Abstract
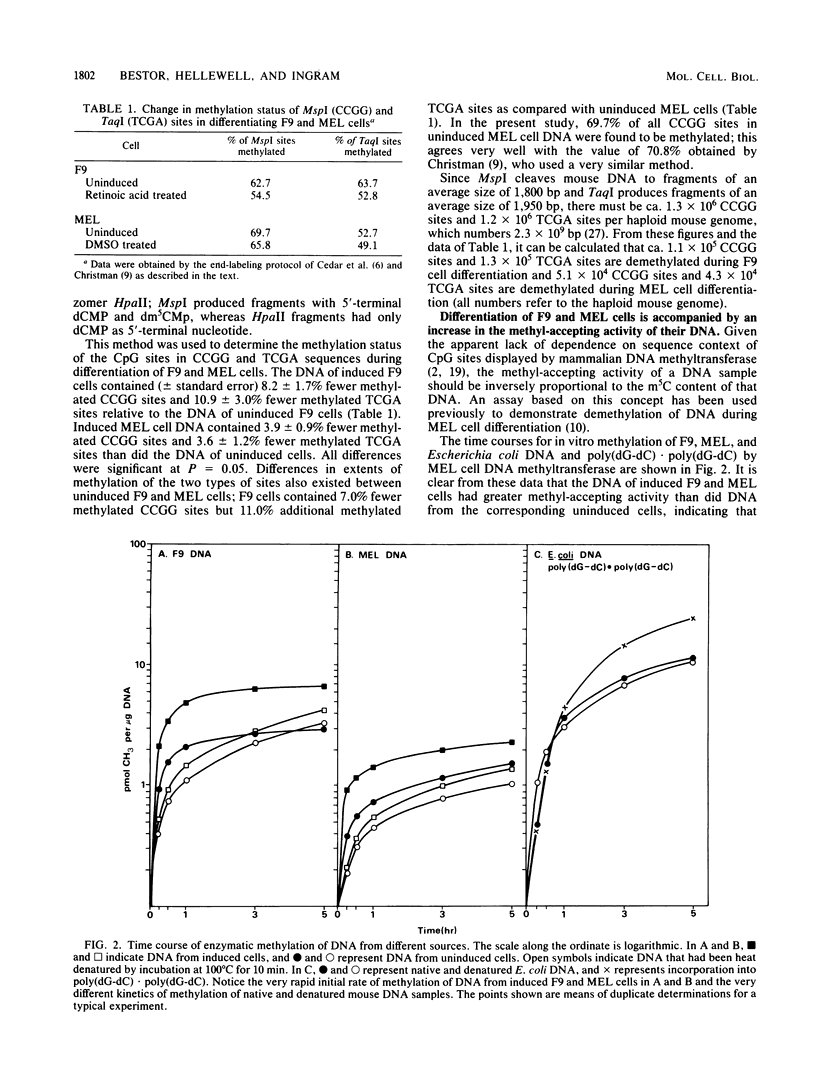
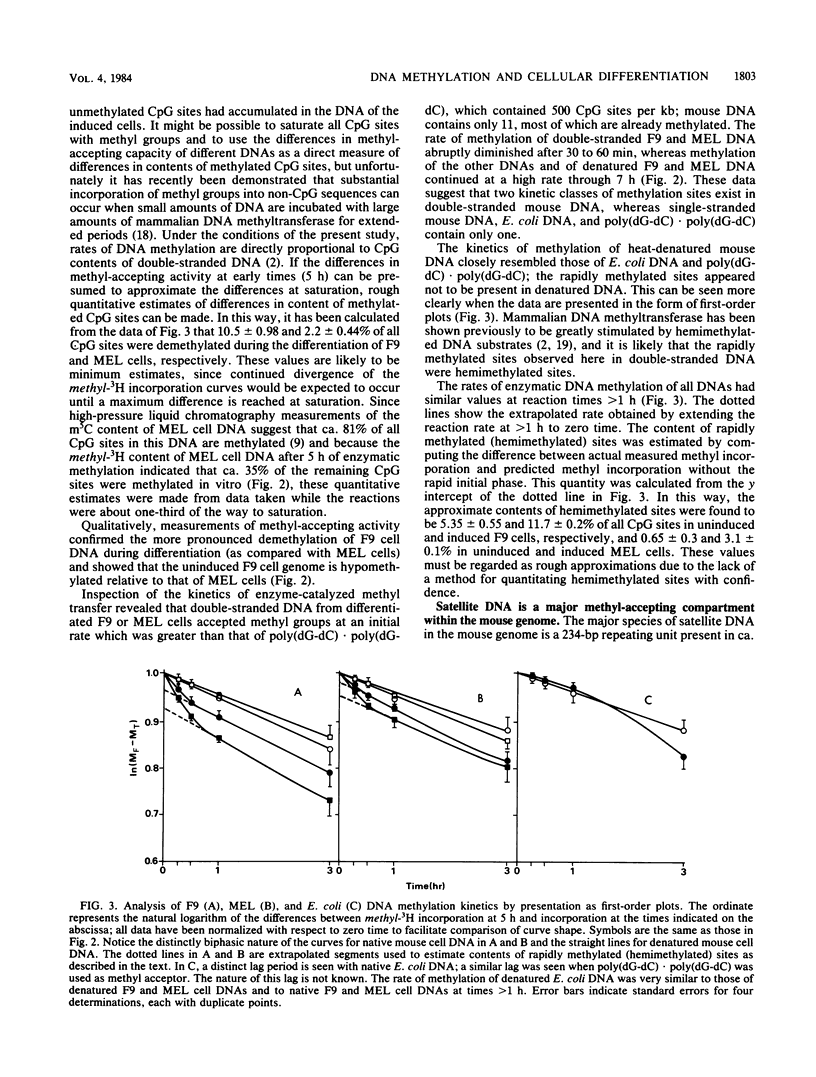
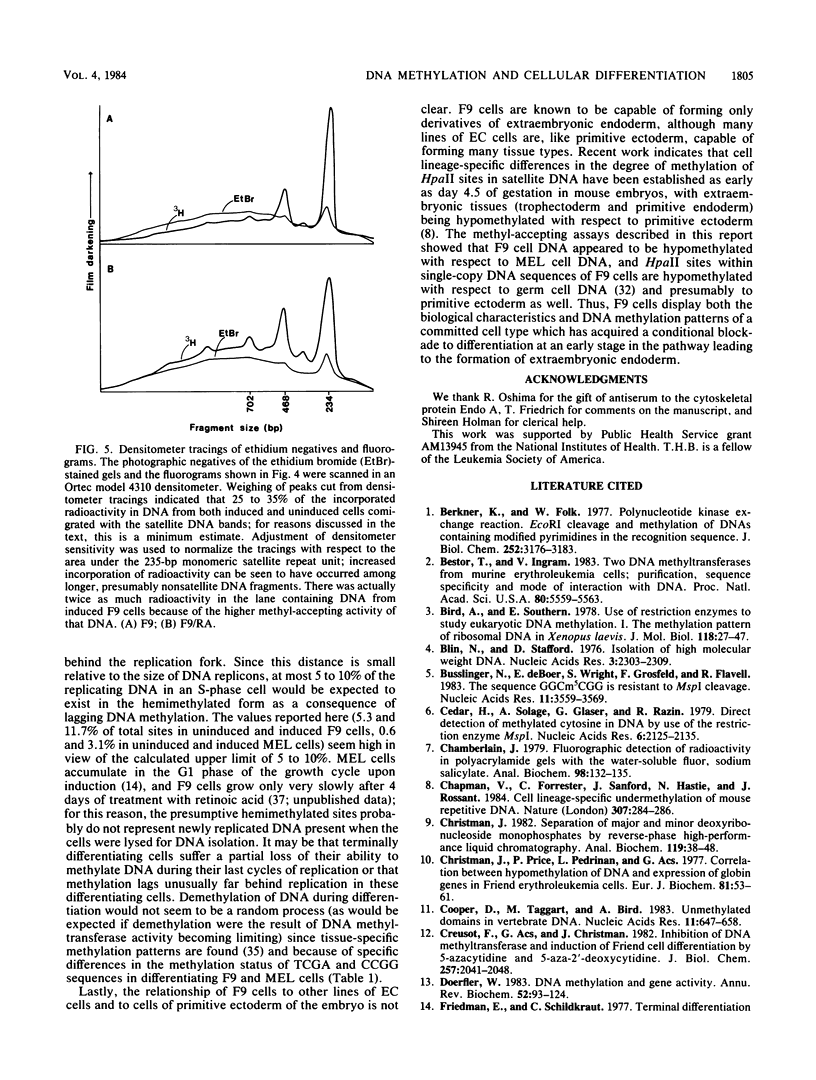
Methyl-accepting assays and a sensitive method for labeling specific CpG sites have been used to show that the DNA of F9 embryonal carcinoma cells decreases in 5-methylcytosine content by ca. 9% during retinoic acid-induced differentiation, whereas the DNA of dimethyl sulfoxide-induced Friend murine erythroleukemia (MEL) cells loses ca. 3.8% of its methyl groups. These values correspond to the demethylation of 2.2 X 10(6) and 0.9 X 10(6) 5'-CpG-3' sites per haploid genome in differentiating F9 and MEL cells, respectively. Fluorography of DNA restriction fragments methylated in vitro and displayed on agarose gels showed that demethylation occurred throughout the genome. In uninduced F9 cells, the sequence TCGA tended to be more heavily methylated than did the sequence CCGG, whereas this tendency was reversed in MEL cells. The kinetics of in vitro DNA methylation reactions catalyzed by MEL cell DNA methyltransferase showed that substantial numbers of hemimethylated sites accumulate in the DNA of terminally differentiating F9 and MEL cells, implying that a partial loss of DNA-methylating activity may accompany terminal differentiation in these two cell types.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berkner K. L., Folk W. R. Polynucleotide kinase exchange reaction: quantitave assay for restriction endonuclease-generated 5'-phosphoroyl termini in DNA. J Biol Chem. 1977 May 25;252(10):3176–3184. [PubMed] [Google Scholar]
- Bestor T. H., Ingram V. M. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5559–5563. doi: 10.1073/pnas.80.18.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. P., Southern E. M. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol. 1978 Jan 5;118(1):27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- Blin N., Stafford D. W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976 Sep;3(9):2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busslinger M., deBoer E., Wright S., Grosveld F. G., Flavell R. A. The sequence GGCmCGG is resistant to MspI cleavage. Nucleic Acids Res. 1983 Jun 11;11(11):3559–3569. doi: 10.1093/nar/11.11.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H., Solage A., Glaser G., Razin A. Direct detection of methylated cytosine in DNA by use of the restriction enzyme MspI. Nucleic Acids Res. 1979;6(6):2125–2132. doi: 10.1093/nar/6.