Abstract
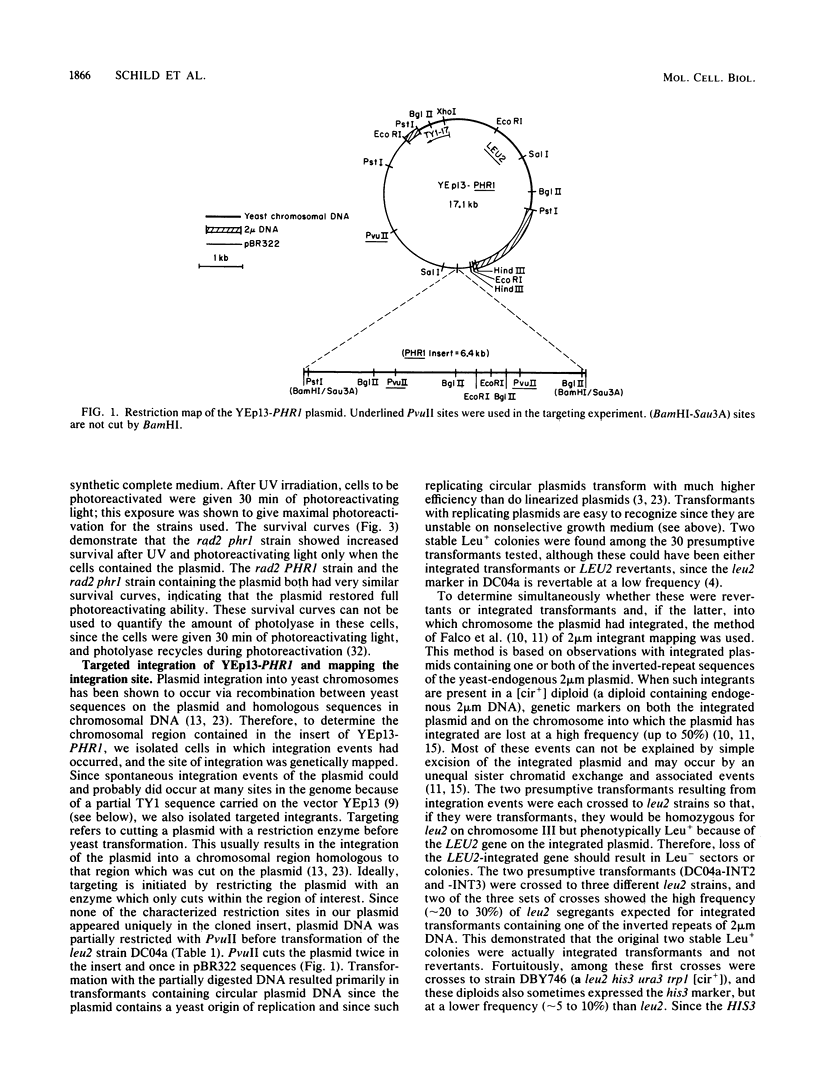
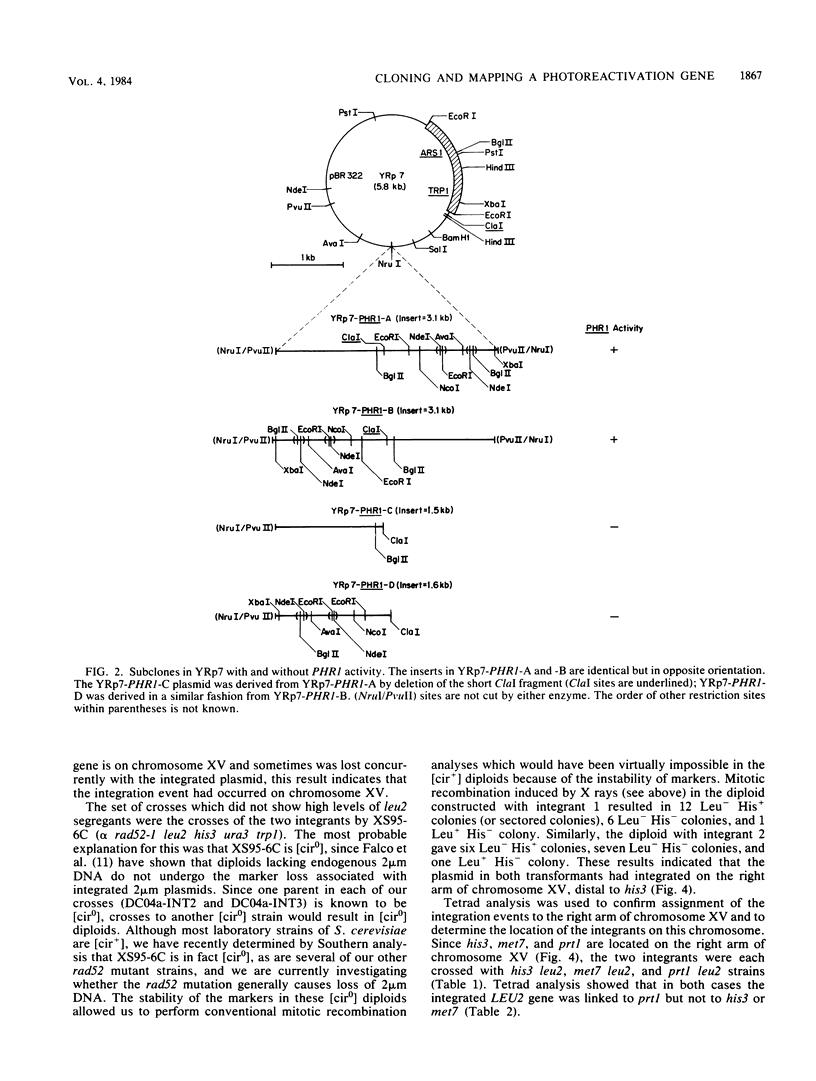
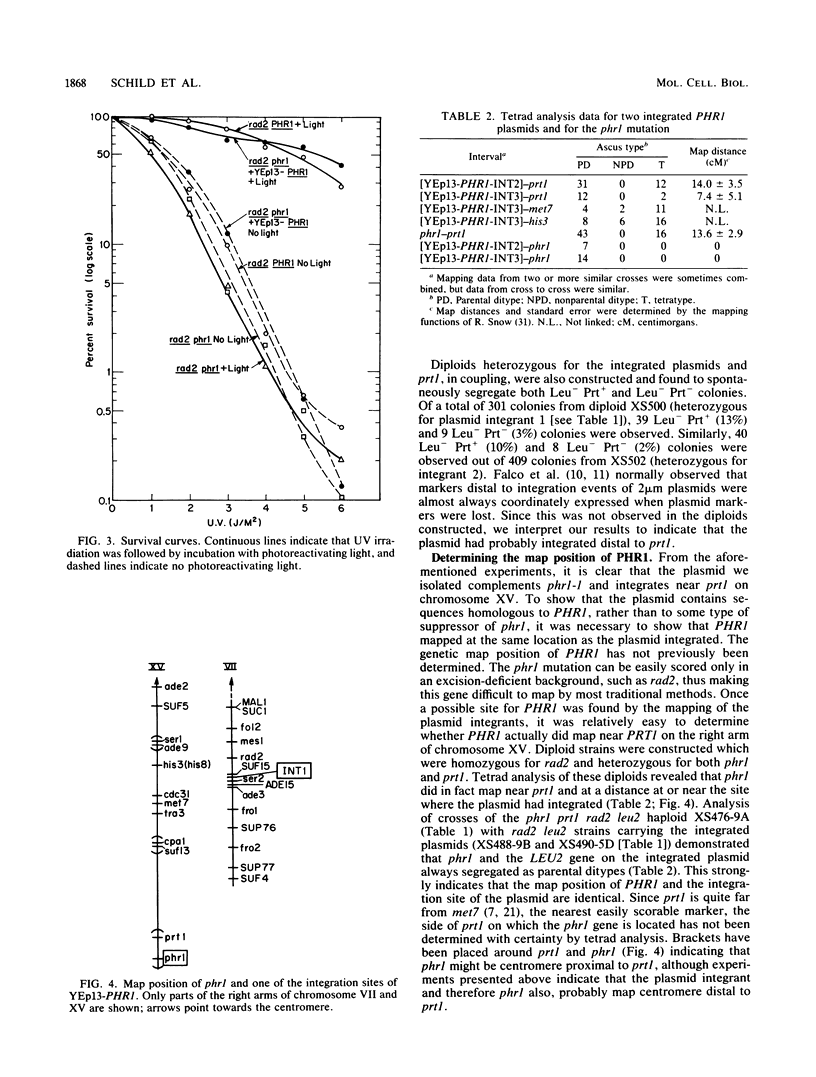
The yeast Saccharomyces cerevisiae, like most organisms, is able to directly repair pyrimidine dimers by using a photoreactivating enzyme and visible light. Cells carrying the phr1 mutation were shown previously to be unable to photoreactivate dimers, but neither the map position nor the primary gene product of the PHR1 gene has been determined. We have cloned this gene and determined its map position. A plasmid containing a 6.4-kilobase yeast DNA insert has been isolated and shown to restore photoreactivation in a phr1 strain. A 3.1-kilobase subclone has also been shown to complement phr1. The original plasmid was targeted to integrate into chromosomal DNA at a site homologous to the insert by cutting within the insert. Two of these integrants have been mapped on the right arm of chromosome XV; the integrants have been further mapped at ca. 13 centimorgans from prt1. It has also been independently determined that phr1 maps at this location. Thus, we have determined the map position of PHR1 and also have shown that the plasmid contains PHR1 rather than a suppressor of the phr1 mutation.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andreadis A., Hsu Y. P., Kohlhaw G. B., Schimmel P. Nucleotide sequence of yeast LEU2 shows 5'-noncoding region has sequences cognate to leucine. Cell. 1982 Dec;31(2 Pt 1):319–325. doi: 10.1016/0092-8674(82)90125-8. [DOI] [PubMed] [Google Scholar]
