Background: Vps9 is a guanine nucleotide exchange factor (GEF) that activates Rab5 paralogs in yeast.
Results: Muk1 has a VPS9 homology domain, exhibits GEF activity against Rab5 paralogs, and is partially redundant with Vps9 in vivo.
Conclusion: Muk1 is a bona fide Rab5 GEF.
Significance: Discovery of a second yeast Rab5 GEF provides a more complete understanding of endosomal traffic.
Keywords: Endocytosis, G Proteins, Guanine Nucleotide Exchange Factor (GEF), Membrane Fusion, Rab, Luciferase Reporter of Intraluminal Deposition (LUCID), MVB, Vps21, Ypt52, Ypt53
Abstract
VPS9 domains can act as guanosine nucleotide exchange factors (GEFs) against small G proteins of the Rab5 family. Saccharomyces cerevisiae vps9Δ mutants have trafficking defects considerably less severe than multiple deletions of the three cognate Rab5 paralogs (Vps21, Ypt52, and Ypt53). Here, we show that Muk1, which also contains a VPS9 domain, acts as a second GEF against Vps21, Ypt52, and Ypt53. Muk1 is partially redundant with Vps9 in vivo, with vps9Δ muk1Δ double mutant cells displaying hypersensitivity to temperature and ionic stress, as well as profound impairments in endocytic and Golgi endosome trafficking, including defects in sorting through the multivesicular body. Cells lacking both Vps9 and Muk1 closely phenocopy double and triple knock-out strains lacking Rab5 paralogs. Microscopy and overexpression experiments demonstrate that Vps9 and Muk1 have distinct localization determinants. These experiments establish Muk1 as the second Rab5 GEF in budding yeast.
Introduction
Rab small G proteins, a subgroup of the Ras superfamily, mark compartmental identity and are key regulators of membrane trafficking. Saccharomyces cerevisiae contains 11 Rabs, and more than 60 have been identified in mammals (1). The Rab5 family coordinates events essential in endolysosomal traffic, including endosome maturation and targeting of Golgi-derived vesicles to early and late endosomes. The Rab7 family resides on late endosomal or vacuolar membranes and controls hetero- and homotypic fusion events at these organelles. Like humans, S. cerevisiae has three Rab5 isoforms (Vps21, Ypt52, and Ypt53) and one Rab7 (Ypt7). Disruption of Vps21 results in obvious endolysosomal trafficking and morphology phenotypes, whereas Ypt52 and Ypt53 are partially redundant with Vps21 (2–6). In addition, recent work implicates Ypt53 in cellular stress responses (3).
Rab signaling is regulated through the binding of GDP and GTP. Rabs adopt an active conformation when GTP-bound, allowing them to bind and coordinate activities of effector molecules that are required for vesicle docking and fusion. Intrinsic rates of Rab GTP hydrolysis and GDP dissociation are slow; Rabs rely on regulatory proteins to catalyze transitions between their active and inactive states. Rab signaling is promoted by guanine nucleotide exchange factors (GEFs)4 that stimulate GDP release, allowing GTP to bind. GTPase-accelerating proteins (GAPs) terminate Rab signaling by triggering GTP hydrolysis. Rabs are anchored to membranes through a pair of prenylated cysteines at their C termini. A chaperone, GDP dissociation inhibitor (GDI), can extract bis-prenylated Rab-GDP from membranes, and can deposit the bound Rabs back onto specific membranes through a mechanism that may be accelerated by GDI displacement factors (7–10).
In many cases, GEFs for small G proteins contain identifiable, evolutionarily conserved catalytic domains. For example, the Sec7 domain acts on the Arf subfamily, the Cdc25 domain acts on the Ras family, and the Dbl homology/pleckstrin homology domain acts on the Rho family of small GTPases (11). Similarly, a large family of Rab GEFs contains the DENN (differentially expressed in normal and neoplasia) domain; however, not all do (12).
The VPS9 domain was identified in S. cerevisiae Vps9 and Homo sapiens Rabex-5; both proteins were shown to stimulate GDP expulsion on G proteins of the Rab5 family (13, 14). The mammalian Rin proteins (Rin1–3) also contain VPS9 domains and act as GEFs against Rab5 isoforms, suggesting that VPS9 domains generally function to stimulate signaling by members of the Rab5 family (15–17). VPS9 domain proteins localize to appropriate membranes by interacting with a variety of targeting determinants. For example, yeast Vps9 has a CUE domain that is believed to bind ubiquitinylated cargo molecules at the endosome surface. Similarly, mammalian Rabex-5 has a ubiquitin-binding domain, and it also interacts with an array of accessory proteins. In contrast, mammalian Rin1 interacts selectively with internalized EGFR receptors to promote their endosomal down-regulation.
At least 22 mammalian proteins have VPS9 domains. Two S. cerevisiae proteins contain intact VPS9 domains: Vps9 and Muk1 (computationally linked to KAP95) (18). Using biochemical and genetic analyses, we now show that Muk1 is a specific GEF for yeast Rab5 proteins and that its in vivo function is partially redundant with the major GEF of the Rab5 family, Vps9.
EXPERIMENTAL PROCEDURES
Cloning and Strain Construction
Strains and plasmids are summarized in Table 1. Generation of DNA cassettes to knock out the MUK1 ORF was performed as described (19), and correct integration was confirmed by genomic PCR mapping. Yeast expression plasmids were generated by gap repair recombination of PCR products into linearized plasmid vectors. GST-tagged Muk1 was inserted into the shuttle vector pFB-HTB-N (20). The resulting plasmid was sequenced and transformed into DH10Bac Escherichia coli, and bacmid DNA was then purified. Bacmid DNA was transfected into Sf9 cells to prepare nuclear polyhedrosis baculovirus stocks. Yeast two-hybrid (Y2H) vectors are as described (21) or were obtained from the Yeast Resource Center. The vectors were sequence-verified prior to use.
TABLE 1.
Strains and plasmids used in this study
| Name | Genotype/description | Reference/source |
|---|---|---|
| S. cerevisiae | ||
| SEY6210 | MATα leu2-3,112 ura3-52 his3-200 trp1-901 lys2-801 suc2-9 | Ref. 26 |
| BHY10 | SEY6210 CPY-Invertase::LEU2 (pBHY11) | Ref. 27 |
| MBY3 | SEY6210 vps4Δ::TRP1 | Ref. 28 |
| GOY223 | BHY10 vps9Δ::HIS3 | Ref. 3 |
| DNY516 | BHY10 muk1Δ::KAN | This study |
| DNY517 | BHY10 vps9Δ::HIS3 muk1Δ::KAN | This study |
| DNY206 | BHY10 vps21Δ::KAN | Ref. 3 |
| DNY471 | BHY10 vps21Δ0 ypt52Δ0 | Ref. 3 |
| E. coli | ||
| TOP10F′ | F′ lacIq, Tn10(TetR) mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (StrR) endA1 nupG | Invitrogen |
| BL21(DE3) | F − ompT gal dcm lon hsdSB (rB − mB −)λ (DE3 [lacI lacUV5-T7 ind1 sam7 nin5]) | Stratagene/Agilent |
| Plasmids | ||
| pRS416 | URA3 AmpR CEN | Ref. 29 |
| pRS426 | URA3 AmpR 2μ | Ref. 29 |
| pDN216 | MUK1 (pRS426) | This study |
| pGO36 | GFP (pRS416) | Ref. 30 |
| pGO45 | GFP-CPS1 (pRS426) | Ref. 30 |
| pDN219 | GFP-MUK1 (pRS416) | This study |
| pDN251 | URA3 LoxP::CEN::LoxP PGK1pr::RLuc 0::FLuc | Ref. 3 |
| pDN252 | PGK1pr::RLuc SNA3-FLuc (pDN251) | Ref. 3 |
| pDN312 | PGK1pr::RLuc STE3-FLuc (pDN251) | This study |
| pDN268 | PGK1pr::RLuc MUP1-FLuc (pDN251) | This study |
| pSna3-GFP | SNA3-GFP (pRS416) | Ref. 31 |
| pCHL642 | URA3 AmpR MUP1-GFP (pRS416) | S. Emr (Cornell) |
| pDN615 | LEU2 AmpR LoxP::CEN::LoxP (pRS415) | This study |
| pDN274.2 | mCherry-VPS9 (pDN615) | This study |
| AMP219 | AmpR GST-VPS21 (pParallel-GST) | Ref. 20 |
| AMP220 | AmpR GST-YPT52 (pParallel-GST) | Ref. 20 |
| AMP217 | AmpR GST-YPT53 (pParallel-GST) | Ref. 20 |
| AMP218 | AmpR GST-YPT7 (pParallel-GST) | Ref. 20 |
| AMP1434 | AmpR GST-MUK1 (pFB HTB) | This study |
| AMP84 | AmpR His6-Gyp1TBC (pET22) | Ref. 20 |
Yeast Two-hybrid Culture and Media
Yeast growth in liquid cultures was monitored using a Bioscreen-C machine (Growth Curves USA). 150-μl cultures of YPD (yeast peptone dextrose) with or without 200 mm CaCl2 at 30 °C were monitored with periodic shaking. Cultures were inoculated at OD600 nm = 0.1 from overnight YPD cultures. Bioscreen-C growth curve data were analyzed using YODA software (22) and plotted using GraphPad Prism 4.0. For solid medium growth assays using limiting dilutions, cells were grown overnight at 30 °C in synthetic medium supplemented with casamino acids to select for retention of the MUK1 overexpression plasmid prior to serial dilution and application of cells to nonselective YPD plates. For Y2H tests, liquid cultures of the bait and prey Y2H library strains were grown in selective medium, then mixed in a 96-well plate, and pinned to YPD plates using a 48-pin manifold. The plates were incubated at 30 °C overnight, and then colonies were replica-plated onto synthetic medium lacking Trp and Leu to select for diploid cells. These plates were incubated at 30 °C for 2 days; then tested for Y2H interactions by replica plating to yeast synthetic medium lacking Trp, Leu, and His; and supplemented with 3 mm 3-amino-1,2,4-triazole. After 5 days at 30 °C, the plates were imaged.
