Background: Interaction with insect midgut receptors is required for Bacillus thuringienesis (Bt) toxicity.
Results: RNAi knockdown of E-cadherin and sodium solute symporter (SSS) genes dramatically decreases Tribolium castaneum (Tc) larval susceptibility to Cry3Ba. A SSS fragment enhances Cry3Ba toxicity.
Conclusion: E-cadherin and SSS but not aminopeptidase N are Cry3Ba receptors in Tc.
Significance: For the first time, SSS was demonstrated as a Bt functional receptor.
Keywords: Cadherins, Host-Pathogen Interactions, Receptors, RNA Interference (RNAi), Transporters
Abstract
Understanding how Bacillus thuringiensis (Bt) toxins interact with proteins in the midgut of susceptible coleopteran insects is crucial to fully explain the molecular bases of Bt specificity and insecticidal action. In this work, aminopeptidase N (TcAPN-I), E-cadherin (TcCad1), and sodium solute symporter (TcSSS) have been identified by ligand blot as putative Cry3Ba toxin-binding proteins in Tribolium castaneum (Tc) larvae. RNA interference knockdown of TcCad1 or TcSSS proteins resulted in decreased susceptibility to Cry3Ba toxin, demonstrating the Cry toxin receptor functionality for these proteins. In contrast, TcAPN-I silencing had no effect on Cry3Ba larval toxicity, suggesting that this protein is not relevant in the Cry3Ba toxin mode of action in Tc. Remarkable features of TcSSS protein were the presence of cadherin repeats in its amino acid sequence and that a TcSSS peptide fragment containing a sequence homologous to a binding epitope found in Manduca sexta and Tenebrio molitor Bt cadherin functional receptors enhanced Cry3Ba toxicity. This is the first time that the involvement of a sodium solute symporter protein as a Bt functional receptor has been demonstrated. The role of this novel receptor in Bt toxicity against coleopteran insects together with the lack of receptor functionality of aminopeptidase N proteins might account for some of the differences in toxin specificity between Lepidoptera and Coleoptera insect orders.
Introduction
The development of improved bioinsecticides based on toxin-producing Bacillus thuringiensis (Bt)3 bacteria requires new toxins with increased activity and wider insecticidal spectrum, as well as more effective toxin delivery methods. Identifying molecules that confer toxin susceptibility in target insects is essential to understand how Bt toxins interact with their hosts, facilitating a more rational design of Bt products. Several membrane components in the insect midgut epithelium capable of binding Bt toxins have been identified; however, not all appear to be functionally relevant (1).
In Lepidoptera, it has been demonstrated that interaction of Cry1 toxins with aminopeptidase N (APN) and cadherin (CAD)-like) midgut proteins (2, 3) is required for toxic action, and alkaline phosphatase and ABC transporter have also been proposed as Cry1 receptors (4, 5). In mosquito larvae, homologous APN, CAD-like, and alkaline phosphatase proteins have been described as Cry11 and Cry4 receptor proteins (6), and in Coleoptera a cadherin-like protein has been demonstrated to act as a Cry3Aa receptor (7). In coleopteran insects, other molecules, such as an ADAM-like metalloprotease (8) and alkaline phosphatase (9), have been proposed as putative Cry receptors. The best characterized Cry receptors, APN and CAD-like proteins, have been unequivocally involved in a Bt mode of action by gene silencing, resulting in a reduced sensitivity to toxin (10, 11).
The APN proteins belong to a family of zinc-binding metalloprotease/peptidase enzymes inserted into the midgut microvillar membrane via a C-terminal glycosylphosphatidylinositol anchor that play an essential role in insect digestion (12). APNs are highly variable in sequence and have been grouped into five phylogenetic classes based on their amino acid sequences (13). They have distinct N- and O-glycosylation patterns, which seem to be determinant for Cry toxin binding (12).
The CAD-like proteins that function as Cry receptors contain several cadherin repeats but, unlike classical cadherins, are not primarily located within adherens junctions involved in cell-cell adhesion but on the apical membrane of midgut columnar epithelial cells (12). In this type of Cry receptors, the membrane-proximal cadherin repeats have been reported to be key interaction sites that mediate toxin susceptibility (14–16).
In Lepidoptera, a sequential toxin interaction with CAD-like and APN proteins in the midgut membrane has been proposed, resulting in toxin recruitment on the membrane surface and conformational changes that promote toxin insertion into the membrane, leading to cell osmotic disequilibrium and eventually to cell death (17).
Through proteomic approaches, other proteins have been identified as Cry binding proteins, although their role as Bt toxin receptors has not been demonstrated yet. Among them are actin and V-ATP-synthase in Lepidoptera (18–20) and Diptera (21), heat shock cognate protein in Lepidoptera (20), and flotillin and prohibitin in Diptera (21). Actin, V-ATP-synthase, and prohibitin have been also identified as Cry binding partners in Coleoptera through targeted mass spectrometry protein analysis.4 Whether these molecules act facilitating the toxic process or contributing to the insect response against Cry toxins remains to be investigated and constitutes an important issue to reveal the entire picture of the Bt mode of action.
The coleopteran model insect Tribolium castaneum (Tc) is a major global pest of stored products for human consumption for which many genetic and genomics tools have been developed (22), so it constitutes an ideal subject for the identification of new biopesticide targets based in Bt. In this work, we carried out receptor binding and ligand blot experiments in Tc larvae with the coleopteran-specific toxin Cry3Ba, previously shown to be active against this insect pest (23). Using LC-MS/MS spectrometry, among other proteins, we have identified as putative Cry3Ba toxin receptors an APN (NP_001164285, in this work denoted as TcAPN-I), an E-cadherin protein (XP_971388, denoted as TcCad1), and a sodium solute symporter protein (EFA03129, denoted as TcSSS) containing cadherin repeats. RNA interference experiments with dsRNA of the corresponding genes demonstrated that TcCad1 and TcSSS proteins were Cry3Ba toxin functional receptors in Tc, whereas TcAPN-I was not related to Cry3Ba susceptibility.
EXPERIMENTAL PROCEDURES
Cry3 Toxin Production
BTS1 and BTS00125L Bt Cry3Aa and Cry3Ba-producing strains were grown in solid sporulation medium (24) at 30 ± 1 °C until complete autolysis. Lysed bacteria were resuspended in 2× PBS (8 mm Na2HPO4, 2 mm KH2PO4, 150 mm NaCl), pH 7.4, and washed twice with 0.02% Triton X-100 in 2× PBS, pH 7.4, and twice with water. Following centrifugation at 6000 × g for 10 min at 4 °C, spore-crystal mixtures were resuspended in water and stored at −20 °C until use. Crystal inclusions were purified from spores by centrifugation in discontinuous sucrose gradients as described in Rausell et al. (25).
Insects
A laboratory colony of Tc founded from Ga-2 strain adults kindly provided by Dr. Beeman (U.S. Department of Agriculture) was used. Insects were reared on whole grain flour with 5% brewer yeast powder at 30 ± 1 °C in darkness.
Tc Brush Border Membrane Vesicle (BBMV) Preparation
Tc BBMV were prepared from 10–14-day-old larvae (after egg laying) according to the method of Wolfesberger et al. (26), as modified by Reuveni and Dunn (27). APN enzyme activity was monitored as described by Hafkenscheid (28) to assess BBMV preparation quality.
