Background: The mechanisms of placental growth factor (PlGF)-mediated blood vessel formation are incompletely understood.
Results: Plasmin cleaves the heparin-binding domain of PlGF-2.
Conclusion: Plasmin regulates PlGF-2/Neuropilin-1-mediated tissue vascularization and growth.
Significance: Plasmin-mediated carboxyl-terminal processing of VEGF family members may be considered as a principal mechanism to regulate their biological activity.
Keywords: Angiogenesis, Plasmin, Protease, Vascular Biology, Vascular Endothelial Growth Factor (VEGF), Mechanism of Growth Factor Regulation, PlGF
Abstract
Placental growth factor (PlGF) is a critical mediator of blood vessel formation, yet mechanisms of its action and regulation are incompletely understood. Here we demonstrate that proteolytic processing regulates the biological activity of PlGF. Specifically, we show that plasmin processing of PlGF-2 yields a protease-resistant core fragment comprising the vascular endothelial growth factor receptor-1 binding site but lacking the carboxyl-terminal domain encoding the heparin-binding domain and an 8-amino acid peptide encoded by exon 7. We have identified plasmin cleavage sites, generated a truncated PlGF118 isoform mimicking plasmin-processed PlGF, and explored its biological function in comparison with that of PlGF-1 and -2. The angiogenic responses induced by the diverse PlGF forms were distinct. Whereas PlGF-2 increased endothelial cell chemotaxis, vascular sprouting, and granulation tissue formation upon skin injury, these activities were abrogated following plasmin digestion. Investigation of PlGF/Neuropilin-1 binding and function suggests a critical role for heparin-binding domain/Neuropilin-1 interaction and its regulation by plasmin processing. Collectively, here we provide new mechanistic insights into the regulation of PlGF-2/Neuropilin-1-mediated tissue vascularization and growth.
Introduction
Placental growth factor (PlGF),2 a member of the VEGF family of growth factors, is a critical regulator of postnatal angiogenesis in various physiological and pathological conditions including repair of soft (1–3) and hard tissues (4), inflammation (5), and cancer (6). The exact molecular mechanisms by which PlGF regulates blood vessel formation are still not completely understood (7). PlGF has been reported to support the formation and maturation of new vessels by direct action on existing endothelial cells but also promotes angiogenesis indirectly by inducing recruitment and survival of other cell types involved in angiogenesis, such as mural cells, monocytes/macrophages, and bone marrow-derived precursor cells (8–10).
Although PlGF and VEGF-A show only a 42% amino acid sequence identity, as well as significant functional differences, PlGF shares remarkable structural similarities with the extensively analyzed family member VEGF-A (11). Comparative structure-function analysis between VEGF-A and PlGF might contribute to advance mechanistic insights into PlGF-mediated activities. Generation of diverse VEGF-A isoforms, which are distinguished by the presence of carboxyl-terminal peptides encoded by exons 6, 7, and 8 of the vegf-a gene, are crucial for the diverse biochemical and functional properties of VEGF-A, including binding to cell surfaces and extracellular matrix components, receptor binding characteristics, endothelial cell adhesion and survival, as well as vascular branch formation (11). Similarly to the generation of VEGF-A isoforms, in humans mRNA splicing of a single plgf gene gives rise to at least four different protein variants (PlGF-1 to -4), with PlGF-1 and -2 being the predominant isoforms expressed and studied so far (12). In mice the plgf gene encodes only the PlGF-2 isoform (13).
PlGF variants are homodimeric molecules, sharing a sequence encoded by exons 1–5 representing the VEGFR-1 binding sites and an 8-amino acid peptide encoded by exon 7. PlGF-1 and -2 differ in the absence or presence of a domain encoded by exon 6, respectively, which exhibits affinity to heparin-Sepharose and is referred to as a heparin-binding domain (HBD) (14). Furthermore, cross-linking experiments and competitive binding studies on endothelial cells indicated that the HBD of PlGF-2 is critical for the interaction with the co-receptor neuropilin-1 (Nrp-1) (15, 16). The biological/functional consequences of PlGF-2 binding with Nrp-1 and/or extracellular matrix components are still unclear. Likewise, functions of the amino acid sequence encoded by exon 7 have not been reported.
Although mechanistically not yet completely understood, binding of VEGF-A165 through its HBD to Nrp-1 was shown to promote VEGF-A165·VEGFR-2 complex formation and to sustain VEGFR-2 signaling and biological activities (17, 18). Furthermore, differential interaction of VEGF-A isoforms with various extracellular matrix components has been intensively analyzed. All VEGF-A isoforms except the short VEGF-A121 and the VEGF-Axxxb forms interact through their HBD encoded by exon 6a and 7 with heparin/heparan sulfate; for VEGF-A165, this feature has been shown to be essential for the establishment of a functional vascular system (19, 20). One could speculate on similar functions for the diverse PlGF isoforms for differential binding to matrix components and/or Nrp-1 interactions.
In addition to differential mRNA splicing, proteolytic processing of long VEGF-A isoforms has been shown to be an important regulatory mechanism to control VEGF-A activities (21). Specifically, we and others have demonstrated the susceptibility of VEGF-A165 to proteolytic cleavage by plasmin and metalloproteinases (21–25). Both proteases generate a protease-resistant core fragment containing the VEGFR binding domain but lacking the carboxyl-terminal domain coding for the HBD and the domain encoded by exon 8. Proteolytic processing of VEGF-A165 substantially reduced and altered the angiogenic response during wound healing and tumor formation (24, 25). Whether the PlGF isoforms display analogous susceptibility to proteases has not been analyzed. Here we investigated the hypothesis that PlGF is a substrate of plasmin and might be regulated in its biological activities by proteolytic processing. We show that PlGF-2 is sensitive to plasmin processing and that truncation of PlGF-2 significantly impacts its HBD/Nrp-1-mediated effects on angiogenesis. Our findings unveil a dual control of PlGF activity by transcriptional regulation and proteolytic mechanisms.
EXPERIMENTAL PROCEDURES
Synthesis and Purification of Recombinant Human PlGF Variants
cDNAs of full-length PlGF-1 (Met-1 to Arg-131), PlGF-2 (Met-1 to Arg-152), and a truncated version PlGF118 (Met-1 to Lys-118) were generated from human placenta and cloned into the eukaryotic expression vector pCEP V149 and pCEP V19. For affinity purification, cDNAs were fused either to an amino-terminal, 8× histidine tag (PlGF118) or an amino-terminal, 8× histidine tag and a carboxyl-terminal, double streptavidine tag (PlGF-1 and PlGF-2). The proteins were produced in human embryonic kidney cells (HEK293 EBNA cells). Recombinant PlGF isoforms were purified to 95% purity by affinity chromatography using Strep-Tactin Superflow Sepharose (PlGF-1 and PlGF-2; IBA BioTAGnology) or immobilized metal ion affinity chromatography using nickel-Sepharose 6 Fast Flow (PlGF118; GE Healthcare). Protein identities were confirmed by peptide mass fingerprinting. Protein concentrations were determined by the BCA assay (Pierce) and ELISA (Quantikine human PlGF; R & D Systems) according to the manufacturer's instructions. As indicated, in some experiments recombinant PlGF proteins were purchased and produced in Sf-9 insect cells (Reliatech).
SDS-PAGE and Immunoblotting
SDS-PAGE was performed following the protocol of Laemmli. To analyze the sensitivity of PlGF isoforms to plasmin, PlGF proteins were incubated with wound exudate, human serum plasmin (0.02 unit/ml or serial dilutions as indicated) (Calbiochem), or α2-antiplasmin (Calbiochem) at 37 °C; at the indicated time points, the reactions were terminated by the addition of reducing or nonreducing Laemmli buffer and heating to 95 °C. The reactions were resolved on a 4–12% reducing Bis-Tris SDS-PAGE gel (NuPAGE; Invitrogen). Integrity of PlGF isoforms was determined by silver staining (SilverQuestTM; Invitrogen) or by detecting immunoreactive products with an anti-human PlGF monoclonal rabbit antibody directed against the first 20 amino-terminal amino acids of human PlGF (Reliatech). Bound primary antibody was detected using an anti-rabbit HRP-conjugated secondary antibody (DAKO A/S). To assess Nrp-1 expression in PAE and PAE/Nrp-1 cells, cell lysates were normalized to 1000 μg of total protein, incubated with concanavalin A-Sepharose beats (Amersham Biosciences) to enrich glycosylated proteins following the manufacturer's instructions; the eluted fraction was analyzed by immunoblotting using a mouse-anti-human Nrp-1 (A12) antibody (Santa Cruz Biotechnology) and an anti-mouse HRP-conjugated secondary antibody (DAKO A/S).
To analyze signal transduction, endothelial cells were starved and incubated with PlGF variants (2.5 nm) in endothelial basal medium-2 (growth factor-free, 0.1% FCS) (Invitrogen), VEGF-A165 (1.8 nm) (produced in Sf9 insect cells; Reliatech), or starvation medium for the indicated time periods. The cells were lysed, and samples were resolved in 4–12% Bis-Tris SDS-PAGE gel and immunoblotted with anti-phospho-focal adhesion kinase (Tyr-576/577), anti-phospho-Akt (Ser-473), anti-phospho-Erk-1/-2 (Thr-202/Tyr-204), anti-total Akt, anti-total Erk-1/-2, anti-β-actin (all Cell Signaling Technologies), anti-phospho-VEGFR-1 (Tyr-1213) (Millipore), anti-total-VEGFR-1 (Millipore), or anti-Nrp-1 (Santa Cruz Biotechnology). Detection was accomplished using the enhanced chemiluminescense Western blot detection system (ECL; Amersham Biosciences). Densitometric analysis of phospho-signal intensity was performed using ImageJ 1.44p software. The signal of pErk, pAkt, or pFAK was normalized to the corresponding total Erk, Akt, or β-actin signal, respectively. Multiple Western blots (≥3) were performed.