Protein Purification
Vps9 and Gyp1–46 were expressed in E. coli and purified as described (23). GST-tagged Rab G-proteins (Vps21, Ypt52, Ypt53, and Ypt7) were purified as described previously (3). GST-Muk1 was expressed in adherent BTI-TN-5B1–4 (Hi-5) cells (21). The cells were lysed by sonication in HEPES lysis buffer (50 mm HEPES, 150 mm NaCl, 5 mm 2-mercaptoethanol, pH 7.4) with protease inhibitors. The clarified lysate was bound to glutathione-Sepharose resin for 4 h at 4 °C. The resin was extensively washed and eluted with 20 mm glutathione in HEPES lysis buffer. Excess glutathione was removed by exchange into HEPES lysis buffer using a PD-10 column (GE Healthcare). GST-Muk1 was then concentrated by ultrafiltration and snap-frozen in liquid nitrogen.
Microscopy
Fluorescence microscopy was performed with an Olympus IX71 light microscope, an EMCCD (Andor Ixon) camera, Pl 60× NA 1.45 or 100× NA 1.3 objectives, and appropriate filter sets. Andor IQ v.6.0.3.62 (Andor Bioimaging, Nottingham, UK) software was used for data collection. ImageJ v.1.45s (J. Rasband, National Institutes of Health), Photoshop v9.0 (Adobe), and Canvas v9.0 (ACD) software were used for contrast and brightness adjustments and for figure layout. For imaging of Mup1-GFP, yeast cells were grown overnight in selective defined medium lacking methionine at 30 °C and diluted to A600 nm 0.2 the following morning. 1.5 ml of culture was harvested at mid-log phase (∼0.5–0.7 OD600 nm) by sedimenting at 3,000 × g for 3 min and suspended in 50 ml of fresh medium lacking Met but containing 50 μm FM 4–64 styryl dye (Invitrogen). The cells were incubated for 15 min at room temperature, centrifuged for 5 min at 3,000 × g, and suspended in 1 ml of fresh medium. This suspension was divided in two with 1 mm methionine added to one portion, and all of the samples were incubated at 30 °C for 1 h. The cells were sedimented at 3,000 × g, resuspended in 50 μl fresh medium with or without Met, and imaged as described above.
GEF Activity Assays
EnzCheck phosphate assay kit (Invitrogen E-6646) was used to measure GEF activity as described previously (23). Briefly, 100-μl aqueous reactions (total) containing reaction buffer (20 mm HEPES·NaOH pH 7.5, 150 mm NaCl, 25 mm MgCl2, 0.375 mm 2-amino-6-mercapto-7-methyl-purine riboside, 0.5 mm GTP, 0.5% (w/v) BSA, and 2 units/ml purine nucleoside phosphorylase), and the indicated Rab at 20 μm were loaded into the wells of a 96-well plate (Corning) and allowed to equilibrate for 10 min. 0.5 μm Gyp1–46 was added and equilibrated for an additional 10 min, followed by the addition of 2 μm GEF. A360 nm was then measured over time at 25–28 °C in a PerkinElmer Life Sciences Victor3 plate reader.
Trafficking Assays
Quantification of carboxypeptidase Y (CPY) secretion was performed using a colorimetric assay as described (24). LUCID assays were performed using a dual luciferase assay system (Promega) as described previously (3). Briefly, the cells were grown overnight at 30 °C in synthetic dropout medium containing 2% dextrose and supplemented with 0.05% (w/v) casamino acids, then diluted, and grown in same medium until log phase. After a 20-min cycloheximide chase (final concentration, 50 μg/ml), ∼0.5 OD600 nm × ml log phase cells were collected by low speed centrifugation, resuspended in 500 μl of lysis buffer, and lysed by vortexing with a slurry of glass beads at room temperature for 15 min. 5-μl aliquots of lysate were analyzed in opaque 96-well plates using a PerkinElmer Life Sciences Victor Light Model 1420 luminometer. FLuc fusions and RLuc were expressed from a single plasmid. Signal from each cargo-FLuc fusion was normalized versus signal from soluble Rluc expressed from the constitutive PGK1 promoter.
LUCID time course monitoring of Mup1-FLuc down-regulation employed cells incubated as described for microscopy experiments (25). Briefly, the cells were grown overnight in synthetic medium (2% glucose) lacking methionine to maximize Mup1 retention at the plasma membrane. Note that media for LUCID assays of Mup1 cannot include casamino acids. A cycloheximide chase was initiated 15 min before harvest and luminometric analysis as described above. The cells were diluted to approximately OD600 nm = ∼0.15 and shaken at 30 °C in low Met medium for 3 h. 1-ml aliquots were then transferred at intervals to culture tubes containing Met at a final concentration of 1 mm. Statistical analyses were performed using GraphPad Prism 4.0.
RESULTS
Growth Phenotypes of Muk1 Deletion and Overexpression in Cells Lacking Vps9
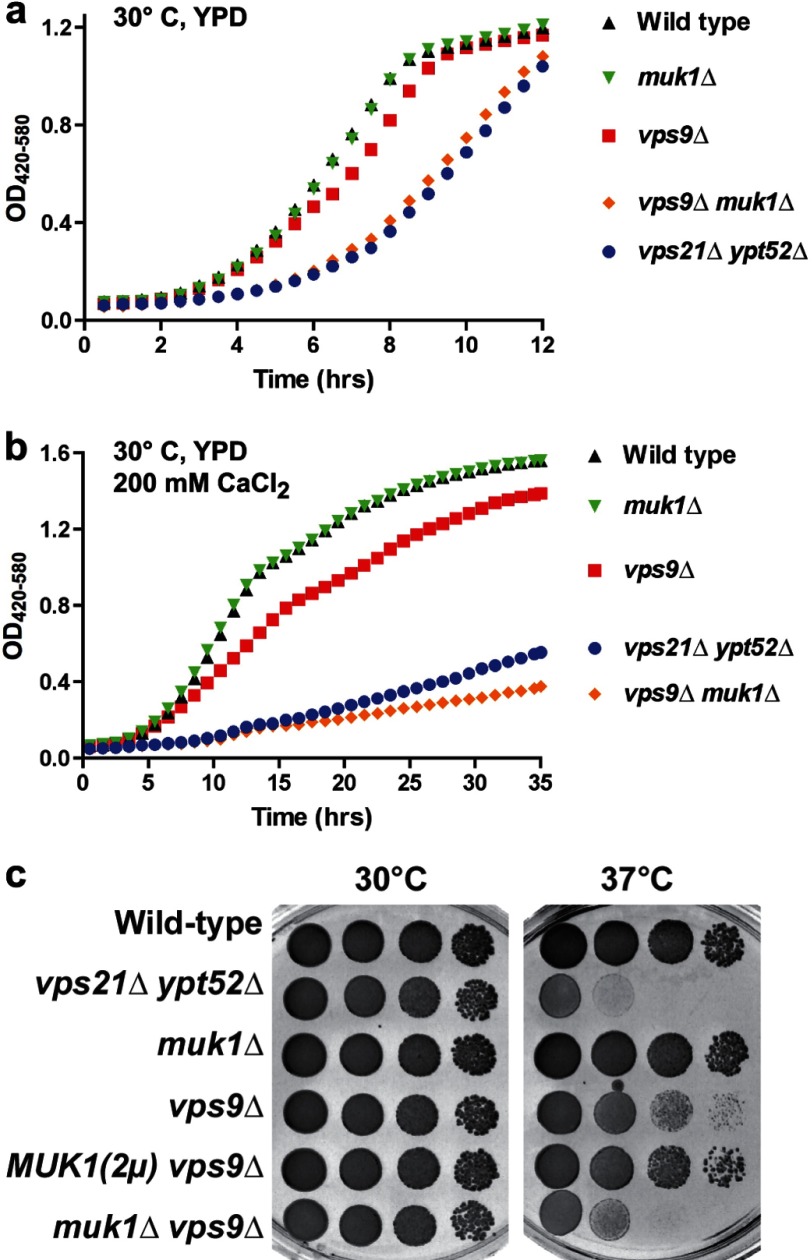
The three Rab5 paralogs of budding yeast (Vps21, Ypt52, and Ypt53) are partially redundant, with single mutants of Vps21 displaying clear sorting and morphology phenotypes that are further enhanced by deletion of Ypt52, Ypt53, or both (3, 4, 6). At 30 °C vps21Δ ypt52Δ double mutants had delayed entry into log phase (Fig. 1a) and had severe growth defects when subjected to ionic or thermal stress (Fig. 1, b and c, and Table 2). Almost identical growth defects were observed with vps21Δ ypt52Δ ypt53Δ triple mutants (3, 6). In marked contrast, single mutants lacking Vps9 grew almost normally when grown on solid or liquid medium (Fig. 1). Together, these results suggested that residual Rab5 signaling occurs in the absence of Vps9.
