Abstract
Polymorphisms of the vitamin D receptor gene (VDR) have been associated inconsistently with various diseases, across populations of diverse origin. The T(f) allele of the functional SNP FokI, in exon 2 of VDR, results in a longer vitamin D receptor protein (VDR) isoform, proposed to be less active. Genetic association of VDR with disease is likely confounded by ethnicity and environmental factors such as plasma 25(OH)D3 status. We hypothesized that VDR expression, VDR level and transactivation of target genes, CAMP and CYP24A1, depend on vitamin D, ethnicity and FokI genotype. Healthy volunteers participated in the study (African, n = 40 and White, n = 20). Plasma 25(OH)D3 levels were quantified by LC-MS and monocytes cultured, with or without 1,25(OH)2D3. Gene expression and protein level was quantified using qRT-PCR and flow cytometry, respectively. Mean plasma 25(OH)D3 status was normal and not significantly different between ethnicities. Neither 25(OH)D3 status nor 1,25(OH)2D3 supplementation significantly influenced expression or level of VDR. Africans had significantly higher mean VDR protein levels (P<0.050), nonetheless transactivated less CAMP expression than Whites. Genotyping the FokI polymorphism by pyrosequencing together with HapMap data, showed a significantly higher (P<0.050) frequency of the CC genotype in Africans than in Whites. FokI genotype, however, did not influence VDR expression or VDR level, but influenced overall transactivation of CAMP and 1,25(OH)2D3-elicited CYP24A1 induction; the latter, interacting with ethnicity. In conclusion, differential VDR expression relates to ethnicity, rather than 25(OH)D3 status and FokI genotype. Instead, VDR transactivation of CAMP is influenced by FokI genotype and, together with ethnicity, influence 1,25(OH)2D3-elicited CYP24A1 expression. Thus, the expression and role of VDR to transactivate target genes is determined not only by genetics, but also by ethnicity and environment involving complex interactions which may confound disease association.
Introduction
The vitamin D receptor (VDR) is a ligand-activated transcription factor that mediates the genomic actions of vitamin D. These actions involve regulation of calcium homeostasis, cell growth and differentiation, detoxification of xenobiotics, and modulation of adaptive and innate immunity; the latter including activation of monocyte-macrophages [1], [2]. 1,25(OH)2D3-bound VDR facilitates heterodimerization with the retinoid X receptor (RXR) and binding to vitamin D response elements (VDREs), essential for transcription of VDR-regulated genes. Thus, 1,25(OH)2D3 availability determines VDR-mediated transactivation of target genes. The genes coding for 1,25(OH)2D3-catabolizing cytochrome P450 enzyme (CYP24A1) and the human cathelicidin antimicrobial protein (CAMP) are examples of 1,25(OH)2D3-regulated target genes. Differential expression of CYP24A1 and CAMP may affect vitamin D status [3] and susceptibility to infectious diseases [4], respectively. Two functional VDREs have been characterized in the promoter of murine CYP24A1 genes [5], [6] and at least two functional VDREs downstream of the human CYP24A1 [7]. CAMP contains at least one identified VDRE in its promoter region [8], and is induced by 1,25(OH)2D3 supplementation in primary keratinocytes, monocytes, and neutrophils [9].
Across populations, single nucleotide polymorphisms (SNPs) in the VDR have been associated inconsistently with diseases of diverse etiology, including tuberculosis (TB), multiple sclerosis, systemic lupus erythematosus (SLE), cirrhosis and various types of cancer [10]. Among VDR SNPs, the functional SNP rs2228570, commonly known as FokI, has been studied extensively. The ancestral f allele (T nucleotide) for this start codon polymorphism, codes for a full length VDR, while the F allele (C nucleotide) results in a three amino acid truncated VDR protein. While it has been shown that the shorter isoform interacts more efficiently with TFIIB [11], [12], reports on the impact of FokI on transactivation are conflicting. Comparing isoforms, Van Etten et al. (2007) found no difference in the transactivation mediated by classical DR3-type VDREs [13], while Alimirah et al. (2011) showed a 1.8 fold higher transactivation of CYP24A1 by the shorter variant, compared to the longer isoform [14].
The impact of debilitating variants on VDR function could be exacerbated by vitamin D deficiency or, alternatively, reduced by adequate vitamin D production or intake. For example, Wilkinson et al. (2000) observed the TT/Tt genotype of the VDR TaqI SNP to be associated with TB in Guajarati Indians living in London, only if vitamin D status was inadequate [15]. The risk of colorectal cancer, the cancer most strongly associated with VDR, more than doubles when individuals carrying the ff genotype of FokI consume a low-calcium or low-fat diet, compared to FF genotypes [16]. Thus, vitamin D status influences the impact of VDR variants on VDR function and associated disease risk. The widely studied association between vitamin D status and disease supports vitamin D deficiency to be involved in impaired immune function. While latitude and consequent UVB intensity influences vitamin D production in the skin [17], a major determinant of vitamin D status is believed to be skin melanin concentration [18]; with individuals with the darker skin type, VI, having notably less vitamin D than those with white skin type I. Similar to VDR variants being associated with disease, vitamin D deficiency have been associated with a higher incidence of TB [19], [20], colorectal cancer [21], [22], [23], cardiovascular disease and SLE [24]. It is uncertain whether the association between vitamin D deficiency and disease prevalence is the cause or effect of disease [20]. Randomized control trials assessing the effect of vitamin D supplementation on disease incidence and prognosis had mixed outcomes. Murdoch et al. (2012) observed no impact of vitamin D supplementation on the incidence of upper respiratory tract infections [25]. Gepner et al. (2012) observed reduced cardiovascular disease risk in postmenopausal women with vitamin D supplementation [26]. Few studies have, however, evaluated whether interactions between genetic variants in the VDR and vitamin D status could confound disease association in diverse populations. In one such a study, Martineau et al. (2011) observed an interaction between vitamin D supplementation and the TaqI VDR polymorphism in TB patients; the tt genotype, but not the Tt/TT, reducing sputum conversion time [27].
We hypothesized that VDR expression, VDR level and transactivation of target genes, CAMP and CYP24A1, depend on a combination of vitamin D, ethnicity and FokI genotype. We assessed the effect of vitamin D, ethnicity and FokI genotype on expression and the functional capabilities of the VDR. Results support that differential VDR expression relates to ethnicity, rather than 25(OH)D3 status and FokI genotype. Instead, VDR transactivation of CAMP is influenced by FokI genotype which, together with ethnicity, influences 1,25(OH)2D3-elicited CYP24A1 expression.
Materials and Methods
Participants and sample collection
Participants were healthy blood donors from the South African National Blood Service (SANBS). Ethical clearance was approved for this study by the Ethics Committees of SANBS and the University of Johannesburg, Faculty of Science. After informed written consent, SANBS collected blood from volunteers by venepuncture and prepared buffy coats. The study included only donors of legal donating age (16 and above), therefore consent from the next of kin, caretakers, or guardians on the behalf of the minors/children participants was not required. Buffy coats, tested to be HIV negative, were supplied anonymously within 24 h of venepuncture. Demographics of the study population are summarized in Table 1.
Table 1. Demographics of the study population.
| Demographic parameters | |
| Number of subjects | 60 |
| Sex | |
| MaleFemale | 2931 |
| Ethnicity | |
| African aWhite | 4020 |
| Mean age (range) | 35 (17–65) |
African donors belonged to any one of the 4 major ethnic groups living in South Africa. These include the Nguni, Sotho, Shangaan-Tsonga and Venda groups. The Nguni group can be subdivided into Zulu and Xhosa. Donors were collected in the Gauteng province of South Africa, residing in the urban region in and around Johannesburg.