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain J. P. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal Biochem. 1979 Sep 15;98(1):132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- Chapman V., Forrester L., Sanford J., Hastie N., Rossant J. Cell lineage-specific undermethylation of mouse repetitive DNA. Nature. 1984 Jan 19;307(5948):284–286. doi: 10.1038/307284a0. [DOI] [PubMed] [Google Scholar]
- Christman J. K., Price P., Pedrinan L., Acs G. Correlation between hypomethylation of DNA and expression of globin genes in Friend erythroleukemia cells. Eur J Biochem. 1977 Nov 15;81(1):53–61. doi: 10.1111/j.1432-1033.1977.tb11926.x. [DOI] [PubMed] [Google Scholar]
- Christman J. K. Separation of major and minor deoxyribonucleoside monophosphates by reverse-phase high-performance liquid chromatography: a simple method applicable to quantitation of methylated nucleotides in DNA. Anal Biochem. 1982 Jan 1;119(1):38–48. doi: 10.1016/0003-2697(82)90662-5. [DOI] [PubMed] [Google Scholar]
- Cooper D. N., Taggart M. H., Bird A. P. Unmethylated domains in vertebrate DNA. Nucleic Acids Res. 1983 Feb 11;11(3):647–658. doi: 10.1093/nar/11.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creusot F., Acs G., Christman J. K. Inhibition of DNA methyltransferase and induction of Friend erythroleukemia cell differentiation by 5-azacytidine and 5-aza-2'-deoxycytidine. J Biol Chem. 1982 Feb 25;257(4):2041–2048. [PubMed] [Google Scholar]
- Doerfler W. DNA methylation and gene activity. Annu Rev Biochem. 1983;52:93–124. doi: 10.1146/annurev.bi.52.070183.000521. [DOI] [PubMed] [Google Scholar]
- Friend C., Scher W., Holland J. G., Sato T. Hemoglobin synthesis in murine virus-induced leukemic cells in vitro: stimulation of erythroid differentiation by dimethyl sulfoxide. Proc Natl Acad Sci U S A. 1971 Feb;68(2):378–382. doi: 10.1073/pnas.68.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Sosa M. A., Midgett R. M., Slagel V. A., Githens S., Kuo K. C., Gehrke C. W., Ehrlich M. Tissue-specific differences in DNA methylation in various mammals. Biochim Biophys Acta. 1983 Jun 24;740(2):212–219. doi: 10.1016/0167-4781(83)90079-9. [DOI] [PubMed] [Google Scholar]