- Boling M. E., Setlow J. K. Photoreactivating enzyme in logarithmic-phase and stationary-phase yeast cells. Biochim Biophys Acta. 1967 Sep 26;145(2):502–505. doi: 10.1016/0005-2787(67)90068-8. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Hicks J. B. Replication and recombination functions associated with the yeast plasmid, 2 mu circle. Cell. 1980 Sep;21(2):501–508. doi: 10.1016/0092-8674(80)90487-0. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Culbertson M. R., Gaber R. F., Cummins C. M. Frameshift suppression in Saccharomyces cerevisiae. V. Isolation and genetic properties of nongroup-specific suppressors. Genetics. 1982 Nov;102(3):361–378. doi: 10.1093/genetics/102.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson M. J., Kingsman S. M., Kingsman A. J. Sequence variation in the LEU2 region of the saccharomyces cerevisiae genome. Gene. 1981 Dec;16(1-3):133–139. doi: 10.1016/0378-1119(81)90069-x. [DOI] [PubMed] [Google Scholar]
- Falco S. C., Botstein D. A rapid chromosome-mapping method for cloned fragments of yeast DNA. Genetics. 1983 Dec;105(4):857–872. doi: 10.1093/genetics/105.4.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco S. C., Li Y., Broach J. R., Botstein D. Genetic properties of chromosomally integrated 2 mu plasmid DNA in yeast. Cell. 1982 Jun;29(2):573–584. doi: 10.1016/0092-8674(82)90173-8. [DOI] [PubMed] [Google Scholar]
- Hicks J. B., Hinnen A., Fink G. R. Properties of yeast transformation. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1305–1313. doi: 10.1101/sqb.1979.043.01.149. [DOI] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston J., Hilger F., Mortimer R. Variation in frequency of transformation by plasmid YRp7 in Saccharomyces cerevisiae. Gene. 1981 Dec;16(1-3):325–329. doi: 10.1016/0378-1119(81)90089-5. [DOI] [PubMed] [Google Scholar]