Plasma vitamin D quantification
Plasma 25-hydroxyvitamin D3 (25(OH)D3) level is currently the best representative measure of vitamin D status; as 1,25(OH)2D3 level is under tight control by parathyroid hormone, calcium and phosphorus levels and kept mostly within reference ranges [28]. The levels of 25(OH)D3 were quantified in plasma by LC-MS at the Department of Chemical Pathology, Faculty of Health Science, University of Witwatersrand. Plasma 25(OH)D3 was extracted according to manufacturer's guidelines using the ClinRep® HPLC Complete 25-OH-Vitamin D2/D3 Kit (RECIPE, Germany). A commercially available internal standard (RECIPE, Germany) was included and LC-MS was performed using a m/z transition of 401>383 for quantification of 25(OH)D3. Serum pools from the Vitamin D External Quality Assessment Scheme (DEQAS, UK) were included in the analysis for quality control purposes.
Monocyte cultures and treatment
Peripheral blood mononuclear cells (PBMCs) were isolated from the buffy coats using a histopaque-1077® gradient (Sigma Aldrich, St Louis, MO). PBMCs were suspended in tissue culture media containing RPMI (GIBCO, Auckland, New Zealand), 10% FCS, 1% Streptomycin and 1% L-glutamine (Highveld Biological, Johannesburg, South Africa). Cells were seeded on a growth area of 189 cm2 per buffy coat and allowed to adhere for 2 h at 37°C, 5% CO2. Adhered monocytes were washed, harvested and reseeded at 10×106 cells per culture dish (60 mm diameter). Cultures were left untreated for 16 h after which they were stimulated for 24 h in the presence or absence of 10 nM 1,25(OH)2D3 (Sigma Aldrich, St Louis, MO). In addition to environmental factors, 25(OH)D3 conversion to the active 1,25(OH)2D3 may be influenced by polymorphisms in, for example, the 1á-hydroxylase gene (CYP27A1) [29]. Treating cells with the active 1,25(OH)2D3 would overcome any genetic variation in this regard and was therefore the metabolite of choice for in vitro supplementation.
qRT-PCR
Expression of VDR and its target genes (CAMP and CYP24A1), were determined by quantitative reverse transcriptase PCR (qRT-PCR). RNA was extracted from monocyte-macrophages with QIAzol® lysis reagent (QIAGEN Sciences, Maryland, USA) according to the manufacturer's guidelines, with the exception that the lysis reagent was increased to 2 ml per 4×106 cells. Extracted RNA was re-dissolved in 25 ìl DEPC-treated water, quantified using Nanodrop spectrophotometry and integrity evaluated using agarose gel electrophoresis. DNA contamination was eliminated using the RQ-1 RNase-free DNase kit (Promega, SA) according to the manufacturer's guidelines. cDNA synthesis was performed using the Tetro cDNA synthesis kit (Bioline, Celtic MolecularDiagnostics, SA). qPCR reactions for each treatment were carried out in duplicate, using Sensimix ™ SYBR No-ROX kit (Bioline, Celtic Diagnostics SA) and the CFX96™ Real-time system, C1000™ Thermal Cycler. Gene normalisation was performed against two stably expressed reference genes: Ubiquitin C (UBC) and tyrosine-3-monooxygenase/tryptophan-5-monooxygenase activation protein, zeta polypeptide (YWHAZ) [30]. Gene expression was quantified using the comparative CT method according to the MIQE guidelines [31], using inter-run calibrators [32] and qBASEPLUS software [33]. Primer sequences used in this study are listed in Table 2.
Table 2. qRT-PCR primer sequences for target and reference gene amplification.
| Gene | Forward primer | Reverse primer |
| VDR | 5′ CTGACCCTGGAGACTTTGAC 3′ | 5′ TTCCTCTGCACTTCCTCATC 3′ |
| CAMP | 5′ GCAGTCACCAGAGGATTGTGAC 3′ | 5′ CACCGCTTCACCAGCCC 3′ |
| CYP24A1 | 5′ ATGAGCACGTTTGGGAGGAT 3′ | 5′ TGCCAGACCTTGGTGTTGAG 3′ |
| UBC | 5′ ATTTGGGTCGCGGTTCTTG 3′ | 5′ TGCCTTGACATTCTCGATGGT 3′ |
| YWHAZ | 5′ ACTTTTGGTACATTGTGGCTTCAA 3′ | 5′ CCGCCAGGACAAACCAGTAT 3′ |
Primer sequences obtained from RTPrimerDB (Bustin et al., 2009; http://medgen.ugent.be/rtprimerdb/)
Flow cytometry
Intracellular VDR protein levels were quantified by flow cytometry in triplicate. Cells (1×106) were permeabilized with 0.2% Triton X-100 (Sigma Aldrich, St Louis, MO) in phosphate buffered saline (PBS). Permeabilized cells were incubated with mouse anti-human IgG2 monoclonal antibody against VDR, purchased from Santa Cruz Biotechnologies, Santa Cruz, CA (20 µg/ml 1% BSA/PBS, 30 min). Unbound primary antibody was washed off (0.2% Triton X-100 in PBS) before the cells were labelled with FITC-conjugated goat anti-mouse-IgG2a secondary antibody (8 µg/ml 1% BSA/PBS) purchased from Santa Cruz Biotechnologies, Santa Cruz, CA. Fluorescence was quantified using a BD FACS ARIA™ Flow Cytometer (excitation: 488 nm, emission: 525 nm). Bead-normalized compensation was performed to control for technical variation in fluorescence readings over time [34].
Genotyping
The FokI polymorphism (rs2228570) in the VDR (chr12:48272895, NCBI dbSNP build 137) was genotyped using pyrosequencing. Genomic DNA was extracted from monocytes using the Nucleon™ BACC2 Genomic DNA Extraction kit at 4°C, according to the manufacturer's instructions (GE Healthcare, Buckinghamshire, UK). Extracted DNA was re-dissolved in 50 µl Tris-EDTA buffer (TE, pH 7.4, Sigma Aldrich, St Louis, MO). Genotyping was outsourced to Epigen DX (MA, USA). HapMap population data were obtained from the International HapMap Project, including HapMap Phase I, II and III samples (release 27) from four populations [35]. These populations include individuals from the Centre d′Etude du Polymorphisme Humain (CEPH) collected in Utah, USA, with ancestry from northern and western Europe (CEU; n = 113); Yoruba in Ibadan, Nigeria (YRI; n = 112); Luhya in Webuye, Kenya (LWK; n = 86) and Maasai in Kinyawa, Kenya (MKK; n = 142).
Statistical Analysis
Statistical analysis was performed using IBM® SPSS® Statistics version 21 for Windows (SPSS Inc., Chicago, Illinois). Gene expression data showed an overall positive skewness and was ln-transformed to obtain normal distribution, meeting the assumption for parametric tests. Two-way ANOVA was used to test whether an interaction exists between treatment and ethnicity or whether these factors have main effects on the data. Pair-wise comparisons of means were computed with the Fisher's least significant difference (LSD) test, with Bonferroni correction for multiple variables. An independent t-test was used to compare plasma 25(OH)D3 levels between Africans and Whites. Chi-square analysis of the frequency distribution of genotypes between populations was performed using Microsoft Excel®.
Data availability
Data presented in this manuscript has been deposited in the Dryad Repository: http://dx.doi.org/10.5061/dryad.12dp5.