FIGURE 1.
A dual GEF deletion mutant phenocopies severe Rab5 deficiency. a and b, growth curves at 30 °C in YPD liquid medium (a) and medium supplemented with 200 mm CaCl2 (b). Data points each represent the mean of eight replicate samples. c, limiting dilution growth assay on YPD agar plates at 30 and 37 °C. MUK1, under native promoter and terminator, was overexpressed on a high copy (2μ) plasmid.
TABLE 2.
Growth rates of yeasts defective in Rab5 signaling.
The values shown are the interval doubling times in min (mean ± S.D., n = 8 each).
| Strain | YPD | YPD + 200 mm CaCl2 |
|---|---|---|
| Wild-type | 88.2 ± 2.3 | 147.5 ± 1.8 |
| vps9Δ | 95.9 ± 2.7 | 220.4 ± 4.6 |
| muk1Δ | 85.3 ± 2.5 | 144.5 ± 2.4 |
| muk1Δ vps9Δ | 125.6 ± 9.4 | 1381.0 ± 251.3 |
| vps21Δ ypt52Δ | 138.2 ± 17.7 | 975.2 ± 199.0 |
Because Muk1 contains a VPS9 domain (32), we hypothesized that Muk1 drives the residual Rab5 signaling presumed to occur in vps9Δ deletion mutants. If this were the case, a vps9Δ muk1Δ double deletion should phenocopy the severe growth and trafficking defects of the vps21Δ ypt52Δ double mutant. As shown in Fig. 1, we observed nearly identical growth delays and defects in both Rab5 null (vps21Δ ypt52Δ) and GEF null (vps9Δ muk1Δ) strains. Moreover, expression of MUK1 from a high copy plasmid largely suppressed the growth defect of a vps9Δ single mutant at 37 °C (Fig. 1c). Hence, MUK1 and VPS9 are partially redundant in conferring cellular tolerance of thermal stress, and loss of both MUK1 and VPS9 phenocopies the growth defects of mutants mostly or completely lacking Rab5.
Muk1 Deletion Exacerbates Golgi-Endosome Traffic Defects in Cells Lacking Vps9
We next asked whether Muk1 and Vps9 cooperate in endolysosomal biogenesis and cargo trafficking. CPY, a soluble vacuolar hydrolase, traffics from the Golgi to the prevacuolar endosome by binding the CPY receptor Vps10, which cycles between these compartments. Perturbation of Vps10 traffic results in CPY secretion, which can be monitored with the chimeric reporter CPY invertase (24). Wild-type cells (Fig. 2) secrete little CPY invertase, vps21Δ or vps9Δ single mutants secrete similar, substantial amounts of CPY invertase, and vps21Δ ypt52Δ double mutants secrete roughly twice as much CPY invertase as either vps21Δ or vps9Δ single mutants (3). Like wild-type cells, muk1Δ single mutants exhibited no detectable CPY invertase mistargeting. In contrast, vps9Δ muk1Δ double mutants mistargeted CPY invertase more severely than vps9Δ single mutants and just as severely as vps21Δ ypt52Δ double mutants (Fig. 2a).
FIGURE 2.
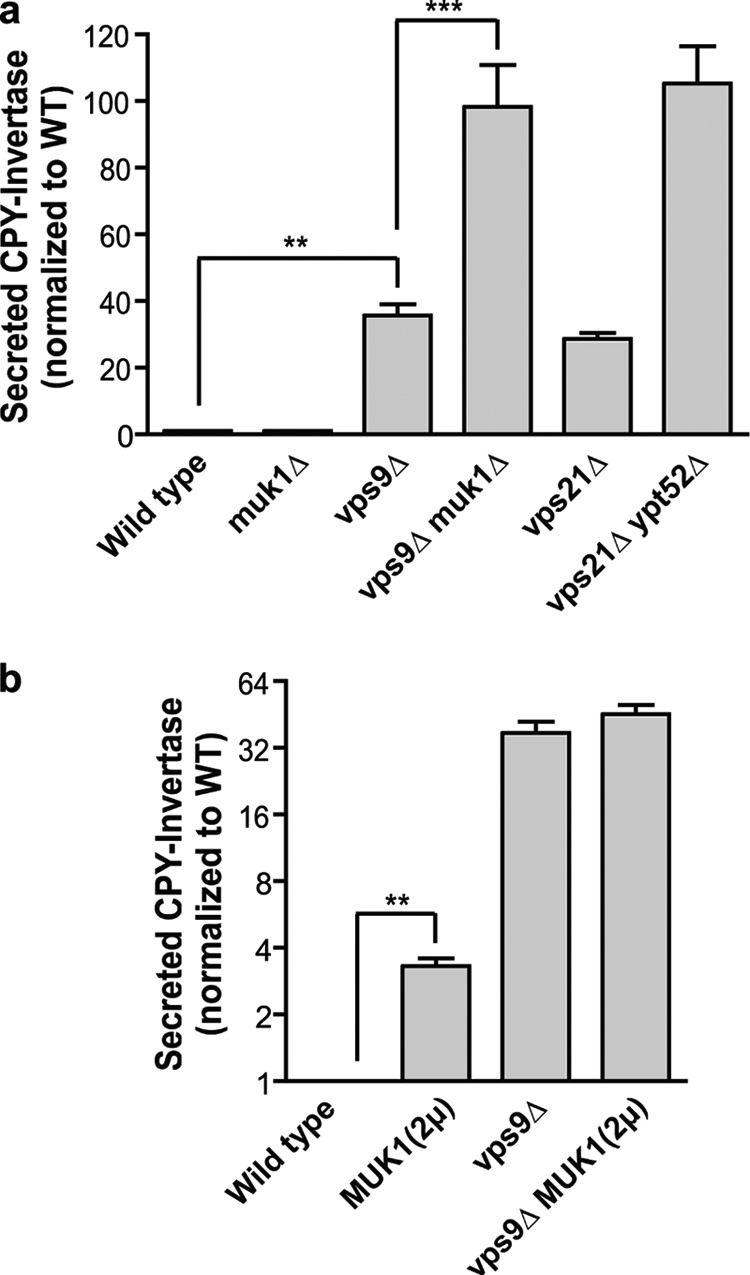
Redundant and nonredundant functions of Vps9 and Muk1 in CPY targeting. a and b, measurement of extracellular CPY invertase normalized to wild type. The bar graphs show the means ± S.E. of four independent experiments. In a, paired one-way analysis of variance: p < 0.0001 overall; ***, p < 0.001; **, p < 0.01. All pairwise comparisons are statistically significant, except wild type versus muk1Δ, vps9Δ versus vps21Δ, and vps21Δ ypt52Δ versus vps9Δ muk1Δ. In b, **, p = 0.0034, paired, two-tailed t test.
Unlike the cell growth assays (Fig. 1c), Muk1 overproduction did not suppress CPY mistargeting in a vps9Δvps21Δ background (Fig. 2b). Moreover, Muk1 overproduction in wild-type cells caused a mild but statistically significant CPY mistargeting phenotype (Fig. 2b). Taken together, these experiments suggest that Vps9 and Muk1 have both overlapping and distinct functions within the Golgi-endosome network.
