Abstract
Seasonal mammals integrate changes in the duration of nocturnal melatonin secretion to drive annual physiologic cycles. Melatonin receptors within the proximal pituitary region, the pars tuberalis (PT), are essential in regulating seasonal neuroendocrine responses. In the ovine PT, melatonin is known to influence acute changes in transcriptional dynamics coupled to the onset (dusk) and offset (dawn) of melatonin secretion, leading to a potential interval-timing mechanism capable of decoding changes in day length (photoperiod). Melatonin offset at dawn is linked to cAMP accumulation, which directly induces transcription of the clock gene Per1. The rise of melatonin at dusk induces a separate and distinct cohort, including the clock-regulated genes Cry1 and Nampt, but little is known of the up-stream mechanisms involved. Here, we used next-generation sequencing of the ovine PT transcriptome at melatonin onset and identified Npas4 as a rapidly induced basic helix-loop-helix Per-Arnt-Sim domain transcription factor. In vivo we show nuclear localization of NPAS4 protein in presumptive melatonin target cells of the PT (α-glycoprotein hormone-expressing cells), whereas in situ hybridization studies identified acute and transient expression in the PT of Npas4 in response to melatonin. In vitro, NPAS4 forms functional dimers with basic helix loop helix-PAS domain cofactors aryl hydrocarbon receptor nuclear translocator (ARNT), ARNT2, and ARNTL, transactivating both Cry1 and Nampt ovine promoter reporters. Using a combination of 5′-deletions and site-directed mutagenesis, we show NPAS4-ARNT transactivation to be codependent upon two conserved central midline elements within the Cry1 promoter. Our data thus reveal NPAS4 as a candidate immediate early-response gene in the ovine PT, driving molecular responses to melatonin.
In seasonal mammals, annual cycles of reproduction, growth, and metabolism are entrained by annual changes in photoperiod (1, 2). Transduction of this photoperiod signal is known to involve the hormone melatonin, which is secreted nocturnally from the pineal gland, the pattern of which is sculpted by seasonal changes in photoperiod/night length (3). In this way, melatonin provides neuroendocrine target sites with an accurate internal hormonal representation of annual photoperiodic changes. Numerous studies have employed timed infusion paradigms in pinealectomized animals to demonstrate that the duration of this nocturnal signal is critical in driving seasonal neuroendocrine reproductive changes, with prolonged daily infusions of long or short durations, mimicking endogenous winter or summer physiologic patterns, respectively, which lead to an induction of an appropriate seasonal physiologic response (reviewed in Ref. 4).
Recent attention has focused on the pars tuberalis (PT) within the pituitary gland as a key site of action of melatonin (3, 5, 6). Here, a dense melatonin receptor (MT1)-rich population of cells are located, and within the PT, summer-like melatonin signals have been shown to regulate seasonal expression of the β-subunit of TSH, activating thyroid hormone-metabolizing pathways in the adjacent hypothalamus and leading to seasonal reproductive responses (reviewed in Refs. 1 and 7). A key current question relates to how changes in duration of the nocturnal melatonin signal are able to convey photoperiodic time within the PT. In one hypothesis, it has been proposed that melatonin drives a daily rhythm of gene expression coincident with dawn or dusk, such that seasonal changes in the duration of melatonin secretion alter the relative phasing of this dawn-dusk rhythmic program (8). Studies in seasonal sheep have provided some evidence in support of such an internal coincidence timer, including the observation that regulation of the core circadian transcription factors Per1 and Cry1 are expressed at dawn and dusk, respectively, with their relative phasing to dawn and dusk controlled by the duration of the duration of the prevailing melatonin signal (9).
Melatonin is known to inhibit cAMP accumulation and derepression of this signaling pathway at dawn (ie, at the time of melatonin decline) induces a surge of intracellular cAMP leading to transcriptional activation of cAMP-regulated genes timed to dawn, including Per1 (10, 11). Rather less is known of the transcriptional program activated at night, coincident with the onset of melatonin secretion. Our studies (6, 12) and those of others (9, 13–15) have already identified a small cluster of melatonin-activated genes, with increased expression in the early night, coincident with the rise in melatonin secretion. These include the clock gene Cry1 but also other circadian-controlled genes such as Nampt and NeuroD1 (6). A particular feature of this melatonin-regulated group of genes is that expression peaks over a relatively long time course, with maximal expression at +3 hours after onset of melatonin rise (6). To date, little is known of up-stream immediate-early response circuits within the PT that may be involved in activation of such a nocturnal transcriptome by melatonin (3).
We describe here experiments in which we use deep sequencing technologies to screen the expressed transcriptome of the ovine PT after the rise in melatonin, from which we have identified the bHLH (basis helix loop helix) DNA-binding transcription factor Npas4 as a candidate immediate-early response gene. We show that this gene exhibits a rapid transient increase in expression in the PT, directly induced by melatonin in the early night phase. We also show that NPAS4 heterodimerizes with a subset of other bHLH transcription factors (ARNT, ARNT2), which are expressed in the PT and transactivate the promoters of known melatonin-responsive genes, and using ovine Cry1 as a model, we mapped NPAS4 responses to two conserved central midline element (CME) binding sites in the proximal promoter region of this circadian clock gene, driving expression. Our data are therefore compatible with the hypothesis that NPAS4 provides the critical upstream element in melatonin action in the early night phase and is a potential key component driving seasonal neuroendocrine responses.
Materials and Methods
Animals
All animal experiments were undertaken in accordance with the Home Office Animals (Scientific Procedures) Act (1986), UK, under a Project License held by A.L.
For studies of acute melatonin-mediated induction of Npas4, 48 Scottish black-face male castrate spring lambs were isolated in October (natural photoperiod, ∼9 h light/15 h dark) and maintained on artificial short photoperiods (SP, 8 h light, 16 h dark) in an indoor facility (Roslin, Edinburgh) for 12 weeks; in early January the animals were transferred to artificial long photoperiods (LP, 16 h light/8 h dark) through symmetrical extension of the light phase by 4 hours at dawn and dusk. After 4 weeks on LP (day 28) at the time of normal lights off (ie, 16 h after lights on) the lighting was left on, and 24 sheep were treated with intradermal implants of melatonin (Regulin; Bestpet Pharmacy, London, UK) or sham treated at Zeitgeber time (ZT) 16 hours (time of normal lights off), as previously described (6). By this means, the precise time of melatonin rise could be regulated in each animal. Animals were then humanely destroyed by overdose of sodium pentobarbital at +1h30, +3h30, +6h30, and +9h30 after implantation (n = 12 [6 melatonin; 6 control per group]) and brains were removed and hypothalamic block with PT attached was frozen in dry ice.
For studies of photoperiodic changes, we used archive PT tissue sections, described earlier (6). Briefly, 48 Soay sheep ewes were housed indoors from early January (natural photoperiod, ∼ 7h light/17 h dark) under SP (8 h light/16 h dark). At week +8, 24 animals were killed at 1 of 6 time points: ZT3h, 7h, 11h, 15h, 19h, 23h (n = 4/time point), where ZT0 = time of lights on and ZT8 = time of lights off.
For RNA-seq experiment 8 male black-face sheep were transferred to an indoor facility in early January (natural photoperiod, ∼7 h light/17 h dark) and acclimated to artificial LP (16 h light/8 h dark) by symmetrical extension of the light phase by 4 hours at dawn and dusk. Twenty eight days after exposure to LP the animals were maintained in constant light. At the time of expected lights off (ZT16), half the cohort received intradermal melatonin implants, and the remaining 4 animals were sham treated as previously described (6). Animals were killed at +1h30 after implantation and PTs were collected and frozen on dry ice.