Results
Ethnicity influences VDR expression and protein level
To determine whether ethnicity or 1,25(OH)2D3 supplementation influence VDR expression and protein level, primary monocyte-macrophage cultures were established from an African and White population and supplemented in vitro with or without 10 nM 1,25(OH)2D3 for 24 h. VDR expression and protein level were quantified and the data was analysed with ethnicity as a fixed factor. Two-way ANOVA revealed a significant main effect of ethnicity on VDR mRNA (P<0.050) and protein level (P<0.001), without treatment interaction (Fig. 1). Fisher's least significant difference (LSD) test showed a significantly higher mean VDR protein level in Africans compared to Whites at basal and control conditions (P<0.050) and in the presence of in vitro 1,25(OH)2D3 supplementation (P = 0.050; Fig. 1B). In vitro 1,25(OH)2D3 supplementation for 24 h did not significantly alter VDR mRNA or protein level compared to the vehicle-treated control irrespective of ethnicity. Post-hoc LSD significance for differences in VDR protein level between Africans and Whites was not maintained after Bonferroni correction.
Figure 1. Ethnicity influenced VDR mRNA and protein level.
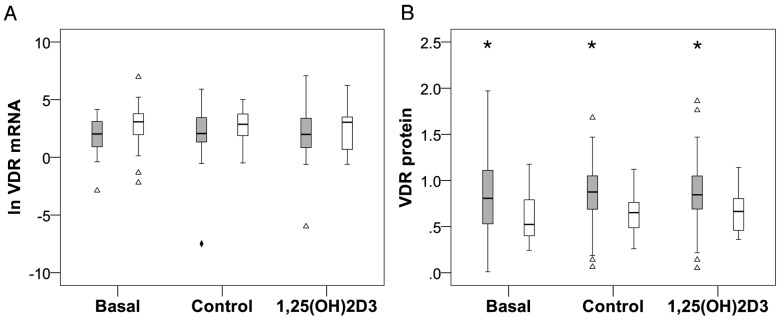
In vitro VDR expression (A) and protein level (B) were quantified in monocyte-macrophages from healthy Africans (grey, n = 40) and Whites (white, n = 20), using RT-qPCR and flow cytometry respectively. Monocytes were analysed directly after isolation (Basal), or cultured for 24 h with 10 nM 1,25(OH)2D3 or vehicle control. Box plots show data distribution: boxes illustrate 50% of cases or interquartile range (IQR, 25th to 75th percentile); horizontal lines, median; whiskers, 1.5 IQR from box, or minimum or maximum values if no case has a value in that range. Africans displayed more variance than Whites as illustrated by the distribution IQRs and outliers (Δ, 1.5 to 3 IQR from box) and extreme outlier (♦, >3 IQR from box). Approximately 95% of the data lie between whiskers. Two-way ANOVA showed an overall, significant main effect for ethnicity; Africans having lower VDR mRNA (P<0.050) but higher protein level (P<0.001) compared to Whites. Fisher's LSD test showed a significantly higher mean VDR protein level in Africans compared to Whites under all conditions (* P<0.050). VDR mRNA data was ln-transformed to meet the assumptions of parametric statistical tests.
VDR-1,25(OH)2D3 transactivation of target gene CAMP, not CYP24A1, was influenced by ethnicity
To evaluate VDR function the mRNA level of VDR target genes, CAMP and CYP24A1, was quantified in response to in vitro 1,25(OH)2D3 supplementation. In vitro 1,25(OH)2D3 supplementation significantly induced CAMP and CYP24A1 gene expression in both Africans (CAMP: P<0.010; CYP24A1: P<0.001) and Whites (CAMP: P<0.050; CYP24A1: P<0.001). Ethnicity had a significant main effect on CAMP (P<0.010), being higher in Whites, but not on CYP24A1 mRNA level (Fig. 2). The extent of CAMP induction by 1,25(OH)2D3 was ethnicity dependent, as pair-wise comparisons (LSD) showed significantly higher mean 1,25(OH)2D3-elicited CAMP expression in Whites compared to Africans (P<0.050). Although the same trend was observed for CYP24A1 expression, the difference was not significant. The significant difference in CAMP levels between Africans and Caucasians was not maintained after Bonferoni correction. Ethnicity-dependent differences in baseline levels of VDR protein (Fig. 1B) prompted the evaluation of transactivation efficiency of VDR (1,25(OH)2D3-elicited target gene expression level/VDR protein level). In the presence of 10 nM 1,25(OH)2D3, VDR transactivation efficiency was marginally lower in Africans than Whites for CAMP (Africans = 1.79; Whites = 2.17) and CYP24A1 (Africans = 3.73; Whites = 4.04) induction.
Figure 2. 1,25(OH)2D3-elicited transactivation of target gene CAMP by VDR is influenced by ethnicity.
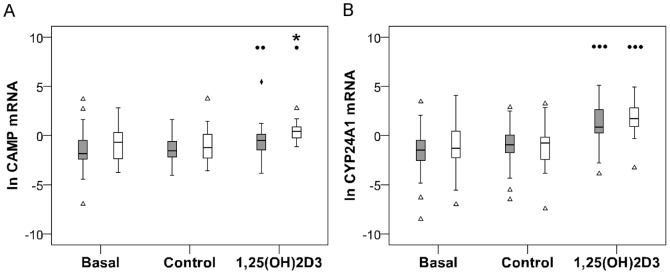
Box plots illustrate expression of VDR target genes, CAMP (A) and CYP24A1 (B). Data is differentiated by ethnicity: Africans (grey, n = 40) and Whites (white, n = 20). Gene expression was quantified in monocyte-macrophages from healthy individuals using RT-qPCR. In vitro 1,25(OH)2D3 supplementation significantly induced CAMP and CYP24A1 expression in both Africans and Whites relative to the vehicle control level (P<.050, P<0.010, P<0.001). Ethnicity had a significant main effect on CAMP (P<0.010), but not CYP24A1 mRNA level. Significantly higher mean CAMP mRNA level was observed in Whites compared to Africans after 1,25(OH)2D3 supplementation (* P<0.050). CAMP and CYP24A1 mRNA data was ln-transformed to meet the assumptions of parametric statistical analysis. Outliers are defined in legend for Fig. 1.
The normal 25(OH)D3 status in Africans and Whites did not influence VDR expression, protein level or function
To determine whether variation in the 25(OH)D3 status of Africans and Whites contributed to the differential VDR function, plasma 25(OH)D3 level was quantified using LC-MS. Overall, the study population had a mean 25(OH)D3 status of 87 nmol/L (Fig. 3). Although Whites (92.9 nmol/L) had a slightly higher mean 25(OH)D3 status compared to Africans (84.4 nmol/L), the difference was not significant. Neither overall nor condition-specific correlation analysis showed any relation between 25(OH)D3 status and in vitro VDR expression, VDR level or VDR function in monocyte-macrophages for Africans and Whites combined or in isolation (data not shown).
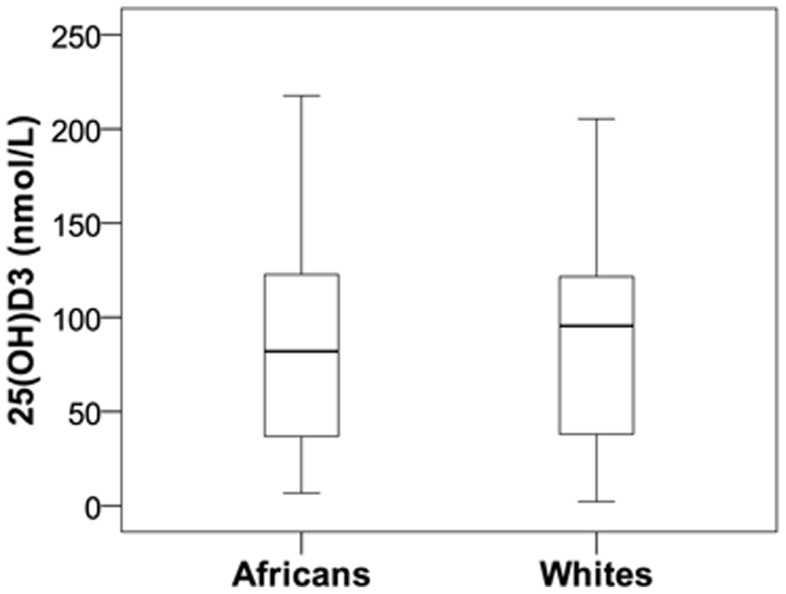
Figure 3. 25(OH)D3 status of Africans and Whites of the Gauteng Province of South Africa is normal and not significantly different.