Mass Spectrometric Analysis
PlGF-2 expressed in HEK293 cells (25 μg in 200 μl of 50 mm Tris-HCl, pH 8) was incubated with nickel-Sepharose beats (15 min at room temperature) to allow binding to the amino-terminal His tag, followed by incubation in plasmin (final concentrations: 0.04 or 0.008 unit/ml in plasmin buffer 50 mm Tris-HCl, pH 8, 37 °C, 5 or 30 min); beats were collected by centrifugation (5 min, 13,500 rpm, 4 °C), and the supernatants were analyzed by LC-MS/MS. LC-MS data were acquired on a Q-TofII quadrupole-TOF mass spectrometer (Micromass) equipped with a Z spray source. Samples were introduced by an Ultimate Nano-LC system (LC Packings) equipped with the Famos autosampler and the Switchos column-switching module. The column setup comprises a 0.3 × 1-mm trapping column and a 0.075 × 150-mm analytical column, both packed with 3-μm Atlantis dC18 (Waters). The samples were diluted 1:10 in 0.1% TFA. A total of 10 μl was injected onto the trap column and desalted for 1 min with 0.1% TFA and a flow rate of 10 μl/min. The 10-port valve switched the trap column into the analytical flow path, and peptides were eluted onto the analytical column by using a gradient of 2% acetonitrile in 0.1% formic acid to 40% acetonitrile in 0.1% formic acid over 65 min and a column flow rate of ∼200 nl/min, resulting from a 1:1,000 split of the 200 μl/min flow delivered by the pump. The electrospray ionization interface comprised an uncoated 10-μm-inner diameter PicoTip spray emitter (New Objective) linked to the HPLC flow path using a 7-μl dead volume stainless steel union mounted onto the PicoTip holder assembly (New Objective). Stable nanospray was established by the application of 1.7- 2.4 kV to the stainless steel union. The data-dependent acquisition of MS and tandem MS (MS/MS) spectra was controlled by the Masslynx 4.0. Survey scans of 1.4 s covered the range from m/z 400 to 1,400. Doubly and triply charged ions rising above a given threshold were selected for MS/MS experiments. In MS/MS mode, the mass range from m/z 40 to 1,400 was scanned in 1.4 s, and four scans were added up for each experiment. Micromass-formatted peak lists were generated from the raw data by using the Proteinlynx software module. A database search using a local installation of MASCOT 1.9 and a custom database containing the sequence of recombinant PIGF-2 was used for a fast identification of PIGF-2-derived peptides. No enzyme specificity was used for the database search. Because it was expected that the sequence stretch of interest contains a pair of oxidized cysteines, cysteine oxidation was allowed as an optional modification. The results reported by the search engine were verified by manual inspection of the deconvoluted spectra.
Surface Plasmon Resonance Spectroscopy
Binding experiments were performed by surface plasmon resonance measurements on a Biacore 3000 instrument (Biacore Inc.) at 25 °C, following a well established procedures (26). Briefly, to assess the binding capacity of the various PlGF isoforms to Nrp-1 in dependence to heparin, a CM5 sensor surface was activated by use of EDC/NHS and coupling buffer in a ratio of 1:1 as activating reagents. The degree of coupling was set to ∼1500 response units. Remaining reactive groups were inactivated with ethanolamine, and PlGF variants were coupled on chip surface at a flow rate of 5 μl/min. The following binding experiments with human Nrp-1 (extracellular domain, Phe-22 to Lys-644; BD Biosciences) as soluble analyte were performed with various concentrations (1–300 nm in HEPES running buffer). The analyte was passed over the sensor chip with a constant flow rate of 30 μl/min for 300 s, and dissociation was measured over 500 s. Between different experimental cycles, the bound proteins were washed from the sensor surface with 2 m NaCl in running buffer. In a second experimental setting, Nrp-1 was coupled to the sensor chip, and the degree of coupling was set to 1500 response units, whereas 1–300 nm of PlGF was used as soluble analyte. Fittings of the data, overlay plots, and calculation of KD values were done with BIAevaluation software 4.1 estimating a 1:1 model.
Binding of PlGF Forms to Heparin
The three proteins were dialyzed against the loading buffer (50 mm NaC1, 20 mm Tris-HCl, pH 7.4), and 1 mg each was applied onto a heparin-Sepharose CL-6B column (GE Healthcare). After five washes with loading buffer, the columns were eluted stepwise with increasing salt concentration (0.1, 0.2, 0.4, 0.5, 0.75, 1, and 2 m NaC1 in 20 mm Tris-HCl, pH 7.4), and the samples were analyzed by SDS-PAGE.
CD Spectroscopy
CD spectra (180–260 nm) were recorded at 100 nm/min using a Jasco-J715 spectropolarimeter and a thermostatted 0.1-cm-path length quartz cell (20 °C, five accumulations each). Data point resolution and bandwidth were set to 0.2 nm, sensitivity to 50 millidegrees. CD spectra of samples containing 5 μm PlGF (PlGF-1, PlGF-2, or PlGF118) in PBS were measured, background-corrected, and converted to mean molar residue ellipticity, and the data were evaluated using J-715 for Windows Standard Analysis software.
Chemotaxis Assay
Chemotaxis assays were performed using a Boyden chamber system with filter inserts of 8-μm pore size (Millipore). Filters were precoated with collagen (BD Biosciences). Medium supplemented with or without PlGF isoforms (2.56 nm) was added into the lower chamber, whereas 2.5 × 105 cells were seeded into the upper chamber. Medium supplemented with 10% FCS or VEGF-A165 (1.79 nm) (Reliatech) served as a positive control. After 4.5 h (37 °C), cells on the lower face were fixed and stained with a variant of Romanowski staining (Quickdiff, Behring) and analyzed by light microscopy; cell number was determined in 10 high power fields per filter (200-fold magnification).
Three-dimensional Spheroid Sprouting Assay
HUVECs (1000 cells/well) were seeded in 20% methylcellulose, 80% growth medium (100 μl) into a 96-roundbottom plate (Nunc) to induce spheroid formation. After 24 h of incubation (37 °C), spheroids were harvested, and 50 spheroids/gel were seeded into 500 μl of a nonpolymerized collagen gel (2 mg/ml) in 24-well plates. Following polymerization, PlGF variants (3.66 nm) or VEGF-A165 (1 nm) as positive control were added to the gel. After 24 h of incubation at 37 °C, 15 randomly chosen spheroids were analyzed for each protein at 100-fold magnification (Nikon bright field microscope). Cumulative sprouting length and number of sprouts/spheroid was measured using ImageJ software.
Animals and Wounding Experiments
C57BLKS/J-m+/+Leprdb/db mice (male, 10–12 weeks of age; Jackson Laboratory) were caged individually under standard pathogen-free conditions, and two independent wound healing experiments were performed as previously described (25). Briefly, the mice were anesthetized, and four full thickness punch biopsy wounds were created on the back of the mouse and treated locally for the following 6 days by either applying PlGF variants (2.5 μm in PBS) or vehicle control (PBS). Solution was allowed to adsorb for at least 1 h before the animal was placed back into its cage. For histological analysis, wound tissues were excised, and the wound area was bisected in caudocranial direction; the tissue was fixed (4% paraformaldehyde) or embedded in OCT compound (Tissue Tek), immediately frozen in liquid nitrogen, and stored at −80 °C. Histological analysis was performed on serial sections (6 μm).
Histology, Immunohistochemistry, and Morphometric Analysis
Cryosections were fixed in acetone, blocked (3% BSA), incubated with antisera against murine CD31 (BD Biosciences), desmin (DakoCytomation), and F4/80 (Dianova); bound primary antibodies were detected using an ALEXA Fluor 488-conjugated polyclonal goat anti-rat antibody and an ALEXA Fluor 594-conjugated polyclonal goat anti-mouse antibody (Invitrogen); nuclei were stained with DAPI. As a control for specificity primary antibodies were omitted and replaced by irrelevant isotype-matched antibodies. Immunofluorescence/immunohistochemical microscopy was conducted at indicated magnifications (Microscope Eclipse 800E; Nikon), and morphometric analysis was performed on digital images as recently described (25).
Human Wound Exudates
Wound exudate was obtained from patients presenting with nonhealing skin ulcers because of venous insufficiency (n = 7) or from patients with normally healing acute cutaneous wounds at indicated time points postinjury (day 2, n = 10; day 7, n = 5; day 14, n = 7; excisional wounds of the lower leg awaiting wound closure by secondary intention); in addition blood serum was obtained from 15 patients. Exudate was harvested and tested for plasmin activity as previously described (23, 26). PlGF content in serum samples or wound exudates was determined by ELISA (Quantikine human PlGF immunoassay; R & D Systems).