For immunohistochemistry and immunofluorescent experiments of LH and α-glycoprotein (GSU) hormone expressing cells, 8 male black-face sheep were transferred to an indoor facility in early January (natural photoperiod, ∼7 h light/17 h dark) and acclimated to artificial SP (8 h light/16 h dark) for 8 weeks. Animals were humanely killed at ZT7, and hypothalamic blocks with PTs attached were fixed using bouins. For immunofluorescent experiments of NPAS4, 4 male and 4 female black-face sheep were transferred to an indoor facility in early November (natural photoperiod, ∼9 h light/15 h dark) and acclimated to artificial SP (8 h light/16 h dark) for 8 weeks. At time of expected lights off (ZT8) animals were maintained in constant light and given intradermal melatonin implants. Animals were humanely killed at +2h after implantation, and hypothalamic blocks were taken with PTs attached. Tissue for immunofluorescent experiments were fixed in neutral-buffered formalin, and those tissues for immunohistochemistry were fixed in bouins.
Melatonin RIA
Melatonin was measured by RIA (16), using a rabbit antimelatonin antiserum (PF1288; P.A.R.I.S., Paris, France) and 2-Iodomelatonin (NEX236050UC; PerkinElmer, Boston, Massachusetts) as tracer. All samples were assayed in a single assay with an intraassay coefficient of variation of 5% and a sensitivity of 5 pg/mL.
RNA preparation
Total RNA was prepared using a Trizol (Invitrogen, Paisley, UK) extraction method as per the manufacturer's protocol. Samples were passed through RNeasy columns (Qiagen, Crawley, UK) to clean up the RNA further. Quality of the RNA was checked on a bioanalyzer (Agilent Technologies, Berkshire, United Kingdom).
Preparation of RNA-seq libraries, sequencing, and analysis
Samples were prepared for mRNA sequencing using 5 μg of total RNA starting material following the Illumina mRNA sequencing 8-sample preparation kit protocol. Resulting libraries were quality checked on an Agilent DNA 1000 bioanalyzer (Agilent Technologies), clustered onto a Single Read flow-cell using the Illumina v2 cluster generation kit at a 4.75 pM concentration, and 36 cycle single-ended sequencing was carried out on the Genome Analyzer IIx using Illumina v3 Sequencing by Synthesis kits (Illumina, Little Chesterford, United Kingdom). The Illumina Genome Analyzer-IIx platform generated 15–18 million RNA-seq tags per sample, each 36 nucleotides in length.
RNA-seq analysis
RNA-seq tags were assigned the reference sheep genome (OARv2.0) using Tophat and Bowtie alignment package (17, 18) by parallel computing using Edinburgh University high performance Computing and Data Facility (ECDF, www.ecdf.ed.ac.uk); unique tags were cross-referenced to sheep gene annotations, and counts of tags were calculated for each gene. Based on this pipeline, about 70 million tags (70% of all tags) were uniquely annotated to known genes. All samples were checked for quality at various steps, and all samples showed greater than 95% correlation with each other. Statistical analysis was then used to identify differentially expressed genes using the edgeR package (19) within Bioconductor. Low RNA-Seq coverage genes have been removed in advance. Statistical significance was set at a false discovery rate of less than 0.01 and greater than a 3-fold-change, P value was set to ≤ .01. Sequence data have been submitted to Ensembl for gene modeling, and experimental data will be made available on short read archive (SRA, http://www.ncbi.nlm.nih.gov/sra) upon completion of the ovine genome.
Immunohistochemistry
Tissues were immersed in 10% neutral-buffered formalin fixative for 24 hours, transferred to 70% ethanol, and then dehydrated and embedded in paraffin wax. Sections (5 μm) were cut, floated onto Superfrost Plus slides (J1800 AMNZ; Thermo Scientific, Rockford, Illinois), dried at 50°C for several hours and then dewaxed and rehydrated. Double immunostaining was carried out for colocalization of αGSU with NPAS4, or the LHβ-subunit. Slides were washed in PBS buffer between treatments. Endogenous peroxidase activity was blocked by incubating sections in 3% (vol/vol) hydrogen peroxidase in methanol for 30 minutes. Sections were blocked using 20% normal goat serum, 5% BSA in PBS (normal goat serum/PBS/5% BSA-blocking buffer) for 30 minutes, and then incubated in a humidified chamber overnight at 4°C in first primary antibody rabbit anti-ovine αGSU (ASMR20 91189, produced by Alan McNeilly) and raised against ovine αGSU (kindly donated by L.E. Reichert, Albany Medical College, New York, New York) (20), diluted at 1:2500 in blocking buffer. Each slide was then incubated with the goat antirabbit peroxidase IgG (ab7171 Abcam, Cambridge, Massachusetts) diluted 1:500, for 1 hour, then followed by trichostatin A (NEL77B001KT, PerkinElmer) diluted 1:50 in kit buffer for 10 minutes in the dark. Slides then were incubated in 0.01 M citrate buffer (2.5 min in boiling solution in the microwave followed by a cool down at room temperature for at least 15 minutes (21). Sections were treated with blocking buffer solution for 30 minutes and incubated overnight at 4°C with the second primary antibody, either rabbit antimouse NPAS4 (NBP1–06574; Novus Biologicals, Littleton, Colorado) diluted 1:2000 or mouse anti-ovine LHβ 518B7 (kindly donated by Dr Jan Roser, University of California, Davis, California). Sections were then incubated with the second secondary antibody goat antirabbit peroxidase IgG (ab7171 Abcam), or goat antimouse peroxidase IgG (ab6823 Abcam) followed by trichostatin A (NEL741B001KT, PerkinElmer), diluted 1:50 in kit buffer for 10 minutes in the dark. Control section were incubated with blocking buffer in place of the first, second primary, or both first and second primary antibodies. Sections were coverslip mounted using PermaFluor mounting medium (TA-030-FM, Thermo Scientific). Images were collected using a LSM 510 Meta microscope.
In situ hybridization (ISH)
Coronal sections (20 μm) from frozen tissues were collected with a cryostat (CM3050s Leica Microsystems, Ltd., Milton Keynes, UK) on polysine slides (VWR International, Lutterworth, UK) and kept at −80°C until ISH. ISH protocols are described elsewhere (6). Probes were hybridized overnight at 60°C on fixed sections, and hybridization signals were visualized on autoradiographic films (Kodak Biomax MR Films, Eastman Kodak, Rochester, New York) after 1-week exposure at −80°C. Signal intensity was quantified by densitometry analysis of autoradiographs using the image-Pro Plus 6.0 software (Media Cybernetics, Inc, Marlow, United Kingdom). Relative optical density in the PT was calculated on each section with a minimum of n = 3 sections per animal and 6 animals per group.