Muk1 Deletion Exacerbates Multivesicular Body Targeting Defects in Cells Lacking Vps9
We recently discovered an absolute requirement for Rab5 signaling in the biogenesis and sorting functions of late endosomal multivesicular bodies (MVBs). vps21Δ ypt52Δ double mutants totally lack organelles with MVB architecture and mistarget cargo that is normally packaged into intraluminal vesicles (ILVs) at the MVB (3). We predicted that if Muk1 operates in concert with Vps9 to activate Rab5, the rate and fidelity of MVB cargo transport should be impaired in vps9Δ muk1Δ double mutants and perhaps in muk1Δ single mutants as well. To test these predictions, we examined four representative transmembrane proteins that are targeted to ILVs: carboxypeptidase S (CPS) and Sna3, which traffic from the late Golgi to the MVB, and Mup1 and Ste3, which traffic from the plasma membrane to the MVB. As the following experiments show, all four MVB cargos are severely missorted in vps9Δ muk1Δ double mutant cells.
CPS targeting was monitored using a GFP-CPS fusion protein. Wild-type cells sort GFP-CPS into ILVs, which deliver GFP-CPS to the vacuole lumen when MVBs fuse with vacuoles. In cells with impaired MVB biogenesis or sorting, GFP-CPS is diverted onto the vacuole limiting membrane or may become trapped in prevacuolar Class E endosomal compartments. Class E compartments appear as large puncta adjacent to the vacuole and are regularly observed in vps4Δ mutants (3). Consistent with the CPY secretion results (Fig. 2), muk1Δ cells correctly targeted CPS to the vacuole lumen, whereas vps9Δ cells exhibited a partial CPS sorting defect (Fig. 3). In muk1Δ vps9Δ double mutants, CPS was mistargeted to the vacuolar limiting membrane and was also observed in prevacuolar endosomal compartments (Fig. 3).
FIGURE 3.
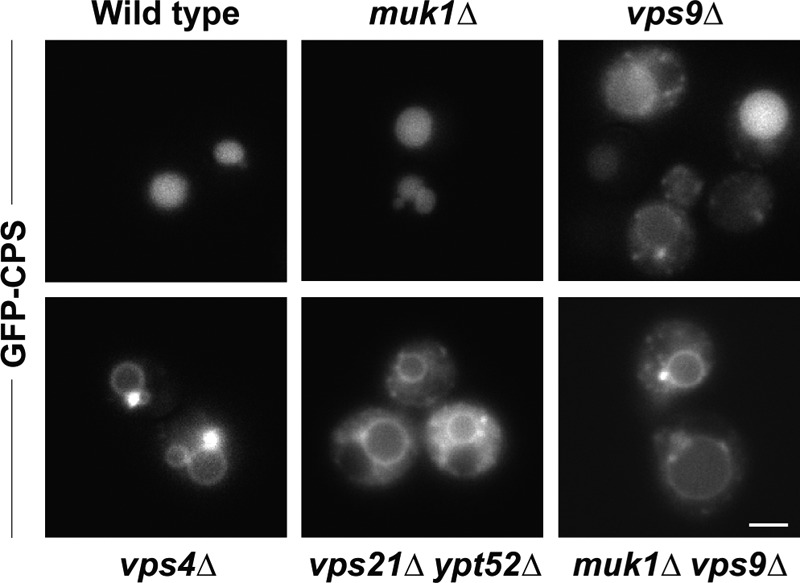
Biosynthetic traffic to the vacuole lumen in GEF null and Rab5-deficient mutants. Fluorescence microscopy of cells expressing GFP-CPS. Note that in contrast to the vps4Δ Class E phenotype of CPS mislocalized to the vacuole limiting membrane and concentrated at large puncta adjacent to the vacuole, cells lacking Rab5 or Rab5 GEF functions display CPS mislocalized to the limiting membrane and distributed throughout the cytoplasm. Scale bar, 2 μm.
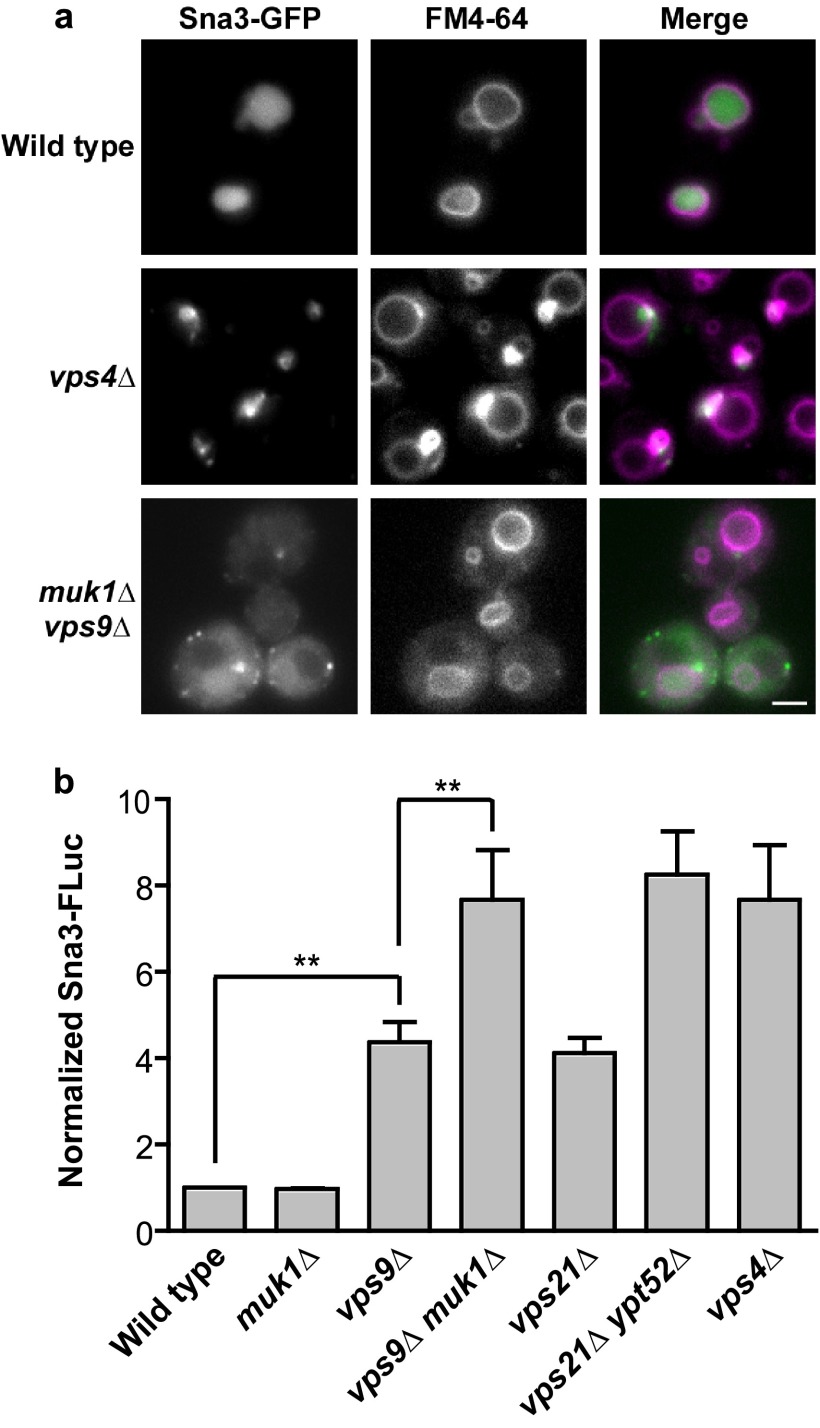
Like CPS, Sna3 traffics from Golgi to MVB and is then deposited into the vacuole lumen. The fidelity of Sna3 luminal targeting is robust compared with other ubiquitin-dependent MVB cargoes, and Sna3 sorting is sometimes used as a proxy for luminal vesicle formation (31, 33–35). We observed Sna3 targeting by both fluorescence microscopy (also employing a vital stain, FM4–64, that labels vacuole membranes and upstream endocytic compartments) and a quantitative assay called LUCID (luciferase reporter of intraluminal deposition), which monitors sequestration of cargo into MVBs (3). LUCID employs a soluble, control Renilla luciferase (RLuc) as well as an ILV cargo (in this case Sna3) fused to firefly luciferase (FLuc) at its cytosolic terminus. The ratio of FLuc to RLuc decreases as internalized Sna3-FLuc is sequestered into ILVs at the MVB (which are subsequently destroyed in the vacuole lumen) and away from its soluble substrate. We observed no discernible missorting of Sna3 in muk1Δ mutant cells, whereas vps9Δ mutants had an intermediate missorting phenotype somewhat more severe than a vps21Δ mutant. The double GEF mutant (vps9Δ muk1Δ) strongly missorted Sna3, on par with the double Rab5 mutant (vps21Δ ypt52Δ).