Binding Assays on Tc BBMV
Purified Cry3Ba toxin was biotinylated using biotinyl-N-hydroxysuccinimide ester (Amersham Biosciences; protein biotinylation module, GE Healthcare) according to the manufacturers' indications. Cry3Ba biotinylated toxin (1.4 nm) was incubated with 10 μg of Tc BBMV in PBS buffer, pH 7.4, 0.1% BSA, for 1 h in the presence or absence of 1000-fold excess unlabeled Cry3Ba or Cry3Aa toxins. Subsequently, unbound toxin was removed by centrifugation (10 min at 14,000 × g), and BBMV with bound toxin were washed twice with the same buffer (100 μl). BBMV were suspended in 15 μl of PBS and the corresponding volume of 8× Laemmli sample loading buffer. Samples were 10% SDS-PAGE electrophoresed and electrotransferred to a nitrocellulose membrane (Hybond ECL; Amersham Biosciences). The biotinylated toxins that were bound to the protein vesicles were visualized by incubating with streptavidin-peroxidase conjugate (Amersham Biosciences; 1:4000 dilution) for 1 h, followed by the addition of Amersham Biosciences ECL Prime Western blotting reagents (GE Healthcare), as recommended by the manufacturers.
Ligand Blot Analysis on Tc BBMV
Tc BBMV proteins (15 μg) were separated in 10% SDS-PAGE and transferred onto a nitrocellulose membrane (Hybond ECL; Amersham Biosciences) that was incubated overnight with PBS buffer, pH 7.4, 3% BSA, 0.1% Tween 20. After washing with PBS buffer, pH 7.4, 2% Tween 20, the membrane was incubated with 10 μg/ml Cry3Ba toxin in PBS buffer, pH 7.4. The blot was then washed twice with PBS buffer, pH 7.4, 2% Tween 20 and incubated with Cry3 rabbit polyclonal antibody (1:10,000) for 1 h. Following two washes with PBS buffer, pH 7.4, 2% Tween 20, the membrane was incubated with alkaline-phosphatase conjugated anti-rabbit secondary antibody (Sigma, 1:30,000), and the immunoreactive proteins were visualized using the ECL detection system Immobilon Western (Millipore). The corresponding bands in a Coomassie-stained gel of Tc BBMV (10% SDS-PAGE) were excised, trypsinized, and analyzed by LC-MS/MS.
Toxicity Assays
Toxicity assays on Tc larvae were performed using preweighed 10 to 14-day-old larvae (after egg laying), fed for 7 days on 20-μl flour discs (20% flour, w/v), prepared as described by Xie et al. (29), containing 12.5 μg/μl Cry3Ba spore-crystal mixture for treatments or water in control assays. The assays were performed in 96-well polystyrene plates (Sterilin, Thermofisher) with one flour disc and one larva per well. Thirty larvae were used in each assay, and at least two replicates were carried out. Mortality was recorded after 7 days under laboratory rearing conditions.
For toxicity assays in gene silencing experiments, 4 days after dsRNA or control buffer injection, larvae were weighed and exposed to 12.5 μg/μl Cry3Ba spore-toxin mixtures in flour discs. Flour discs prepared with water were used as controls of the toxicity assays.
Toxicity assays with a 29-mer peptide containing amino acids 1110–1138 of TcSSS protein (PepTcSSS peptide, Ac-KVDAAGSATVELKDTIELITILTPKLTFT-NH2) were carried out exposing groups of 20 larvae of 0.33–0.50-mg weight per larva to flour discs containing 8.8 μg/μl Cry3Ba spore-crystal mixture with or without 10 μg/μl peptide (final concentration). As a control, larvae were challenged with PepTcSSS peptide alone.
RNAi
RNA isolation and cDNA synthesis were performed as described before (23), using 10–14-day-old larvae (after egg laying). cDNA was used as template for PCR amplification using Prime Star polymerase (Takara) and specific primers generated from TcAPN-I, two other APN proteins (EEZ99297, denoted as TcAPN-II, and EEZ99296, denoted as TcAPN-III), TcCad1 and TcSSS protein NCBI gene sequences, containing a T7 polymerase promoter sequence at their 5′ end (see Table 1). The PCR products (1 μg) were used for in vitro transcription to prepare dsRNA using Ambion MEGAscript T7 kit (Applied Biosystems, Austin, TX) according to the manufacturer's protocol. Purified dsRNA was stored at −20 °C until injected into Tc larvae.
TABLE 1.
Primers used in qRT-PCR to analyze the expression of genes corresponding to TcCad1, TcSSS, TcAPN-I, TcAPN-II, and TcAPN-III proteins and to generate dsRNA in RNAi experiments
| Accession number and name | Sequence |
|---|---|
| XP_971388 | |
| TcCad1 RNAi Fw | 5′-GAATTGTAATACGACTCACTATAGGGAACTGACCAAATCACCTTCG-3′ |
| TcCad1 RNAi Rv | 5′-GAATTGTAATACGACTCACTATAGGTGTCTCCAACATCTTTATCGGT-3′ |
| TcCad1 qRT-PCR Fw | 5′-AACAACCCGAGTGGCGAAT-3′ |
| TcCad1 qRT-PCR Rv | 5′-TCTGCCATTGATGAGTTCTTGGT-3′ |
| EFA03129 | |
| TcSSS RNAi Fw | 5′-GAATTGTAATACGACTCACTATAGGCTTCCAAACTTACAGTAAAACTAG-3′ |
| TcSSS RNAi Rv | 5′-GAATTGTAATACGACTCACTATAGGAACACTGATAAGATATTGGTCC-3′ |
| TcSSS qRT-PCR Fw | 5′-AAACCGCGATTCTGGTAAACC-3′ |
| TcSSS qRT-PCR Rv | 5′-TGACCGAATTGTGGTATGGTGAT-3′ |
| NP_001164285 | |
| TcAPN-I RNAi Fw | 5′-GAATTGTAATACGACTCACTATAGGAGTCCACGATGTTTCTAGAGC-3′ |
| TcAPN-I RNAi Rv | 5′-GAATTGTAATACGACTCACTATAGGCAGTCAGTATGTTCAACGTCAG-3′ |
| TcAPN-I qRT-PCR Fw | 5′-CAAGTGGCGGTCCCAGAT-3′ |
| TcAPN-I qRT-PCR Rv | 5′-TCAACAATCCCCAATTTTCCA-3′ |
| EEZ99297 | |
| TcAPN-II RNAi Fw | 5′-GAATTGTAATACGACTCACTATAGGCGATAACAAGTTTGGTAACACG-3′ |
| TcAPN-II RNAi Rv | 5′-GAATTGTAATACGACTCACTATAGGAGATATTTGTTAAGTACGGCTTC-3′ |
| TcAPN-II qRT-PCR Fw | 5′-CGCCCTATTCGCCACTGA-3′ |
| TcAPN-II qRT-PCR Rv | 5′-CGACTATTTGTGCCCGGTTT-3′ |
| EEZ99296 | |
| TcAPN-III RNAi Fw | 5′-GAATTGTAATACGACTCACTATAGGCTGAAGTACTACTAGCAGCGG-3′ |
| TcAPN-III RNAi Rv | 5′-GAATTGTAATACGACTCACTATAGGCATCGCTTGCTAGAACAGG-3′ |
| TcAPN-III qRT-PCR Fw | 5′-CACCTGGTCTGTTCACGAAATG-3′ |
| TcAPN-III qRT-PCR Rv | 5′-CCCAACTTTTTCGGCCAAT-3′ |
| EFA04159 | |
| TcRPS18 qRT-PCR Fw | 5′-TGATGGCAAACGCAAAGTCA-3′ |
| TcRPS18 qRT-PCR Rv | 5′-TCGGCCGACACCTTTGA-3′ |
Larvae were anesthetized for 5 min on ice before ventral injection of ∼0.2 μl of 1 μg/μl dsRNA in injection buffer (1.4 mm NaCl, 0.07 mm Na2HPO4, 0.03 mm KH2PO4, 4 mm KCl), using thin wall capillars (World Precision Instruments) in a microinjection system (Narishige). Control larvae were injected with injection buffer. Following injection, larvae were grown under standard rearing conditions.