Plasmids
For riboprobes and coding sequences cloning, 1.5 μg of ovine PT RNA was reverse transcribed into cDNA. A 339-bp fragment of the ovine NPAS4 transcript was cloned in pGEM-T easy (Promega, Southampton, United Kingdom). Primers and cDNA sequences are deposited in GenBank under the following accession number: HQ224964. The coding sequence of ovine Npas4 (GenBank accession no. HF912412) was amplified by PCR with the following primers: forward, 5′-TCA TCG ATT CCA CCA TGT ACC GCT CCA CCA AGG G-3′; reverse, 5′-CAA TCG ATT GAA ACG TTG GTT CCC CTC CAC TTC-3′ in a vector containing 6× Myc tags (pCS2MT, http://sitemaker.umich.edu/dlturner.vectors). The ovine Arnt, Arntl, and Arnt2 coding sequences were also amplified by PCR with a His-tagged reverse primer and cloned in pCS2 using ClaI/XhoI sites (GenBank accession nos. HF912413, HF912415, and HF912414, respectively). Primers were as followed: Arnt forward, 5′-GAA TCG ATT CCA CCA TGG CGG CGA CTA CTG CTA A-3′; Arnt reverse, 5′-AGG CTC GAG GTG GTG ATG GTG ATG ATG TTC TGA AAA GGA GGG AAA CAT AGT TAG GC-3′; Arntl forward, 5′-GAA TCG ATT CCA CCA TGG CCG ACC AGA GAA TGGAC-3′; Arntl reverse, 5′-AGG CTC GAG GTG GTG ATG GTG ATG ATG CAG TGG CCA CGG CAA GTC AC-3′; Arnt2 forward, 5′-GAA TCG ATT CCA CCA TGG CAA CCC CGG CGG CCG-3′, Arnt2 reverse, 5′-AGG CTC GAG GTG GTG ATG GTG ATG ATG CTC CGA AAA CGG CGG GAA CAT G-3′.
The 1902-bp ovine pGL3-Cry1 promoter construct, which includes 101 bp downstream of transcription start site (TSS) was a kind gift of Professor David Hazlerigg (22) (GenBank accession no. EF651797). Subsequent 5′-deletions were performed by PCR to produce a 313-bp, 188-bp, and 130-bp fragments, which all had the same 3′-end, 97 bp downstream of TSS. The 2239-bp ovine Nampt promoter was amplified by PCR from an ovine bacterial artificial chromosome (BAC CH243–9F3; BACPAC; Oakland, California), using primers designed using bovine genomic data (ENSBTAG0000001550: forward, 5′-TCC TGC ATT AGC AGT CTG ATA CCC-3′; reverse, 5′-CGC TGG CAA TCT AGG GAC TC-3′): The obtained fragment was purified and submitted to another PCR using primers containing the appropriate restriction site for cloning in the luciferase pGL3 vector (Promega).
All site-directed mutagenesis was conducted using QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies, Berkshire, United Kingdom) according to the manufacturer's instructions. Mutation of putative CMEs was done in accordance with previous CME ablation studies (23, 26).
All PCRs were performed using Phusion High-Fidelity DNA polymerase (Thermo Fisher Scientific, Cheshire, United Kingdom). Original sequence fidelity was confirmed by comparison with reference sheep genome (OARv2.0). The fidelity of subclones and confirmation of site-directed mutagenesis was checked through sequencing and alignment to parent molecule.
Cell culture and transfections
COS7 cells were grown in DMEM high glucose + l-Glutamine, sodium pyruvate, sodium bicarbonate (Sigma-Aldrich, Poole, UK), supplemented with 10% fetal bovine serum (Sigma-Aldrich) and 1% penicillin streptomycin solution (Sigma-Aldrich), in 5% CO2 atmosphere at 37°C. Cells were seeded at 5.104 cells per well/mL in 24-well plates for luciferase assays or at 15.104 cells per well/3 mL in 6-well plates for Western blots. Cells were left to attach for 24 h before transfecting with Genejuice (Merck Chemical Ltd, Nottingham, United Kingdom) with a total amount of 250 ng or 1 μg of DNA per well (for luciferase assays and Western blots, respectively).
Luciferase assays
Cells were washed in PBS and then lysed using 150 μL of 1× passive lysis buffer (Promega), 24 hours after transfection. The dual luciferase reporter assay system was used to assess bioluminescence of lysate following the manufacturer's guidelines (Promega) using an Orion L microplate luminometer (Berthold, Hertfordshire, United Kingdom). Three to 5 technical repeats were carried out for each assay, with 4 replicates per group.
Protein harvest and Western blots
Cells were trypsinized for 5 minutes and harvested in 300 μL of lysis buffer (1% Triton X, 150 mM NaCl, 1 mM Tris-HCl [pH 7.4], 1 mM EDTA [pH 8], 1 mM EGTA, 0.5% Nonidet P-40). Bicinchoninic acid assays were carried out to quantify the total amount of protein in each group. Samples were boiled for 5 minutes with Laemmli sample buffer (Bio-Rad, Hertfordshire, United Kingdom) and stored on ice before being loaded onto a precast 4%–15% TGX gel (Bio-Rad). Gels were run at 120 V for 1.5 hours and then transferred via iBlot system to a nitrocellulose membrane (Life Technologies, Paisley, United Kingdom). Membranes were blocked in 5% milk solution in TBS-T (TBS/0.05% Tween 20) for 1 hour before overnight incubation with α-Myc (Caltag-Medsystems Ltd, 562, 1:20 000) or α-His (Genscript, A00186, 1:250) at 4°C. Membranes were then washed 5 times for 5 minutes with TBS-T and then incubated with horseradish peroxidase-linked mouse or rabbit α-IgG (raised in donkey, GE Healthcare, Berkshire, United Kingdom) at 1:20 000 for 1 hour at room temperature. After secondary antibody incubation the membranes were washed again 5 times for 5 minutes with TBS-T. Membranes were briefly washed with TBS after which immunoreactive proteins were visualized using ECL advance chemiluminescent kit (GE Healthcare, Berkshire, United Kingdom) and Kodak MR film (Eastman Kodak, Rochester, New York).
Statistical analyses
Differences between groups were analyzed with GraphPad Prism 5 software (GraphPad Software, Inc, San Diego, California) using 1- or 2-way ANOVA followed by Bonferroni post hoc test.
Results
RNA-seq reveals Npas4 as a melatonin-induced gene in the PT
For sheep housed in constant light at the time of normal lights off, melatonin implants induced circulating levels of the hormone equivalent to normal nighttime concentrations (16) whereas control implant-free animals maintained basal levels throughout the night (Supplemental Figure 1 published on The Endocrine Society's Journals Online web site at http://mend.endojournals.org). RNA-seq measures revealed a 38.8-fold induction of Npas4 mRNA within 1.5 hours within the ovine PT, after melatonin implant insertion (Table 1). Other known melatonin-regulated genes (Cry1 and NeuroD1) also exhibited a significant but lower amplitude increases in expression (Table 1). Candidate transcriptional partners for NPAS4 (ARNT, ARNT2, and ARNTL) were expressed in the PT but did not show any significant change in expression after melatonin treatment (Table 1).
Table 1.
RNA-seq Values for 8 Genes in the Ovine PT 1.5 h after Exposure to Melatonin in Early Night in LP-Housed Animals
| Gene Name | Gene ID | FC |
|---|---|---|
| Gadd45g | 3172 | 39.1 |
| Npas4 | 24718 | 38.8 |
| Nppc | 8974 | 27.3 |
| Drd1 | 8906 | 20.9 |
| Neurod2 | 22180 | 19.9 |
| Neurod1 | 7892 | 18.9 |
| MIR7–1 | 10103 | 12.9 |
| MIR1179 | 9908 | 12.4 |
| Cry1 | 8105 | 10.3 |
| Gtf2a1 | 17155 | 9.0 |
| Nlrp12 | 21562 | 5.0 |
| Fosl2 | 23262 | 4.5 |
| Ier5 | 21722 | 4.0 |
| Mterfd3 | 16759 | 3.7 |
| Six2 | 20158 | 3.2 |
| NPAS4 partners, not significantly melatonin regulated | Gene ID | FC |
| Arnt | 8860 | 1.05 |
| Arnt2 | 22664 | 1.07 |
| Arntl | 23327 | 1.03 |
Significantly induced genes have a false discovery rate of ≤ 0.01, P values were set ≤ .01. Gene identifications (IDs) are accession numbers for short-repeat archive database. FC, fold change over nonimplanted animals.
