090026000586 |
FA |
FA |
| AO090038000086 cluster |
AO090038000086 |
AO090038000078 - AO090038000086 |
AO090038000078 - AO090038000092 |
- |
n/a |
n/a |
| AO090038000098 cluster |
AO090038000098 |
- |
AO090038000092 - AO090038000101 |
AO090038000098 - AO090038000100 |
ECS |
ECS |
| AO090038000149 cluster |
AO090038000149 |
AO090038000144 - AO090038000149 |
AO090038000139 - AO090038000156 |
AO090038000145 - AO090038000151 |
IGD |
IGD |
| AO090038000210 cluster |
AO090038000210 |
- |
AO090038000205 - AO090038000220 |
AO090038000210 - AO090038000214 |
FA |
ECS, IGD |
| AO090038000390 cluster |
AO090038000390 |
AO090038000390 - AO090038000399 |
AO090038000385 - AO090038000399 |
AO090038000390 - AO090038000392 |
ECS |
FA |
| AO090038000488 cluster |
AO090038000488 |
- |
AO090038000480 - AO090038000492 |
AO090038000486 - AO090038000490 |
FA |
IGD |
| AO090038000543 cluster |
AO090038000543 |
AO090038000543 - AO090038000557 |
AO090038000540 - AO090038000558 |
AO090038000543 - AO090038000550 |
ECS, IGD |
FA |
| AO090102000166 cluster |
AO090102000166 |
AO090102000166 - AO090102000169 |
AO090102000157 - AO090102000173 |
AO090102000165 - AO090102000170 |
FA |
FA |
| AO090102000322 cluster |
AO090102000322 |
AO090102000316 - AO090102000322 |
AO090102000318 - AO090102000324 |
AO090102000317 - AO090102000322 |
IGD |
ECS, IGD |
| AO090102000338 cluster |
AO090102000338 |
AO090102000329 - AO090102000338 |
AO090102000331 - AO090102000342 |
AO090102000336 - AO090102000338 |
FA |
ECS, IGD |
| AO090102000465 cluster |
AO090102000465 |
AO090102000457 - AO090102000466 |
AO090102000457 - AO090012001037 |
AO090102000464 - AO090102000466 |
FA |
ECS, IGD |
| AO090103000167 cluster |
AO090103000167 |
AO090103000165 - AO090103000179 |
AO090103000159 - AO090103000177 |
AO090103000165 - AO090103000170 |
ECS,IGD |
ECS |
| AO090103000224 cluster |
AO090103000224 |
AO090103000222 - AO090103000227 |
AO090103000215 - AO090103000231 |
AO090103000220 - AO090103000226 |
ECS, FA |
ECS |
| AO090103000355 cluster |
AO090103000355 |
AO090103000355 - AO090103000366 |
AO090103000347 - AO090103000364 |
AO090103000352 - AO090103000361 |
FA |
FA |
| AO090113000209 cluster |
AO090113000209 |
AO090113000209 - AO090113000204 |
AO090113000208 - AO090113000199 |
- |
n/a |
n/a |
| AO090120000024 cluster |
AO090120000024 |
AO090120000024 - AO090120000020 |
AO090120000033 - AO090120000013 |
AO090120000024 - AO090120000022 |
ECS |
FA, ECS |
| AO090124000040 cluster |
AO090124000040 |
- |
AO090124000035 - AO090124000048 |
AO090124000035 - AO090124000040 |
ECS, FA |
ECS |
| AO090206000074 cluster |
AO090206000074 |
AO090206000075 - AO090206000074 |
AO090206000082 - AO090206000067 |
AO090206000074 - AO090206000072 |
ECS, IGD |
ECS, IGD |
| AO090701000530 cluster |
AO090701000530 |
AO090701000525 - AO090701000543 |
AO090701000525 - AO090701000539 |
AO090701000525 - AO090701000530 |
ECS, IGD |
ECS, IGD |
| AO090701000600 cluster |
AO090701000600 |
AO090701000600 - AO090701000603 |
AO090701000912 - AO090701000604 |
AO090701000597 - AO090701000600 |
ECS |
ECS, IGD |
| AO090701000826 cluster |
AO090701000826 |
AO090701000826 - AO090701000833 |
AO090701000817 - AO090701000917 |
AO090701000824 - AO090701000838 |
ECS, IGD |
ECS |
| Non-PKS/NRPS cluster 1 |
n/a |
- |
AO090120000058 - AO090120000068 |
- |
n/a |
n/a |
| Non-PKS/NRPS cluster 2 | n/a | - | AO090113000164 - AO090113000175 | - | n/a | n/a |
* Experimentally characterized; Abbreviations: ECS, End of cluster synteny; IGD, Increase in intergenic distance; FA, Change in functional annotation; ED, Experimentally determined.
Table 8.
Number of gene clusters predicted by SMURF, antiSMASH, or manual and experimental methods
| SMURF | antiSMASH | Manual | Experimental | Total clusters predicted* | |
|---|---|---|---|---|---|
|
A. nidulans |
49 |
66 |
63 |
9 |
71 |
|
A. fumigatus |
33 |
38 |
29 |
5 |
39 |
|
A. niger |
79 |
70 |
66 |
0 |
81 |
| A. oryzae | 57 | 73 | 62 | 2 | 75 |
* The number for total clusters predicted is the sum total of non-redundant clusters predicted by any method for a species. Manual boundary predictions include those that were experimentally detemined and published, or were made by evaluating published expression profile data (M. Andersen et al, 2013), cluster synteny among Aspergillus strains/species, changes in functional annotation and/or increases in intergenic distance, in that order of relative weight.
Figure 1.