Quantitative Real Time PCR
TcCad1, TcAPN-I, TcAPN-II, TcAPN-III, and TcSSS transcript levels were evaluated 8 days after dsRNA injection by quantitative real time PCR. qRT-PCR amplification was performed on a StepOnePlus Real-Time PCR system (Applied Biosystems) thermocycler, following the manufacturer's recommendations, using Power SYBR Green PCR Master Mix (Applied Biosystems), 100 ng of cDNA, and gene-specific forward and reverse primers (Table 1), designed with Primer Express software (Applied Biosystems). For each sample, two biological replicates were analyzed using the mean values of three technical replicates. Gene expression was normalized using RPS18 (ribosomal protein S18, EFA04159) expression as an endogenous control (30) (primers included in Table 1). The data were analyzed by Student's t test for statistically significant differences (p < 0.05). To analyze the expression of TcCad1, TcAPN-I, and TcSSS genes during larval development, qRT-PCR amplification was performed as described above on RNA obtained from larvae of different weight within a range of 0.32–1.8 mg.
RESULTS
Aminopeptidase N, E-cadherin, and Sodium Solute Symporter Are Putative Cry3Ba Toxin-binding Proteins in Tc
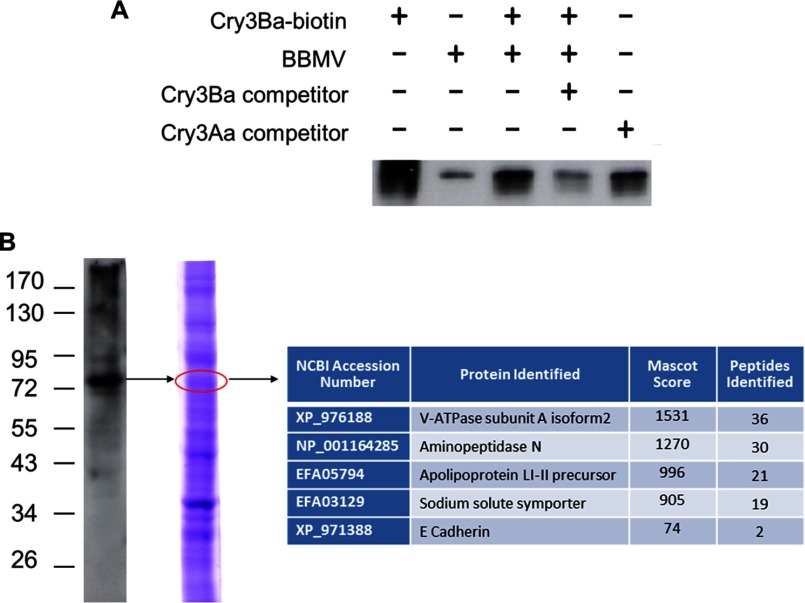
Purified Cry3Ba toxin was biotin-labeled and homologous, and heterologous competition binding assays were performed on BBMV of Tc (Fig. 1A). Binding experiments showed that Cry3Ba toxin was able to bind to Tc BBMV, and this interaction was specific because it was competed with 1000-fold excess of unlabeled Cry3Ba toxin but not with 1000-fold unlabeled Cry3Aa.
FIGURE 1.
Cry3Ba toxin binds TcAPN-I, TcCad1, and TcSSS. A, competition binding assays on Tc BBMV. Biotin-labeled Cry3Ba toxin was incubated with Tc BBMV in the absence or presence of an excess of unlabeled toxin competitor. After 1 h of incubation, unbound toxins were removed by centrifugation, and vesicles containing bound toxins were loaded onto a SDS-PAGE and blotted to a nitrocellulose membrane. Labeled proteins were visualized by means of streptavidin-peroxidase conjugate. B, Cry3Ba toxin ligand blot of Tc BBMV proteins immunodetected with anti-Cry3 antibody is shown on the left. The arrow points to the band at ∼75 kDa that was excised for LC-MS/MS analysis from the corresponding Coomassie-stained gel shown on the right. The most relevant identifications obtained after searching the NCBInr database (taxonomic restriction to Tc) with peptide mass fingerprinting data using Mascot (MatrixScience) are shown in the box.
Tc BBMV proteins were blotted onto a nitrocellulose membrane, and binding of proteins to the Cry3Ba toxin was visualized by immunodetection of the bound toxin. The major protein band recognized by this toxin was ∼75 kDa (Fig. 1B). The corresponding band in a Coomassie-stained gel of Tc BBMV (10% SDS-PAGE) was not among the most intense bands in the gel, indicating that the ligand blotting was performed in optimal conditions so that nonimmunoreactive proteins present in high concentration did not adsorb the primary antibody nonspecifically. The 75-kDa band excised from the Coomassie-stained gel was analyzed by LC-MS/MS, and the NCBInr database (taxonomic restriction to Tc) was searched with peptide mass fingerprinting data using Mascot (MatrixScience) to establish the best protein matches (a score value higher than 57 was considered a significant hit). The most relevant identifications are shown in Fig. 1B. The highest Mascot score corresponded to a predicted protein similar to V-ATPase subunit A isoform 2 (XP_976188), which has been reported to bind Cry toxins in other insects (19–21). Among the identified proteins in the 75-kDa band, we detected aminopeptidase N and E-cadherin that have been demonstrated to act as Cry toxin receptors (12), and with a high Mascot score, the reported Cry binding protein apolipoprotein LI-II precursor (31) and a novel putative Cry receptor, the sodium solute symporter protein, were also found (Fig. 1B).
To demonstrate whether these proteins act as functional Bt receptors in Tc midgut epithelial membrane, we used RNAi to assess the effect of expression knockdown of the corresponding genes in Cry3Ba toxin insecticidal activity against Tc larvae. Because it has been described that V-ATPase silencing compromises Tc viability (32), and apolipoprotein has been proposed to be involved in toxin sequestration by a coagulation reaction inside the gut lumen (31), we focused on gene silencing of aminopeptidase N NP_001164285 (in this work denoted as APN-I), E-cadherin XP_971388 (TcCad1), and the sodium solute symporter protein EFA03129 (TcSSS).