NPAS4 is quickly and transiently induced in the sheep PT in vivo
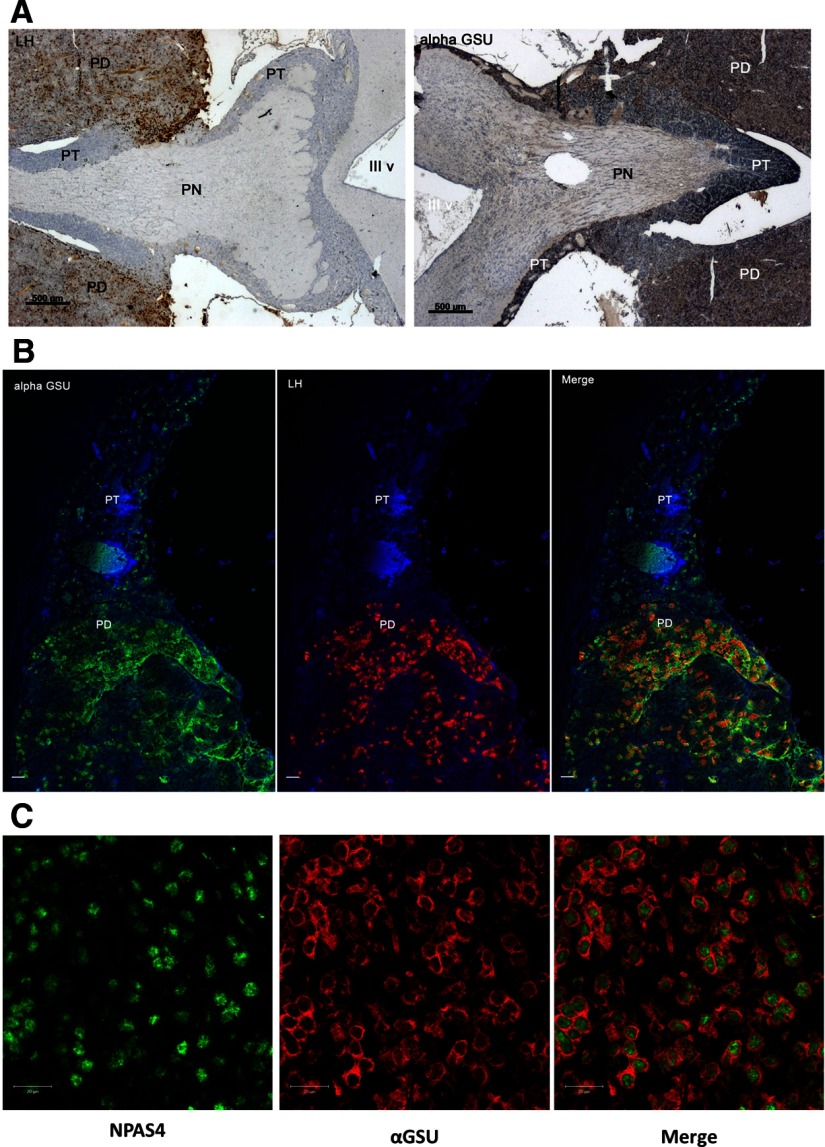
We undertook quantification of Npas4 expression in the PT, using ISH. In animals housed on LP conditions, we saw activation of Npas4 transcription in the PT (Figure 1A) with maximal induction at +1h30 after melatonin implant insertion, (Figure 1A, black bars) and a rapid decline to levels similar to those in untreated controls (Figure 1A, white bars) at +3h30, +6h30, and +9h30 after melatonin treatment. In order to confirm whether this activation was photoperiod specific, we also screened archival sections (6) from PT tissue collected from animals over a full light/dark cycle housed in SP conditions. This also revealed a rapid induction at the early night onset, with a decline in expression over a time course similar to that seen on LP conditions (Figure 1, A and B). Thus, Npas4 is induced in response to melatonin rise on both long and short photoperiods. We identified a population of αGSU expressing cells in the ovine PT but (in contrast to PD) were unable to detect any presumptive gonodotrophs within this region, based on colocalization with LHβ (Figure 2, A and B). Subsequent immunofluorescent analysis of NPAS4 in the PT showed that the protein coexpressed in αGSU-expressing cells (Figure 2C), with strong nuclear and weak cytoplasmic expression for NPAS4 and exclusively cytoplasmic staining for αGSU. The suitability of the antimouse NPAS4 antibody for ovine tissue was checked by comparing the protein sequence used as an antigenic region, and this is highly conserved across species (Supplemental Figure 2a). Further, we repeated the immunostaining studies with two additional antihuman NPAS4 antibodies in PT sections (both male and female), and this revealed an identical staining pattern (Supplemental Table 1 and Supplemental Figure 2b).
Figure 1.
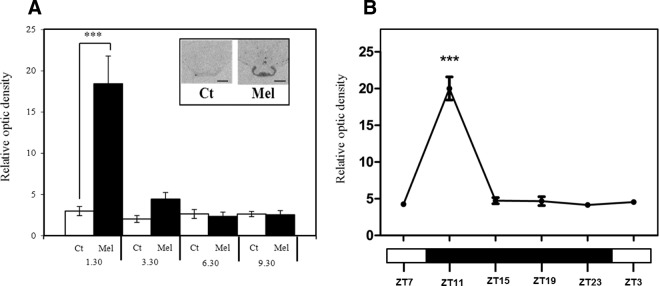
Npas4 mRNA Is quickly and Transiently Induced in Sheep after Melatonin Treatment and in SP Conditions. A, Quantitative ISH for Npas4 in the sheep PT housed in LPs (16 h light:8 h dark) after 1h30, 3h30, 6h30, and 9h30 melatonin (mel) or sham (Ct) treatment. Insert, representative autoradiographs after 1h30 treatment in melatonin and control group; scale bar, 200 μm. B, Quantitative ISH for Npas4 in the sheep PT every 4 hours in SPs (8 h light:16 h dark). Data show the mean relative optic density across 3 sections for each animal (n = 6 LP; n = 4 SP). Statistical differences between groups were analyzed after 2-way ANOVA followed by Bonferroni's post hoc test (***, P < .001).
Figure 2.
NPAS4 Is Detected in Nuclei of αGSU Expressing Ovine PT Cells. A, Immunohistochemistry, showing LH and αGSU expression in ovine PT and pars distalis (PD). Scale bars, 500 μm. B, Double immunofluorescence, showing expression of LH, αGSU, and merged images in ovine PT and PD. Scale bars, 20 μm. C, Double immunofluorescence, showing expression of αGSU and NPAS4 in ovine PT. Scale bars, 20 μm. PN, pars nervosa; III v, third ventricle.