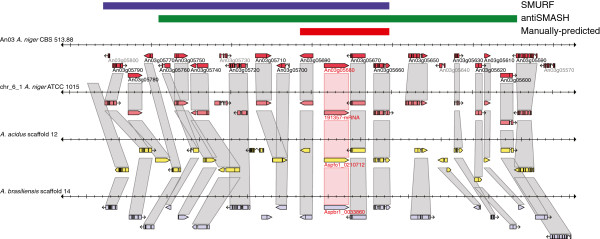
Genomic context of the predicted An03g05680 cluster of A. niger viewed with the Sybil multiple genome browser. Boundary predictions for A. niger CBS 513.88 species identifies predicted clusters in A. niger ATCC 1015, A. acidus and A. brasiliensis by matching orthologous protein clusters in Sybil. The red bar delineates the manually predicted cluster boundary based on cluster synteny between 2 A. niger strains and 2 additional Aspergillus species. The blue bar indicates the extent of the SMURF cluster prediction and the green bar indicates the antiSMASH-predicted boundaries.
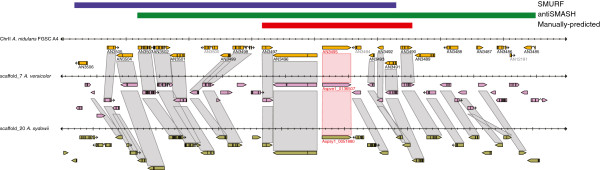
Andersen et al.[16] recently reported another strategy of identifying the extent of secondary metabolite gene cluster boundaries. Their method uses genome-wide microarray expression studies from A. nidulans to identify coregulated genes surrounding secondary metabolite gene cluster backbone enzymes. Since secondary metabolite gene clusters often show cryptic expression under many laboratory growth conditions, this study generated expression data from cultures grown on a wide variety of media (to maximize the possibility of expression), and combined these data with previously generated expression data to analyze a superset of 44 expression conditions [16]. Their analysis produced a list of 53 predicted secondary metabolite gene clusters of A. nidulans, some of which show clear patterns of coregulated expression while some of the expressed backbone enzymes showed no correlation with adjacent genes. Five of these were DTS-based gene clusters not identified by the SMURF or antiSMASH algorithms. These data have been curated at AspGD and were used as a criterion for our manual cluster boundary predictions (see below). An example of the inpA- and inpB-containing gene cluster determined by this criterion is shown in Figure 2. The gene clusters of A. nidulans with all of the boundary predictions made with ‘expression pattern’ as the primary evidence are listed in Table 4. The total number of boundaries predicted using this criterion is summarized in Table 9.
Figure 2.
A. nidulans AN3497 gene cluster predicted based of gene expression analysis of Andersen et al. 2013. Red bar indicates manually predicted cluster boundary (AN3490-AN3497) based on expression pattern and aligned with orthologous clusters of A. versicolor and A. sydowii. Blue bar indicates SMURF boundary prediction (AN3491-AN3506) and green bar indicates the antiSMASH-predicted boundary (AN3485-AN3503).
Table 9.
Summary of primary criteria used for making manual secondary metabolite gene cluster boundary predictions
| ED | EP | ECS | FA | IGD | |
|---|---|---|---|---|---|
|
A. nidulans |
24 (18%) |
38 (29%) |
47 (36%) |
17 (13%) |
6 (4%) |
|
A. fumigatus |
10 (15%) |
n/a |
39 (57%) |
7 (10%) |
12 (18%) |
|
A. niger |
0 (0%) |
n/a |
129 (98%) |
2 (<2%) |
1 (<1%) |
| A. oryzae | 8 (6%) | n/a | 90 (73%) | 17 (14%) | 9 (7%) |
Abbreviations: ED, Experimentally determined; EP, Published expression pattern (M. Anderson et al, 2013); ECS, End of cluster synteny; FA, Change in functional annotation; IGD, Increase in intergenic distance; n/a, not applicable.
To generate a high-quality set of candidate secondary metabolite biosynthetic gene clusters, we used SMURF and antiSMASH as the source of cluster predictions, along with manually predicted DTS clusters and then manually refined the gene cluster boundaries. Manual cluster boundary annotations (Tables 4, 5, 6, 7 and Additional files 2, 3, 4, 5) were made based on several criteria: published experimental data (including gene expression studies), synteny between clustered genes among different species indicated by the presence of conserved gene cluster boundaries (Figure 1), functional annotation of predicted genes within and adjacent to clusters and increases in intergenic distance between boundary genes and adjacent genes, which we frequently observed (Figure 3). We determined that gene clusters tend to be conserved between species and that breaks in cluster synteny frequently indicate a cluster boundary. To the best of our knowledge, no gene cluster prediction algorithm or research group has used genomic comparisons between species for large-scale cluster predictions. We used the Sybil viewer [51], which displays alignments of orthologous genes across multiple species in their genomic context, to manually examine potential boundaries and to compare synteny between clusters of different species and/or strains (Figure 1) and the adjacent syntenic regions outside each predicted cluster. The genome sequence is available for two strains each of A. fumigatus (Af293 and A1163) and A. niger (CBS513.88 and ATCC 1015), which allowed us to consider cluster synteny, which approached 100%, between these strains in addition to the orthology between Aspergillus species.
Figure 3.