Sna3 is also strongly missorted in vps4Δ mutant cells. Like Rab5-deficient cells (3), vps9Δ muk1Δ double mutant cells accumulated Sna3 in puncta dispersed throughout the cytoplasm rather than in a Class E prevacuolar compartment (Fig. 4). Similarly, in several previous studies (33, 35–37) mistargeted Sna3 did not accumulate at the vacuolar limiting membrane, as was the case with mistargeted CPS (Fig. 3). In contrast, when multiple Sna3 sorting motifs are simultaneously disrupted, including multiple cytosolic lysines and tyrosines, Sna3 does accumulate at the vacuole limiting membrane (37), which might indicate that mistargeted Sna3 can be retrieved from the vacuole. Our findings suggest that vps9Δ muk1Δ double mutants have a defect distinct, and probably upstream, of the Class E compartment formed in cells that lack Vps4 or other proteins needed for ILV biogenesis (3).
FIGURE 4.
Biosynthetic traffic to the vacuole lumen in GEF null and Rab5-deficient mutants. a, fluorescence microscopy of cells expressing Sna3-GFP (green) and stained with the endocytic tracer dye FM4–64 (purple) that stains vacuole membranes. Note that although vps4Δ Class E mutants concentrate Sna3 at FM 4–64-stained perivacuolar puncta, muk1Δ vps9Δ cells mislocalize Sna3 at puncta throughout the cytoplasm. Scale bar, 2 μm. b, bar graphs show means ± S.E. of five independent LUCID assays of Sna3-FLuc. Paired one-way analysis of variance: p < 0.0001 overall; **, p < 0.01. All pairwise comparisons are statistically significant, except wild type versus muk1Δ, vps9Δ versus vps21Δ, vps21Δ ypt52Δ versus vps9Δ muk1Δ, vps4Δ versus vps21Δ ypt52Δ, and vps4Δ versus vps9Δ muk1Δ.
Mup1, a high affinity Met transporter, localizes to the plasma membrane in Met-limited medium. At higher levels of extracellular Met, Mup1 is rapidly endocytosed and trafficked through the MVB en route to the vacuole lumen (25, 33). As shown in Fig. 5a, Mup1 localized to the plasma membrane in both vps9Δ and muk1Δ single mutants, with a small fraction accumulating in puncta. As with wild-type cells, Met exposure resulted in rapid and complete Mup1 internalization in a muk1Δ single mutant, whereas a vps9Δ mutant displayed residual Mup1 at the plasma membrane. Double mutant vps9Δ muk1Δ cells correctly targeted Mup1 to the plasma membrane and were competent to internalize Mup1 when exposed to Met. However, subsequent Mup1 trafficking through the endolysosomal system was severely disrupted in the double mutant, with Mup1 trapped in both intracellular punctate structures and on the vacuolar limiting membrane. There was no detectable Mup1 in the vacuole lumen (Fig. 5a).
FIGURE 5.
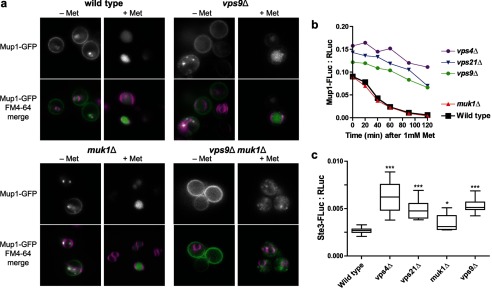
Endocytic traffic to the vacuole lumen in GEF null and Rab5-deficient mutants. a, Mup1-GFP (green) was monitored by fluorescence microscopy in cells labeled with FM4–64 (purple) both in the absence of methionine (− Met) and 1 h after the addition of 1 mm methionine (+ Met) to stimulate internalization of Mup1-GFP from the plasma membrane and transport to the vacuole lumen. b, time course LUCID analysis of Mup1-FLuc down-regulation after addition of 1 mm methionine. Points represent means of two duplicate samples from a representative experiment. c, LUCID analysis of Ste3-FLuc. The box plot summarizes 10 biological replicates pooled across three independent experiments. One-way analysis of variance: p < 0.0001 overall; ***, p < 0.001; *, p < 0.05 compared with wild type. b and c, Mup1-FLuc and Ste3-FLuc were each normalized to a cytosolic RLuc loading control expressed from the same plasmid.
To quantify the kinetics of Mup1 delivery into MVBs, we used a Mup1-LUCID reporter. We observed normal Mup1 trafficking in wild-type and muk1Δ single mutant cells. In contrast, vps9Δ, vps21Δ, or vps4Δ mutants had both elevated steady-state levels of Mup1 in the absence of Met, and severe defects in Mup1 targeting to the MVB when Met was added to the medium (Fig. 5b). Double mutant vps21Δ ypt52Δ and vps9Δ muk1Δ cells were not analyzed in LUCID experiments because of their extremely slow growth in Met-limited synthetic medium.
Ste3 is a G protein-coupled receptor at the plasma membrane where it binds yeast a-factor to initiate the mating response. In the absence of mating factor Ste3 is constitutively internalized, packaged into ILVs at the MVB, and routed to the vacuole for degradation (38). Cells without Vps4 entirely lack MVB intraluminal vesicles (30, 39). Using Ste3-LUCID, vps4Δ mutants accumulated substantially more Ste3 than wild-type cells, whereas vps9Δ and vps21Δ mutants had somewhat less severe Ste3 accumulation. Cells lacking Muk1 had a small but statistically significant accumulation of Ste3 relative to wild-type cells (Fig. 5c). These results indicate that endocytic traffic to the vacuole generally tolerates loss of Muk1 without strong defects but that Muk1 may contribute to efficient endocytic down-regulation or recycling of Ste3.
Muk1 Is a GEF for Rab5 Paralogs
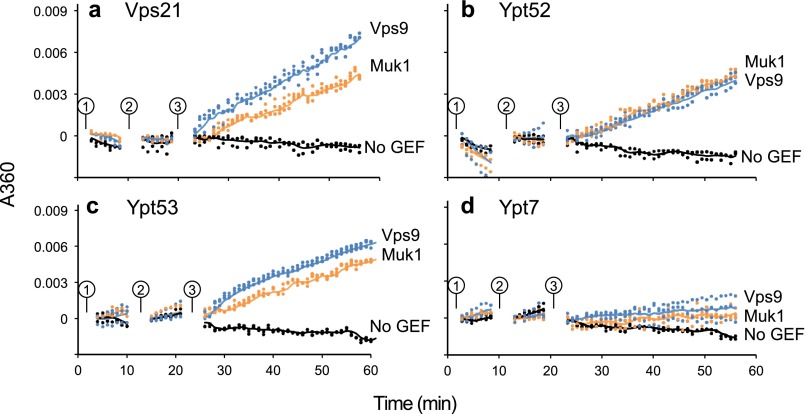
We next assayed the GEF activities of purified recombinant Vps9 and Muk1 for various endolysosomal Rabs. In the presence of Gyp1TBC, a hydrolysis-accelerating GAP, Vps21-GTP, and several other yeast Rabs undergo a single turnover of GTP hydrolysis (3, 23, 40, 41). In the presence of GTP and a GEF such as Vps9, GDP is exchanged, and additional cycles of GTP hydrolysis can then occur (23). In this reaction system, we measure GTP hydrolysis through evolution of inorganic phosphate, using a real time spectrophotometric assay based on purine nucleoside phosphorylase and 2-amino-6-mercapto-7-methyl-purine riboside, a small molecule reporter that changes its UV absorbance when phosphorylated by purine nucleoside phosphorylase (41, 42).
As expected (Fig. 6), Vps9 stimulated multiple cycles of GTP hydrolysis by Vps21, Ypt52, and Ypt53, but not on Ypt7, the vacuolar Rab. Muk1 exhibited a pattern of GEF activity similar to Vps9, stimulating multiple cycle GTP hydrolysis on Vps21, Ypt52, and Yp53, but not on Ypt7. In combination with the genetic experiments described above, these experiments establish Muk1 as an authentic GEF with selectivity for Rab5 paralogs.
FIGURE 6.
GEF activities of purified Vps9 and Muk1. Coupled GEF-GAP phosphate release assay with Vps9 (blue), Muk1 (orange), or no GEF (black) against Vps21 (a), Ypt52 (b), Ypt53 (c), and Ypt7 (d). Individual points show data from triplicate samples in a representative experiment. The lines show smoothed means. The reactions were monitored following Rab and reaction buffer for 10 min (circle 1), addition of GAP for 10 min (circle 2), and addition of GEF for 1 h (circle 3).