NPAS4 preferentially cooperates with ARNT and ARNT2 to activate Cry1 and Nampt promoter transcriptional activity in vitro
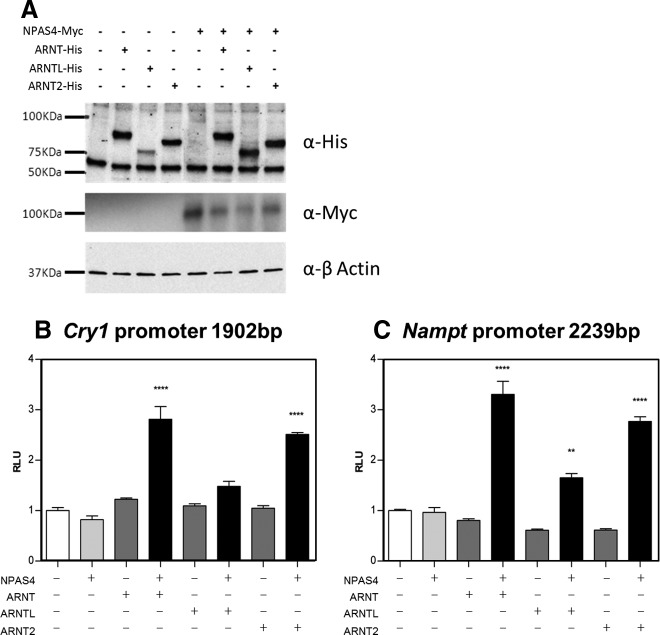
NPAS4 is known to interact with other PAS domain proteins to actively up-regulate the expression of its target genes, and studies by Ooe et al (23, 24) and Pruunsild et al (25) have identified members of the aryl hydrocarbon receptor nuclear translocator (ARNT) family as cofactors. Accordingly, we cloned full-length ovine NPAS4, ARNT, ARNTL, and ARNT2 in either Myc- or His-tagged expression vectors (NPAS4-Myc, ARNT-His, ARNTL-His, ARNT2-His) and characterized their effects on expression of the ovine Cry1 promoter using a luciferase reporter assay in COS7 cells. Appropriate expression of each protein in COS7 cells was confirmed by Western blot, using antibodies against Myc and His tags (Figure 3A). Luciferase assays revealed that single transfection of NPAS4, ARNT, ARNTL, or ARNT2 individually did not increase Cry1 promoter activity. However, when combined, NPAS4/ARNT and NPAS4/ARNT2 significantly activated Cry1 (Figure 3B). We had earlier identified nicotinamide phosophoribosyltransfersase (Nampt) as a candidate melatonin-regulated gene (6). We therefore also cloned a 2.2-Kb fragment of the ovine Nampt promoter, which revealed similar but lower amplitude activation by NPAS4/ARNTL (Figure 3C).
Figure 3.
NPAS4 Activates Cry1 and Nampt Promoter Activity in the Presence of Heterodimeric Partner Proteins. A, Western blots on total proteins from COS7 cells transfected with empty Myc and/or His expression vectors and vectors expressing the full-length ovine NPAS4-Myc, ARNT-His, ARNTL-His, or ARNT2-His. B and C, In vitro transfection of COS7 cells with a fragment of the ovine Cry1 1902 bp (B, −1805 +97 relative to TSS) or Nampt 2239bp (C, −1845 +394 relative to putative TSS) promoter driving the expression of the luciferase gene, together with empty Myc and/or His expression vectors and vectors expressing the full-length ovine NPAS4, ARNT, ARNTL, and/or ARNT2. Results show relative biolumnescence (RLU), after 24 hours, in each group (n = 4 replicates per group) normalized to control group (empty vectors), from 1 of 3 representative experiments. Statistical differences between groups were analyzed after 1-way ANOVA followed by Bonferrroni's post hoc test (**, P ≤ .01; ****, P ≤ .0001 against control group).
NPAS4 heterodimers transactivate Cry1 activity through two conserved CMEs
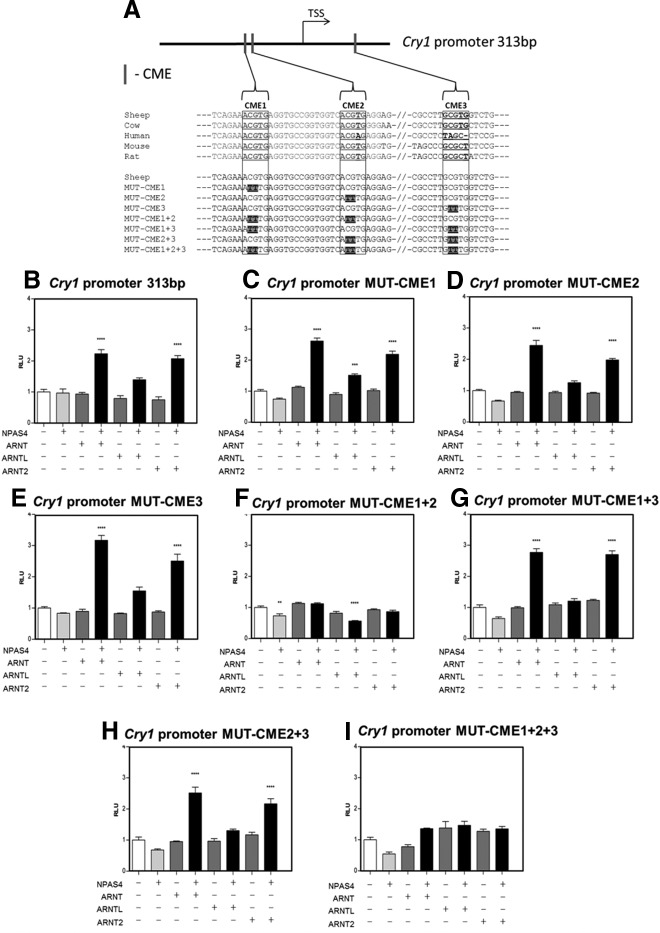
To determine the critical region essential to the NPAS4 heterodimer-mediated transcriptional activation of the Cry1 promoter, we carried out a 5′-truncation of the Cry1-luc construct to obtain a 313-bp fragment showing strong interspecies conservation (Figure 3A). This construct behaved similarly to the 1.9-kb long promoter, with significant induction when NPAS4 was cotransfected, either with ARNT or ARNT2 (Figure 3B).
NPAS4/ARNT dimers have been shown to activate transcription of target genes through DNA binding via their bHLH domain on CME (central nervous system midline element sequence) and mutation of the CGTG half-site (the ARNT binding site) to TTTG abolished the NPAS4/ARNT dependent effect (23, 26). Three putative CMEs were identified within the 313-bp region of the ovine promoter (Figure 3A). Each of these sites was mutated individually but did not affect the NPAS4 heterodimer-mediated activation of the Cry1 promoter (Figure 3, C–E). Similarly, double mutation of CME1+3 or CME2+3 did not impede NPAS4 heterodimeric activation (Figure 3, G and H). However, double mutation of CME1+2 or triple mutation of CME1+2+3 completely removed the transactivating potential of NPAS4 heterodimers (Figure 3, F and I).
In parallel, we characterized the critical Cry1 promoter region essential for regulation by NPAS4 heterodimers by 5′-truncation of the 313-bp construct and investigated responses of a 188-bp and 130-bp long fragment region (Figure 4A). Interestingly, 5′-deletion of the region containing CME1 and -2 does not appear to effect Cry1 promoter transactivation (Supplemental Figure 3, A and B). These studies (Supplemental Figure 3, C and D) revealed that NPAS4 heterodimer-mediated responses of the Cry1 promoter 5′ deleted from position −91 (relative to TSS) requires a 58-bp region located a few base pairs upstream of the TSS. Within this region, we undertook serial mutations, one or two nucleotides at a time, to identify a potential response element specific for NPAS4 heterodimers (Supplemental Figure 3E). Using this strategy, two critical sites (encompassing sites 2, 3 and 5–7) were identified in which mutations impaired (by 50% on their own and 100% when combined) the NPAS4/ARNT-mediated transcriptional activation of the Cry1-luc (188-bp) promoter construct (Supplemental Figure 3F). Predictive transcription factor-binding site analysis (AliBaba 2.1, http://www.gene-regulation.com/pub/programs.html) identified these regions as a putative CCAAT box and WT1 binding sites (Supplemental Figure 3G).
Figure 4.