RNA Interference Knockdown of TcCad1 or TcSSS Proteins Resulted in Decreased Susceptibility to Cry3Ba Toxin
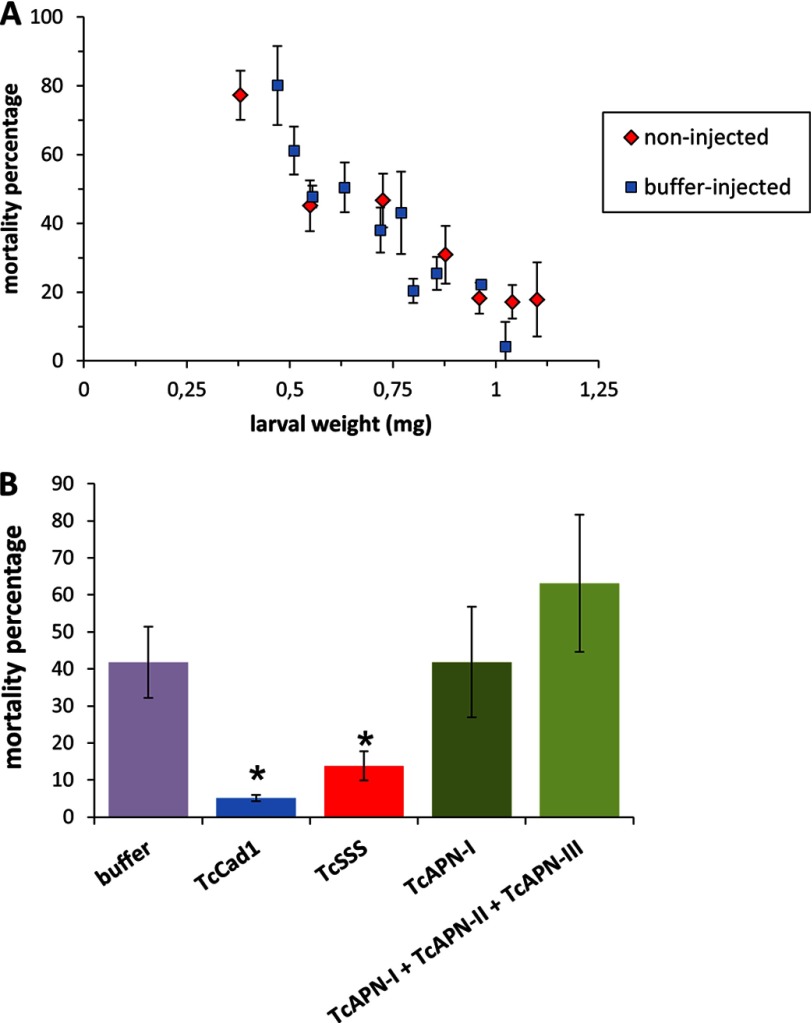
To choose the appropriate larval size for silencing, we obtained the transcription profile of the TcAPN-I, TcCad1, and TcSSS protein genes in different Tc larval developmental stages using qRT-PCR (Fig. 2A), with RPS18 mRNA as an internal control. TcCad1 and TcSSS showed similar expression profile. In contrast, TcAPN-I displayed a completely different pattern. The optimal larval weight range that allowed toxicity assays to be performed following silencing, so that pupation is not reached during bioassay time course, was 0.25–0.80 mg. Because within that range the abundance of TcCad1 and TcSSS transcripts showed a maximum in larvae of 0.64-mg weight, we selected this larval stage to examine mRNA levels of the corresponding transcripts by qRT-PCR, in dsRNA-injected larvae and control (buffer-injected) larvae (Fig. 2A). Because the TcAPN-I expression profile remained stable during the analyzed larval weight range, we selected the same larval stage as in TcCad1 and TcSSS to assess gene knockdown efficiency (Fig. 2A). Additionally, we performed a gene silencing experiment injecting a mixture of dsRNAs of TcAPN-I and the closely related APN EEZ99297 (in this work denoted as TcAPN-II) and APN EEZ99296 (in this work denoted as TcAPN-III) (Fig. 2B). As shown in Fig. 2C, following dsRNA injection, reductions of 80.5, 76.8, and 84.2% compared with control buffer-injected larvae were observed in TcCad1, TcSSS, and TcAPN-I transcript abundance, respectively. In the triple silenced larvae, reductions of 84.7, 37.7, and 90.9% compared with control buffer-injected larvae were observed in TcAPN-I, TcAPN-II, and TcAPN-III transcripts, respectively (Fig. 2D). Single or multiple gene silencing did not cause larval mortality in any of the assayed genes, suggesting that these genes' normal expression must not be essential for Tc larvae viability.
FIGURE 2.
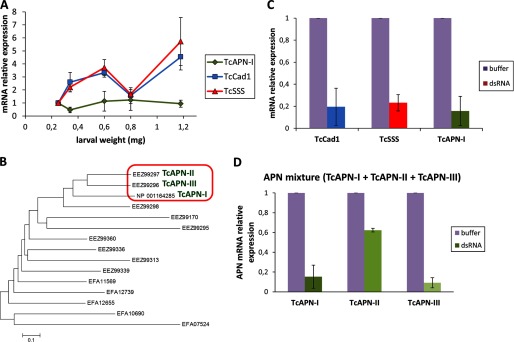
TcAPN-I, TcCad1, and TcSSS mRNA expression is reduced in response to injection of dsRNA. A, qRT-PCR analysis of TcAPN-I, TcCad1, and TcSSS mRNA expression levels in Tc larvae of different weight. RPS18 mRNA abundance was used to normalize gene expression. B, unrooted phylogenetic tree generated with Mega 5 (46) of 15 Tc aminopeptidases N and aminopeptidases-like amino acid sequences. The neighbor-joining method (47) for reconstructing the phylogenetic tree was used. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (48) and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated. C, gene silencing in Tc larvae in response to injection of TcAPN-I or TcCad1 or TcSSS dsRNA. The relative amount of Tc gene transcripts estimated by qRT-PCR in buffer-injected control larvae and silenced larvae was compared, normalized to the expression of RPS18 gene. The statistical significance of the gene expression between the two samples was evaluated using Student's t test, and significant knockdown was observed (p < 0.05) for all genes. The error bars represent standard errors of the mean of two biological samples and three technical replicates each. D, gene silencing in Tc larvae in response to injection of a mixture of TcAPN-I and TcAPN-II and TcAPN-III dsRNA. The relative amount of each individual Tc gene transcript corresponding to TcAPN-I, TcAPN-II, or TcAPN-III estimated by qRT-PCR in buffer-injected control larvae, and silenced larvae was compared normalized to the expression of RPS18 gene. The statistical significance of the gene expression between the two samples was evaluated using Student's t test, and significant knockdown was observed (p < 0.05) for all genes. The error bars represent the standard error of the mean of two biological samples and three technical replicates each.
Prior to toxicity assays in silenced larvae, a control assay was performed to investigate the influence of injection on Cry3Ba toxicity. Tc larvae of different weight were injected with injection buffer and 4 days post-injection were fed with flour discs containing Cry3Ba spore-crystal mixture. Mortality was recorded after 7 days and compared with that of noninjected larvae (Fig. 3A). In both cases, increased larval weight correlated with lower Cry3Ba mortality showing similar mortality profiles, indicating that injection had no effect on the mortality caused by Cry3Ba toxin.
FIGURE 3.