The mean plasma 25(OH)D3 status of Africans (84.4 nmol/L, n = 30) and Whites (92.4 nmol/L, n = 14), quantified using LC-MS, was normal according to the IOM recommendations and not significantly different between the two ethnic groups.
The impact of increasing in vitro 1,25(OH)2D3 supplementation on CAMP expression
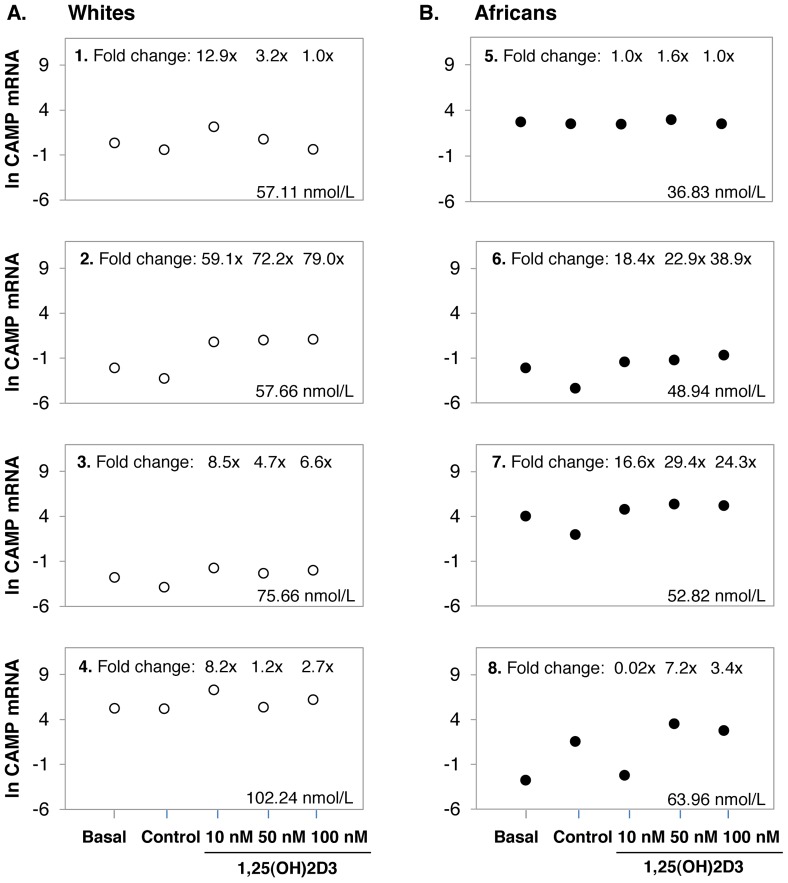
To determine whether the effect of ethnicity on in vitro VDR expression, VDR protein level and function would be influenced by increasing concentrations of 1,25(OH)2D3, a dose response was performed in an independent study in Africans (n = 4) and Whites (n = 4). Considering the mean of only 4 replicates per group, there was no differential response between Africans and Whites, as observed in the larger study, likely due to the notable inter-individual variation in responses, eminent throughout the study (Fig. 4, data not shown for VDR and CYP24A1). However, the trend for higher CAMP in Whites and lower CAMP in Africans at 10 nM, appeared to be reversed at higher levels of 1,25(OH)2D3 supplementation (50 nM and 100 nM).The notable inter-individual variation observed in the larger study prompted us to consider individual responses in the dose response analysis (Fig. 5). In all four White individuals 10 nM 1,25(OH)2D3 increased CAMP expression, which was only true in the case of two of the four African individuals. The two Africans lacking a response at 10 nM (individual 5 and 8) increased CAMP expression at 50 nM 1,25(OH)2D3, but not at 100 nM. In 75% of Whites responding at 10 nM (individual 1, 3 and 4), and 75% of Africans responding at 50 nM (individual 5, 7 and 8), increased 1,25(OH)2D3 concentration was not necessarily beneficial in terms of CAMP expression, and in some cases reduced CAMP expression. For the 8 individuals used in the dose-response study, the mean plasma 25(OH)D3 status was not significantly different between Africans and Whites and did not correlate with VDR mRNA level, VDR protein level or function in any condition.
Figure 4. The impact of increasing in vitro 1,25(OH)2D3 supplementation on CAMP expression.

Error-bar plots illustrate the mean level of CAMP expression. Data is differentiated by ethnicity: Africans (black, n = 4) and Whites (white, n = 4). Gene expression was quantified in monocyte-macrophages from healthy individuals using RT-qPCR before (basal) and after 24 h of in vitro 1,25(OH)2D3 supplementation at increasing concentrations (10 nM, 50 nM, and 100 nM). The trend for higher CAMP in Whites and lower CAMP in Africans at 10 nM, appears to be reversed at higher levels of 1,25(OH)2D3 supplementation (50 nM and 100 nM). Error bars display the LSD for each data set.
Figure 5. Individual-specific response to increasing in vitro 1,25(OH)2D3 supplementation is 25(OH)D3 status-independent.
Dots illustrate the mean level of CAMP expression per individual for Whites (A, 1–4) and Africans (B, 5–8). The 25(OH)D3 status, as well as the 1,25(OH)2D3-mediated fold change in expression level relative to the control is shown for each individual (untransformed data). Gene expression was quantified in monocyte-macrophages from healthy individuals using RT-qPCR before (basal) and after 24 h of in vitro 1,25(OH)2D3 supplementation at increasing concentrations (10 nM, 50 nM, and 100 nM). Plasma 25(OH)D3 status was quantified using LC-MS. A trend towards higher CAMP level is present at 10 nM 1,25(OH)2D3 for Whites, while Africans showed marginally increased CAMP expression at 50 nM 1,25(OH)2D3 concentrations. No trend between plasma 25(OH)D3 status and in vitro response to increasing concentrations of 1,25(OH)2D3 is present.
FokI genotype distribution differs between Africans and Whites
The CC genotype of the FokI SNP is a functional coding variant in the VDR that could potentially contribute to differential VDR function in Africans and Whites. We evaluated the FokI genotype distribution between African and White individuals in the South African cohort (Table 3) and in African and White populations from the International HapMap Project (Table 4). In the South African cohort, the frequency for the CC FokI genotype was significantly higher in Africans compared to Whites (Table 3, P<0.050). Similarly, African populations from the International HapMap Project (YRI, LWK and MKK) had a significantly higher frequency for the CC genotype than Whites of Western-European descent (CEU; Table 4, P<0.001). Moreover, no significant difference in FokI genotype distribution was observed between YRI, LWK and MKK (Table 4).