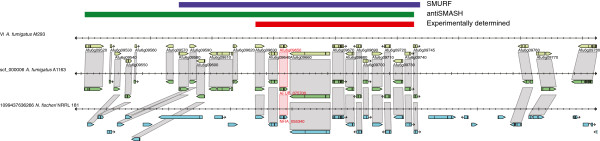
Conserved cluster synteny between the gliotoxin cluster of A. fumigatus and the orthologous cluster of Neosartorya fischeri. The predicted gene cluster is indicated with a red bar. The left border of the Afu6g09650 cluster shows a small increase in intergenic distance while the right border shows a large change in intergenic distance. Both borders are examples of interspecies cluster synteny. Red bar indicates experimentally determined cluster boundary (Afu6g09630 - Afu6g09740). Blue bar indicates SMURF boundary prediction (Afu6g09580 - Afu6g09740) and green bar indicates the antiSMASH-predicted boundary (Afu6g09520 - Afu6g09745).
AspGD displays and provides sequence resources for 15 Aspergillus genomes and related species. A given genome is typically particularly closely related to that of one or two of the other species; the A. fumigatus genome best matches that of Neosartorya fischeri (see Sybil syntenic genomic context in Additional file 3), A. niger best matches A. acidus and A. brasiliensis (Additional file 4) and A. oryzae best matches A. flavus (Additional file 5). Unlike A. fumigatus, A. niger and A. oryzae, A. nidulans lacks such a closely related species in AspGD with sufficient synteny to enable routine use of cluster orthology in boundary determination. Therefore, we used other criteria such as published gene expression patterns [16], increases in intergenic distance and changes from secondary metabolism-related gene annotations to non-secondary metabolism-related gene annotations (described below) for making these predictions in A. nidulans (Figure 1). The numbers of manually predicted gene clusters in each of these additional species, determined by observing breaks in gene cluster synteny (see Methods), are summarized in Table 9.
In some cases, the functional annotation of the putative gene cluster members was informative in predicting cluster boundaries, especially for A. nidulans, which often lacked cluster synteny with other species present in AspGD. In addition to genes encoding the core backbone enzymes, clusters typically include one or more acyl transferase, oxidoreductase, hydrolase, cytochrome P450, transmembrane transporter and a transcription factor. We manually inspected each cluster and the genomic region surrounding it; changes in functional annotations from typical secondary metabolism annotations to annotations atypical of secondary metabolic processes were frequently observed upon traversing a cluster boundary (Additional files 2, 3, 4, 5) and this was used as an additional criterion for boundary prediction, especially in cases where inter- or intra-species clustering or published gene expression data were not available. In some instances, genes with functional annotations unrelated to secondary metabolism are embedded within a cluster. For example, A. nidulans bglD (AN7915) encodes a glucosidase present in the F9775 biosynthetic gene cluster (Additional file 2). In a cclAΔ strain background in which histone 3 lysine 4 methylation is impaired, the expression of cryptic secondary metabolite clusters, such as F9775, is activated [52]. The activation of bglD expression was observed along with other genes in the F9775 cluster and based on this pattern of coregulation, bglD is included as a member of this cluster [52]. It is unclear, however, whether bglD actually plays a role in F9775 biosynthesis. The gene encoding translation elongation factor 1 gamma, stcT, is a member of the ST gene cluster (stc) of A. nidulans. Its inclusion in the stc cluster was based on its pattern of coregulation with 24 other genes, some of which have experimentally determined roles in A. nidulans ST biosynthesis, or are orthologous to A. parasiticus proteins involved in AF production, for which ST is a precursor [46]. We also observed a gene, AN2546, that is expressed, and is predicted to encode a glycosylphosphatidylinositol (GPI)-anchored protein [53], located in the emericellamide cluster (Additional file 2); however, an AN2546 deletion strain still produces emericellamide, thus its inclusion in the cluster is based on its genomic location and expression pattern rather than function. These examples indicate that some genes are located within clusters and yet may not contribute to secondary metabolite production. The frequency and significance of unrelated genes that have become incorporated into a secondary metabolism gene cluster remains unclear; experimental verification is needed to further assess these. In cases where the cluster synteny data were compelling, cluster synteny was given higher precedence than functional annotation in the delineation of the cluster boundaries.
Increases in the distance between predicted boundary genes and the gene directly adjacent to a boundary (which we refer to as intergenic distance) were frequently observed. An example with a large intergenic distance at the right boundary is shown in the A. fumigatus gliotoxin (gli) cluster (Figure 3). However, we found that more subtle increases in intergenic distance were only somewhat reliable when compared to boundaries with experimental evidence. We therefore only based a cluster boundary prediction on an increase in intergenic distance in a small number of cases where no other data were available (Table 9).
Discussion
AspGD provides high-quality manual and computational gene structure and function annotations for A. nidulans, A. fumigatus, A. niger and A. oryzae, along with sequence analysis and visualization resources for these and additional Aspergilli and related species. Among fungal databases, AspGD is the only resource performing comprehensive manual literature curation for Aspergillus species. AspGD contains curated data covering the entire corpus of experimental literature for A. nidulans, A. fumigatus, A. niger and A. oryzae, with phenotype and GO annotations for every gene described in the literature for these species, including those related to secondary metabolism. The direct, manual curation of genes from the literature forms the basis for the computational annotations at AspGD. This information, collected in a centralized, freely accessible resource, provides an indispensible resource for scientific information for researchers.
During the course of curation, we identified gaps in the set of GO terms that were available in the Biological Process branch of the ontology. To improve the GO annotations for secondary metabolite biosynthetic genes, we added new, more specific BP terms to the GO and used these new terms for direct annotation of Aspergillus genes. These terms include the specific secondary metabolite in each GO term name. Because ‘secondary metabolic process’ (GO:0019748) and ‘regulation of secondary metabolite biosynthetic process’ (GO:0043455) map to different branches in the GO hierarchy, complete annotation of transcriptional regulators of secondary metabolite biosynthetic gene clusters, such as laeA, requires an additional annotation to the regulatory term that we also added for each secondary metabolite.