Statistical Analysis
The data were summarized as means ± S.E. Location differences were assessed by analysis of variance and Tukey post hoc tests for pairwise comparisons. Thus, the type I error is strongly controlled, at least pertaining to specific families of (pairwise) hypotheses. Moreover, in case the S.E. clearly increased with the mean (based on Bartlett's test for equal variances), a log transformation was applied to stabilize the variance. Statistical significance (adjusted for multiplicity) was graded according to <0.05 (*), <0.01 (**), or <0.001 (***). All statistical analyses were performed using GraphPad Prism 5 (GraphPad Software Inc.).
RESULTS
PlGF Is Cleaved by Plasmin
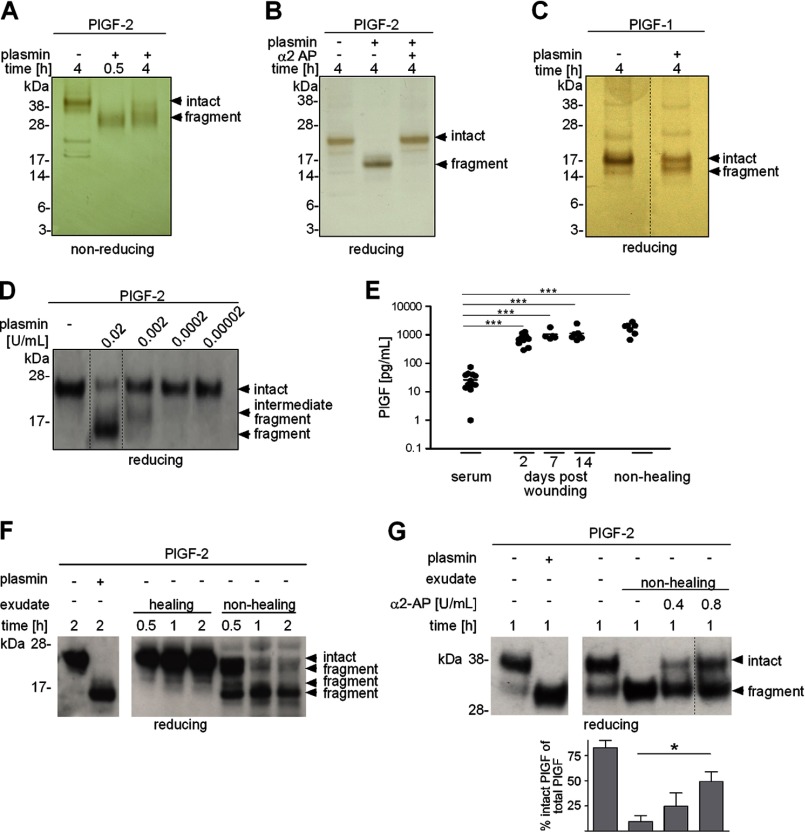
To investigate the hypothesis that PlGF might be a substrate for the serine protease plasmin, PlGF-1 and PlGF-2 were incubated with plasmin and samples were subjected to gel electrophoresis and analyzed by silver staining (Fig. 1, A–C) and Western blotting (Fig. 1D). In nonreducing conditions, dimeric PlGF-2 migrated with a molecular mass of ∼42 kDa (Fig. 1A). Extended incubation of PlGF-2 with plasmin for 4 h resulted in its fragmentation and formation of a protease-resistant cleavage product of ∼32 kDa. In reducing conditions, the PlGF-2 monomer exhibited an electrophoretic mobility shift from 23 to ∼16 kDa following plasmin-mediated digestion (Fig. 1B). Cleavage products migrating below 16 kDa could not be detected. Plasmin-mediated fragmentation of PlGF-2 was inhibited by the plasmin inhibitor α2-antiplasmin (Fig. 1B). In reducing conditions, monomeric PlGF-1 migrated with a molecular mass of 17 kDa, with a shift of ∼1–2 kDa following plasmin digestion (Fig. 1C). The generation of a major protease-resistant fragment with similar electrophoretic mobility for PlGF-1 and -2, as well as the detection of this protease-resistant 16 kDa fragment with an antibody raised against the amino-terminal domain of PlGF, suggests carboxyl-terminal cleavage in both proteins (Fig. 1D). A dose-response analysis indicated that plasmin-mediated cleavage of PlGF-2 occurred in multiple stages (Fig. 1D).
FIGURE 1.
PlGF is cleaved by plasmin. A–D, PlGF-1 and -2 were incubated with plasmin and α2-antiplasmin (α2 AP) for different time periods, subjected to nonreducing or reducing gel electrophoresis, and analyzed by silver staining (A–C) and Western blotting (D). E, quantification (ELISA) of endogenous PlGF protein in exudates obtained from healing or nonhealing human wounds and blood serum; each dot represents a sample of a different patient. F, representative Western blot analysis of PlGF-2 incubated without/with plasmin or spiked into wound exudates. G, shown is a representative Western blot analysis of PlGF-2 incubated without/with plasmin or spiked into wound exudates preincubated without/with α2-AP and the mean fraction of intact/fragmented PlGF-2 ± S.E. of multiple blots (n = 3). A–F, PlGF proteins were expressed in Sf9 cells; G, PlGF-2 protein was expressed in HEK293 cells.
To examine whether PlGF proteolysis might be a relevant biological event in the human system, we determined the concentration and integrity of PlGF in exudates obtained from normal healing or nonhealing chronic human wounds. Wound exudate is the interstitial fluid of injured tissue that reflects the metabolic condition of the wound microenvironment and has been proven to be useful for identification of factors involved in wound healing physiology and pathology (27, 28). As quantified by ELISA, in exudates obtained from both healing and nonhealing wounds, the mean concentration of PlGF was significantly increased (at least 25-fold) as compared with the mean concentration of plasma serum levels of 26 pg/ml, arguing for local PlGF expression at the wound site (Fig. 1E). PlGF concentrations were similar in healing and nonhealing wounds. To analyze the integrity of PlGF protein in the wound environment, recombinant PlGF-2 was spiked into wound exudates obtained from normal healing or nonhealing wounds, and samples were subjected to Western blot analysis in reducing conditions (Fig. 1F). Whereas in exudates obtained from healing wounds to large extent PlGF-2 maintained its stability, in exudates obtained from nonhealing wounds, PlGF-2 underwent a gradual degradation resulting in the formation of a protease-resistant fragment with an approximate molecular mass of 16 kDa after 2 h of incubation with exact the same electrophoretic mobility as plasmin-digested PlGF-2 (Fig. 1F). Fragmentation of PlGF-2 (produced in human embryonic kidney 293 EBNA cells) in exudates obtained from nonhealing wounds could be partially rescued by preincubation with α2-antiplasmin (Fig. 1G). As shown previously (27, 28), in this study plasmin activity was significantly increased in exudates obtained from chronic versus healing wounds (plasmin activity in chronic exudates (n = 3) mean 27 milliunits/ml ± S.D. 7.8, in healing exudates (n = 4) mean 17 units/ml ± S.D. 5.7; p = 0.05). Collectively, these findings suggest that proteolytic processing of the carboxyl terminus in PlGF occurs during tissue repair and is differentially regulated in healing and nonhealing conditions.
Processing of PlGF-2 by Plasmin Results in Cleavage of the HBD
To identify the plasmin cleavage sites in PlGF-2, we used LC-MS/MS analysis. Consistent with the Western blot analysis that suggested a multistage cleavage event, LC-MS/MS analysis revealed several fragments that indicate cleavage sites within the HBD at residues Arg-151, Lys-136, Arg-134, Lys-131, Lys-127, and Lys-118 (Fig. 2). Extended plasmin digestion therefore results in a complete degradation of the HBD, with Lys-118 representing position P1 of the cleavage site closest to the amino terminus, and the plasmin-resistant amino-terminal fragment PlGF1–118 representing the VEGFR-1-binding domain. Although we did not subject plasmin-digested PlGF-1 fragments to LC-MS/MS analysis, several arguments suggest that also in PlGF-1 the position Lys-118 is the plasmin cleavage site closest to the amino terminus: first, the cleavage site Lys-118 to Met-119 is localized within the domain encoded by exon 5, which is equivalent in PlGF-1 and -2; second, cleavage at position Lys-118 would be consistent with the electrophoretic mobility shift of 1–2 kDa under reducing conditions following plasmin processing of PlGF-1 (Fig. 1C). Notably, the plasmin cleavage site for VEGF-A165 has also been mapped within exon 5 (Fig. 2A) (22, 23).
FIGURE 2.
Identification of plasmin cleavage sites in PlGF-2 by LC-MS/MS analysis. PlGF-2 (expressed in HEK293 cells) was incubated with plasmin, and peptide fragments were analyzed by LC-MS/MS. A, schematic presentation of plasmin cleavage sites (arrowheads) in PlGF-2 based on identified fragments; the HBD in PlGF-2 and VEGF-A165 is illustrated in gray; alignment of plasmin cleavage sites closest to the amino termini of VEGFA165, PlGF-1, and PlGF-2. B, absolute and relative deviation between expected and found molecular mass of PlGF-2 peptide fragments identified by LC-MS/MS.