- Gama-Sosa M. A., Wang R. Y., Kuo K. C., Gehrke C. W., Ehrlich M. The 5-methylcytosine content of highly repeated sequences in human DNA. Nucleic Acids Res. 1983 May 25;11(10):3087–3095. doi: 10.1093/nar/11.10.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graessmann M., Graessmann A., Wagner H., Werner E., Simon D. Complete DNA methylation does not prevent polyoma and simian virus 40 virus early gene expression. Proc Natl Acad Sci U S A. 1983 Nov;80(21):6470–6474. doi: 10.1073/pnas.80.21.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y., Cedar H., Razin A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982 Feb 18;295(5850):620–622. doi: 10.1038/295620a0. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y., Szyf M., Cedar H., Razin A. Methylation of replicating and post-replicated mouse L-cell DNA. Proc Natl Acad Sci U S A. 1983 Aug;80(16):4919–4921. doi: 10.1073/pnas.80.16.4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigh L. S., Owens B. B., Hellewell O. S., Ingram V. M. DNA methylation in chicken alpha-globin gene expression. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5332–5336. doi: 10.1073/pnas.79.17.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörz W., Altenburger W. Nucleotide sequence of mouse satellite DNA. Nucleic Acids Res. 1981 Feb 11;9(3):683–696. doi: 10.1093/nar/9.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörz W., Zachau H. G. Characterization of distinct segments in mouse satellite DNA by restriction nucleases. Eur J Biochem. 1977 Mar 1;73(2):383–392. doi: 10.1111/j.1432-1033.1977.tb11329.x. [DOI] [PubMed] [Google Scholar]
- Kapp L. N., Painter R. B. DNA replication fork movement rates in mammalian cells. Int Rev Cytol. 1982;80:1–25. doi: 10.1016/s0074-7696(08)60365-4. [DOI] [PubMed] [Google Scholar]
- Keshet E., Cedar H. Effect of CpG methylation on Msp I. Nucleic Acids Res. 1983 Jun 11;11(11):3571–3580. doi: 10.1093/nar/11.11.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur J Biochem. 1975 Aug 15;56(2):335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
- McClelland M., Ivarie R. Asymmetrical distribution of CpG in an 'average' mammalian gene. Nucleic Acids Res. 1982 Dec 11;10(23):7865–7877. doi: 10.1093/nar/10.23.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M. The effect of site specific methylation on restriction endonuclease cleavage (update). Nucleic Acids Res. 1983 Jan 11;11(1):r169–r173. doi: 10.1093/nar/11.1.235-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveh-Many T., Cedar H. Topographical distribution of 5-methylcytosine in animal and plant DNA. Mol Cell Biol. 1982 Jul;2(7):758–762. doi: 10.1128/mcb.2.7.