Mutational Analysis of CMEs of the 313-bp Ovine Cry1 Promoter. A, Localization of the 3 putative CMEs (N(G/A)CGTG, CME1, CME2 and CME3) identified in the conserved 313-bp proximal promoter region (position −216 +97 relative to TSS) of the ovine Cry1 promoter (TSS). Nucleotide sequence alignment and site-directed mutagenesis alterations shown below. B–I, In vitro transfection of COS7 cells with a 313-bp fragment of the ovine Cry1 promoter driving the expression of the luciferase gene, together with empty Myc and/or His expression vectors and vectors expressing the full-length ovine NPAS4, ARNT, ARNTL, and/or ARNT2. Results show relative biolumnescence (RLU) after 24 hours, in each group (n = 4 replicates per group) normalized to control group (empty vectors), from 1 of 3 representative experiments. Statistical differences between groups were analyzed after 1-way ANOVA followed by Bonferroni's post hoc test (**, P ≤ .01; ***, P ≤ .001; ****, P ≤ .0001 against control group).
NPAS4 and ARNT cellular localization in COS7 cells
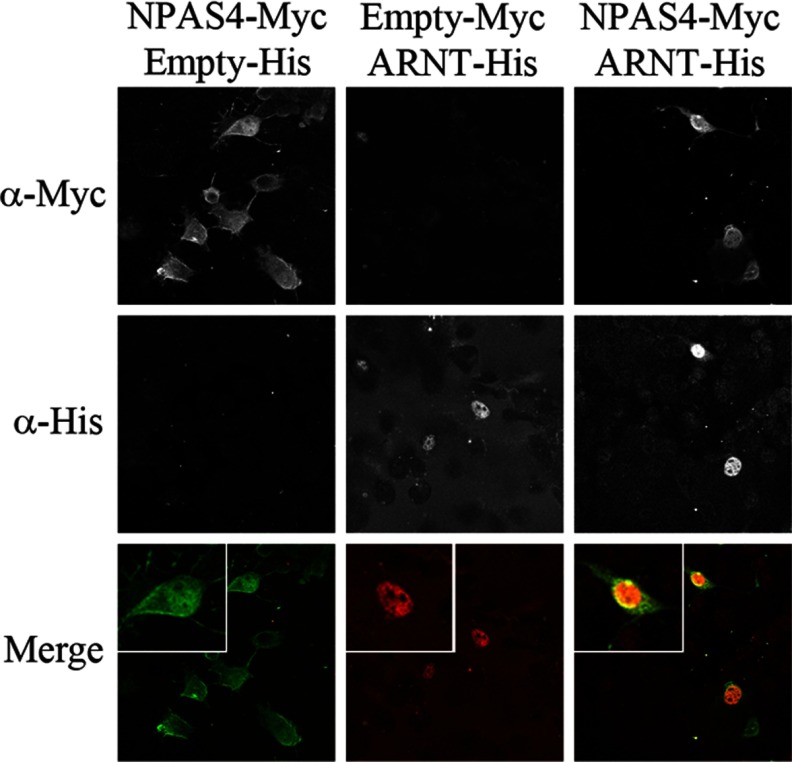
We characterized NPAS4 and ARNT cellular localization in transfected COS7 cells (Figure 5) using anti-Myc and anti-His tag antibodies. As previously described, ARNT expression is nuclear (27); interestingly, NPAS4 shows both cytoplasmic and nuclear localization, as seen in the ovine PT (Figure 2c and Supplemental Figure 2b). Previous studies have shown heterologous expression of NPAS4 to localize in the nucleus in COS7 cells (28). Here the additional localization of some NPAS4 in the cytoplasm may be suggestive of disparities in the biology of ovine and murine proteins, although this would be expected for a putative transcription factor that does not possess a nuclear localization signal. Coexpression of ARNT and NPAS4 revealed nuclear colocalization indicative of their transactivation potential as heterodimeric transcription factors.
Figure 5.
Cellular Localization of NPAS4 and ARNT in Transfected COS7 Cells. Immunocytochemistry of COS7 cells overexpressing NPAS4-Myc or/and ARNT-His. Images show staining obtained with the anti-Myc only (top panels), anti-His only (middle panels), and combined (bottom panels).
Discussion
The transduction of photoperiodic information in seasonal mammals is known to involve the PT. Melatonin has been shown to suppress cAMP activity within this tissue, and the decline in melatonin at dawn is associated with a deinhibition and rise in intracellular cAMP, directly stimulating induction of cAMP-responsive genes, including the circadian clock gene Per1 (10, 11). The mechanisms involved in the nocturnal activation of an additional cohort of genes induced by melatonin, which includes the clock gene Cry1, regulated via an MT1 receptor-dependent mechanism (6, 10, 30–32), are less well understood. MT1 signaling has been associated with Gαi, Gαq/11, and βγ G protein subunits; notably, RGS4 (a regulator of G protein signaling) is induced by melatonin and demonstrated to modulate MT1 signaling in vitro (33–35). However, none of these interactions have been successfully linked to the mechanism underpinning melatonin-driven gene activation. Seasonal changes in the phase relationship between gene cohorts, driven by changes in duration of the melatonin signal and timed to the onset and offset of secretion, have been proposed as a potential mechanism whereby a photoperiodic transcriptional circuit is activated (8). Within all other mammalian tissues and cell types, PER and CRY are normally coexpressed in a similar phase relationship, and heterodimeric repression by these two proteins is considered to provide rhythmic negative feedback to the core circadian clock (36). Melatonin action on the PT may represent a special case in which the relative phasing of these two components changes with season.
We screened the ovine PT for rapid and acute alterations in gene expression in response to melatonin and identified Npas4 as a candidate immediate-response gene to melatonin. NPAS4 is a member of the PAS-domain family, which includes the circadian clock regulators CLOCK and NPAS2. Our work now identifies NPAS4 as a key element in seasonal timing. PAS domain proteins trigger specific transcriptional programs upon various stimuli, with the example of CLOCK, HIF1α, and AHR sensing changes in light, oxygen, or toxicants, respectively, in the environment (37). Like other members of the PAS family, NPAS4 can interact with other PAS domain proteins such as ARNT to convey an immediate response to a specific signal through active transcriptional induction of its target genes.
Npas4 is primarily expressed in the brain, notably in the hippocampus and the habenula (24, 28), structures that also coexpress melatonin receptors (38–40). The gene has previously been described as an activity-dependent transcription factor, quickly induced upon synaptic activity, stress response, or drug challenge, with a major role in learning, memory, and conditional fear responses (23, 25, 41–43). It is also very rapidly induced after a variety of different stimuli (43), with induction in the hippocampal CA3 region in as little as 5 minutes after contextual learning, where it is thought to be essential for memory formation (42). Within the hippocampus, Bdnf (brain-derived neurotrophic factor) has been described as a key target gene for NPAS4 (25), but although the gene is expressed in the PT, in contrast to the hippocampus, we did not observe induction with melatonin or altered pattern of expression across the light-dark cycle on either short or long photoperiods (data not shown). Thus, the repertoire of downstream NPAS4 targets may be tissue and cell type specific.
We detected rapid and PT-specific activation of NPAS4 mRNA, both by RNA-seq and ISH methods. Our data also reveal that strong nuclear and weak cytoplasmic NPAS4 is widespread in αGSU (common subunit for TSH)-expressing cells of the ovine PT. The use of αGSU as a marker for PT thyrotrophs was justified by IHC and immunofluorescence, which showed that αGSU colocalized with LHβ in the pars distalis gonadotrophs, but not in the PT. Thus αGSU was used to mark presumptive thyrotrophs of the ovine PT as shown previously in the pars distalis of both mice and human pituitaries (20). The TSH-immunoreactive cells of the PT are known to colocalize the MT1 receptor in the rat (44), and our data are therefore consistent with the hypothesis that TSH-expressing cells of the PT are the primary target for melatonin regulation of NPAS4. Npas4 may be a strongly conserved component in melatonin signaling in the PT, because it has previously been identified by microarray experiments in melatonin-proficient strains of mice, with up-regulation in the PT at night, at the time of maximal melatonin secretion, and with greatly reduced expression in mice bearing a knockout for the MT1 receptor (15).