Muk1 and Vps9 Localization
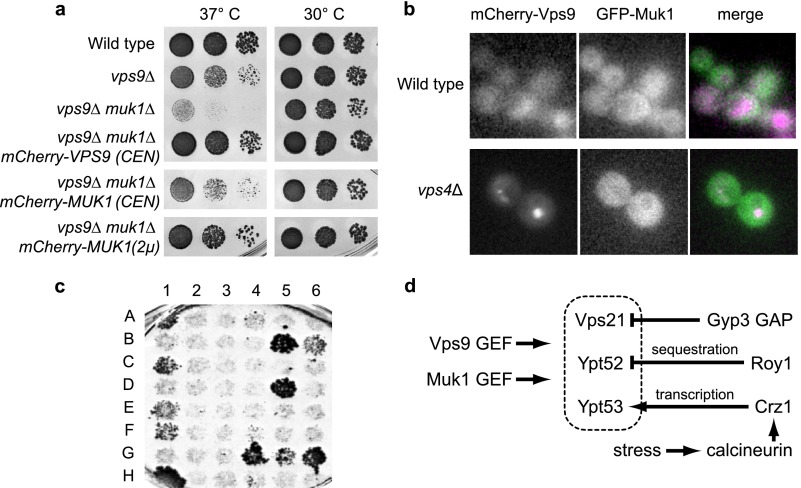
To compare the localization of Vps9 and Muk1, we expressed GFP-Muk1 and mCherry-Vps9 fusion proteins. The tagged proteins were functional, as evidenced by their ability to rescue the heat stress sensitivity phenotype of muk1Δ vps9Δ double mutant cells (Fig. 7a). When observed in live cells, Vps9 is predominantly cytosolic with no discernable localization to specific subcellular structures. However, in a vps4Δ mutant, mCherry-Vps9 localizes to the prevacuolar Class E compartment, which is also highly enriched with Vps21 (36). We therefore compared the localization of tagged Vps9 and Muk1 in wild-type and vps4Δ mutant cells. Both GFP-Muk1 and mCherry-Vps9 were predominantly cytosolic in wild-type cells, but only mCherry-Vps9 redistributed to Class E compartment punctae in a vps4Δ mutant (Fig. 7b).
FIGURE 7.
Muk1 and Vps9 localization, Muk1 interactions, and summary of Rab5 regulation in yeast. a, chimeric Muk1 and Vps9 constructs rescue heat sensitivity phenotypes of muk1Δ vps9Δ cells. Limiting dilution growth assay on YPD agar plates at 30 and 37 °C. Plasmid-borne chimeras were expressed using low copy (CEN) or high copy (2μ) plasmids. b, GFP-Muk1 (green) and mCherry-Vps9 (purple) were coexpressed using low copy plasmids in the indicated strains and imaged. Both GFP-Muk1 and mCherry-Vps9 appear cytosolic in wild-type cells. GFP-Muk1 remains cytosolic in vps4Δ, whereas mCherry-Vps9 accumulates at Class E compartments. c, Y2H interactions of Muk1. Muk1 was screened against potential interactors using a mating procedure as described under “Experimental Procedures.” The Y2H plates were scored for growth on 3 mm 3-amino-1,2,4-triazole after 5 days at 30 °C. The interactors and plate coordinates are summarized in Table 3. d, working model for regulation of signaling by the yeast Rab5 paralogs. See “Discussion” for details.
To explore possible binding partners and help determine where Muk1 may be functioning, we assayed Muk1 against a host of endocytic and other trafficking proteins using yeast two-hybrid tests (Fig. 7c and Table 3). As expected, Muk1 interacted with Vps21. Interactions with Ypt52 and Ypt53 were not detected; however, our conditions for two-hybrid (single-copy vectors and 3-amino-1,2,4-triazole on the test plates) are relatively stringent. We detected two-hybrid interactions with multiple trafficking proteins including Ypt6, a Rab that controls Golgi endosome transport, Exo84, a subunit of the plasma membrane Exocyst tethering complex, and weak interactions with the vacuolar Rab Ypt7 and its GEF Mon1 (43). Notably, all of these candidate interactors reside within the peripheral zones that delineate the boundaries of the Rab5 signaling domain: the plasma membrane, the vacuole, and the late Golgi. In addition, although Vps9 and mammalian Rabex-5 both comprise ubiquitin-binding domains, Muk1 lacks motifs implicated in ubiquitin binding. We conclude that although Muk1 and Vps9 are both able to catalyze nucleotide exchange with all three yeast Rab5 proteins, the two yeast Rab5 GEFs possess distinct localization determinants and probably operate at different subcellular locations.
TABLE 3.
Muk1 Y2H results
Y2H bait plasmid Muk1 was mated with a Y2H prey library; representative data are shown in Fig. 7C. The plates were evaluated after 5 days at 30 °C on synthetic medium containing 3 mm 3-amino-1,2,4-triazole and Ade and lacking Leu and Trp.
| Score | Location | Protein | |
|---|---|---|---|
| + | A | 1 | Ypt7 |
| A | 2 | Vps39 | |
| A | 3 | Vps8 | |
| A | 4 | Vps19 | |
| A | 5 | Vps11 | |
| A | 6 | Vps16 | |
| B | 1 | Vps18 | |
| B | 2 | Vps41 | |
| B | 3 | Vps45 | |
| B | 4 | Vps33 | |
| ++ | B | 5 | Vps21 |
| + | B | 6 | Vps21 S21N |
| + | C | 1 | Vps21 Q66L |
| C | 2 | Pep12 | |
| C | 3 | Gyp7 | |
| C | 4 | Gyp1 | |
| C | 5 | Ypt1 | |
| C | 6 | Sna3 | |
| D | 1 | Ccz1 | |
| D | 2 | Arf1 | |
| D | 3 | Arf2 | |
| D | 4 | Sec1 | |
| ++ | D | 5 | Mon1 |
| D | 6 | Yif1 | |
| + | E | 1 | Yip1 |
| E | 2 | Yip2 | |
| E | 3 | Yip3 | |
| E | 4 | Yip4 | |
| E | 5 | Yip5 | |
| E | 6 | Sec4 | |
| + | F | 1 | Ypt6 |
| F | 2 | Ypt10 | |
| F | 3 | Ypt11 | |
| F | 4 | Ypt31 | |
| F | 5 | Ypt32 | |
| F | 6 | Ypt53 | |
| G | 1 | Vps9 | |
| G | 2 | Ubi4 | |
| G | 3 | Ypt52 | |
| ++ | G | 4 | Ady3 |
| ++ | G | 5 | Bzz1 |
| ++ | G | 6 | Exo84 |
| ++ | H | 1 | Gcd7 |
| H | 2 | Vps35 | |
| H | 3 | Vps5 | |
| H | 4 | Muk1 | |
| H | 5 | YML002W | |
| H | 6 | Control | |
DISCUSSION
VPS9 has been identified in several protein sorting screens, but MUK1 was not identified in these screens (26, 44, 45). Although high-throughput protein interaction screens and our yeast two-hybrid experiments suggest physical interactions between Muk1 and several other trafficking proteins (46–48), the role of Muk1 in the endocytic network has not been examined until very recently. Deletion of Vps9 causes endolysosomal protein sorting and morphology defects (49). These phenotypes were much less severe than combined deletion of the three Rab5 paralogs on which Vps9 acts, suggesting the presence of an additional Rab5 GEF. A combination of protein trafficking assays and in vitro biochemistry now establish Muk1 as the second Rab5 GEF. While the present report was in preparation, Cabrera et al. (2) reported parallel experiments independently showing that Muk1 has a Rab5 GEF activity.
Only one phenotypic defect arising from Muk1 deletion has so far been detected, a small but reproducible increase in the level of Ste3 in LUCID experiments (Fig. 5c). This may reflect increased endocytic recycling of Ste3 to the plasma membrane, inefficient Ste3 transport into the vacuole lumen through the MVB pathway, or both. The traffic of other cargo molecules (CPY, CPS, Sna3, and Mup1) was not detectably impaired in muk1Δ single mutants. In contrast, muk1Δ vps9Δ double mutants had synthetic growth and trafficking defects stronger than either single mutant and similar to those of double and triple Rab5 knock-out cells. Cabrera et al. (2) also observed that muk1Δ mutants had no overt phenotypes but that cells lacking both Vps9 and Muk1 showed prominent drug sensitivity and failed to properly localize subunits of the endosomal tethering and fusion machinery. Together these results indicate functional redundancy of Muk1 with Vps9 and help explain why Muk1 eluded genetic identification. Overexpression of Muk1 increased CPY secretion in wild-type cells (Fig. 2b), demonstrating that normal Golgi endosome cycling of the CPY receptor Vps10 is perturbed by high levels of Muk1.