Knockdown of TcCad1 or TcSSS enhances survival of Cry3Ba toxin challenged T. castaneum larvae. A, mortality percentage following Cry3Ba spore-crystal treatment on either noninjected or buffer-injected Tc larvae of different weight. For each experiment, mortality was recorded using 30 Tc larvae. The error bars represent standard error of the mean of at least two biological replicates. B, mortality percentage following Cry3Ba spore-crystal treatment on either buffer-injected (control) or dsRNA-injected larvae. Mortality experiments were performed using 30 Tc larvae. The error bars represent standard error of the mean of two biological samples and three technical replicates each. Asterisks indicate that the mortality increase observed upon Bt treatments in silenced larvae with respect to buffer-injected larvae was statistically significant using Student's t test (p < 0.05).
We next carried out toxicity assays with Cry3Ba toxin on dsRNA-injected and buffer-injected larvae in a weight range of 0.64–0.79 mg (Fig. 3B). TcCad1 and TcSSS dsRNA treatments led to statistically significant mortality decrease compared with buffer-injected larvae (∼87.8 and 67.0% mortality decreases, respectively) (Student's t test, p < 0.05), consistent with a role of both proteins as Cry3Ba toxin functional receptors. In contrast, TcAPN-I dsRNA and a mixture of TcAPN-I, TcAPN-II, and TcAPN-III dsRNA did not show significant mortality differences when compared with buffer-injected larvae (Student's t test, p > 0.05) (Fig. 3B). The results demonstrated that TcCad1 and TcSSS but not aminopeptidase N proteins are Cry3Ba receptors in Tc.
Cry3Ba Toxicity Is Enhanced by a TcSSS Peptide Fragment Containing a Putative Binding Epitope Found in Other Bt Cadherin Functional Receptors
PROSITE patterns database was searched with Motif Scan to identify known motifs in TcSSS and TcCad1 sequences, and the corresponding domain figures were generated using MyDomains-Image Creator PROSITE tool (Fig. 4A). Similar to the CAD-like functional Bt receptor in Tenebrio molitor (TmCad1, ABL86001) (7), in the TcCad1sequence, extracellular, transmembrane, and intracellular domains were found using the TMHMM server. In the extracellular domain, a signal sequence (using SignalP server) (33) and 12 extracellular repeat domains were predicted. TcSSS contains the common topological motif of an arrangement of 11 transmembrane domains present in the sodium solute symporter family (34) and 3 predicted cadherin repeats in the extracellular domain. TcSSS and TcCad1 sequences were scanned to identify conserved patterns using the PRATT 2.1 tool that allows discovering patterns of the PROSITE database conserved in sets of unaligned protein sequences (35). Fig. 4A shows the Clustal alignment corresponding to the PRATT identified pattern in TcSSS and TcCad1 sequences. This alignment was then used to identify the previously described cadherin Bt toxin binding epitopes in Manduca sexta CAD-like protein (1416GVLTLNIQ1423) and in T. molitor TmCad1 (1359GDITINFE1366) (7, 36). Sequence alignments of M. sexta and T. molitor Cry receptor binding regions showed a high degree of similarity to the TcSSS 1115GSATVELK1122 sequence corresponding to the homology region in TcCad1 identified using PRATT (Fig. 4B).
FIGURE 4.
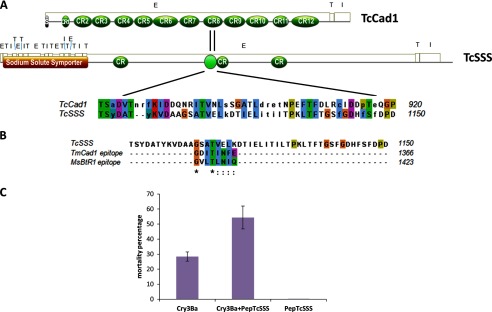
TcSSS contains cadherin repeats and a putative binding epitope homologous to other Bt cadherin functional receptors, which enhances Cry3Ba toxicity in Tc larvae. A, schematic representation of TcCad1 and TcSSS receptors secondary structure obtained with Motif Scan showing the Clustal alignment corresponding to an identified pattern in TcSSS and TcCad1 sequences using PRATT 2.1 (33). The extracellular (E), transmembrane (T), and intracellular (I) domains and numbered cadherin repeat regions (CR1–CR12) are illustrated. B, clustal alignment of the previously described cadherin Bt toxin binding epitopes in M. sexta MsBtR1 (1416GVLTLNIQ1423) and in T. molitor TmCad1 (1359GDITINFE1366) (7, 36) and residues 1103–1150 of the TcSSS sequence corresponding to the homology region in TcCad1 identified using PRATT. In this TcSSS fragment, a putative binding epitope (1115GSATVELK1122) was found. C, enhancement of Cry3Ba toxicity to Tc larvae by a 29-mer peptide (PepTcSSS) spanning amino acids 1110–1138, containing the identified putative binding epitope in TcSSS protein.
It has been demonstrated that peptides containing the above mentioned binding epitopes of M. sexta and T. molitor enhanced the activity of Cry1A and Cry3Aa toxins in different lepidopteran and coleopteran larvae, respectively (36, 37). Therefore, consistent with TcSSS being a functional receptor in Tc, it would be expected that a peptide containing the homologous fragment identified in the TcSSS protein would also enhance the toxicity of Cry3Ba against Tc larvae. A 29-mer peptide Ac-1110KVDAAGSATVELKDTIELITILTPKLTFT1138-NH2 (PepTcSSS) containing the putative binding epitope in TcSSS was synthesized and, when used in bioassays, significantly enhanced Cry3Ba toxicity against Tc larvae, resulting in 54.4% mortality in contrast to 28.4% mortality with Cry3Ba toxin alone (Fig. 4C). As shown in Fig. 4C, PepTcSSS itself did not cause larval mortality.
DISCUSSION
It is well known that Bt toxins insecticidal activity relies on the interaction with midgut epithelial membrane components of the target insect. In the case of Cry3Ba toxin, reported active against Tc (23), as expected, the toxin specifically bound to Tc larvae BBMV (Fig. 1A). By ligand blot analysis, a protein band of 75 kDa was mainly recognized by Cry3Ba toxin and several putative binding proteins were identified using mass spectrometry (Fig. 1B). The considered genuine Cry receptors APN and CAD-like proteins were found among them and also other proteins reported as Cry binding proteins, such as V-ATPase (19–21), with the highest score, and apolipoprotein LI-II precursor (31) (Fig. 1B). Additionally, a novel Cry binding protein, TcSSS, was identified with a high score (Fig. 1B).
A substantial amount of evidence suggests that toxin-binding APNs found in various insect larvae act as Bt receptors (12). It has been proposed that the main significance of toxin binding to these proteins might be to concentrate the prepore toxin structure at the cell membrane surface prior to membrane insertion (17). However, our RNAi results with TcAPN-I in Tc did not support a role for this protein in Cry3Ba toxicity because no change in mortality was observed in Tc silenced larvae treated with Cry3Ba toxin relative to nonsilenced larvae challenged with the toxin. Failure to demonstrate the receptor function of TcAPN-I made us consider the possibility that in Tc larvae lacking TcAPN-I gene expression, other APN proteins might functionally replace it. We then performed a multiple gene silencing experiment in which TcAPN-I and the closely related TcAPN-II and TcAPN-III genes were simultaneously knockdown. Following Cry3Ba intoxication in Tc triple silenced larvae, no mortality decrease was observed relative to control larvae, ruling out the involvement of these APN proteins in Cry3Ba toxin action in Tc. Although not statistically significant, Cry3Ba intoxicated triple silenced larvae showed a mortality increase relative to intoxicated control larvae, probably evidencing the detrimental effect of the multiple silencing. Because the effectiveness of TcAPN-II silencing (37.7%) was significantly lower than that of the other two Tc APNs (84.7% for TcAPN-I and 90.9% for TcAPN-III) (Fig. 2D), it is not possible to infer whether all three APN proteins equally contributed to this effect.