Our in vitro studies suggest that NPAS4 preferentially acts in cooperation with ARNT or ARNT2 to stimulate the activity of Cry1 and Nampt promoters. The cofactor ARNT has been previously shown to act as a homodimer (45), but our studies suggest that ARNT acts primarily as a heterodimer in driving Cry1 and Nampt expression. Yeast two-hybrid experiments have also shown interactions between NPAS4 and ARNTL (BMAL1) (23), but we revealed a lower efficiency of action, when compared with NPAS4/ARNT or NPAS4/ARNT2. NPAS4 heterodimers have been shown to transactivate gene transcription through a Per-Arnt-Sim-response element domain and a response element similar to a CME (23, 25). Although lacking a Per-Arnt-Sim-response element, three putative CME were identified in the conserved 313-bp Cry1 promoter. Our site-directed mutagenesis studies showed that whereas no single site could sufficiently account for the action of the target gene, mutation of two closely associated sites (CME1+2) or a triple mutation of all three sites (CME1+2+3) completely removed NPAS4 heterodimer-mediated transactivation. Interestingly, however, 5′-truncation of the 313-bp Cry1 promoter to produce a 188-bp promoter region (removing CME 1 and 2) resulted in an NPAS4 response. Specific mutations within the proximal promoter region of the 188-bp Cry1 promoter revealed critical sequences affecting the NPAS4/ARNT-dependent response are likely to be binding sites for factors essential to the initiation of the transcription. It is possible that NPAS4 heterodimers may also potentiate basal transcription as cofactors, recruited by other transcription factors essential for RNA polymerase II-driven transcription. Indeed, chromatin immunoprecipitation-seq data in mice showed that NPAS4 colocalizes with RNA polymerase II on promoters, implying that NPAS4 is involved as a cofactor in the recruitment of the preinitiation complex and general transcriptional activation (42). The paradoxical recovery of NPAS4 activation of the minimal promoter (Cry1 188bp promoter, lacking CME1 and 2) could be due to loss of binding site(s) for a repressor complex. Thus the conserved blocks 1 and 2 (Supplemental Figure 3) are critical for NPAS4-mediated activation of the Cry1 promoter.
To convey photoperiodic information the PT has to accurately track the onset of the dark phase, and these signals therefore need to exhibit rapid onset and precision, matching the sharp rise in cAMP activation at dawn. Because our data reveal that Npas4 is activated by melatonin on both short and long photoperiods, we propose that this member of the bHLH-PAS transcription factor family acts as the dark-onset switch, triggering the initiation of a downstream transcriptional program within this key photoperiodic tissue. Our studies also demonstrate that NPAS4, previously considered as a brain-specific gene primarily regulating neural plasticity and memory formation (42, 43, 48), may also serve an important role in melatonin-regulated timing circuits within the pituitary gland.
Supplementary Material
Acknowledgments
We thank Beverley Bright and Ben Saer for laboratory technical assistance, and Joan Docherty, Marjorie Thompson, and John Hogg for assistance with animal work at the Marshall Building, University of Edinburgh; Professor David Hazlerigg, University of Aberdeen, for kind gift of a Cry1 plasmid, and an anonymous referee for helpful suggestions on an earlier draft.
This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC, Grant BB G003033 A.L. and J.D.) and BB/J004235/1 and BB/J004316/1 (to D.B.).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- ARNT
- aryl hydrocarbon receptor nuclear translocator
- bHLH
- basic helix loop helix
- CME
- central midline element
- GSU
- α-glycoprotein
- ISH
- in situ hybridization
- LP
- long photoperiod
- PT
- pars tuberalis
- SP
- short photoperiod
- TBS-T
- Tris-biuffered saline/0.05% Tween 20
- ZT
- Zeitgeber time.
References
- 1. Hazlerigg D, Loudon A. New insights into ancient seasonal life timers. Curr Biol. 2008;18:R795–R804 [DOI] [PubMed] [Google Scholar]
- 2. Lincoln GA, Short RV. Seasonal breeding: nature's contraceptive. Recent Prog Horm Res. 1980;36:1–52 [DOI] [PubMed] [Google Scholar]
- 3. Dupré SM. Encoding and decoding photoperiod in the mammalian pars tuberalis. Neuroendocrinology. 2011;94:101–112 [DOI] [PubMed] [Google Scholar]
- 4. Bartness TJ, Powers JB, Hastings MH, Bittman EL, Goldman BD. The timed infusion paradigm for melatonin delivery: what has it taught us about the melatonin signal, its reception, and the photoperiodic control of seasonal responses? J Pineal Res. 1993;15:161–190 [DOI] [PubMed] [Google Scholar]
- 5. Hanon EA, Lincoln GA, Fustin JM, et al. Ancestral TSH mechanism signals summer in a photoperiodic mammal. Curr Biol. 2008;18:1147–1152 [DOI] [PubMed] [Google Scholar]
- 6. Dupré SM, Burt DW, Talbot R, et al. Identification of melatonin-regulated genes in the ovine pituitary pars tuberalis, a target site for seasonal hormone control. Endocrinology. 2008;149:5527–5539 [DOI] [PubMed] [Google Scholar]
- 7. Yoshimura T. Neuroendocrine mechanism of seasonal reproduction in birds and mammals. Anim Sci J. 2010;81(4):403–410 [DOI] [PubMed] [Google Scholar]
- 8. Lincoln GA, Andersson H, Loudon A. Clock genes in calendar cells as the basis of annual timekeeping in mammals—a unifying hypothesis. J Endocrinol. 2003;179(1):1–13 [DOI] [PubMed] [Google Scholar]
- 9. Lincoln G, Messager S, Andersson H, Hazlerigg D. Temporal expression of seven clock genes in the suprachiasmatic nucleus and the pars tuberalis of the sheep: evidence for an internal coincidence timer. Proc Natl Acad Sci U S A. 2002;99:13890–13895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Messager S, Ross AW, Barrett P, Morgan PJ. Decoding photoperiodic time through Per1 and ICER gene amplitude. Proc Natl Acad Sci U S A. 1999;96:9938–9943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morgan PJ, Ross AW, Graham ES, Adam C, Messager S, Barrett P. oPer1 is an early response gene under photoperiodic regulation in the ovine pars tuberalis. J Neuroendocrinol. 1998;10:319–323 [DOI] [PubMed] [Google Scholar]
- 12. Dupré SM, Miedzinska K, Duval CV, et al. Identification of Eya3 and TAC1 as long-day signals in the sheep pituitary. Curr Biol. 2010;20:829–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dardente H, Menet JS, Poirel VJ, et al. Melatonin induces Cry1 expression in the pars tuberalis of the rat. Brain Res Mol Brain Res. 2003;114:101–106 [DOI] [PubMed] [Google Scholar]
- 14. Fischer C, Christ E, Korf HW, von Gall C. Tafa-3 encoding for a secretory peptide is expressed in the mouse pars tuberalis and is affected by melatonin 1 receptor deficiency. Gen Comp Endocrinol. 2012;177:98–103 [DOI] [PubMed] [Google Scholar]
- 15. Unfried C, Burbach G, Korf HW, von Gall C. Melatonin receptor 1-dependent gene expression in the mouse pars tuberalis as revealed by cDNA microarray analysis and in situ hybridization. J Pineal Res. 2010;48:148–156 [DOI] [PubMed] [Google Scholar]