- MacQuillan A. M., Herman A., Coberly J. S., Green G. A second photoreactivation-deficient mutation in Saccharomyces cerevisiae. Photochem Photobiol. 1981 Dec;34(6):673–677. [PubMed] [Google Scholar]
- Mortimer R. K., Schild D. Genetic map of Saccharomyces cerevisiae. Microbiol Rev. 1980 Dec;44(4):519–571. doi: 10.1128/mr.44.4.519-571.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. A., Tatchell K. The structure of transposable yeast mating type loci. Cell. 1980 Mar;19(3):753–764. doi: 10.1016/s0092-8674(80)80051-1. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick M. A. A photoreactivationless mutant of Saccharomyces cerevisiae. Photochem Photobiol. 1969 Apr;9(4):307–312. doi: 10.1111/j.1751-1097.1969.tb07294.x. [DOI] [PubMed] [Google Scholar]
- Resnick M. A., Setlow J. K. Photoreactivation and gene dosage in yeast. J Bacteriol. 1972 Mar;109(3):1307–1309. doi: 10.1128/jb.109.3.1307-1309.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar G. B., Smith F. W., Sancar A. Identification and amplification of the E. coli phr gene product. Nucleic Acids Res. 1983 Oct 11;11(19):6667–6678. doi: 10.1093/nar/11.19.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild D., Ananthaswamy H. N., Mortimer R. K. An endomitotic effect of a cell cycle mutation of Saccharomyces cerevisiae. Genetics. 1981 Mar-Apr;97(3-4):551–562. doi: 10.1093/genetics/97.3-4.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow R. Maximum likelihood estimation of linkage and interference from tetrad data. Genetics. 1979 May;92(1):231–245. doi: 10.1093/genetics/92.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiberg J. S. Floating-loop replica-plating device. J Appl Bacteriol. 1977 Jun;42(3):433–436. doi: 10.1111/j.1365-2672.1977.tb00712.x. [DOI] [PubMed] [Google Scholar]