GO annotations facilitate predictions of gene function across multiple species and, as part of this project, we used orthology relationships between experimentally characterized A. nidulans, A. fumigatus, A. niger and A. oryzae genes to provide orthology-based GO predictions for the unannotated secondary metabolism-related genes in AspGD. The prediction and complete cataloging of these candidate secondary metabolism-related genes will facilitate future experimental studies and, ultimately, the identification of all secondary metabolites and the corresponding secondary metabolism genes in Aspergillus and other species.
The SMURF and antiSMASH algorithms are efficient at predicting gene clusters on the basis of the presence of certain canonical backbone enzymes; however, disparities between boundaries predicted by these methods became obvious when the clusters predicted by each method were aligned. While there was an extensive overlap between the two sets of identified clusters, in most cases the cluster boundaries predicted by SMURF and antiSMASH were different, requiring manual refinement.
The data analysis of Andersen et al.[16] used a clustering matrix to identify superclusters, defined as clusters with similar expression, independent of chromosomal location, that are predicted to participate in cross-chemistry between clusters to synthesize a single secondary metabolite. They identified seven superclusters of A. nidulans. Two known meroterpenoid clusters that exhibit cross-chemistry, and are located on separate chromosomes, are the austinol (aus) clusters involved in the synthesis of austinol and dehydroaustinol [31,37]. The biosynthesis of prenyl xanthones in A. nidulans is dependent on three separate gene clusters [36]. This was apparent because the mdpG gene cluster was shown to be required for the synthesis of the anthraquinone emodin, monodictyphenone, and related compounds. Emodin and monodictyphenone are precursors of prenyl xanthones and the mdpG cluster lacked a prenyltransferase, required for prenyl xanthone synthesis [36]. A search of the A. nidulans genome for prenyltransferases that may participate in prenyl xanthone synthesis predicts seven prenyltransferases. Two strains (ΔxptA and ΔxptB) with mutated prenyltransferase genes at chromosomal locations distant from the mdpG cluster, have been described as being defective in prenyl xanthone synthesis. Therefore, while a total of 266 unique clusters were identified in our analysis, published data indicate that some of these clusters may function as superclusters that display cross-chemistry synthesis of a single secondary metabolite or group of related secondary metabolites [16,31,36].
Our manual annotation of secondary metabolite gene clusters in four Aspergillus species complements the computational prediction methods for identifying fungal secondary metabolites and the genes responsible for their biosynthesis. Implicit in our interspecies cluster synteny analysis is the prediction of secondary metabolite gene clusters orthologous to those in our curated species. For example, A. nidulans gene clusters most closely matched those in A. versicolor, thus identifying several new predicted A. versicolor gene clusters by orthology and interspecies cluster synteny with the predicted A. nidulans clusters (Additional file 2).
Conclusions
These new curated data, based on both computational analysis and manual evaluation of the Aspergillus genomes, provide researchers with a comprehensive set of annotated secondary metabolite gene clusters and a comprehensive functional annotation of the secondary metabolite gene products within AspGD. We anticipate that these new data will promote research in this important and complex area of Aspergillus biology.
Methods
Generation of new GO terms
The Gene Ontology Consortium requires that any compounds within BP term names in the GO be cataloged in the Chemical Entities of Biological Interest (ChEBI) database (http://www.ebi.ac.uk/chebi/). To enable the creation of the new GO terms, we first requested and were assigned ChEBI identifiers for all secondary metabolites recorded in AspGD. Once ChEBI term identifiers were assigned, the relevant GO terms were requested from the GO Consortium through TermGenie (http://go.termgenie.org/) for biosynthetic process, metabolic process and catabolic process terms for each new secondary metabolic process term and regulation of secondary metabolic process term (Additional file 1).
Orthologous protein predictions
Jaccard-clustering, which groups together highly similar proteins within a genome of interest, was used to make ortholog predictions between the Aspergillus species and is described in detail at http://sybil.sourceforge.net/documentation.html#jaccard. Briefly, the first step of this algorithm identifies highly similar proteins within each genome of interest. The resulting groups (“clusters”) from multiple genomes are themselves grouped in the second step to form orthologous groups (“Jaccard Orthologous Clusters”). The corresponding genes can be subsequently analyzed in their genomic context to visually identify conserved synteny blocks that are displayed in the Sybil genome viewer (aspgd.broadinstitute.org). The ortholog predictions for all AspGD species are available for download at http://www.aspergillusgenome.org/download/homology/orthologs/. Orthologous protein predictions between Saccharomyces cerevisiae, Schizosaccharomyces pombe and the Aspergillus protein sets were made by pair-wise comparisons using the InParanoid software [54]. InParanoid was chosen based on compatibility with the existing ortholog analysis pipeline at AspGD, and comparable accuracy when compared with alternative methods [55]. Stringent cutoffs were used: BLOSUM80 and an InParanoid score of 100% (parameters: -F \“m S\” -M BLOSUM80). The data from this comparison are available for download at (http://www.aspergillusgenome.org/download/homology/).