Table 3. Frequency distribution of genotypes for the Fok I SNP differs between Africans and Whites from the South African study population.
| Ethnicity | Genotype frequency distribution n (%) | χ2 | df | P-value | |
| CC | CT/TT | ||||
| African | 26 (68.4) | 12 (31.4) | 5.182 | 1 | <0.050 |
| White | 7 (36.8) | 12 (63.2) | |||
Table 4. Frequency distribution of genotypes for the Fok I SNP differs between Africans and Whites, but not between Africans, from the International HapMap Project.
| Ethnicity a | Genotype frequency distribution n (%) | χ2 | df | P-value | |
| CC | CT/TT | ||||
| YRICEU | 73 (65.2)42 (37.2) | 39 (34.8)71 (62.8) | 17.6 | 1 | <0.001 |
| LWKCEU | 62 (72.1)42 (37.2) | 24 (27.9)71 (62.8) | 22.6 | 1 | <0.001 |
| MKKCEU | 88 (62.0)42 (37.2) | 54 (38.0)71 (62.8) | 12.2 | 1 | <0.001 |
| YRILWKMKK | 73 (65.2)62 (72.0)88 (62.0) | 39 (34.8)24 (27.9)54 (38.0) | 2.456 | 2 | >0.050 |
HapMap population data were obtained from the International HapMap Project, including all HapMap Phase I, II and III samples from four populations: Individuals from the Centre d′Etude du Polymorphisme Humain (CEPH) collected in Utah, USA, with ancestry from northern and western Europe (CEU, n = 113); Yoruba in Ibadan, Nigeria (YRI, n = 112); Luhya in Webuye, Kenya (LWK, n = 86) and Maasai in Kinyawa, Kenya (MKK, n = 142).
FokI influences VDR function, but not expression
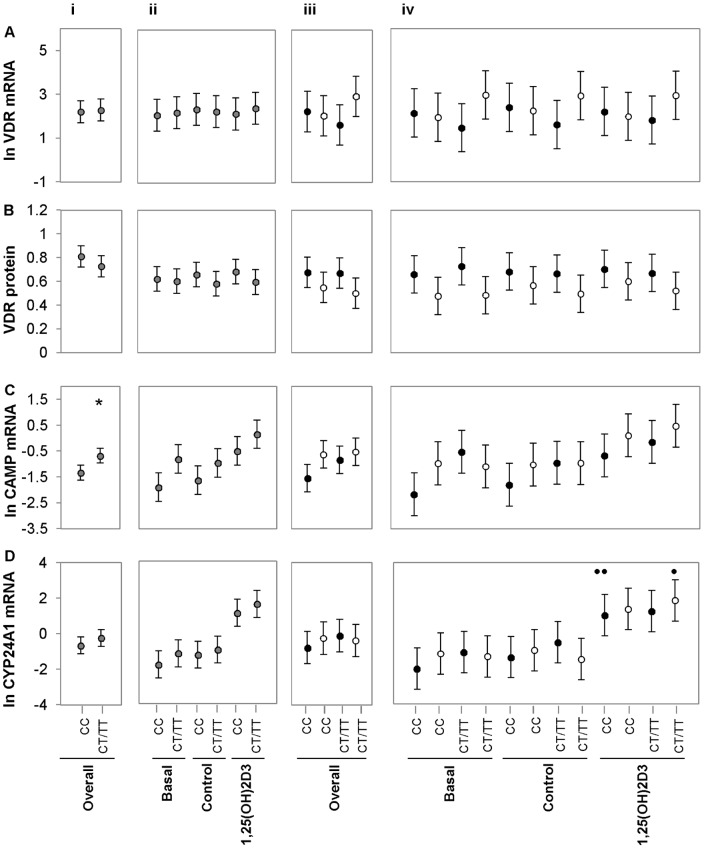
To determine whether the FokI genotype influenced VDR expression or function, individuals were genotyped for FokI and data analysed based on CC and CT/TT genotype. No significant difference was observed between genotype CC and CT/TT regarding level of VDR expression (Fig. 6A) or VDR protein (Fig. 6B) level. The CT/TT genotype showed significantly higher overall levels of CAMP mRNA compared to the CC genotype (Fig. 6C i, P<0.050). This difference in CAMP level between genotypes was not seen at any individual treatment (Fig 6C ii) or between ethnic groups, whether overall (Fig 6C iii) or as defined by treatment (Fig. 6C iv). Furthermore, 1,25(OH)2D3-elicited induction of CYP24A1 expression was influenced by FokI genotype, showing interaction with ethnicity (Fig 6D iv). In Africans (Fig 6D iv, black dots), 1,25(OH)2D3-elicited induction of CYP24A1 mRNA was significant for only the CC genotype (P<0.010, n = 26). In contrast, in Whites (Fig 6D iv. White dots), 1,25(OH)2D3-elicited induction of CYP24A1 mRNA was significant with the CT/TT genotype (P<0.050, n = 12) but not with the CC genotype (n = 7).
Figure 6. FokI influence VDR function, but not expression.
Error-bar plots illustrate the mean level of VDR expression (A), VDR protein (B), CAMP expression (C) and CYP24A1 expression (D) differentiated by FokI genotype (CC and CT/TT). Data was analysed combining ethnicity (i and ii, grey dots, n = 57) or differentiating ethnicity (iii and iv, Africans [n = 38], black dots and Whites [n = 19], white dots). Data was further analysed combining (overall, i and iii) or separating treatments (ii and iv). VDR expression and protein level was not significantly influenced by FokI genotype. Combining ethnicity and treatment (overall), CAMP mRNA level was significantly higher in the CT/TT genotypes compared to the CC genotype (* P<0.050). 1,25(OH)2D3-elicited induction of CYP24A1 mRNA was significant in Africans with the CC genotype (P<0.010, n = 26) and in Whites with the CT/TT genotypes (P<0.050, n = 12). Error bars display the LSD for each data set with Bonferroni correction. All significances indicated withstood Bonferroni correction.
Discussion
VDR target gene expression is modulated by 1,25(OH)2D3 [8], [36] and thought to be affected by variant VDR isoforms generated by FokI genotypes [14]. Here we assessed the combined effect of vitamin D, ethnicity and FokI genotype on expression and the functional capabilities of the VDR. Results support that differential VDR expression relates to ethnicity, to a lesser extent to vitamin D status, but not FokI genotype. Instead, VDR transactivation of CAMP is influenced by FokI genotype and, together with ethnicity, influence 1,25(OH)2D3-elicited CYP24A1 expression. Our results support a complex interaction between FokI, ethnicity and 1,25(OH)2D3-elicited VDR transactivation capacity of certain target genes, which may explain inconsistent genetic association of VDR with disease.
The inconsistent association between VDR variants and vitamin D level with diseases in diverse populations led us to investigate the possibility that VDR expression, VDR protein level and function may differ between ethnicities. Our results illustrate that ethnicity had a significant main effect on VDR expression and protein level, trending towards a higher VDR mRNA level in Whites and a significantly higher basal and control VDR protein level in Africans. This inverse relationship between gene expression and protein level suggests differential post-transcriptional regulation between the two ethnicities, and that the dynamics of VDR mRNA translation may differ between Africans and Whites. Despite having lower VDR levels than Africans, Whites produced a significantly higher level of CAMP mRNA than Africans in response to in vitro 1,25(OH)2D3 supplementation. Increasing the concentrations of in vitro 1,25(OH)2D3 supplementation further illustrated that the response to 1,25(OH)2D3 supplementation is individual-specific and did not correlate with 25(OH)D3 status. While more 1,25(OH)2D3 seemed beneficial for CAMP expression in the Africans that did not respond to 10 nM supplementation, it did not hold any additional benefit for individuals that did. This suggests that optimal 1,25(OH)2D3 level for VDR function may be individual-specific and that the efficiency of VDR function differs between ethnicities. Furthermore, the relationship between VDR protein level and VDR function observed within these results supports the complex link suggested between regulatory processes and overall phenotype [37]. Ethnicity-dependent VDR activity, as reflected in CAMP gene expression, may explain differential disease predisposition, as illustrated by the higher prevalence of TB [38] and colorectal cancer [39] in Africans compared to Whites. Thus, ethnicity influences VDR expression, VDR protein level and CAMP gene transactivation.