As suggested by the presence of a VPS9 domain, Muk1 has nucleotide exchange activity against the three endocytic Rab5 paralogs (Vps21, Ypt52, and Ypt53) but is unable to catalyze exchange with Ypt7, the vacuolar Rab. Cabrera et al. (2) similarly reported that Muk1 functions as a Rab GEF against Vps21 and Ypt52. A minor point of divergence is our finding that Vps9 and Muk1 stimulate comparable levels of nucleotide exchange on Ypt53. We note that different biochemical assays were employed in these studies. We assayed multiple turnover hydrolysis of unmodified GTP, whereas Cabrera et al. monitored ejection of a Rab-bound fluorescent GDP analog.
Vps9 and Muk1 are mainly cytosolic in wild-type cells. However, in vps4Δ cells, Vps9 becomes trapped at Class E compartments, whereas Muk1 does not, demonstrating that Muk1 and Vps9 have distinct localization determinants and underscoring the possible functional specialization of these GEFs (Fig. 7a).
GEF localization is a critical determinant of Rab localization and activity (50). Mammalian Rabex-5, a VPS9-type GEF, is targeted to membranes by its effector, Rabaptin-5, which in turn interacts with proteins at Golgi, early, and late endosomal membranes (51–55). In contrast Rin1, which also has a VPS9 domain, specifically localizes to internalized EGF receptor, which may ensure priority transit for receptor down-regulation (56). Yeast Vps9 binds monoubiquitylated cargo, which is hypothesized to localize Vps9 within the yeast endosomal network (57). It is becoming clear that VPS9 GEF family members share a conserved catalytic domain but that localization of individual GEFs to Rab5-positive membranes occurs through distinct modes of spatial and temporal regulation. Further investigation into these varied mechanisms of GEF localization will clarify how diverse Rab5-dependent trafficking pathways are regulated both in yeast and higher eukaryotes.
In the GEF cascade model proposed by Novick and co-workers (58), GEF localization is the key contributor to sequential Rab activation during compartmental maturation. In the simplest form of this model, activated Rab “A” recruits a downstream GEF for Rab “B” as the membrane matures. Because Vps9 and Muk1 have distinct localization determinants but similar substrate specificities, it will be important to elucidate the normal order and locations of action of these GEFs. Yeast two-hybrid interactions between Muk1 and both Ypt6 and Ypt7 (Table 3) are consistent with the cascade model.
Several studies have shown that even in the relatively simple yeast system, Rab5 signaling is subject to intricate regulation (Fig. 7b). The three yeast Rab5 isoforms share overlapping capacities to support endolysosomal biogenesis and cargo transport (2–6), differentially control effector recruitment (2), and differ in their intrinsic ability to mediate vesicle tethering (23). Ypt52 is typically held in an inactive state by Roy1 (repressor of ypt52) (4), and Ypt53 expression is induced upon cellular stress as part of the Crz1-calcineurin regulon (3). Moreover, Vps21 signaling is spatially and temporally restricted GAP Gyp3/Msb3 (3, 59). It will be important to understand how these interactions govern specific cellular processes including stress responses (3), organelle inheritance (60), and metabolic regulation (61).
Acknowledgments
We thank R. Plemel for essential technical assistance, M. Kaeberlein for access to the BioScreen machine, and V. MacKay and members of the Merz lab for critical comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM077349.
- GEF
- guanine nucleotide exchange factor
- GAP
- GTPase-accelerating protein
- MVB
- multivesicular body
- Y2H
- yeast two-hybrid
- CPY
- carboxypeptidase Y
- ILV
- intraluminal vesicle
- CPS
- carboxypeptidase S
- FLuc
- firefly luciferase
- RLuc
- Renilla luciferase
- LUCID
- luciferase reporter of intraluminal deposition.
REFERENCES
- 1. Barr F., Lambright D. G. (2010) Rab GEFs and GAPs. Curr. Opin. Cell Biol. 22, 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cabrera M., Arlt H., Epp N., Lachmann J., Griffith J., Perz A., Reggiori F., Ungermann C. (2013) Functional separation of endosomal fusion factors and the class C core vacuole/endosome tethering (CORVET) complex in endosome biogenesis. J. Biol. Chem. 288, 5166–5175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nickerson D. P., Russell M. R., Lo S. Y., Chapin H. C., Milnes J. M., Merz A. J. (2012) Termination of isoform-selective Vps21/Rab5 signaling at endolysosomal organelles by Msb3/Gyp3. Traffic 13, 1411–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu Y., Nakatsukasa K., Kotera M., Kanada A., Nishimura T., Kishi T., Mimura S., Kamura T. (2011) Non-SCF-type F-box protein Roy1/Ymr258c interacts with a Rab5-like GTPase Ypt52 and inhibits Ypt52 function. Mol. Biol. Cell 22, 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Singer-Krüger B., Stenmark H., Zerial M. (1995) Yeast Ypt51p and mammalian Rab5. Counterparts with similar function in the early endocytic pathway. J. Cell Sci. 108, 3509–3521 [DOI] [PubMed] [Google Scholar]
- 6. Singer-Krüger B., Stenmark H., Düsterhöft A., Philippsen P., Yoo J. S., Gallwitz D., Zerial M. (1994) Role of three rab5-like GTPases, Ypt51p, Ypt52p, and Ypt53p, in the endocytic and vacuolar protein sorting pathways of yeast. J. Cell Biol. 125, 283–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sivars U., Aivazian D., Pfeffer S. R. (2003) Yip3 catalyses the dissociation of endosomal Rab-GDI complexes. Nature 425, 856–859 [DOI] [PubMed] [Google Scholar]
- 8. Abdul-Ghani M., Gougeon P. Y., Prosser D. C., Da-Silva L. F., Ngsee J. K. (2001) PRA isoforms are targeted to distinct membrane compartments. J. Biol. Chem. 276, 6225–6233 [DOI] [PubMed] [Google Scholar]
- 9. Collins R. N. (2003) “Getting it on.” GDI displacement and small GTPase membrane recruitment. Mol. Cell 12, 1064–1066 [DOI] [PubMed] [Google Scholar]
- 10. Barrowman J., Novick P. (2003) Three Yips for Rab recruitment. Nat. Cell Biol. 5, 955–956 [DOI] [PubMed] [Google Scholar]
- 11. Cherfils J., Chardin P. (1999) GEFs. Structural basis for their activation of small GTP-binding proteins. Trends Biochem. Sci 24, 306–311 [DOI] [PubMed] [Google Scholar]
- 12. Yoshimura S., Gerondopoulos A., Linford A., Rigden D. J., Barr F. A. (2010) Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J. Cell Biol. 191, 367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hama H., Tall G. G., Horazdovsky B. F. (1999) Vps9p is a guanine nucleotide exchange factor involved in vesicle-mediated vacuolar protein transport. J. Biol. Chem. 274, 15284–15291 [DOI] [PubMed] [Google Scholar]
- 14. Horiuchi H., Lippé R., McBride H. M., Rubino M., Woodman P., Stenmark H., Rybin V., Wilm M., Ashman K., Mann M., Zerial M. (1997) A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell 90, 1149–1159 [DOI] [PubMed] [Google Scholar]
- 15. Tall G. G., Barbieri M. A., Stahl P. D., Horazdovsky B. F. (2001) Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev. Cell 1, 73–82 [DOI] [PubMed] [Google Scholar]
- 16. Saito K., Murai J., Kajiho H., Kontani K., Kurosu H., Katada T. (2002) A novel binding protein composed of homophilic tetramer exhibits unique properties for the small GTPase Rab5. J. Biol. Chem. 277, 3412–3418 [DOI] [PubMed] [Google Scholar]
- 17. Kajiho H., Saito K., Tsujita K., Kontani K., Araki Y., Kurosu H., Katada T. (2003) RIN3. A novel Rab5 GEF interacting with amphiphysin II involved in the early endocytic pathway. J. Cell Sci. 116, 4159–4168 [DOI] [PubMed] [Google Scholar]
- 18. Samanta M. P., Liang S. (2003) Predicting protein functions from redundancies in large-scale protein interaction networks. Proc. Natl. Acad. Sci. U.S.A. 100, 12579–12583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gueldener U., Heinisch J., Koehler G. J., Voss D., Hegemann J. H. (2002) A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 30, e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brett C. L., Plemel R. L., Lobinger B. T., Vignali M., Fields S., Merz A. J. (2008) Efficient termination of vacuolar Rab GTPase signaling requires coordinated action by a GAP and a protein kinase. J. Cell Biol. 182, 1141–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Plemel R. L., Lobingier B. T., Brett C. L., Angers C. G., Nickerson D. P., Paulsel A., Sprague D., Merz A. J. (2011) Subunit organization and Rab interactions of Vps-C protein complexes that control endolysosomal membrane traffic. Mol. Biol. Cell 22, 1353–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Olsen B., Murakami C. J., Kaeberlein M. (2010) YODA. Software to facilitate high-throughput analysis of chronological life span, growth rate, and survival in budding yeast. BMC Bioinformatics 11, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lo S. Y., Brett C. L., Plemel R. L., Vignali M., Fields S., Gonen T., Merz A. J. (2012) Intrinsic tethering activity of endosomal Rab proteins. Nat. Struct. Mol. Biol. 19, 40–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Darsow T., Odorizzi G., Emr S. D. (2000) Invertase fusion proteins for analysis of protein trafficking in yeast. Methods Enzymol. 327, 95–106 [DOI] [PubMed] [Google Scholar]