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima R. G. Identification and immunoprecipitation of cytoskeletal proteins from murine extra-embryonic endodermal cells. J Biol Chem. 1981 Aug 10;256(15):8124–8133. [PubMed] [Google Scholar]
- Rahe B., Erickson R. P., Quinto M. Methylation of unique sequence DNA during spermatogenesis in mice. Nucleic Acids Res. 1983 Nov 25;11(22):7947–7959. doi: 10.1093/nar/11.22.7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy E. P. Nucleotide sequence analysis of the T24 human bladder carcinoma oncogene. Science. 1983 Jun 3;220(4601):1061–1063. doi: 10.1126/science.6844927. [DOI] [PubMed] [Google Scholar]
- Romanov G. A., Vanyushin B. F. Methylation of reiterated sequences in mammalian DNAs. Effects of the tissue type, age, malignancy and hormonal induction. Biochim Biophys Acta. 1981 Apr 27;653(2):204–218. doi: 10.1016/0005-2787(81)90156-8. [DOI] [PubMed] [Google Scholar]
- Sano H., Sager R. Tissue specificity and clustering of methylated cystosines in bovine satellite I DNA. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3584–3588. doi: 10.1073/pnas.79.11.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer J., Roberts-Ems J., Luthardt F. W., Riggs A. D. Methylation of DNA in mouse early embryos, teratocarcinoma cells and adult tissues of mouse and rabbit. Nucleic Acids Res. 1979 Dec 20;7(8):2369–2385. doi: 10.1093/nar/7.8.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland S., Mahdavi V. The induction of differentiation in teratocarcinoma stem cells by retinoic acid. Cell. 1978 Oct;15(2):393–403. doi: 10.1016/0092-8674(78)90008-9. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Levy J., Terada M., Breslow R., Rifkind R. A., Marks P. A. Induction of erythroid differentiation in murine virus infected eythroleukemia cells by highly polar compounds. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1003–1006. doi: 10.1073/pnas.72.3.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. DNA methylation at a CCGG sequence in the large intron of the rabbit beta-globin gene: tissue-specific variations. Nucleic Acids Res. 1978 Dec;5(12):4631–4634. doi: 10.1093/nar/5.12.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]