Orthology- and domain-based GO transfer
To augment the annotations for all genes, including secondary metabolism related genes, we used manual and domain-based GO annotations to annotate the predicted orthologs that lacked direct experimental characterization. Ortholog predictions for A. nidulans, A. fumigatus, A. niger and A. oryzae were made based on the characterized proteins of S. cerevisiae, S. pombe and the other Aspergillus species in AspGD. Candidate GO annotations to be used as the basis for these inferences are limited to those with experimental evidence, that is, with evidence codes of IDA (Inferred from Direct Assay), IPI (Inferred from Physical Interaction), IGI (Inferred from Genetic Interaction) or IMP (Inferred from Mutant Phenotype). Annotations that are themselves predicted in S. cerevisiae, S. pombe or in Aspergillus, either based on sequence similarity or by some other methods, are excluded from this group to avoid transitive propagation of predictions. Also excluded from the predicted annotation set are annotations that are redundant with existing, manually curated annotations or those that assign a related but less specific GO term. The orthology-based GO assignments are given the evidence code IEA (Inferred from Electronic Annotation) and displayed with the source species and name of the gene from which they were derived, along with a hyperlink to the appropriate gene page at AspGD, SGD or PomBase. The new annotations that have been manually assigned or electronically transferred from S. cerevisiae and S. pombe to A. nidulans, A. fumigatus, A. niger and A. oryzae, and between the Aspergillus species are summarized in Table 3.
Domain-based GO transfers were assigned to a lower precedence than orthology-based transfers. IprScan predicts InterPro domains based on protein sequences [56]. The Interpro2go mapping file (http://www.ebi.ac.uk/interpro) was used to map GO annotations to genes with the corresponding domain predictions. A domain-based GO prediction was made only if it was not redundant with an existing manually-curated or orthology-based GO term, or one of its parental terms, that was already assigned to an orthologous protein.
Finally, descriptions for genes lacking manual or GO-based annotations were constructed from the manual GO terms assigned to characterized orthologs. GO annotations were included with the following precedence: BP, followed by MF, and then CC. For genes that lacked experimental characterization and characterized orthologs, but had functionally characterized InterPro domains, descriptions were generated from the domain-based GO annotations. The same precedence rules applied as to the descriptions generated using orthology-based GO information. For genes that lacked experimental characterization and characterized orthologs, and without functionally characterized InterPro domains, but had uncharacterized orthologs, the descriptions simply list the orthology relationship because no inferred GO information was available.
Secondary metabolic gene cluster analysis and annotation
The pre-computed results file (smurf_output_precomputed_08.13.08.zip) was downloaded from the SMURF website (http://jcvi.org/smurf/index.php). Version 1.2.1 of the antiSMASH program [39] was downloaded from (http://antismash.secondarymetabolites.org/) and run locally on the chromosome and/or contig sequences of A. nidulans FGSC A4, A. fumigatus Af293, A. niger CBS 513.88 and A. oryzae RIB40. Details of the parameters the antiSMASH program uses to predict boundaries are in described in Medema et al. 1998 [39] and those for SMURF are described in Khaldi et al. 2010 [38]. The secondary metabolic gene clusters predicted by these programs were manually analyzed and annotated using functional data available for each gene in AspGD. Cluster membership was determined based on physical proximity of candidate genes to cluster backbone genes. Adjacent genes were added to the cluster if they had functional annotations common to known secondary metabolism genes. In cases where backbone genes had Jaccard orthologs in other species (see above), we required orthology between all other cluster members. Confirmation of orthology between clusters was facilitated by use of the Sybil multiple genome browser which can be used to evaluate synteny between species. We visually evaluated synteny by examining whether a gene that was putatively in a cluster had orthologs in the other species – where a gene in the species in which the cluster was identified no longer had orthologs in the other species that were adjacent, we inferred a break in synteny. Cluster boundaries were also determined by changes in common functional annotation, or by an increase in intergenic distances. tRNAs and other non-coding RNAs were excluded in cluster boundary analysis. Annotated images of the orthologous gene clusters are included in Additional files 2, 3, 4, 5.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DOI, MBA and MSS designed the project, DOI wrote the manuscript, GS, JRW, MBA and MSS edited the manuscript, DOI and MSS analyzed the data, DOI and MSS annotated the data, JB, GC, PS and FW performed bioinformatics analysis of the data. All authors read and approved the final manuscript.
Supplementary Material
Contains a table listing all GO terms available from the GO Consortium describing fungal secondary metabolic processes as of December 2012.
Contains a table listing the manually annotated gene clusters predicted by SMURF and antiSMASH for A. nidulans.
Contains a table listing manually annotated gene clusters predicted by SMURF and antiSMASH for A. fumigatus.
A table listing the manually annotated gene clusters predicted by SMURF and antiSMASH for A. niger.
A table listing manually annotated gene clusters predicted by SMURF and antiSMASH for A. oryzae.
Contributor Information
Diane O Inglis, Email: dinglis@stanford.edu.
Jonathan Binkley, Email: jpbinkley@stanford.edu.
Marek S Skrzypek, Email: marek.skrzypek@stanford.edu.
Martha B Arnaud, Email: arnaudm@stanford.edu.
Gustavo C Cerqueira, Email: gustavo@broadinstitute.org.
Prachi Shah, Email: prachi@stanford.edu.
Farrell Wymore, Email: wymore@stanford.edu.
Jennifer R Wortman, Email: jwortman@broadinstitute.org.
Gavin Sherlock, Email: gsherloc@stanford.edu.
Acknowledgements
The authors would like to thank Gail Binkley for the AspGD Oracle Database administration, Stuart Miyasato and Matt Simison for the AspGD database software and hardware maintenance and the editors at CheBI and the GO Consortium. We would also like to thank Vinita Joardar at JCVI for providing an updated set of A. oryzae secondary metabolite gene cluster predictions. This work was supported by the National Institute of Allergy and Infectious Diseases at the US National Institutes of Health [R01 AI077599 to GS and JW].