- 25. Menant A., Barbey R., Thomas D. (2006) Substrate-mediated remodeling of methionine transport by multiple ubiquitin-dependent mechanisms in yeast cells. EMBO J. 25, 4436–4447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. (1988) Protein sorting in Saccharomyces cerevisiae. Isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8, 4936–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Horazdovsky B. F., Busch G. R., Emr S. D. (1994) VPS21 encodes a rab5-like GTP-binding protein that is required for the sorting of yeast vacuolar proteins. EMBO J. 13, 1297–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Babst M., Sato T. K., Banta L. M., Emr S. D. (1997) Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16, 1820–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sikorski R. S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Odorizzi G., Babst M., Emr S. D. (1998) Fab1p PtdIns3P 5-kinase function essential for protein sorting in the multivesicular body. Cell 95, 847–858 [DOI] [PubMed] [Google Scholar]
- 31. Reggiori F., Pelham H. R. (2001) Sorting of proteins into multivesicular bodies. Ubiquitin-dependent and -independent targeting. EMBO J. 20, 5176–5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carney D. S., Davies B. A., Horazdovsky B. F. (2006) Vps9 domain-containing proteins. Activators of Rab5 GTPases from yeast to neurons. Trends Cell Biol. 16, 27–35 [DOI] [PubMed] [Google Scholar]
- 33. MacDonald C., Stringer D. K., Piper R. C. (2012) Sna3 is an Rsp5 adaptor protein that relies on ubiquitination for its MVB sorting. Traffic 13, 586–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stawiecka-Mirota M., Pokrzywa W., Morvan J., Zoladek T., Haguenauer-Tsapis R., Urban-Grimal D., Morsomme P. (2007) Targeting of Sna3p to the endosomal pathway depends on its interaction with Rsp5p and multivesicular body sorting on its ubiquitylation. Traffic 8, 1280–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McNatt M. W., McKittrick I., West M., Odorizzi G. (2007) Direct binding to Rsp5 mediates ubiquitin-independent sorting of Sna3 via the multivesicular body pathway. Mol. Biol. Cell 18, 697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Russell M. R., Shideler T., Nickerson D. P., West M., Odorizzi G. (2012) Class E compartments form in response to ESCRT dysfunction in yeast due to hyperactivity of the Vps21 Rab GTPase. J. Cell Sci. 125, 5208–5220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Oestreich A. J., Aboian M., Lee J., Azmi I., Payne J., Issaka R., Davies B. A., Katzmann D. J. (2007) Characterization of multiple multivesicular body sorting determinants within Sna3. A role for the ubiquitin ligase Rsp5. Mol. Biol. Cell 18, 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen L., Davis N. G. (2000) Recycling of the yeast a-factor receptor. J. Cell Biol. 151, 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nickerson D. P., West M., Odorizzi G. (2006) Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J. Cell Biol. 175, 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Albert S., Gallwitz D. (2000) Msb4p, a protein involved in Cdc42p-dependent organization of the actin cytoskeleton, is a Ypt/Rab-specific GAP. Biol. Chem. 381, 453–456 [DOI] [PubMed] [Google Scholar]
- 41. Pan X., Eathiraj S., Munson M., Lambright D. G. (2006) TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature 442, 303–306 [DOI] [PubMed] [Google Scholar]
- 42. Webb M. R. (1992) A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc. Natl. Acad. Sci. U.S.A. 89, 4884–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nordmann M., Cabrera M., Perz A., Bröcker C., Ostrowicz C., Engelbrecht-Vandré S., Ungermann C. (2010) The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr. Biol. 20, 1654–1659 [DOI] [PubMed] [Google Scholar]
- 44. Banta L. M., Robinson J. S., Klionsky D. J., Emr S. D. (1988) Organelle assembly in yeast. Characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J. Cell Biol. 107, 1369–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Raymond C. K., Howald-Stevenson I., Vater C. A., Stevens T. H. (1992) Morphological classification of the yeast vacuolar protein sorting mutants. Evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 3, 1389–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Uetz P., Giot L., Cagney G., Mansfield T. A., Judson R. S., Knight J. R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Qureshi-Emili A., Li Y., Godwin B., Conover D., Kalbfleisch T., Vijayadamodar G., Yang M., Johnston M., Fields S., Rothberg J. M. (2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627 [DOI] [PubMed] [Google Scholar]
- 47. Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. U.S.A. 98, 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yu H., Braun P., Yildirim M. A., Lemmens I., Venkatesan K., Sahalie J., Hirozane-Kishikawa T., Gebreab F., Li N., Simonis N., Hao T., Rual J. F., Dricot A., Vazquez A., Murray R. R., Simon C., Tardivo L., Tam S., Svrzikapa N., Fan C., de Smet A. S., Motyl A., Hudson M. E., Park J., Xin X., Cusick M. E., Moore T., Boone C., Snyder M., Roth F. P., Barabási A. L., Tavernier J., Hill D. E., Vidal M. (2008) High-quality binary protein interaction map of the yeast interactome network. Science 322, 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Burd C. G., Mustol P. A., Schu P. V., Emr S. D. (1996) A yeast protein related to a mammalian Ras-binding protein, Vps9p, is required for localization of vacuolar proteins. Mol. Cell. Biol. 16, 2369–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Blümer J., Rey J., Dehmelt L., Mazel T., Wu Y. W., Bastiaens P., Goody R. S., Itzen A. (2013) RabGEFs are a major determinant for specific Rab membrane targeting. J. Cell Biol. 200, 287–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hirst J., Lui W. W., Bright N. A., Totty N., Seaman M. N., Robinson M. S. (2000) A family of proteins with γ-adaptin and VHS domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome. J. Cell Biol. 149, 67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mattera R., Arighi C. N., Lodge R., Zerial M., Bonifacino J. S. (2003) Divalent interaction of the GGAs with the Rabaptin-5-Rabex-5 complex. EMBO J. 22, 78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shiba Y., Takatsu H., Shin H. W., Nakayama K. (2002) γ-Adaptin interacts directly with Rabaptin-5 through its ear domain. J. Biochem. 131, 327–336 [DOI] [PubMed] [Google Scholar]
- 54. Zhu Y., Doray B., Poussu A., Lehto V. P., Kornfeld S. (2001) Binding of GGA2 to the lysosomal enzyme sorting motif of the mannose 6-phosphate receptor. Science 292, 1716–1718 [DOI] [PubMed] [Google Scholar]
- 55. McBride H. M., Rybin V., Murphy C., Giner A., Teasdale R., Zerial M. (1999) Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell 98, 377–386 [DOI] [PubMed] [Google Scholar]
- 56. Barbieri M. A., Kong C., Chen P. I., Horazdovsky B. F., Stahl P. D. (2003) The SRC homology 2 domain of Rin1 mediates its binding to the epidermal growth factor receptor and regulates receptor endocytosis. J. Biol. Chem. 278, 32027–32036 [DOI] [PubMed] [Google Scholar]
- 57. Davies B. A., Topp J. D., Sfeir A. J., Katzmann D. J., Carney D. S., Tall G. G., Friedberg A. S., Deng L., Chen Z., Horazdovsky B. F. (2003) Vps9p CUE domain ubiquitin binding is required for efficient endocytic protein traffic. J. Biol. Chem. 278, 19826–19833 [DOI] [PubMed] [Google Scholar]
- 58. Mizuno-Yamasaki E., Medkova M., Coleman J., Novick P. (2010) Phosphatidylinositol 4-phosphate controls both membrane recruitment and a regulatory switch of the Rab GEF Sec2p. Dev. Cell 18, 828–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lachmann J., Barr F. A., Ungermann C. (2012) The Msb3/Gyp3 GAP controls the activity of the Rab GTPases Vps21 and Ypt7 at endosomes and vacuoles. Mol. Biol. Cell 23, 2516–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Weisman L. S. (2006) Organelles on the move. Insights from yeast vacuole inheritance. Nat. Rev. Mol. Cell Biol. 7, 243–252 [DOI] [PubMed] [Google Scholar]
- 61. Bridges D., Fisher K., Zolov S. N., Xiong T., Inoki K., Weisman L. S., Saltiel A. R. (2012) Rab5 proteins regulate activation and localization of target of rapamycin complex 1. J. Biol. Chem. 287, 20913–20921 [DOI] [PMC free article] [PubMed] [Google Scholar]