Plasmin Cleavage Attenuates PlGF-2-mediated VEGFR-1 Phosphorylation
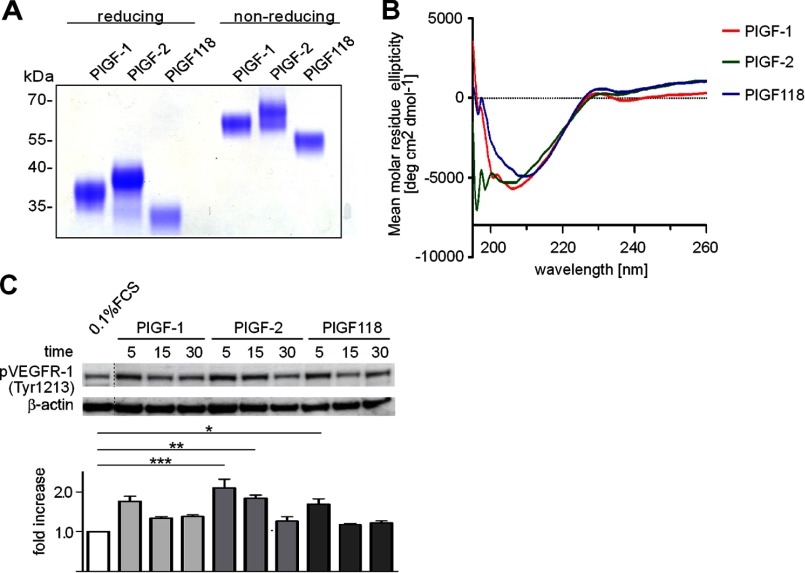
To evaluate the functional impact of plasmin-mediated carboxyl-terminal processing of PlGF, we generated constructs of PlGF-1, PlGF-2, and a truncated isoform that mimicked the plasmin-resistant PlGF fragment (PlGF118). These constructs were used to produce recombinant proteins in HEK293 EBNA cells. To assess proper folding of the PlGF variants, we analyzed dimerization and secondary structure properties by SDS-PAGE analysis and CD spectroscopy. In nonreducing conditions, PlGF-2, PlGF-1, and PlGF118 migrated with a molecular mass of ∼65–68, 58–60, and 54–55 kDa, respectively (Fig. 3A). In reducing conditions, monomers of PlGF-2, PlGF-1, and PlGF118 exhibited a molecular mass of ∼38–39, 36–37, and 32–33 kDa, respectively (Fig. 3A). Differences in electrophoretic mobility of recombinant proteins depicted in Figs. 1 and 3 are due to differences in posttranslational modifications (e.g., glycosylation) and histidine/double streptavidine tag expression. All isoforms showed a comparable degree in their ellipticity, indicating that loss of the carboxyl terminus does not affect the secondary structure of PlGF isoforms (Fig. 3B). As revealed by Western blot analyses, all isoforms resulted in Tyr-1213 phosphorylation of VEGFR-1 when compared with stimulation with 0.1% FCS, although weaker in PlGF-1 and PlGF118 when compared with PlGF-2 (Fig. 3C).
FIGURE 3.
PlGF isoforms expressed in HEK293 cells are biologically active. A and B, SDS-PAGE and Coomassie stain (A) and CD spectroscopy (B) of PlGF isoforms expressed in HEK293 cells. C, stimulation of HUVECs with PlGF isoforms (2.5 nm) and analysis of VEGFR-1 phosphorylation (Tyr-1213); the degree of phosphorylation was determined by densitometric analysis of phospho-signal intensity and normalized to the corresponding β-actin signal.
Plasmin Cleavage Attenuates PlGF-2-mediated Endothelial Cell Chemotaxis and Vascular Sprouting
To analyze the functional impact of plasmin processing for PlGF, the chemotactic activity of PlGF variants on endothelial cells was analyzed in a Boyden chamber assay. Previous studies reported on equal chemotactic potency of PlGF-1 and -2 (produced in Sf9 insect cells) in bovine aortic arch-derived endothelial cells (16). Here we used porcine aortic endothelial (PAE) cells stably transfected with the human Nrp-1 receptor (PAE/Nrp-1) or untransfected cells lacking Nrp-1 expression (Fig. 4A) to examine the effect of diverse PlGF variants. Nrp-1 transfected PAE cells treated with PlGF-2 (expressed in Sf9 insect cells) exhibited at least a 2-fold higher chemotactic response as compared with cells treated with PlGF-1 or control cells (Fig. 4B). Preincubation of PlGF-2 with plasmin resulted in a significant loss of its chemotactic activity that was partly rescued by α2-antiplasmin. Both PlGF-1 and PlGF-2 were unable to induce chemotaxis in untransfected cells lacking Nrp-1 expression (Fig. 4B). Repeating the experiment with PlGF isoforms produced in HEK293 cells and extending the analysis on HUVEC substantiated the critical role of the HBD for endothelial cell chemotaxis. In both PAE/Nrp-1 and HUVECs, chemotactic activity was significantly increased in response to PlGF-2 as compared with PlGF-1 and PlGF118 (Fig. 4, C and D).
FIGURE 4.
Plasmin cleavage attenuates PlGF-2-mediated endothelial cell chemotaxis and vascular sprouting. A, concanavalin A pulldown and Western blot analysis of Nrp-1 expression in untransfected or human Nrp-1-transfected PAE cells. B–D, analysis of chemotactic activity of PlGF isoforms in a Boyden chamber assay; PAE, PAE/Nrp-1, or HUVECs were stimulated with PlGF isoforms (produced in Sf9 insect cells) with or without plasmin and/or α2-antiplasmin (B) and PlGF isoforms (expressed in HEK293 cells) (C and D); samples were analyzed in triplicate in at least three independent experiments, and the results are shown ± S.E. (n ≥ 3); hpf, high power field. E, representative images of HUVEC spheroids embedded in collagen gels and stimulated with various PlGF isoforms (3.66 nm) (expressed in HEK293 cells), VEGF-A165 (1 nm) (positive control), or 0.1% FCS (negative control) for 24 h. F, quantitative analysis of cumulative sprout length/spheroid and number of sprouts/spheroid; 15 spheroids/condition were analyzed in three independent experiments; the results are shown ± S.E. Scale bars, 100 μm.
We next tested the role of plasmin processing of PlGF in endothelial sprout formation using a three-dimensional spheroid outgrowth assay in type collagen I gels. Both the cumulative sprouting length (reflecting sprout outgrowth) and the number of sprouts (reflecting sprout induction) per spheroid were significantly increased by PlGF-2-treated cultures as compared with controls (Fig. 4, E and F). Although PlGF-1 and PlGF118 also revealed stimulating activity on sprout formation when compared with controls, the overall response was weaker when compared with PlGF-2, and the response on sprout induction did not reach statistical significance when compared with controls. Of note, after 24 h of treatment, all PlGF-treated spheroids revealed a greater stability as compared with control cultures, suggesting that PlGF acts as a survival factor for endothelial cells, independently of the HBD and/or the carboxyl-terminal domain encoded by exon 7.
The HBD of PlGF-2 Is Critical for Its Repair-promoting Activities in Diabetic Mice
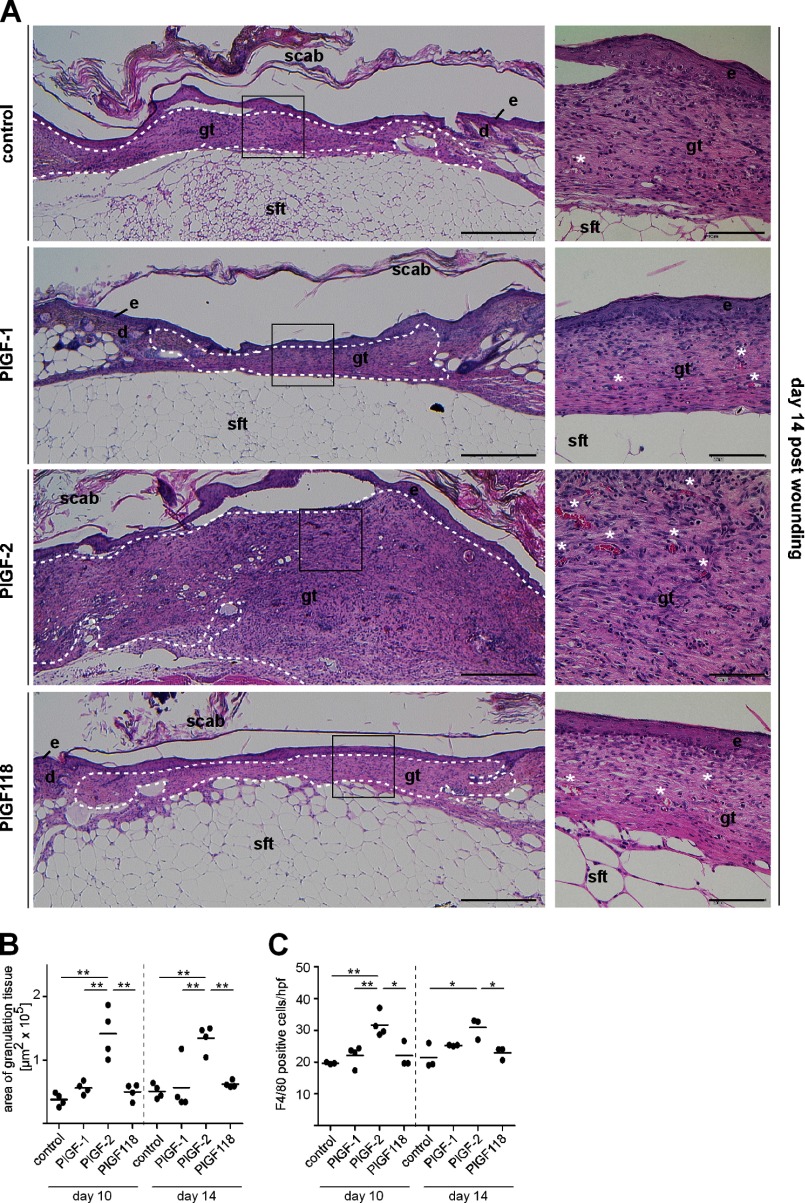
Further exploration of biological effects of plasmin processing of PlGF was performed in an in vivo wound healing assay. Vascular growth is profoundly involved in effective tissue repair and has been shown to be significantly attenuated in preclinical models of impaired healing such as diabetic mice and chronic skin ulcers in humans (23, 25). Full thickness excisional skin wounds were created on the back of diabetic mice (db/db mice), and the repair response was characterized following local treatment with repetitive applications of PlGF isoforms (2.5 μm) or vehicle control. Quantitative analysis of granulation tissue formation (hematoxylin- and eosin-stained wound sections), macrophages (F4/80 positive cells), vascular density, and perivascular cells (CD31/desmin double staining) present in granulation tissue showed that in PlGF-2-treated wounds, the amount (Fig. 5), cellularity, and vascularization of granulation tissue (Fig. 6) were significantly increased at days 10 and 14 postinjury, as compared with PlGF-1-, PlGF118-, or vehicle-treated wounds. Granulation tissue formation and vascularization were also increased in response to PlGF-1 and PlGF118 as compared with vehicle controls; however, these effects were minor and did not reach statistical significance. Furthermore, whereas in PlGF-1- and PlGF118-treated wounds, the vascular density decreased toward day 14 postinjury, the increased vascular density in PlGF-2-treated wounds sustained until day 14. The ratio of desmin/CD31-double-stained vascular structures within the granulation tissue was significantly increased in response to PlGF-2 as compared with PlGF-1-, PlGF118-, and vehicle-treated wounds at days 10 and 14. These findings indicate an enhanced coverage of vascular structures by perivascular cells in PlGF-2-treated wounds and suggest increased stability/maturation of PlGF-2-induced blood vessels at the wound site (Fig. 6B). Wound closure kinetics was not significantly altered in PlGF-treated wounds at days 10 and 14 postinjury (data not shown).