References
- Bhetariya PJ, Madan T, Basir SF, Varma A, Usha SP. Allergens/Antigens, toxins and polyketides of important Aspergillus species. Indian J Clin Biochem. 2011;26:104–119. doi: 10.1007/s12291-011-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlfs M, Albert M, Keller NP, Kempken F. Secondary chemicals protect mould from fungivory. Biol Lett. 2007;3:523–525. doi: 10.1098/rsbl.2007.0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCabe AP, van Liempt H, Palissa H, Unkles SE, Riach MB, Pfeifer E, von Döhren H, Kinghorn JR. Delta-(L-alpha-aminoadipyl)-L-cysteinyl-D-valine synthetase from Aspergillus nidulans. Molecular characterization of the acvA gene encoding the first enzyme of the penicillin biosynthetic pathway. J Biol Chem. 1991;266:12646–12654. [PubMed] [Google Scholar]
- MacCabe AP, Riach MB, Unkles SE, Kinghorn JR. The Aspergillus nidulans npeA locus consists of three contiguous genes required for penicillin biosynthesis. EMBO J. 1990;9:279–287. doi: 10.1002/j.1460-2075.1990.tb08106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón D, Carramolino L, Patiño C, Sánchez F, Peñalva MA. Cloning and characterization of the isopenicillin N synthetase gene mediating the formation of the beta-lactam ring in Aspergillus nidulans. Gene. 1987;57:171–181. doi: 10.1016/0378-1119(87)90120-X. [DOI] [PubMed] [Google Scholar]
- Yu JH, Leonard TJ. Sterigmatocystin biosynthesis in Aspergillus nidulans requires a novel type I polyketide synthase. J Bacteriol. 1995;177:4792–4800. doi: 10.1128/jb.177.16.4792-4800.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller NP, Segner S, Bhatnagar D, Adams TH. stcS, a putative P-450 monooxygenase, is required for the conversion of versicolorin A to sterigmatocystin in Aspergillus nidulans. Appl Environ Microbiol. 1995;61:3628–3632. doi: 10.1128/aem.61.10.3628-3632.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelkar HS, Keller NP, Adams TH. Aspergillus nidulans stcP encodes an O-methyltransferase that is required for sterigmatocystin biosynthesis. Appl Environ Microbiol. 1996;62:4296–4298. doi: 10.1128/aem.62.11.4296-4298.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butchko RA, Adams TH, Keller NP. Aspergillus nidulans mutants defective in stc gene cluster regulation. Genetics. 1999;153:715–720. doi: 10.1093/genetics/153.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelkar HS, Skloss TW, Haw JF, Keller NP, Adams TH. Aspergillus nidulans stcL encodes a putative cytochrome P-450 monooxygenase required for bisfuran desaturation during aflatoxin/sterigmatocystin biosynthesis. J Biol Chem. 1997;272:1589–1594. doi: 10.1074/jbc.272.3.1589. [DOI] [PubMed] [Google Scholar]
- Luque MI, Rodríguez A, Andrade MJ, Martín A, Córdoba JJ. Development of a PCR protocol to detect aflatoxigenic molds in food products. J Food Prot. 2012;75:85–89. doi: 10.4315/0362-028X.JFP-11-268. [DOI] [PubMed] [Google Scholar]
- Kupfahl C, Michalka A, Lass-Flörl C, Fischer G, Haase G, Ruppert T, Geginat G, Hof H. Gliotoxin production by clinical and environmental Aspergillus fumigatus strains. Int J Med Microbiol. 2008;298:319–327. doi: 10.1016/j.ijmm.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Lewis RE, Wiederhold NP, Lionakis MS, Prince RA, Kontoyiannis DP. Frequency and species distribution of gliotoxin-producing Aspergillus isolates recovered from patients at a tertiary-care cancer center. J Clin Microbiol. 2005;43:6120–6122. doi: 10.1128/JCM.43.12.6120-6122.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton CO, Bouzani M, Loeffler J, Rogers TR. Direct interaction studies between Aspergillus fumigatus and human immune cells; what have we learned about pathogenicity and host immunity? Front Microbiol. 2012;3:413. doi: 10.3389/fmicb.2012.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf DH, Heinekamp T, Remme N, Hortschansky P, Brakhage AA, Hertweck C. Biosynthesis and function of gliotoxin in Aspergillus fumigatus. Appl Microbiol Biotechnol. 2012;93:467–472. doi: 10.1007/s00253-011-3689-1. [DOI] [PubMed] [Google Scholar]
- Andersen MR, Nielsen JB, Klitgaard A, Petersen LM, Zachariasen M, Hansen TJ, Blicher LH, Gotfredsen CH, Larsen TO, Nielsen KF, Mortensen UH. Accurate prediction of secondary metabolite gene clusters in filamentous fungi. Proc Natl Acad Sci USA. 2013;110:E99–E107. doi: 10.1073/pnas.1205532110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez JF, Somoza AD, Keller NP, Wang CC. Advances in Aspergillus secondary metabolite research in the post-genomic era. Nat Prod Rep. 2012;29:351–371. doi: 10.1039/c2np00084a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhired S, Weber M, Kempf-Sontag A, Keller NP, Hoffmeister D. Accurate prediction of the Aspergillus nidulans terrequinone gene cluster boundaries using the transcriptional regulator LaeA. Fungal Genet Biol. 2007;44:1134–1145. doi: 10.1016/j.fgb.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Perrin RM, Federova ND, Bok JW, Cramer RA, Wortman JR, Kim HS, Nierman WC, Keller NP. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 2007;3:523–525. doi: 10.1371/journal.ppat.0030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JM, Keller NP. Secondary metabolism in fungi: does chromosomal location matter? Curr Opin Microbiol. 