Although target-gene transactivation was directly influenced by availability of 1,25(OH)2D3 [40], in vitro 1,25(OH)2D3 supplementation had no significant effect on VDR expression. While Zella et al. (2010) showed that 1,25(OH)2D3 induced the accumulation of VDR in osteocarcinoma cells [41], Adams et al. (2010) and Selvaraj et al. (2009), found that neither in vitro 25(OH)D3 nor 1,25(OH)2D3 induced VDR expression in monocyte-macrophages of healthy individuals, respectively [42], [43]. Combined, the results suggest that while 1,25(OH)2D3 modulates the innate immune response via the VDR, baseline VDR mRNA and protein level are tightly regulated and not influenced by in vitro 1,25(OH)2D3 supplementation or plasma 25(OH)D3 level in primary monocyte-macrophages of individuals within normal range for 25(OH)D3 status. The effect of 1,25(OH)2D3 on VDR expression and protein level may therefore depend on cell type. Thus, the influence of 1,25(OH)2D3 on VDR expression and VDR protein level in monocyte-macrophages may depend on other confounding factors.
Furthermore, we found that the mean plasma level of 25(OH)D3 was not significantly different between Africans and Whites and that both ethnicities had, on average, a normal sufficient 25(OH)D3 status according to the Institute of Medicine (IOM) recommendations (>50 nmol/L). In contrast, an American based study found 25(OH)D3 status to be lower in Africans than Whites, and that Africans are vitamin D deficient [44], [45]; which may relate to the higher latitude and reduced UVB intensity in North America. A South African study conducted in the Western Cape showed a high prevalence of vitamin D deficiency among Africans, latently infected with M. tuberculosis [20]. While latency may influence 25(OH)D3 status, vitamin D synthesis is also compromised during the rainy winter months in Cape Town (Latitude 33° S); but not significantly altered throughout the year in individuals living in sunshine-rich Johannesburg (Latitude 26° S) [46]. The lack of disparity in 25(OH)D3 status between Africans and Whites in the current study may relate to the higher latitude, Highveld summer rainfall and sunny winter climate. A more recent study conducted in Africa, revealed that the mean serum 25(OH)D concentration of two African populations living in Kenya (Maasai) and Tanzania (Hadzabe) was relatively high. Both populations had normal mean serum 25(OH)D concentrations of 119 nmol/L and 109 nmol/L, respectively [47]. Combined, these findings suggest that 25(OH)D3 status is dependent on latitude and climate, with the influence of skin type evident only at less favorable climates. Furthermore, the similar level observed between Africans and Whites in the current study population indicates that, when sufficient, plasma 25(OH)D3 status may not be the determining factor in differential VDR expression, VDR protein level and function in healthy individuals. The 25(OH)D3 status did not correlate with VDR expression, VDR protein level or function in our study population. In agreement, Hendrickson et al. (2011) and Adams et al. (2009) found no association of plasma 25(OH)D3 status with VDR and CAMP expression, respectively [48], [42]. It has been suggested however that plasma 1,25(OH)2D3 status, independent of 25(OH)D3, may differ between individuals. For example, in the case of extra-renal hydroxylation by activated macrophages, where genetic variation in the VDR may influence 1,25(OH)2D3 production and ultimately plasma status [49]. Thus it is possible that 1,25(OH)2D3 status may have differed between Africans and Whites in the current study and may have influenced differential VDR expression, VDR protein level and function.
The genotype distribution of the FokI SNP was significantly different between the two ethnicities in our study, with the CC genotype present in 68% of Africans compared to the 37% in Whites. A similar distribution was observed for HapMap populations. This ethnicity difference in FokI genotype distribution did not appear to influence differential VDR expression or VDR protein level between ethnicities. The lack of a significant effect on VDR expression and protein level was not entirely unexpected, as FokI is a coding-region SNP which would affect protein function, unlike regulatory-region SNPs. Similarly, Selvaraj et al. (2009) found no significant difference in VDR protein level between variant FokI genotypes in both healthy Indian controls and pulmonary TB patients [43]. SNPs in the 3′ end of the VDR, together with a variable poly(A) microsatellite have, been shown to influence VDR mRNA stability [50], [51]. These variables may be responsible for the inverse relation between VDR mRNA and protein level observed in ethnicities of our cohort. Combined, these results suggest that variant genotypes of FokI do not influence VDR expression or level, while ethnicity does, implicating environment or other genetic factors.
While the FokI genotype did not affect VDR gene regulation in our cohort, it had a target-gene specific effect on VDR function. This finding agreed with the coding, functional nature of FokI; with previous work supporting a more robust transactivation capacity for the shorter VDR isoform (C nucleotide), as shown in CYP24A1 reporter gene constructs [11], [50], [14]. Van Etten et al. (2007) however, found no difference in transactivational capacity between the two alleles using a similar reporter gene assay [13]. In our study, the only evidence for higher activity of the C allele was the significant 1,25(OH)2D3-dependent induction of CYP24A1 in Africans for CC homozygotes, but not for those of CT/TT genotype. In contrast, CC homozygosity significantly hampered CAMP transactivation, irrespective of ethnicity or condition, and lacked significant 1,25(OH)2D3-elicited induction of CYP24A1 in Whites. This suggests that FokI genotype influences VDR transactivation capacity in a target-gene dependent manner. It is possible that the long and short VDR isoforms interact differently with VDRE's in different target genes. This target-gene specific interaction between VDR and VDREs may further be influenced by SNPs in the recognition elements. A low frequency SNP in African-Americans in the VDRE of the CYP24A1 promoter for example, has been shown to decrease the ability of the VDR to bind to and induce expression of the gene [52]. It is thus possible that VDRE SNPs may differentially influence transactivation of specific target genes. Although the cohort we investigated is small, the functional analysis was done under circumstances closer to normal physiology than reporter assays, and the results suggest that FokI not only has a target-gene specific effect on VDR function, but also interacts with ethnicity and 1,25(OH)2D3.
Considering that the ancestral allele of FokI is the T nucleotide [53], we propose that the interaction between genotype and ethnicity regarding 1,25(OH)2D3-elicited induction of CYP24A1 reflects natural selection. As early homonins with dark pigmented skin migrated North, their pigmentation decreased as an adaptation to synthesize sufficient vitamin D at higher latitude and limited UVB radiation. Modern humans in middle and East Africa however, were exposed to high intensity UV radiation and adapted by increasing skin pigmentation [54]. Despite their dark skin, individuals in Central and East Africa may still produce relatively high levels of 25(OH)D [45]. The more active 1,25(OH)2D3 breakdown in Africans, facilitated by CYP24A1, to regulate high baseline 25(OH)D level provides some support for this hypothesis. Thus, natural selection of the CC genotype of the VDR, associated with reduced CAMP induction and increased 1,25(OH)2D3-elicited CYP24A1 transactivation, was favoured in Africans. Unfortunately, CC likely conferred increased susceptibility to infection with pathogens such as Mycobacterium tuberculosis, brought to Africa in the late 1600's to early 1700's through colonisation and trade with the East [55]. Decreased CAMP expression in Africans may contribute to the infectious disease burden of South Africa; currently ranked globally with the third highest TB burden [56]. Based on the fact that CAMP is a 1,25(OH)2D3-inducible gene, it is expected that 1,25(OH)2D3 supplementation would be beneficial in TB prevention and treatment. However, our results suggest that the CC genotype associated with Africans may moderate CAMP induction through 1,25(OH)2D3-elicitated CYP24A1 induction and consequent 1,25(OH)2D3 catabolism. Thus, the efficacy of 1,25(OH)2D3-elicitation and subsequent application in therapy should be considered in the context of ethnicity-dependent variables.