FIGURE 5.
The HBD promotes PlGF-2-induced granulation tissue formation in diabetic mice. A, representative hematoxylin and eosin staining of PlGF-treated wound tissues of diabetic mice day 14 postinjury. The right panels present the boxed areas in the left panels at higher magnification. The dashed line outlines area of granulation tissue. e, epidermis; d, dermis; gt, granulation tissue; sft, subcutaneous fat tissue. White asterisks indicate blood vessels. Scale bars, 500 μm (left panel) and 100 μm (right panel). B and C, morphometric analysis of area of granulation tissue (B) and quantification of F4/80+-stained cells (C) within a defined area within the granulation tissue of PlGF-treated wound tissues at days 10 and 14 postinjury. Each dot represents a different wound tissue. PlGF proteins were expressed in HEK293 cells. hpf, high power field.
FIGURE 6.
The HBD promotes PlGF-2-induced wound angiogenesis and recruitment of perivascular cells. A, representative immunohistochemical staining for endothelial cells (CD31) and perivascular cells (desmin) of PlGF-treated wound tissues in diabetic mice at day 14 postinjury. B, quantification of the area within the granulation tissue that stained positive for CD31 or desmin at days 10 and 14 postinjury. Coverage of vessels with pericytes was quantified by calculating the ratio of desmin/CD31 double stained area. The dotted line indicates epidermal-dermal junction. e, epidermis. Scale bars, 100 μm. PlGF proteins were expressed in HEK293 cells.
Plasmin Processing of PlGF-2 Abrogates Its Binding Capacity to Heparin and Neuropilin-1
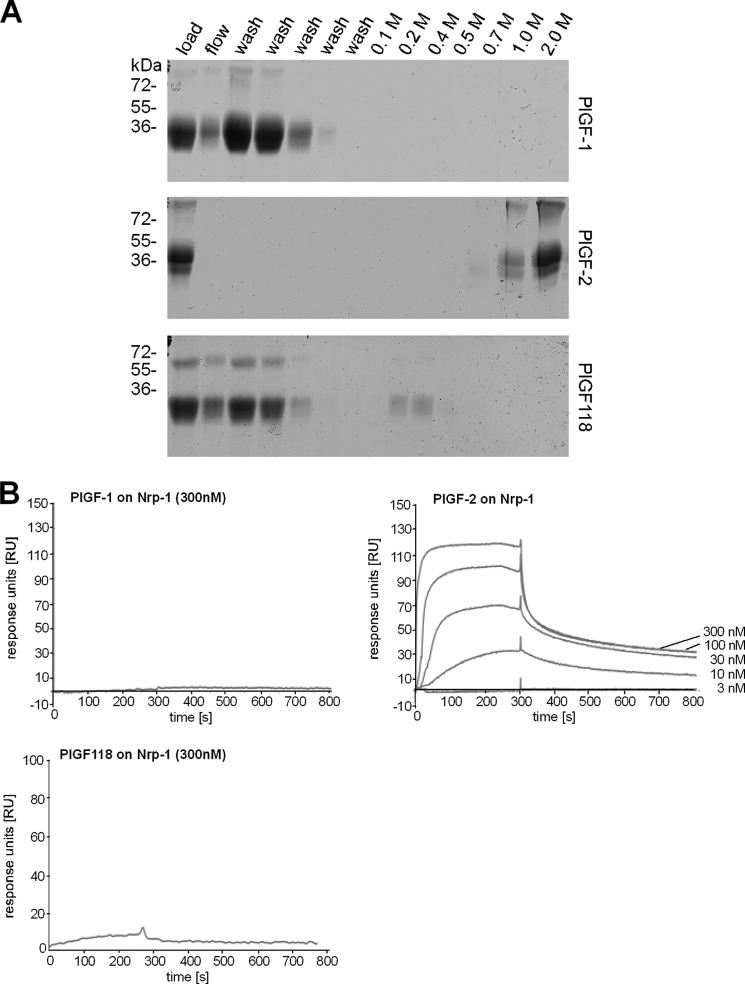
Next we evaluated the impact of plasmin processing upon the binding capacity of PlGF to heparin and to the co-receptor Nrp-1. As shown by affinity purification, PlGF-2 binds strongly to heparin (Fig. 7A), confirming earlier studies that reported binding of PlGF-2 to heparin-Sepharose (14). Importantly, PlGF-1 and PlGF118 do not interact with heparin (Fig. 7A). As revealed by plasmon resonance spectroscopy, binding of PlGF-2 to the immobilized Nrp-1 extracellular domain was clearly concentration-dependent, and the calculated KD was 0.1 μm (Fig. 7B). Plasmin digestion of PlGF-2 abrogated interaction with Nrp-1. PlGF-1 did not bind to Nrp-1. Binding characteristics for Nrp-1 were confirmed for all PlGF variants by inverted settings using Nrp-1 as soluble analyte (data not shown).
FIGURE 7.
Binding analysis of PlGF isoforms to heparin or Nrp-1. A, to test whether PlGF-1, PlGF-2, or PlGF118 binds to heparin, the proteins (expressed in HEK293 cells) were dialyzed against 50 mm NaCl and applied to heparin-Sepharose column. Only PlGF-2 bound to heparin and was eluted with 1–2 M NaCl. B, Nrp-1 was immobilized on sensor chips, and binding sensorgrams were recorded for PlGF variants as soluble analyte; different concentrations of PlGF proteins (3–300 nm) were monitored by measuring the variation in the plasmon resonance angle as function of time and described as response units (RU). The curves are shown in ascending order depending on the concentration of the analyte.
The HBD Differentially Regulates PlGF-2-induced Phosphorylation of the Focal Adhesion Kinase but Does Not Modulate Erk-1/-2 or Akt-mediated Signaling
We further explored downstream pathways that might mediate the differential effects in response to plasmin processed PlGF-2. We focused the analysis on signaling molecules previously shown to be critically involved in PlGF-mediated cell functions. Stimulation of HUVECs with either PlGF-2 or PlGF118 resulted in rapid phosphorylation of Erk-1/-2 and Akt, which was similar in signal intensity for both molecules (Fig. 8A). Also, overexpression of Nrp-1 in PAE cells did not alter the signal strength for Erk-1/-2 and Akt activation in response to PlGF-2 (Fig. 8B). Based on these findings, it seems unlikely that Erk/Akt activation is involved in differential endothelial cell activities in response to PlGF-2 and PlGF118 stimulation. We then investigated the involvement of FAK signaling in response to PlGF stimulation. Whereas stimulation with PlGF-2 provoked an increased and sustained phosphorylation signal of FAK, the activation signal was substantially weaker in response to PlGF118 (Fig. 8C). Furthermore, PlGF-2-mediated activation of FAK was pronounced in PAE-Nrp-1 transfected cells as compared with cells lacking Nrp-1. Together, our findings propose a critical role for PlGF-2/Nrp-1 interaction for FAK activation.
FIGURE 8.