2010;13:431–436. doi: 10.1016/j.mib.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim FY, Hou Y, Chen Y, Oh JH, Lee I, Bugni TS, Keller NP. Genome-based cluster deletion reveals an endocrocin biosynthetic pathway in Aspergillus fumigatus. Appl Environ Microbiol. 2012;78:4117–4125. doi: 10.1128/AEM.07710-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YM, Szewczyk E, Nayak T, Davidson AD, Sanchez JF, Lo HC, Ho WY, Simityan H, Kuo E, Praseuth A, Watanabe K, Oakley BR, Wang CC. Molecular genetic mining of the Aspergillus secondary metabolome: discovery of the emericellamide biosynthetic pathway. Chem Biol. 2008;15:527–532. doi: 10.1016/j.chembiol.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja M, Chiang YM, Chang SL, Praseuth MB, Entwistle R, Sanchez JF, Lo HC, Yeh HH, Oakley BR, Wang CC. Illuminating the diversity of aromatic polyketide synthases in Aspergillus nidulans. J Am Chem Soc. 2012;134:8212–8221. doi: 10.1021/ja3016395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa T, Ishiuchi K, Praseuth A, Noguchi H, Hotta K, Watanabe K. Overexpressing transcriptional regulator in Aspergillus oryzae activates a silent biosynthetic pathway to produce a novel polyketide. ChemBioChem. 2012;13:855–861. doi: 10.1002/cbic.201200107. [DOI] [PubMed] [Google Scholar]
- Arnaud MB, Chibucos MC, Costanzo MC, Crabtree J, Inglis DO, Lotia A, Orvis J, Shah P, Skrzypek MS, Binkley G, Miyasato SR, Wortman JR, Sherlock G. The Aspergillus Genome Database, a curated comparative genomics resource for gene, protein and sequence information for the Aspergillus research community. Nucleic Acids Res. 2010;38:D420–427. doi: 10.1093/nar/gkp751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud MB, Cerqueira GC, Inglis DO, Skrzypek MS, Binkley J, Chibucos MC, Crabtree J, Howarth C, Orvis J, Shah P, Wymore F, Binkley G, Miyasato SR, Simison M, Sherlock G, Wortman JR. The Aspergillus Genome Database (AspGD): recent developments in comprehensive multispecies curation, comparative genomics and community resources. Nucleic Acids Res. 2012;40:D653–659. doi: 10.1093/nar/gkr875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. Gene Ontology Annotations and Resources. Nucleic Acids Res. 2012;41:D530–535. doi: 10.1093/nar/gks1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, Drabkin H, Eppig JT, Hill DP, Ni L, Ringwald M, Balakrishnan R, Cherry JM, Christie KR, Costanzo MC, Dwight SS, Engel S, Fisk DG, Hirschman JE, Hong EL, Nash RS, Sethuraman A, Theesfeld CL, Botstein D, Dolinski K, Feierbach B, Berardini T, Mundodi S, Rhee SY, Apweiler R, Barrell D, Camon E, Dimmer E, Lee V, Chisholm R, Gaudet P, Kibbe W, Kishore R, Schwarz EM, Sternberg P, Gwinn M, Hannick L, Wortman J, Berriman M, Wood V, de la Cruz N, Tonellato P, Jaiswal P, Seigfried T, White R. Gene Ontology Consortium. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodiyar VK, Hill DP, Howe D, Berardini TZ, Tweedie S, Talmud PJ, Breckenridge R, Bhattarcharya S, Riley P, Scambler P, Lovering RC. The representation of heart development in the gene ontology. Dev Biol. 2011;354:9–17. doi: 10.1016/j.ydbio.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk E, Chiang YM, Oakley CE, Davidson AD, Wang CC, Oakley BR. Identification and characterization of the asperthecin gene cluster of Aspergillus nidulans. Appl Environ Microbiol. 2008;74:7607–7612. doi: 10.1128/AEM.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo HC, Entwistle R, Guo CJ, Ahuja M, Szewczyk E, Hung JH, Chiang YM, Oakley BR, Wang CC. Two separate gene clusters encode the biosynthetic pathway for the meroterpenoids austinol and dehydroaustinol in Aspergillus nidulans. J Am Chem Soc. 2012;134:4709–4720. doi: 10.1021/ja209809t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez-Fernández O, Trigos A, Ramos-Balderas JL, Viniegra-González G, Deising HB, Aguirre J. Phosphopantetheinyl transferase CfwA/NpgA is required for Aspergillus nidulans secondary metabolism and asexual development. Eukaryot Cell. 2007;6:710–720. doi: 10.1128/EC.00362-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames BD, Haynes SW, Gao X, Evans BS, Kelleher NL, Tang Y, Walsh CT. Complexity generation in fungal peptidyl alkaloid biosynthesis: oxidation of fumiquinazoline A to the heptacyclic hemiaminal fumiquinazoline C by the flavoenzyme Af12070 from Aspergillus fumigatus. Biochemistry. 2011;50:8756–8769. doi: 10.1021/bi201302w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez JF, Chiang YM, Szewczyk E, Davidson AD, Ahuja M, Elizabeth Oakley C, Woo Bok J, Keller N, Oakley BR, Wang CC. Molecular genetic analysis of the orsellinic acid/F9775 gene cluster of Aspergillus nidulans. Mol Biosyst. 2010;6:587–593. doi: 10.1039/b904541d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiya S, Grundmann A, Li X, Li SM, Turner G. Identification of a hybrid PKS/NRPS required for pseurotin A biosynthesis in the human pathogen Aspergillus fumigatus. ChemBioChem. 2007;8:1736–1743. doi: 10.1002/cbic.200700202. [DOI] [PubMed] [Google Scholar]