Functional studies have turned midgut CAD-like proteins into one of the most likely Cry toxin receptor molecules in lepidopteran, dipteran, and coleopteran larvae (7, 12, 38). It has been proposed that they play the role of the first receptor of Cry toxins, binding toxin monomer and facilitating further processing required for the prepore oligomer formation (17). In the beetle T. molitor, lower expression of the CAD-like gene TmCad1 in larvae directly correlated with survival on Cry3Aa-treated diet, demonstrating the functional role of this protein as Cry3Aa receptor (7). In this report we have demonstrated that the TmCad1 ortholog CAD-like protein TcCad1, identified as a Cry3Ba binding protein in Tc larvae, also acts as a functional Cry3Ba receptor in this insect. Tc silenced larvae intoxicated with a Cry3Ba spore-crystal mixture concentration that corresponds approximately to LC50 in nonsilenced larvae showed a dramatic decrease in susceptibility because 95 ± 1% larvae survived. This result adds to those of others highlighting the critical role of CAD-like receptors in Bt mode of action.
In this work, we have demonstrated that the novel Cry3Ba binding protein TcSSS also functions as a toxin receptor in Tc larvae. As in the case of TcCad1, reduction of TcSSS expression by RNA interference resulted in increased larval survival on a Cry3Ba spore-crystal-treated diet (86 ± 4% Tc larvae survived when treated with a concentration that corresponds approximately to LC50). Interestingly, the TcCad1 and TcSSS proteins that act as functional Cry3Ba receptors in Tc larvae exhibited the same gene expression profile during larval development (Fig. 2A) and have cadherin repeats (Fig. 4A), supporting that CAD-like proteins are relevant to Cry3Ba toxicity in Tc. Consistent with the role of TcSSS as a CAD-like Cry receptor, Cry3Ba toxicity was enhanced by a TcSSS peptide fragment containing a putative binding epitope found in other Bt cadherin functional receptors, such as in M. sexta and T. molitor CAD-like proteins.
These features, while suggesting a parallel role of TcCad1 and TcSSS mode of action, are in contrast with the Cry3Ba mortality data obtained in silenced larvae. If both proteins were fully redundant in their participation in Cry3Ba toxic action, the dramatic decrease in mortality observed after toxin challenge in larvae in which each of the corresponding genes were independently knocked down would not be expected (Fig. 3B). The results are more in accordance with a complementary function of both receptor molecules in a stepwise mechanism of action.
SSSs are Na+-dependent transport proteins responsible for the absorption of nutrients, vitamins, osmolytes, and ions across the plasma membrane of pro- and eukaryotic cells (34). SSS proteins constitute a family belonging to the amino acid-polyamine-organocation superfamily Transporter Classification Database (TCDB) (39), which is composed of 11 families (40). Members of the SSS family generally share a core topology characterized by two inverted structural repeats of five transmembrane α-helices each containing the binding site for substrate and ion, and a periplasmic N terminus and a cytoplasmic C terminus.
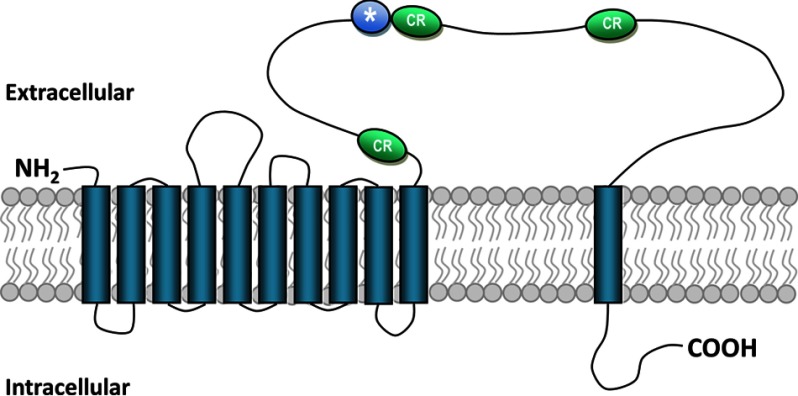
Very recently, the functionality for ATP-binding cassette transporter subfamily C member 2 (ABCC2) as Cry1 toxin receptor has been reported in Bombyx mori larvae (5), in accord with the role of this protein in the mechanism of Cry1 toxin resistance (41, 42). Remarkably, B. mori ABCC2 protein (42) structurally resembles TcSSS protein in that both are multimembrane-spanning transporters (Fig. 5).
FIGURE 5.
Schematic structure of the T. castaneum TcSSS transporter. Using TMHMM version 2.0, 11 transmembrane domains were predicted based on the amino acid sequence of T. castaneum EFA03129. In the schematic diagram, cadherin repeat regions (CR) and localization of the TcSSS fragment (asterisk) containing the region homologous to cadherin binding epitopes in M. sexta MsBtR1 and T. molitor TmCad1 (7, 36) are also illustrated.
There are two main classes of active transmembrane transport systems: primary active transporters, such as ABC proteins, which rely on ATP hydrolysis to actively pump their substrates across membranes; and secondary active transporters, such as SSS, in which transport is driven by proton or sodium transmembrane gradients (43). The common element of primary and secondary transporters is that transporter-mediated movement of solutes across membranes involves the alternating access mechanism proposed by Jardetzky (44), in which transport occurs by binding of substrate to the “open-to-in” state followed by isomerization of the transporter to the “open-to-out” state, allowing the release of the substrate to the cytoplasm. In relation to Bt mode of action, it has been proposed that toxin oligomer insertion might be coupled to the transport cycle of the ABCC2 protein (45).
The biological role of the TcSSS protein is not known, but results of the RNAi experiments together with its functional similarity to ABCC2 transporter, suggest that this novel Cry3Ba binding protein could also be implicated in Cry3Ba toxin insertion. Alternatively, because of the essential role of ion gradients in active transport in almost any cell type, natural products and toxins that collapse the ion gradients across cellular membranes are poisons, and therefore the interaction of Cry3Ba toxin with the TcSSS protein might result in enhanced activity caused by transporter-related toxicity. That could also be the case of the V-ATPase subunit A isoform 2, identified in this work as a putative Cry3Ba binding protein (Fig. 1B), that couples the energy of ATP hydrolysis to proton transport across intracellular and plasma membranes of eukaryotic cells.