The HBD promotes PlGF-2-induced phosphorylation of focal adhesion kinase but is dispensable for Erk-1/-2 or Akt-mediated signaling. A–C, Western blot analysis of signal transduction in HUVECs (A), PAE/Nrp-1 (B and C), or PAE cells stimulated with PlGF-2, PlGF118, or VEGF-A165 (C). The samples were subjected to SDS-PAGE, and phosphorylation was analyzed by antibodies directed against phospho-Erk-1/-2 (Thr-202/Tyr-204), phospho-Akt (Ser-473), or phospho-FAK (Tyr-576/Tyr-577). Antibodies directed against total proteins or β-actin served as loading control and were used to determine the increase over negative control. Shown are representative Western blots and the mean signal intensity ± S.E. of multiple blots (n ≥ 3); PlGF proteins were expressed in HEK293 cells. D, model for PlGF-mediated activities in relation to amino acid sequences encoded by exons 6 and 7. Left panel, PlGF-mediated phosphorylation of VEGFR-1 and downstream activation of Erk and Akt is independent of exon 6 and 7; middle panel, exon 6 in PlGF-2 is essential for binding to heparin/HS or Nrp-1; right panel, it is hypothesized that Nrp-1/heparin binding promotes the interaction/clustering of PlGF-2 with VEGFR-1, which then together with integrin activation leads to enhanced FAK activation and pro-angiogenic responses.
DISCUSSION
In this study, we demonstrated that PlGF-1 and -2 are substrates for plasmin and that plasmin-catalyzed cleavage results in loss of the carboxyl-terminal domain in both proteins. Plasmin-mediated processing of PlGF-2 significantly decreased its binding to heparin and Nrp-1 and substantially attenuated its pro-angiogenic activities. Our findings provide novel mechanistic insights into the regulation of PlGF-2/Nrp-1 interactions by proteolytic processing and important biological consequences of this event for tissue vascularization and growth.
We and other investigators previously demonstrated the critical role of plasmin processing of VEGF-A165 for its biological behavior (21, 22, 25). In addition, plasmin-mediated processing has been reported to be critical for VEGF-C and -D activation, which are primarily involved in lymphangiogenesis (29). Here we show that kinetics and structural consequences of plasmin-mediated processing of PlGF-2 are similar to plasmin-mediated cleavage of VEGF-A165. Both proteins reveal gradual degradation following exposure to plasmin, with the highest proteolytic sensitivity within their HBD localized at the carboxyl terminus. In both proteins, the cleavage site closest to the amino terminus is localized within the domain encoded by exon 5, in close proximity to the cysteine residues critical for the formation of intrachain disulfide bonds. Thus, for VEGF-A165 and PlGF-2, plasmin-mediated cleavage yields an amino-terminal homodimeric VEGF-A110 and PlGF118, respectively, comprising the VEGFR-binding domain and lacking the carboxyl-terminal domains. Plasmin processing apparently does not substantially affect folding and dimerization of either protease-resistant fragment, which are still capable of activating VEGFRs, although at different levels. Therefore, we propose to consider the carboxyl-terminal processing of these two VEGF family members by plasmin as a principal mechanism to regulate their biological activity.
Plasmin processing revealed different functional consequences for PlGF-1 and -2. Whereas plasmin digestion of PlGF-2 significantly attenuated endothelial cell chemotaxis, vascular sprouting, and granulation tissue formation during tissue repair, biological responses to PlGF-1 were similar to PlGF118- or vehicle control-stimulated cells/tissues. Therefore, the effects of plasmin digestion on the biological activities of PlGF-1 appear to be minimal. These findings strongly indicate that most of the PlGF-2-stimulated activities we observed in our studies are dependent upon the HBD encoded by exon 6.
To examine the molecular mechanisms underlying the loss of PlGF-2-mediated responses following plasmin digestion, we investigated several potential mechanisms. First, we assessed whether endothelial cell stimulation with different PlGF variants leads to differences in VEGFR-1 tyrosine phosphorylation and activation of known downstream mediators. PlGF is capable of signaling directly through VEGFR-1, and several VEGFR-1 tyrosine phosphorylation sites in endothelial cells have been reported to be activated upon stimulation with PlGF-2 (8, 30). However, we could not find a systematic analysis in the literature investigating the pattern and function of different tyrosine phosphorylation sites of VEGFR-1 in response to diverse PlGF isoforms. Here we show that following stimulation of HUVECs, all PlGF variants, although weaker in the absence of sequences encoded by exon 6 and/or 7, had the potency to phosphorylate Tyr-1213 and to equally activate downstream mediators including Erk and Akt. Therefore, this pathway is unlikely to explain the different biological activities that we found in response to various PlGF isoforms. However, these results on downstream signaling support the observation that all PlGF variants mediated stability of spheroid structures when compared with vehicle control and indicate that PlGF-mediated endothelial cell survival is independent of the HBD and/or the carboxyl-terminal domain encoded by exon 7 as well as binding to Nrp-1 and/or heparin. Furthermore, our findings suggest that PlGF-2-mediated activation of Erk-1/-2 is independent of Nrp-1 binding, which is paralleled by recent reports demonstrating that VEGF-A165-mediated activation of Erk-1/-2 is independent of Nrp-1 (31). At this stage we cannot exclude that other VEGFR-1 tyrosine residues than Tyr-1213 might be differentially activated by different PlGF variants, and this aspect requires more detailed examination in future studies.
Second, we investigated functional consequences of differential binding of PlGF variants to the co-receptor Nrp-1. The exact mechanism by which Nrp-1 acts on endothelial cell functions is still not resolved, and so far most of the functional studies were performed with VEGF-A165 (32, 33). Based on those studies, it was proposed that Nrp-1 acts as a co-receptor for VEGF-A165, enhancing VEGF binding to VEGFR-2 and thus increasing VEGFR signaling. More recent studies indicated that rather than affecting VEGFR-2 activity directly, Nrp-1 is likely to modulate signaling kinetics and the activation of specific signal output via VEGFR-2 (17). The short carboxyl-terminal sequence of VEGF-A encoded by exon 8a (KPRR) was identified as a critical sequence to direct binding and specificity toward Nrp-1 (17, 34–37). Interestingly, against common knowledge, recently it was shown that also VEGF-A121 binds directly to Nrp-1 (38) through exon 8a (39). In contrast to the situation with VEGF-A165, VEGF-A121 was not able to bridge VEGFR-2/Nrp-1 interaction and to induce the formation of a VEGFR-2·Nrp-1 complex (38). The understanding for PlGF/Nrp-1 interaction is so far based on earlier cross-linking experiments and competitive binding analysis on endothelial cells that identified the HBD of PlGF-2 as an epitope for the Nrp-1b1b2 domain, whereas PlGF-1 revealed no binding to Nrp-1 (15, 16). Interestingly, the short carboxyl-terminal sequence of PlGF-1 and -2 encoded by exon 7 has high sequence homology with exon 8a of VEGF-A. In light of the critical role of exon 8a of the vegf-a gene for Nrp-1 binding (17, 34–37), we hypothesized whether similarly to VEGF-A121, also PlGF-1 might be able to bind to Nrp-1. Using surface plasmon resonance spectroscopy, here we demonstrated that PlGF-2 but not PlGF-1 specifically and strongly binds to Nrp-1. Furthermore, we show that plasmin processing of PlGF-2 completely abrogates binding of PlGF-2 to Nrp-1. Thereby, our findings support earlier cross-linking experiments and corroborate the critical role of exon 6 and not exon 7 as a Nrp-1 binding sequence in PlGF-2. Given the sequence homology between the basic peptides encoded by exons 8a and 7 of vegf-a and plgf, respectively, we believe this is an interesting finding unraveling a potential difference in binding of PlGF and VEGF-A to Nrp-1 and merits further investigation in future studies.
Our findings indicate a critical role of PlGF-2/Nrp-1 binding for PlGF-2 function. FAK activation is closely linked to cell survival and migration in endothelial cells (40). Interestingly, a recent report demonstrated the critical role of VEGF-A165 binding to Nrp-1 for FAK activation, endothelial cell migration, and tube formation (31). We hypothesized that increased FAK activation in response to PlGF-2/Nrp-1 stimulation mediates the findings of superior PlGF-2 pro-angiogenic activities over those of PlGF-1 or PlGF118. Here we substantiated this hypothesis by showing that phosphorylation of FAK was increased and sustained by PlGF-2 when compared with PlGF118. Furthermore, this effect was enhanced in Nrp-1-transfected PAE cells when compared with cells lacking Nrp-1 expression. Thus, our findings indicate an important role for PlGF-2 binding to Nrp-1 in pathways controlling actin cytoskeletal rearrangements critical for cell migration and vessel growth.
Third, differential interaction of VEGF-A isoforms with diverse components of the extracellular matrix, in particular glycosaminoglycans, has been shown to be critical for VEGF-A-mediated biological activities (21). Here we provide evidence that PlGF-2, but neither PlGF-1 nor PlGF118, binds strongly to heparin and that this interaction is entirely abrogated by plasmin processing. These findings substantiate earlier studies demonstrating high affinity of PlGF-2 to heparin-Sepharose (14). At this stage we can only speculate on potential functions of PlGF-2/heparin binding for angiogenic processes. As reported for VEGF-A165, PlGF-2/heparin interactions might allow for the formation of growth factor gradients that are critical for vascular sprouting and/or modulate Nrp-1-mediated activities (40, 41).