- Sanchez JF, Entwistle R, Hung JH, Yaegashi J, Jain S, Chiang YM, Wang CC, Oakley BR. Genome-based deletion analysis reveals the prenyl xanthone biosynthesis pathway in Aspergillus nidulans. J Am Chem Soc. 2011;133:4010–4017. doi: 10.1021/ja1096682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen ML, Nielsen JB, Rank C, Klejnstrup ML, Holm DK, Brogaard KH, Hansen BG, Frisvad JC, Larsen TO, Mortensen UH. A genome-wide polyketide synthase deletion library uncovers novel genetic links to polyketides and meroterpenoids in Aspergillus nidulans. FEMS Microbiol Lett. 2011;321:157–166. doi: 10.1111/j.1574-6968.2011.02327.x. [DOI] [PubMed] [Google Scholar]
- Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 2010;47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39:W339–346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YM, Szewczyk E, Davidson AD, Keller N, Oakley BR, Wang CC. A gene cluster containing two fungal polyketide synthases encodes the biosynthetic pathway for a polyketide, asperfuranone, in Aspergillus nidulans. J Am Chem Soc. 2009;13:2965–2970. doi: 10.1021/ja8088185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann S, Schümann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat Chem Biol. 2007;3:213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- Gerke J, Bayram O, Feussner K, Landesfeind M, Shelest E, Feussner I, Braus GH. Breaking the silence: protein stabilization uncovers silenced biosynthetic gene clusters in the fungus Aspergillus nidulans. Appl Environ Microbiol. 2012;78:8234–8244. doi: 10.1128/AEM.01808-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann S, Funk AN, Scherlach K, Schroeckh V, Shelest E, Horn U, Hertweck C, Brakhage AA. Activation of a silent fungal polyketide biosynthesis pathway through regulatory cross talk with a cryptic nonribosomal peptide synthetase gene cluster. Appl Environ Microbiol. 2010;76:8143–8149. doi: 10.1128/AEM.00683-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YM, Szewczyk E, Davidson AD, Entwistle R, Keller NP, Wang CC, Oakley BR. Characterization of the Aspergillus nidulans monodictyphenone gene cluster. Appl Environ Microbiol. 2010;76:2067–2074. doi: 10.1128/AEM.02187-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J. Clusters of genes for the biosynthesis of antibiotics: regulatory genes and overproduction of pharmaceuticals. J Ind Microbiol. 1992;9:73–90. doi: 10.1007/BF01569737. [DOI] [PubMed] [Google Scholar]
- Brown DW, Yu JH, Kelkar HS, Fernandes M, Nesbitt TC, Keller NP, Adams TH, Leonard TJ. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc Natl Acad Sci USA. 1996;93:1418–1422. doi: 10.1073/pnas.93.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok JW, Hoffmeister D, Maggio-Hall LA, Murillo R, Glasner JD, Keller NP. Genomic mining for Aspergillus natural products. Chem Biol. 2006;13:31–37. doi: 10.1016/j.chembiol.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Robinson SL, Panaccione DG. Chemotypic and genotypic diversity in the ergot alkaloid pathway of Aspergillus fumigatus. Mycologia. 2012;104:804–812. doi: 10.3852/11-310. [DOI] [PubMed] [Google Scholar]
- Maiya S, Grundmann A, Li SM, Turner G. The fumitremorgin gene cluster of Aspergillus fumigatus: identification of a gene encoding brevianamide F synthetase. ChemBioChem. 2006;7:1062–1069. doi: 10.1002/cbic.200600003. [DOI] [PubMed] [Google Scholar]
- Gardiner DM, Howlett BJ. Bioinformatic and expression analysis of the putative gliotoxin biosynthetic gene cluster of Aspergillus fumigatus. FEMS Microbiol Lett. 2005;248:241–248. doi: 10.1016/j.femsle.2005.05.046. [DOI] [PubMed] [Google Scholar]
- Crabtree J, Angiuoli SV, Wortman JR, White OR. Sybil: methods and software for multiple genome comparison and visualization. Meth Mol Biol. 2007;408:93–108. doi: 10.1007/978-1-59745-547-3_6. [DOI] [PubMed] [Google Scholar]
- Bok JW, Chiang YM, Szewczyk E, Reyes-Dominguez Y, Davidson AD, Sanchez JF, Lo HC, Watanabe K, Strauss J, Oakley BR, Wang CC, Keller NP. Chromatin-level regulation of biosynthetic gene clusters. Nat Chem Biol. 2009;5:462–464. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot PW, Brandt BW, Horiuchi H, Ram AF, de Koster CG, Klis FM. Comprehensive genomic analysis of cell wall genes in Aspergillus nidulans. Fungal Genet Biol. 2009;46:S72–81. doi: 10.1016/j.fgb.2008.07.022. [DOI] [PubMed] [Google Scholar]
- Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- Altenhoff AM, Dessimoz C. Phylogenetic and Functional Assessment of Orthologs Inference Projects and Methods. PLoS Comput Biol. 2009;5:e1000262. doi: 10.1371/journal.pcbi.1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Contains a table listing all GO terms available from the GO Consortium describing fungal secondary metabolic processes as of December 2012.
Contains a table listing the manually annotated gene clusters predicted by SMURF and antiSMASH for A. nidulans.
Contains a table listing manually annotated gene clusters predicted by SMURF and antiSMASH for A. fumigatus.
A table listing the manually annotated gene clusters predicted by SMURF and antiSMASH for A. niger.
A table listing manually annotated gene clusters predicted by SMURF and antiSMASH for A. oryzae.