- 16. Almeida OF, Lincoln GA. Photoperiodic regulation of reproductive activity in the ram: evidence for the involvement of circadian rhythms in melatonin and prolactin secretion. Biol Reprod. 1982;27(5):1062–1075 [DOI] [PubMed] [Google Scholar]
- 17. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature Methods 4;. 2012;9(4):357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Robinson MD, McCarthy DJ, Smyth GK. EdgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 1;. 2010;26(1):139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pope C, McNeilly JR, Coutts S, Millar M, Anderson RA, McNeilly AS. Gonadotrope and thyrotrope development in the human and mouse anterior pituitary gland. Dev Biol. 2006;297(1):172–181 [DOI] [PubMed] [Google Scholar]
- 21. Tóth ZE, Mezey E. Simultaneous visualization of multiple antigens with tyramide signal amplification using antibodies from the same species. J Histochem Cytochem. 2007;55(6):545–554 [DOI] [PubMed] [Google Scholar]
- 22. Fustin JM, O'Neill JS, Hastings MH, Hazlerigg DG, Dardente H. Cry1 circadian phase in vitro: wrapped up with an E-box. J Biol Rhythms. 2009;24(1):16–24 [DOI] [PubMed] [Google Scholar]
- 23. Ooe N, Motonaga K, Kobayashi K, Saito K, Kaneko H. Functional characterization of basic helix-loop-helix-PAS type transcription factor NXF in vivo: putative involvement in an “on demand” neuroprotection system. J Biol Chem. 2009;284:1057–1063 [DOI] [PubMed] [Google Scholar]
- 24. Ooe N, Saito K, Mikami N, Nakatuka I, Kaneko H. Identification of a novel basic helix-loop-helix-PAS factor, NXF, reveals a Sim2 competitive, positive regulatory role in dendritic-cytoskeleton modulator drebrin gene expression. Mol Cell Biol. 2004;24:608–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pruunsild P, Sepp M, Orav E, Koppel I, Timmusk T. Identification of cis-elements and transcription factors regulating neuronal activity-dependent transcription of human BDNF gene. J Neurosci. 2011;31:3295–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wharton KA, Jr., Franks RG, Kasai Y, Crews ST. Control of CNS midline transcription by asymmetric E-box-like elements: similarity to xenobiotic responsive regulation. Development. 1994;120:3563–3569 [DOI] [PubMed] [Google Scholar]
- 27. Hord NG, Perdew GH. Physicochemical and immunocytochemical analysis of the aryl hydrocarbon receptor nuclear translocator: characterization of two monoclonal antibodies to the aryl hydrocarbon receptor nuclear translocator. Mol Pharmacol. 1994;46(4):618–626 [PubMed] [Google Scholar]
- 28. Moser M, Knoth R, Bode C, Patterson C. LE-PAS, a novel Arnt-dependent HLH-PAS protein, is expressed in limbic tissues and transactivates the CNS midline enhancer element. Brain Res Mol Brain Res. 2004;128(2):141–149 [DOI] [PubMed] [Google Scholar]
- 29. Jin X, von Gall C, Pieschl RL, et al. Targeted disruption of the mouse Mel(1b) melatonin receptor. Mol Cell Biol. 2003;23:1054–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lincoln GA, Johnston JD, Andersson H, Wagner G, Hazlerigg DG. Photorefractoriness in mammals: dissociating a seasonal timer from the circadian-based photoperiod response. Endocrinology. 2005;146:3782–3790 [DOI] [PubMed] [Google Scholar]
- 31. Weaver DR, Liu C, Reppert SM. Nature's knockout: the Mel1b receptor is not necessary for reproductive and circadian responses to melatonin atonin in Siberian hamsters. Mol Endocrinol. 1996;10:1478–1487 [DOI] [PubMed] [Google Scholar]
- 32. Yasuo S, Yoshimura T, Ebihara S, Korf HW. Melatonin transmits photoperiodic signals through the MT1 melatonin receptor. J Neurosci. 2009;29:2885–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brydon L, Roka F, Petit L, et al. Dual signaling of human Mel1a melatonin receptors via G(i2), G(i3), and G(q/11) proteins. Mol Endocrinol. 1999;13(12):2025–2038 [DOI] [PubMed] [Google Scholar]
- 34. Witt-Enderby PA, MacKenzie RS, McKeon RM, Carroll EA, Bordt SL, Melan MA. Melatonin induction of filamentous structures in non-neuronal cells that is dependent on expression of the human mt1 melatonin receptor. Cell Motil Cytoskeleton. 2000;46(1):28–42 [DOI] [PubMed] [Google Scholar]
- 35. Dupré SM, Dardente H, Birnie MJ, Loudon AS, Lincoln GA, Hazlerigg DG. Evidence for RGS4 modulation of melatonin and thyrotrophin signalling pathways in the pars tuberalis. J Neuroendocrinol. 2011;23(8):725–732 [DOI] [PubMed] [Google Scholar]
- 36. Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418(6901):935–941 [DOI] [PubMed] [Google Scholar]
- 37. Crews ST, Fan CM. Remembrance of things PAS: regulation of development by bHLH-PAS proteins. Curr Opin Genet Dev. 1999;9:580–587 [DOI] [PubMed] [Google Scholar]
- 38. Bittman EL, Weaver DR. The distribution of melatonin binding sites in neuroendocrine tissues of the ewe. Biol Reprod. 1990;43(6):986–993 [DOI] [PubMed] [Google Scholar]
- 39. de Reviers MM, Tillet Y, Pelletier J. Melatonin binding sites in the brain of sheep exposed to light or pinealectomized. Neurosci Lett. 1991;121(1–2):17–20 [DOI] [PubMed] [Google Scholar]
- 40. Pandi-Perumal SR, Trakht I, Srinivasan V, et al. Physiological effects of melatonin: role of melatonin receptors and signal transduction pathways. Prog Neurobiol. 2008;85(3):335–353 [DOI] [PubMed] [Google Scholar]
- 41. Lin Y, Bloodgood BL, Hauser JL, et al. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature. 2008;455(7217):1198–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ramamoorthi K, Fropf R, Belfort GM, et al. Npas4 regulates a transcriptional program in CA3 required for contextual memory formation. Science. 2011;334:1669–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ploski JE, Monsey MS, Nguyen T, DiLeone RJ, Schafe GE. The neuronal PAS domain protein 4 (Npas4) is required for new and reactivated fear memories. PLoS One. 2011;6:e23760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Klosen P, Bienvenu C, Demarteau O, et al. The mt1 melatonin receptor and RORbeta receptor are co-localized in specific TSH-immunoreactive cells in the pars tuberalis of the rat pituitary. J Histochem Cytochem. 2002;50(12):1647–1657 [DOI] [PubMed] [Google Scholar]
- 45. Arpiainen S, Lämsä V, Pelkonen O, Yim SH, Gonzalez FJ, Hakkola J. Aryl hydrocarbon receptor nuclear translocator and upstream stimulatory factor regulate Cytochrome P450 2a5 transcription through a common E-box site. J Mol Biol. 2007;369(3):640–652 [DOI] [PubMed] [Google Scholar]
- 46. Dolfini D, Gatta R, Mantovani R. NF-Y and the transcriptional activation of CCAAT promoters. Crit Rev Biochem Mol Biol. 2012;47:29–49 [DOI] [PubMed] [Google Scholar]
- 47. Philipsen S, Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 1999;27(15):2991–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Maya-Vetencourt JF, Tiraboschi E, Greco D, et al. Experience-dependent expression of NPAS4 regulates plasticity in adult visual cortex. J Physiol. 2012;590:4777–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.