Finally, our findings raise the general question on the biological role of PlGF-1 per se and on the dual generation of a short PlGF isoform lacking the HBD by either transcriptional control and/or proteolysis. Contrary to the generation of diverse PlGF isoforms by differential mRNA splicing in humans, in mice PlGF-2 is the only isoform identified so far (13). Hence, our findings suggest that in mice, modification of PlGF-2 activity by proteolytic processing provides a unique mechanism to control activities mediated by the carboxyl-terminal domain. In humans, next to mRNA splicing of the plgf gene, proteolytic processing of PlGF isoforms provides an additional mechanism to control processes mediated by the carboxyl-terminal domain. Our findings indicate that the impact of proteolytic processing, specifically on PlGF-2, becomes particularly critical in conditions of high/uncontrolled proteolytic activity, e.g., in degenerative conditions such as chronic wounds associated with diabetes mellitus, age, and vascular disease. In fact, here we showed that proteolytic processing of PlGF is increased in human chronic venous ulcers when compared with healing wounds. As previously shown for VEGF-A165 (25), this process might contribute to attenuated angiogenesis and tissue growth, which is a hallmark of chronic skin ulcers, the most common cause of impaired healing conditions in humans.
The findings reported here might be of relevance for the development of PlGF-based concepts of therapeutic vascular growth in the field of regenerative medicine. Current molecular/cellular based therapeutic angiogenesis trials showed only limited beneficial effects for some patients with ischemic diseases, and there is a need for the development of alternative treatment strategies (42). In preclinical studies, PlGF has been shown to be a potent inducer of tissue vascularization (1, 3, 9, 43), often without the side effects reported for VEGF-A therapy. Therefore, PlGF alone or in combination with other pro-angiogenic factors has been proposed for angiogenic therapies. Based on our findings, we propose the use of the PlGF-2 isoform because of its superior activity, when compared with PlGF-1. Moreover, in conditions of unrestrained inflammation with unbalanced proteases, we suggest protecting PlGF-2 from proteolytic processing to maintain its integrity and biological activity.
Acknowledgments
We thank M. Klagsbrun for the Nrp-1-transfected PAE cells. We are grateful to Mats Paulsson, Raimund Wagener, Herbert Weich, Stephanie Traub, and Thomas Krieg for helpful discussions. We thank Raphael Reuten for help with the synthesis of recombinant proteins. Expert technical assistance by Michael Piekarek is gratefully acknowledged.
This work was supported by Grant Angioscaff NMP-LA-2008-214402 (to S. A. E., M. E., and J. A. H.) from the European Union and Deutsche Forschungsgemeinschaft Grants SFB 829 (to S. A. E., P. K., and M. K.) and SFB 832 (to M. K.).
- PlGF
- placental growth factor
- FAK
- focal adhesion kinase
- HBD
- heparin-binding domain
- HEK cells
- human embryonic kidney cells
- HUVEC
- human umbilical vein endothelial cell
- LC-MS/MS
- liquid chromatography tandem mass spectrometry
- Nrp-1
- neuropilin-1
- PAE cells
- porcine aortic endothelial cells
- VEGFR
- vascular endothelial growth factor receptor.
REFERENCES
- 1. Cianfarani F., Zambruno G., Brogelli L., Sera F., Lacal P. M., Pesce M., Capogrossi M. C., Failla C. M., Napolitano M., Odorisio T. (2006) Placenta growth factor in diabetic wound healing. Altered expression and therapeutic potential. Am. J. Pathol. 169, 1167–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gargioli C., Coletta M., De Grandis F., Cannata S. M., Cossu G. (2008) PlGF-MMP-9-expressing cells restore microcirculation and efficacy of cell therapy in aged dystrophic muscle. Nat. Med. 14, 973–978 [DOI] [PubMed] [Google Scholar]
- 3. Iwasaki H., Kawamoto A., Tjwa M., Horii M., Hayashi S., Oyamada A., Matsumoto T., Suehiro S., Carmeliet P., Asahara T. (2011) PlGF repairs myocardial ischemia through mechanisms of angiogenesis, cardioprotection and recruitment of myo-angiogenic competent marrow progenitors. PLoS One 6, e24872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maes C., Coenegrachts L., Stockmans I., Daci E., Luttun A., Petryk A., Gopalakrishnan R., Moermans K., Smets N., Verfaillie C. M., Carmeliet P., Bouillon R., Carmeliet G. (2006) Placental growth factor mediates mesenchymal cell development, cartilage turnover, and bone remodeling during fracture repair. J. Clin. Invest. 116, 1230–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oura H., Bertoncini J., Velasco P., Brown L. F., Carmeliet P., Detmar M. (2003) A critical role of placental growth factor in the induction of inflammation and edema formation. Blood 101, 560–567 [DOI] [PubMed] [Google Scholar]
- 6. Van de Veire S., Stalmans I., Heindryckx F., Oura H., Tijeras-Raballand A., Schmidt T., Loges S., Albrecht I., Jonckx B., Vinckier S., Van Steenkiste C., Tugues S., Rolny C., De Mol M., Dettori D., Hainaud P., Coenegrachts L., Contreres J. O., Van Bergen T., Cuervo H., Xiao W. H., Le Henaff C., Buysschaert I., Kharabi Masouleh B., Geerts A., Schomber T., Bonnin P., Lambert V., Haustraete J., Zacchigna S., Rakic J. M., Jiménez W., Noël A., Giacca M., Colle I., Foidart J. M., Tobelem G., Morales-Ruiz M., Vilar J., Maxwell P., Vinores S. A., Carmeliet G., Dewerchin M., Claesson-Welsh L., Dupuy E., Van Vlierberghe H., Christofori G., Mazzone M., Detmar M., Collen D., Carmeliet P. (2010) Further pharmacological and genetic evidence for the efficacy of PlGF inhibition in cancer and eye disease. Cell 141, 178–190 [DOI] [PubMed] [Google Scholar]
- 7. Fischer C., Mazzone M., Jonckx B., Carmeliet P. (2008) FLT1 and its ligands VEGFB and PlGF. Drug targets for anti-angiogenic therapy? Nat. Rev. Cancer 8, 942–956 [DOI] [PubMed] [Google Scholar]
- 8. Autiero M., Waltenberger J., Communi D., Kranz A., Moons L., Lambrechts D., Kroll J., Plaisance S., De Mol M., Bono F., Kliche S., Fellbrich G., Ballmer-Hofer K., Maglione D., Mayr-Beyrle U., Dewerchin M., Dombrowski S., Stanimirovic D., Van Hummelen P., Dehio C., Hicklin D. J., Persico G., Herbert J. M., Communi D., Shibuya M., Collen D., Conway E. M., Carmeliet P. (2003) Role of PlGF in the intra- and intermolecular cross talk between the VEGF receptors Flt1 and Flk1. Nat. Med. 9, 936–943 [DOI] [PubMed] [Google Scholar]
- 9. Carmeliet P., Moons L., Luttun A., Vincenti V., Compernolle V., De Mol M., Wu Y., Bono F., Devy L., Beck H., Scholz D., Acker T., DiPalma T., Dewerchin M., Noel A., Stalmans I., Barra A., Blacher S., VandenDriessche T., Ponten A., Eriksson U., Plate K. H., Foidart J. M., Schaper W., Charnock-Jones D. S., Hicklin D. J., Herbert J. M., Collen D., Persico M. G. (2001) Synergism between vascular endothelial growth factor and placental growth factor contributes to angiogenesis and plasma extravasation in pathological conditions. Nat. Med. 7, 575–583 [DOI] [PubMed] [Google Scholar]
- 10. Hattori K., Heissig B., Wu Y., Dias S., Tejada R., Ferris B., Hicklin D. J., Zhu Z., Bohlen P., Witte L., Hendrikx J., Hackett N. R., Crystal R. G., Moore M. A., Werb Z., Lyden D., Rafii S. (2002) Placental growth factor reconstitutes hematopoiesis by recruiting VEGFR1 + stem cells from bone-marrow microenvironment. Nat. Med. 8, 841–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ferrara N. (2004) Vascular endothelial growth factor. Basic science and clinical progress. Endocr. Rev. 25, 581–611 [DOI] [PubMed] [Google Scholar]
- 12. Maglione D., Guerriero V., Viglietto G., Ferraro M. G., Aprelikova O., Alitalo K., Del Vecchio S., Lei K. J., Chou J. Y., Persico M. G. (1993) Two alternative mRNAs coding for the angiogenic factor, placenta growth factor (PlGF), are transcribed from a single gene of chromosome 14. Oncogene 8, 925–931 [PubMed] [Google Scholar]
- 13. DiPalma T., Tucci M., Russo G., Maglione D., Lago C. T., Romano A., Saccone S., Della Valle G., De Gregorio L., Dragani T. A., Viglietto G., Persico M. G. (1996) The placenta growth factor gene of the mouse. Mamm. Genome 7, 6–12 [DOI] [PubMed] [Google Scholar]