Our data suggest a complex mechanism underlying the toxicity process of Cry3Ba toxin in Tc; therefore, more research is needed to understand its mode of action. In contrast with the Bt mode of action described for lepidopteran insects, in which APN plays a relevant role in toxicity, in coleopteran insects APN protein has never been involved in Bt mode of action, and our results support the possibility that these proteins might not act as a Cry functional receptor in this insect order. On the other hand, TcCad1 and TcSSS, and probably other midgut proteins identified in this work, are determinant in coleopteran-specific Cry toxicity. The model coleopteran insect Tc represents an ideal experimental subject to obtain a complete picture of the complexity of Bt interactions and the molecular bases of insect toxin specificity.
Acknowledgments
We thank Dr. Martin Klingler (Department of Biology, Developmental Biology Unit, University of Erlangen-Nuremberg) for kindly sharing expertise on Tc RNAi experiments and the Genomics Facility of SCSIE (University of Valencia) and Centro de Investigación Príncipe Felipe (Valencia, Spain) for technical support.
This work was supported by Ministerio de Ciencia e Innovación Grants BIO 2007-67860 and AGL2010-22300-C03-03
M. D. Real and C. Rausell, unpublished results.
- Bt
- B. thuringiensis
- APN
- aminopeptidase N
- CAD
- cadherin
- Tc
- T. castaneum
- SSS
- sodium solute symporter
- dsRNA
- double-stranded RNA
- BBMV
- brush border membrane vesicle
- qRT-PCR
- quantitative RT-PCR.
REFERENCES
- 1. Crickmore N. (2005) Using worms to better understand how Bacillus thuringiensis kills insects. Trends Microbiol. 13, 347–350 [DOI] [PubMed] [Google Scholar]
- 2. Knight P. J., Crickmore N., Ellar D. J. (1994) The receptor for Bacillus thuringiensis CrylA(c) δ-endotoxin in the brush border membrane of the lepidopteran Manduca sexta is aminopeptidase N. Mol. Microbiol. 11, 429–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vadlamudi R. K., Weber E., Ji I., Ji T. H., Bulla L. A., Jr. (1995) Cloning and expression of a receptor for an insecticidal toxin of Bacillus thuringiensis. J. Biol. Chem. 270, 5490–5494 [DOI] [PubMed] [Google Scholar]
- 4. Jurat-Fuentes J. L., Adang M. J. (2004) Characterization of a Cry1Ac-receptor alkaline phosphatase in susceptible and resistant Heliothis virescens larvae. Eur. J. Biochem. 271, 3127–3135 [DOI] [PubMed] [Google Scholar]
- 5. Tanaka S., Miyamoto K., Noda H., Jurat-Fuentes J. L., Yoshizawa Y., Endo H., Sato R. (2013) The ATP-binding cassette transporter subfamily C member 2 in Bombyx mori larvae is a functional receptor for Cry toxins from Bacillus thuringiensis. FEBS J. 280, 1782–1794 [DOI] [PubMed] [Google Scholar]
- 6. Likitvivatanavong S., Chen J., Bravo A., Soberón M., Gill S. S. (2011) Cadherin, alkaline phosphatase, and aminopeptidase N as receptors of Cry11Ba toxin from Bacillus thuringiensis subsp. jegathesan in Aedes aegypti. Appl. Environ. Microbiol. 77, 24–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fabrick J., Oppert C., Lorenzen M. D., Morris K., Oppert B., Jurat-Fuentes J. L. (2009) A novel Tenebrio molitor cadherin is a functional receptor for Bacillus thuringiensis Cry3Aa toxin. J. Biol. Chem. 284, 18401–18410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ochoa-Campuzano C., Real M. D., Martínez-Ramírez A. C., Bravo A., Rausell C. (2007) An ADAM metalloprotease is a Cry3Aa Bacillus thuringiensis toxin receptor. Biochem. Biophys. Res. Commun. 362, 437–442 [DOI] [PubMed] [Google Scholar]
- 9. Martins E. S., Monnerat R. G., Queiroz P. R., Dumas V. F., Braz S. V., de Souza Aguiar R. W., Gomes A. C., Sánchez J., Bravo A., Ribeiro B. M. (2010) Midgut GPI-anchored proteins with alkaline phosphatase activity from the cotton boll weevil (Anthonomus grandis) are putative receptors for the Cry1B protein of Bacillus thuringiensis. Insect Biochem. Mol. Biol. 40, 138–145 [DOI] [PubMed] [Google Scholar]
- 10. Rajagopal R., Sivakumar S., Agrawal N., Malhotra P., Bhatnagar R. K. (2002) Silencing of midgut aminopeptidase N of Spodoptera litura by double-stranded RNA establishes its role as Bacillus thuringiensis toxin receptor. J. Biol. Chem. 277, 46849–46851 [DOI] [PubMed] [Google Scholar]
- 11. Soberón M., Pardo-López L., López I., Gómez I., Tabashnik B. E., Bravo A. (2007) Engineering modified Bt toxins to counter insect resistance. Science 318, 1640–1642 [DOI] [PubMed] [Google Scholar]
- 12. Pigott C. R., Ellar D. J. (2007) Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiol. Mol. Biol. Rev. 71, 255–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Herrero S., Gechev T., Bakker P. L., Moar W. J., de Maagd R. A. (2005) Bacillus thuringiensis Cry1Ca-resistant Spodoptera exigua lacks expression of one of four aminopeptidase N genes. BMC Genomics 6, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gómez I., Oltean D. I., Gill S. S., Bravo A., Soberón M. (2001) Mapping the epitope in cadherin-like receptors involved in Bacillus thuringiensis Cry1A toxin interaction using phage display. J. Biol. Chem. 276, 28906–28912 [DOI] [PubMed] [Google Scholar]
- 15. Dorsch J. A., Candas M., Griko N. B., Maaty W. S., Midboe E. G., Vadlamudi R. K., Bulla L. A., Jr. (2002) Cry1A toxins of Bacillus thuringiensis bind specifically to a region adjacent to the membrane-proximal extracellular domain of BT-R1 in Manduca sexta. Involvement of a cadherin in the entomopathogenicity of Bacillus thuringiensis. Insect Biochem. Mol. Biol. 32, 1025–1036 [DOI] [PubMed] [Google Scholar]
- 16. Hua G., Jurat-Fuentes J. L., Adang M. J. (2004) Bt-R1a extracelular cadherin repeat 12 mediates Bacillus thuringiensis Cry1Ab binding and cytotoxicity. J. Biol. Chem. 279, 28051–28056 [DOI] [PubMed] [Google Scholar]
- 17. Bravo A., Gómez I., Conde J., Muñoz-Garay C., Sánchez J., Miranda R., Zhuang M., Gill S. S., Soberón M. (2004) Oligomerization triggers differential binding of a pore-forming toxin to a different receptor leading to efficient interaction with membrane microdomains. Biochim. Biophys. Acta 1667, 38–46 [DOI] [PubMed] [Google Scholar]
- 18. McNall R. J., Adang M. J. (2003) Identification of novel Bacillus thuringiensis Cry1Ac binding proteins in Manduca sexta midgut through proteomic analysis. Insect Biochem. Mol. Biol. 33, 999–1010 [DOI] [PubMed] [Google Scholar]
- 19. Krishnamoorthy M., Jurat-Fuentes J. L., McNall R. J., Andacht T., Adang M. J. (2007) Identification of novel Cry1Ac binding proteins in midgut membranes from Heliothis virescens using proteomic analyses. Insect Biochem. Mol. Biol. 37, 189–201 [DOI] [PubMed] [Google Scholar]