Taken together, differential VDR expression relates to ethnicity rather than 25(OH)D3 status and FokI genotype, while VDR activity, specifically CAMP and 1,25(OH)2D3-elicited CYP24A1 transactivation relates to FokI genotype, interacting with ethnicity in the latter case. Contrary to conclusions of previous studies that the CC genotype of FokI results in higher VDR transactivation capacity, our data suggests that FokI genotype influences VDR transactivation capacity in a target-gene dependent manner. Although vitamin D is essential for VDR function, it is likely not the sole contributing factor in VDR-related disease susceptibility, as both the level and activity of the VDR differ between populations. Thus, the expression and role of VDR in target gene transactivation is determined not only by genetics, but also by ethnicity and environment involving complex interactions which may confound disease association. With current literature supporting the long-held belief that genetic variants in the VDR-pathway may be key in the association between vitamin D status and disease susceptibility, future work should evaluate the combined contribution of multiple factors, including environment, in various ethnic groups.
Acknowledgments
We would like to acknowledge the South African National Blood Service for providing blood products for the study as well as Tracy Ferrao (Chemical Pathology, University of Witwatersrand Medical School) for assistance in quantification of 25(OH)D3. We acknowledge Brandon R. Jones and Tenielle M. Cooke for assistance in the dose-response study.
Funding Statement
This study was supported by funding from the National Research Foundation (NRF, www.nrf.ac.za, Grant No 81774) and Cancer Association of South Africa (CANSA, www.cansa.org.za, Research Grant L. Bornman). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Baeke F, Takiishi T, Korf H, Gysemans C, Mathieu C (2011) Vitamin D: modulator of the immune system. Curr Opin Pharmacol 10: 482–496. [DOI] [PubMed] [Google Scholar]
- 2. Haussler MR, Whitfield GK, Kaneko I, Haussler CA, Hsieh D, et al. (2013) Molecular Mechanisms of Vitamin D Action. Calcif Tissue Int 92: 77–98. [DOI] [PubMed] [Google Scholar]
- 3. Wang TJ, Zhang F, Richards JB, Kestenbaum B, van Meurs JB, et al. (2010) Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet 7;376(9736): 180–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Campbell GR, Spector SA (2012) Vitamin D inhibits human immunodeficiency virus type 1 and Mycobacterium tuberculosis infection in macrophages through the induction of autophagy. PLoS Pathog 8: e1002689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zierold C, Darwish HM, DeLuca HF (1995) Two Vitamin D Response Elements Function in the Rat 1,25-dihydroxyvitamin D 24-Hydroxylase. J Biol Chem 270(4): 1675–78. [DOI] [PubMed] [Google Scholar]
- 6. Kim S, Shevde NK, Pike JW (2005) 1,25-Dihydroxyvitamin D3 stimulates cyclic vitamin D receptor/retinoid X receptor DNA-binding, co-activator recruitment, and histone acetylation in intact osteoblasts. J Bone Miner Res 20: 305–17. [DOI] [PubMed] [Google Scholar]
- 7. Meyer MB, Goetsch PD, Pike JW (2010) A downstream intergenic cluster of regulatory enhancers contributes to the induction of CYP24A1 expression by 1alpha,25–dihydroxyvitamin D3 . J Biol Chem 285(20): 15599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gombart AF, Borregaard N, Koeffler HP (2005) Human cathelicidin antimicrobial peptide (CAMP) gene is a direct target of the vitamin D receptor and is strongly up-regulated in myeloid cells by 1,25-dihydroxyvitamin D3 . FASEB J 19: 1067–77. [DOI] [PubMed] [Google Scholar]
- 9. Wang TT, Nestel FP, Bourdeau V, Nagai Y, Wang Q, et al. (2004) Cutting edge: 1,25-dihydroxyvitamin D3 is a direct inducer of antimicrobial peptide gene expression. J Immunol 173: 2909–12. [DOI] [PubMed] [Google Scholar]
- 10. Valdivielso JM, Fernandez E (2060) Vitamin D receptor polymorphisms and diseases. Clinica Chimica Acta 371: 1–12. [DOI] [PubMed] [Google Scholar]
- 11. Arai H, Miyamoto K, Taketani Y, Yamamoto H, Iemori Y, et al. (1997) A vitamin D receptor gene polymorphism in the translation initiation codon: effect on protein activity and relation to bone mineral density in Japanese women. J Bone Miner Res 12: 915–921. [DOI] [PubMed] [Google Scholar]
- 12. Jurutka PW, Remus LS, Whitfield GK, Thompson PD, Hsieh JC, et al. (2000) The polymorphic N terminus in human vitamin D receptor isoforms influences transcriptional activity by modulating interaction with transcription factor IIB. Mol Endocrinol 14: 401–20. [DOI] [PubMed] [Google Scholar]
- 13. van Etten E, Verlinden L, Giulietti A, Ramos-Lopez E, Branisteanu DD, et al. (2007) The vitamin D receptor gene FokI polymorphism: functional impact on the immune system. Eur J Immunol 37: 395–405. [DOI] [PubMed] [Google Scholar]
- 14. Alimirah F, Peng X, Murillo G, Mehta RG (2011) Functional Significance of Vitamin D Receptor FokI Polymorphism in Human Breast Cancer Cells. PLoS One 6: e16024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wilkinson RJ, Llewelyn M, Toossi Z, Patel P, Pasvol G, et al. (2000) Influence of vitamin D deficiency and vitamin D receptor polymorphisms on tuberculosis among Gujarati Asians in west London: a case-control study. Lancet 355: 618–21. [DOI] [PubMed] [Google Scholar]
- 16. Wong H, Seow A, Arakawa K, Lee H, Yu MC, et al. (2003) Vitamin D receptor start codon polymorphism and colorectal cancer risk: effect modification by dietary calcium and fat in Singapore Chinese. Carcinogenesis 24: 1091–5. [DOI] [PubMed] [Google Scholar]
- 17. Norman AW (1998) Sunlight, season, skin pigmentation, vitamin D, and 25-hydroxyvitamin D: integral components of the vitamin D endocrine system. Am J Clin Nutr 67: 1108–10. [DOI] [PubMed] [Google Scholar]
- 18. Hagenau T, Vest R, Gissel TN, Poulsen CS, Erlandsen M, et al. (2009) Global vitamin D levels in relation to age, gender, skin pigmentation and latitude: an ecologic meta-regression analysis. Osteoporos Int 20: 133–40. [DOI] [PubMed] [Google Scholar]
- 19. Gibney KB, MacGregor L, Leder K, Torresi J, Marshall C, et al. (2008) Vitamin D deficiency is associated with tuberculosis and latent tuberculosis infection in immigrants from sub-Saharan Africa. Clin Infect Dis 46: 443–6. [DOI] [PubMed] [Google Scholar]
- 20. Martineau AR, Nhamoyebonde S, Oni T, Rangakad MX, Maraisd S, et al. (2011) Reciprocal seasonal variation in vitamin D status and tuberculosis notifications in Cape Town, South Africa. PNAS USA 108: 19013–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Buttigleiro C, Monagheddu C, Petroni P, Saini A, Dogliotti L, et al. (2011) Prognostic role of vitamin D supplementation efficacy of vitamin D supplementation in cancer patients: A systematic review. The Oncologist 16: 1215–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Luong K, Nguyen LTH (2010) The beneficial role of vitamin D and its analogs in cancer treatment and prevention. Crit Rev Oncol Hematol 73: 192–201. [DOI] [PubMed] [Google Scholar]