- 14. Hauser S., Weich H. A. (1993) A heparin-binding form of placenta growth factor (PlGF-2) is expressed in human umbilical vein endothelial cells and in placenta. Growth Factors 9, 259–268 [DOI] [PubMed] [Google Scholar]
- 15. Mamluk R., Gechtman Z., Kutcher M. E., Gasiunas N., Gallagher J., Klagsbrun M. (2002) Neuropilin-1 binds vascular endothelial growth factor 165, placenta growth factor-2, and heparin via its b1b2 domain. J. Biol. Chem. 277, 24818–34825 [DOI] [PubMed] [Google Scholar]
- 16. Migdal M., Huppertz B., Tessler S., Comforti A., Shibuya M., Reich R., Baumann H., Neufeld G. (1998) Neuropilin-1 is a placenta growth factor-2 receptor. J. Biol. Chem. 273, 22272–22278 [DOI] [PubMed] [Google Scholar]
- 17. Kawamura H., Li X., Goishi K., van Meeteren L. A., Jakobsson L., Cébe-Suarez S., Shimizu A., Edholm D., Ballmer-Hofer K., Kjellén L., Klagsbrun M., Claesson-Welsh L. (2008) Neuropilin-1 in regulation of VEGF-induced activation of p38MAPK and endothelial cell organization. Blood 112, 3638–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Soker S., Fidder H., Neufeld G., Klagsbrun M. (1996) Characterization of novel vascular endothelial growth factor (VEGF) receptors on tumor cells that bind VEGF165 via its exon 7-encoded domain. J. Biol. Chem. 271, 5761–5767 [DOI] [PubMed] [Google Scholar]
- 19. Gerhardt H., Golding M., Fruttiger M., Ruhrberg C., Lundkvist A., Abramsson A., Jeltsch M., Mitchell C., Alitalo K., Shima D., Betsholtz C. (2003) VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J. Cell Biol. 161, 1163–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ruhrberg C., Gerhardt H., Golding M., Watson R., Ioannidou S., Fujisawa H., Betsholtz C., Shima D. T. (2002) Spatially restricted patterning cues provided by heparin-binding VEGF-A control blood vessel branching morphogenesis. Genes Dev. 16, 2684–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Houck K. A., Leung D. W., Rowland A. M., Winer J., Ferrara N. (1992) Dual regulation of vascular endothelial growth factor bioavailability by genetic and proteolytic mechanisms. J. Biol. Chem. 267, 26031–26037 [PubMed] [Google Scholar]
- 22. Keyt B. A., Berleau L. T., Nguyen H. V., Chen H., Heinsohn H., Vandlen R., Ferrara N. (1996) The carboxyl-terminal domain (111–165) of vascular endothelial growth factor is critical for its mitogenic potency. J. Biol. Chem. 271, 7788–7795 [DOI] [PubMed] [Google Scholar]
- 23. Lauer G., Sollberg S., Cole M., Flamme I., Stürzebecher J., Mann K., Krieg T., Eming S. A. (2000) Expression and proteolysis of vascular endothelial growth factor is increased in chronic wounds. J. Invest. Dermatol. 115, 12–18 [DOI] [PubMed] [Google Scholar]
- 24. Lee S., Jilani S. M., Nikolova G. V., Carpizo D., Iruela-Arispe M. L. (2005) Processing of VEGF-A by matrix metalloproteinases regulates bioavailability and vascular patterning in tumors. J. Cell Biol. 169, 681–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roth D., Piekarek M., Paulsson M., Christ H., Bloch W., Krieg T., Davidson J. M., Eming S. A. (2006) Plasmin modulates vascular endothelial growth factor-A-mediated angiogenesis during wound repair. Am. J. Pathol. 168, 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ricard-Blum S., Féraud O., Lortat-Jacob H., Rencurosi A., Fukai N., Dkhissi F., Vittet D., Imberty A., Olsen B. R., van der Rest M. (2004) Characterization of endostatin binding to heparin and heparan sulfate by surface plasmon resonance and molecular modeling. J. Biol. Chem. 279, 2927–2936 [DOI] [PubMed] [Google Scholar]
- 27. Eming S. A., Smola-Hess S., Kurschat P., Hirche D., Krieg T., Smola H. (2006) A novel property of povidon-iodine. Inhibition of excessive protease levels in chronic non-healing wounds. J. Invest. Dermatol. 126, 2731–2733 [DOI] [PubMed] [Google Scholar]
- 28. Palolahti M., Lauharanta J., Stephens R. W., Kuusela P., Vaheri A. (1993) Proteolytic activity in leg ulcer exudate. Exp. Dermatol. 2, 29–37 [DOI] [PubMed] [Google Scholar]
- 29. Joukov V., Sorsa T., Kumar V., Jeltsch M., Claesson-Welsh L., Cao Y., Saksela O., Kalkkinen N., Alitalo K. (1997) Proteolytic processing regulates receptor specificity and activity of VEGF-C. EMBO J. 16, 3898–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sawano A., Takahashi T., Yamaguchi S., Aonuma M., Shibuya M. (1996) Flt-1 but not KDR/Flk-1 tyrosine kinase is a receptor for placenta growth factor, which is related to vascular endothelial growth factor. Cell Growth Differ. 7, 213–221 [PubMed] [Google Scholar]
- 31. Herzog B., Pellet-Many C., Britton G., Hartzoulakis B., Zachary I. C. (2011) VEGF binding to NRP1 is essential for VEGF stimulation of endothelial cell migration, complex formation between NRP1 and VEGFR2, and signaling via FAK Tyr407 phosphorylation. Mol. Biol. Cell 22, 2766–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zachary I. C. (2011) How neuropilin-1 regulates receptor tyrosine kinase signalling. The knowns and known unknowns. Biochem. Soc. Trans. 39, 1583–1591 [DOI] [PubMed] [Google Scholar]
- 33. Grünewald F. S., Prota A. E., Giese A., Ballmer-Hofer K. (2010) Structure-function analysis of VEGF receptor activation and the role of coreceptors in angiogenic signaling. Biochim. Biophys. Acta. 1804, 567–580 [DOI] [PubMed] [Google Scholar]
- 34. Cébe-Suarez S., Grünewald F. S., Jaussi R., Li X., Claesson-Welsh L., Spillmann D., Mercer A. A., Prota A. E., Ballmer-Hofer K. (2008) Orf virus VEGF-E NZ2 promotes paracellular NRP-1/VEGFR-2 coreceptor assembly via the peptide RPPR. FASEB J. 22, 3078–3086 [DOI] [PubMed] [Google Scholar]
- 35. Vander Kooi C. W., Jusino M. A., Perman B., Neau D. B., Bellamy H. D., Leahy D. J. (2007) Structural basis for ligand and heparin binding to neuropilin B domains. Proc. Natl. Acad. Sci. U.S.A. 104, 6152–6157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Parker M. W., Xu P., Li X., Vander Kooi C. W. (2012) Structural basis for selective vascular endothelial growth factor-A (VEGF-A) binding to neuropilin-1. J. Biol. Chem. 287, 11082–11089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ballmer-Hofer K., Andersson A. E., Ratcliffe L. E., Berger P. (2011) Neuropilin-1 promotes VEGFR-2 trafficking through Rab11 vesicles thereby specifying signal output. Blood 118, 816–826 [DOI] [PubMed] [Google Scholar]
- 38. Pan Q., Chathery Y., Wu Y., Rathore N., Tong R. K., Peale F., Bagri A., Tessier-Lavigne M., Koch A. W., Watts R. J. (2007) Neuropilin-1 binds to VEGF121 and regulates endothelial cell migration and sprouting. J. Biol. Chem. 282, 24049–24056 [DOI] [PubMed] [Google Scholar]
- 39. von Wronski M. A., Raju N., Pillai R., Bogdan N. J., Marinelli E. R., Nanjappan P., Ramalingam K., Arunachalam T., Eaton S., Linder K. E., Yan F., Pochon S., Tweedle M. F., Nunn A. D. (2006) Tuftsin binds neuropilin-1 through a sequence similar to that encoded by exon 8 of vascular endothelial growth factor. J. Biol. Chem. 281, 5702–5710 [DOI] [PubMed] [Google Scholar]
- 40. Abu-Ghazaleh R., Kabir J., Jia H., Lobo M., Zachary I. (2001) Src mediates stimulation by vascular endothelial growth factor of the phosphorylation of focal adhesion kinase at tyrosine 861, and migration and anti-apoptosis in endothelial cells. Biochem. J. 360, 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shintani Y., Takashima S., Asano Y., Kato H., Liao Y., Yamazaki S., Tsukamoto O., Seguchi O., Yamamoto H., Fukushima T., Sugahara K., Kitakaze M., Hori M. (2006) Glycosaminoglycan modification of neuropilin-1 modulates VEGFR2 signaling. EMBO J. 25, 3045–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ylä-Herttuala S., Alitalo K. (2003) Gene transfer as a tool to induce therapeutic vascular growth. Nat. Med. 9, 694–701 [DOI] [PubMed] [Google Scholar]
- 43. Luttun A., Tjwa M., Moons L., Wu Y., Angelillo-Scherrer A., Liao F., Nagy J. A., Hooper A., Priller J., De Klerck B., Compernolle V., Daci E., Bohlen P., Dewerchin M., Herbert J. M., Fava R., Matthys P., Carmeliet G., Collen D., Dvorak H. F., Hicklin D. J., Carmeliet P. (2002) Revascularization of ischemic tissues by PlGF treatment, and inhibition of tumor angiogenesis, arthritis and atherosclerosis by anti-Flt1. Nat. Med. 8, 831–840 [DOI] [PubMed] [Google Scholar]