- 20. Chen L. Z., Liang G. M., Zhang J., Wu K. M., Guo Y. Y., Rector B. G. (2010) Proteomic analysis of novel Cry1Ac binding proteins in Helicoverpa armigera (Hübner). Arch. Insect Biochem. Physiol. 73, 61–73 [DOI] [PubMed] [Google Scholar]
- 21. Bayyareddy K., Andacht T. M., Abdullah M. A., Adang M. J. (2009) Proteomic identification of Bacillus thuringiensis subsp. israeliensis toxin Cry4Ba binding proteins in midgut membranes from Aedes (Stegomyia) aegypti Linnaeus (Diptera, Culicidae) larvae. Insect Biochem. Mol. Biol. 39, 279–286 [DOI] [PubMed] [Google Scholar]
- 22. Morris K., Lorenzen M. D., Hiromasa Y., Tomich J. M., Oppert C. (2009) Tribolium castaneum larval gut transcriptome and proteome. A resource for the study of the coleopteran gut. J. Proteome Res. 8, 3889–3898 [DOI] [PubMed] [Google Scholar]
- 23. Contreras E., Rausell C., Real M. D. (2013) Proteome response of Tribolium castaneum larvae to Bacillus thuringiensis toxin producing strains. PLoS One 8, e55330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stewart G. S., Johnstone K., Hagelberg E., Ellar D. J. (1981) Commitment of bacterial spores to germinate. A measure of the trigger reaction. Biochem. J. 198, 101–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rausell C., García-Robles I., Sánchez J., Muñoz-Garay C., Martínez-Ramírez A. C., Real M. D., Bravo A. (2004) Role of toxin activation on binding and pore formation activity of the Bacillus thuringiensis Cry3 toxins in membranes of Leptinotarsa decemlineata (Say). Biochim. Biophys. Acta 1660, 99–105 [DOI] [PubMed] [Google Scholar]
- 26. Wolfersberger M., Lüthy P., Maurer A., Parenti P., Sacchi F. V., Giordana B., Hanozet G. M. (1987) Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae). Comp. Biochem. Physiol. 86A, 301–308 [Google Scholar]
- 27. Reuveni M., Dunn P. E. (1991) Differential inhibition by Bacillus thuringiensis delta endotoxin of leucine and aspartic uptake in BBMV from midgut of Manduca sexta. Biochem. Biophys. Res. Commun. 181, 1089–1093 [DOI] [PubMed] [Google Scholar]
- 28. Hafkenscheid J. C. (1984) Aminopeptidases and amino acid arylamidases. In Methods of Enzymatic Analysis (Bergmeyer H. U., Bergmeyer J., Grassl M., eds) Vol. V, 3rd Ed., pp. 2–34, Verlag Chemie, Weinheim [Google Scholar]
- 29. Xie Y. S., Bodnaryk R. P., Fields P. G. (1996) A rapid and simple flour-disk bioassay for testing substances active against stored-product insects. Can. Entomol. 128, 865–875 [Google Scholar]
- 30. Lord J. C., Hartzer K., Toutges M., Oppert B. (2010) Evaluation of quantitative PCR reference genes for gene expression studies in Tribolium castaneum after fungal challenge. J. Microbiol. Methods 80, 219–221 [DOI] [PubMed] [Google Scholar]
- 31. Ma G., Rahman M. M., Grant W., Schmidt O., Asgari S. (2012) Insect tolerance to the crystal toxins Cry1Ac and Cry2Ab is mediated by the binding of monomeric toxin to lipophorin glycolipids causing oligomerization and sequestration reactions. Dev. Comp. Immunol. 37, 184–192 [DOI] [PubMed] [Google Scholar]
- 32. Whyard S., Singh A. D., Wong S. (2009) Ingested double-stranded RNAs can act as species-specific insecticides. Insect. Biochem. Mol. Biol. 39, 824–832 [DOI] [PubMed] [Google Scholar]
- 33. Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011) SignalP 4.0. Discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 34. Jung H. (2002) The sodium/substrate symporter family. Structural and functional features. FEBS Lett. 529, 73–77 [DOI] [PubMed] [Google Scholar]
- 35. Jonassen I., Collins J. F., Higgins D. (1995) Finding flexible patterns in unaligned protein sequences. Protein Sci. 4, 1587–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen J., Hua G., Jurat-Fuentes J. L., Abdullah M. A., Adang M. J. (2007) Synergism of Bacillus thuringiensis toxins by a fragment of a toxin-binding cadherin. Proc. Natl. Acad. Sci. U.S.A. 104, 13901–13906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gao Y., Jurat-Fuentes J. L., Oppert B., Fabrick J. A., Liu C., Gao J., Lei Z. (2011) Increased toxicity of Bacillus thuringiensis Cry3Aa against Crioceris quatuordecimpunctata, Phaedon brassicae and Colaphellus bowringi by a Tenebrio molitor cadherin fragment. Pest Manag. Sci. 67, 1076–1081 [DOI] [PubMed] [Google Scholar]
- 38. Chen J., Aimanova K. G., Fernandez L. E., Bravo A., Soberon M., Gill S. S. (2009) Aedes aegypti cadherin serves as a putative receptor of the Cry11Aa toxin from Bacillus thuringiensis subsp. israelensis. Biochem. J. 424, 191–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saier M. H., Jr., Yen M. R., Noto K., Tamang D. G., Elkan C. (2009) The Transporter Classification Database. Recent advances. Nucleic Acids Res. 37, 274–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wong F. H., Chen J. S., Reddy V., Day J. L., Shlykov M. A., Wakabayashi S. T., Saier M. H., Jr. (2012) The amino acid-polyamine-organocation superfamily. J. Mol. Microbiol. Biotechnol. 22, 105–113 [DOI] [PubMed] [Google Scholar]
- 41. Gahan L. J., Pauchet Y., Vogel H., Heckel D. G. (2010) An ABC transporter mutation is correlated with insect resistance to Bacillus thuringiensis Cry1Ac toxin. PLoS Genet. 6, e1001248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Atsumi S., Miyamoto K., Yamamoto K., Narukawa J., Kawai S., Sezutsu H., Kobayashi I., Uchino K., Tamura T., Mita K., Kadono-Okuda K., Wada S., Kanda K., Goldsmith M. R., Noda H. (2012) A single amino acid mutation in an ABC transporter gene causes resistance to Bt toxin Cry1Ab in the silkworm, Bombyx mori. Proc. Natl. Acad. Sci. U.S.A. 109, E1591–E1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Saier M. H., Jr. (2000) A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64, 354–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jardetzky O. (1966) Simple allosteric model for membrane pumps. Nature 211, 969–970 [DOI] [PubMed] [Google Scholar]
- 45. Heckel D. G. (2012) Learning the ABCs of Bt. ABC transporters and insect resistance to Bacillus thuringiensis provide clues to a crucial step in toxin mode of action. Pestic. Biochem. Physiol. 104, 103–110 [Google Scholar]
- 46. Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011) MEGA5. Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Saitou N., Nei M. (1987) The neighbor-joining method. A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 48. Zuckerkandl E., Pauling L. (1965) Evolutionary divergence and convergence in proteins. In Evolving Genes and Proteins (Bryson V., Vogel H. J., eds) pp. 97–166, Academic Press, New York [Google Scholar]