- 23. Parisi E, Rene JM, Cardus A, Valcheva P, Pinol-Felis C, et al. (2008) Vitamin D receptor levels in colorectal cancer: Possible role of BsmI polymorphism. J Steroid Biochem Mol Biol 111: 87–90. [DOI] [PubMed] [Google Scholar]
- 24.Sumethkul K, Boonyaratavej S, Kitumnuaypong T, Angthararuk S, Cheewasat P, et al.. (2012) The predictive factors of low serum 25-hydroxyvitamin D and vitamin D deficiency in patients with systemic lupus erythematosus. Rheumatol Int [Epub ahead of print]. [DOI] [PubMed]
- 25. Murdoch DR, Slow S, Chambers ST, Jennings LC, Stewart AW, et al. (2012) Effect of vitamin D3 supplementation on upper respiratory tract infections in healthy adults: the VIDARIS randomized controlled trial. JAMA 308: 1333–9. [DOI] [PubMed] [Google Scholar]
- 26. Gepner AD, Ramamurthy R, Krueger DC, Korcarz CE, Binkley N, et al. (2012) A prospective randomized controlled trial of the effects of vitamin D supplementation on cardiovascular disease risk. PLoS One 7: e36617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martineau AR, Timms PM, Bothamley GH, Hanifa Y, Islam K, et al. (2011) High-dose vitamin D(3) during intensive-phase antimicrobial treatment of pulmonary tuberculosis: a double-blind randomised controlled trial. Lancet 377: 242–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carter GD (2011) Accuracy of 25-hydroxyvitamin D assays: confronting the issues. Curr Drug Targets 12: 19–28. [DOI] [PubMed] [Google Scholar]
- 29. Berry D, Hyppönen E (2011) Determinants of vitamin D status: focus on genetic variations. Curr Opin Nephrol 20: 331–6. [DOI] [PubMed] [Google Scholar]
- 30. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. (2002) Accurate normalization of real-time quantitative RT–PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, et al. (2009) The MIQE Guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55: 611–22. [DOI] [PubMed] [Google Scholar]
- 32. Vermeulen J, Pattyn F, De Preter K, Vercruysse L, Derveaux S, et al. (2009) External oligonucleotide standards enable cross laboratory comparison and exchange of real-time quantitative PCR data. Nucleic Acids Res 37: e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dendrou CA, Fung E, Esposito L, Todd JA, Wicker LS, et al.. (2009) Fluorescence Intensity Normalisation: Correcting for Time Effects in Large-Scale Flow Cytometric Analysis. Adv Bioinformatics 476106. [DOI] [PMC free article] [PubMed]
- 35.International HapMap Project (2013). Retrieved from http://hapmap.ncbi.nlm.nih.gov on 15 January 2013.
- 36. Chen KS, DeLuca HF (1995) Cloning of the human 1 alpha,25-dihydroxyvitamin D-3 24-hydroxylase gene promoter and identification of two vitamin D-responsive elements. Biochim Biophys Acta 1263: 1–9. [DOI] [PubMed] [Google Scholar]
- 37. Milner J, Trujillo EB, Kaefer CM, Ross S. Nutrigenomics. In: National Research Council (US) Committee on Advances in Collecting, Utilizing Biological Indicators, Genetic Information in Social Science Surveys; Weinstein M, Vaupel JW, Wachter KW, editors. Biosocial Surveys. Washington (DC): National Academies Press (US) 2008: 14 Available from: http://www.ncbi.nlm.nih.gov/books/NBK62425/. [Google Scholar]
- 38. Stead WW, Senner JW, Reddick WT, Lofgren JP (1990) Racial differences in susceptibility to infection by Mycobacterium tuberculosis. N Engl J Med 322: 422–7. [DOI] [PubMed] [Google Scholar]
- 39. Ashktorab H, Nguza B, Fatemi M, Nouraie M, Smoot DT, et al. (2011) Case-control study of vitamin D, dickkopf homolog 1 (DKK1) gene methylation, VDR gene polymorphism and the risk of colon adenoma in African Americans. PLoS One 6: e25314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lui PT Stenger, S. Huiying L, Wenzel L, Tan BH, et al. (2006) Toll-Like receptor triggering of vitamin D-mediated human antimicrobial response. Science 311: 1770–3. [DOI] [PubMed] [Google Scholar]
- 41. Zella LA, Meyer MB, Nerenz RD, Lee SM, Martowicz ML, et al. (2010) Multifunctional Enhancers Regulate Mouse and Human Vitamin D Receptor Gene Transcription Molecular Endocrinology. 24: 128–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Adams JS, Ren S, Liu PT, Chun RF, Lagishetty V, et al. (2009) Vitamin d-directed rheostatic regulation of monocyte antibacterial responses. J Immunol 182: 4289–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Selvaraj P, Anand SP, Harishankar M, Alagarasu K (2009) Plasma 1,25 Dihydroxy Vitamin D3 Level and Expression of Vitamin D Receptor and Cathelicidin in Pulmonary Tuberculosis. J Clin Immunol 29: 470–8. [DOI] [PubMed] [Google Scholar]
- 44. Harris SS, Dawson-Hughes B (1998) Seasonal changes in plasma 25-hydroxyvitamin D concentrations of young American black and white women. Am J of Clin Nutr 67: 1232–6. [DOI] [PubMed] [Google Scholar]
- 45. Dawson-Hughes B (2004) Racial/ethnic considerations in making recommendations for vitamin D for adult and elderly men and women. Am J Clin Nutr 80: 1763S–6S. [DOI] [PubMed] [Google Scholar]
- 46. Pettifor JM, Moodley GP, Hough FS, Koch H, Chen T, et al. (1996) The effect of season and latitude on in vitro vitamin D formation by sunlight in South Africa. S Afr Med J 86: 1270–2. [PubMed] [Google Scholar]
- 47. Luxwolda MF, Kuipers RS, Kema IP, Janneke Dijck-Brouwer DA, et al. (2012) Traditionally living populations in East Africa have a mean serum 25-hydroxyvitamin D concentration of 115 nmol/l. Br J Nutr 108: 1557–61. [DOI] [PubMed] [Google Scholar]
- 48.Hendrickson WK, Flavin R, Kasperzyk JL, Fiorentino M, Fang F, et al.. (2011) Vitamin D Receptor Protein Expression in Tumor Tissue and Prostate Cancer Progression. J Clin Oncol: 2378–85. [DOI] [PMC free article] [PubMed]
- 49. Lips P (2007) Relative value of 25(OH)D and 1,25(OH)2D measurements. J Bone Miner Res 22: 1668–71. [DOI] [PubMed] [Google Scholar]
- 50. Whitfield GK, Remus LS, Jurutka PW, Zitzer H, Oza AK, et al. (2001) Functionally relevant polymorphisms in the human nuclear vitamin D receptor gene. Mol Cell Endocrinol 177: 145–59. [DOI] [PubMed] [Google Scholar]
- 51. Uitterlinden AG, Fang Y, van Meurs JBJ, Pols HAP, van Leeuwen JPTM (2004) Genetics and biology of vitamin D receptor polymorphisms. Gene 338: 143–56. [DOI] [PubMed] [Google Scholar]
- 52. Roff A, Carastro LM, Wilson RT (2008) A novel SNP in a vitamin D response element of the CYP24A1 promoter reduces protein binding, transactivation, and gene expression. J. Steroid Biochem. Mol. Biol 112: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.dbSNP Short Genetic Variations (2013). Retrieved from http://www.ncbi.nlm.nih.gov/projects/SNP/ on 10 January 2013.
- 54. Jablonski NG, Chaplin G (2010) Human skin pigmentation as an adaptation to UV radiation. PNAS 107: 8962–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Oswald NC (1946) Pulmonary tuberculosis in African native troops. Thorax 1: 100–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.WHO Global TB Report (2012). Retrieved from www.who.int on 2 February 2013.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data presented in this manuscript has been deposited in the Dryad Repository: http://dx.doi.org/10.5061/dryad.12dp5.