Background: HAT1 is involved in homologous recombination repair.
Results: HAT1 facilitates the incorporation of H4K5/K12-acetylated H3.3 at double-strand break sites through its HIRA-dependent histone turnover activity, thereby promoting efficient homologous recombination process.
Conclusion: HAT1 is a key regulator of homologous recombination repair.
Significance: This work provides a mechanistic insight into the regulation of histone dynamics by HAT1.
Keywords: DNA Damage Response, DNA Repair, Histone Acetylase, Histone Chaperone, Histone Modification
Abstract
Faithful repair of DNA double-strand breaks is vital to the maintenance of genome integrity and proper cell functions. Histone modifications, such as reversible acetylation, phosphorylation, methylation, and ubiquitination, which collectively contribute to the establishment of distinct chromatin states, play important roles in the recruitment of repair factors to the sites of double-strand breaks. Here we report that histone acetyltransferase 1 (HAT1), a classical B type histone acetyltransferase responsible for acetylating the N-terminal tail of newly synthesized histone H4 in the cytoplasm, is a key regulator of DNA repair by homologous recombination in the nucleus. We found that HAT1 is required for the incorporation of H4K5/K12-acetylated H3.3 at sites of double-strand breaks through its HIRA-dependent histone turnover activity. Incorporated histones with specific chemical modifications facilitate subsequent recruitment of RAD51, a key repair factor in mammalian cells, to promote efficient homologous recombination. Significantly, depletion of HAT1 sensitized cells to DNA damage compromised the global chromatin structure, inhibited cell proliferation, and induced cell apoptosis. Our experiments uncovered a role for HAT1 in DNA repair in higher eukaryotic organisms and provide a mechanistic insight into the regulation of histone dynamics by HAT1.
Introduction
The maintenance of genome integrity is vital to the proper cell functions. Faithful repair of DNA damage is thus essential for the suppression of genetic instability and tumorigenesis. Eukaryotic cells have evolved two conserved mechanisms to detect and repair DNA double-strand breaks (DSBs)2: homologous recombination (HR) repairs the break using genetic information that is retrieved from an undamaged sister chromatid or chromosomal homologue, whereas non-homologous end joining (NHEJ) involves direct ligations of DNA ends. Characterization of the factors and pathways governing DSB repair is important to the understanding of the mutagenesis processes during oncogenic events and to the development of proper cancer therapies.
Histone modifications, such as reversible acetylation, phosphorylation, methylation, ubiquitination, and ADP-ribosylation, play important roles in the promotion of DSB repairs through creating specific interaction platforms for regulatory proteins and complexes (1–5). Additionally, incorporation of variant histones could affect DSB repairs by influencing chromatin configurations; thus, the recognition by downstream effectors.
Histone turnover, referring to the replacement of “old” histones by “new” histones in a replication-independent manner, does not involve obvious changes in nucleosome occupancy; instead, it affects the composition of histone marks and thereby the epigenetic mechanisms of chromatin-based events such as gene regulation and DNA repair (6–8). A well known example of histone turnover activity is the yeast Swr1p ATPase complex, which catalyzes the replacement of conventional histone H2A with histone H2A.Z variant in nucleosome arrays (9, 10). Coupling of histone acetylation with histone exchange activity at the site of damaged DNA has been observed with the Drosophila Tip60 complex (11), suggesting that histone exchange factors can play an important role during DNA repair process.
HAT1 is the first histone acetyltransferase identified and is highly conserved from yeast to human (12, 13, 64). HAT1 has been described as a classical B type HAT, which can only acetylate the N-terminal tail of newly synthesized histone H4 but not nucleosomal histones. On the contrary, type A histone acetyltransferases are nuclear enzymes capable of acetylating histones that have already been packaged into the chromatin. Evidence from yeast model show that HAT1 forms the NuB4 complex with HAT2 (human RbAp46 homolog), Hif1p (HAT1-interacting factor 1, a histone chaperone that selectively interacts with histones H3), and histone tetramer in nuclei and plays an important role in de novo chromatin assembly during replication. The NuB4 complex has also been shown to regulate HR repair of DSBs by participating in repair-linked chromatin reassembly through its B-HAT activity (14–16). Although HAT1 has long been thought to play a role in DNA repair, it has been mostly studied in yeast cells; whether and how HAT1 participates in the DNA repair process in mammalian cells are largely unknown.
In current study we found that, in addition to a role of HAT1 in post-repair chromatin reassembly, HAT1 has a direct role in HR repair in human cells. HAT1 facilitates the enrichment of H4K5/K12-acetylated H3.3 (H3.3-H4K5/12ac) to the DSB sites through its HIRA-dependent histone turnover activity, thereby marking the damaged area for subsequent recruitment of key repair factor RAD51 to promote efficient HR process.
EXPERIMENTAL PROCEDURES
Cells, Plasmids, Antibodies, and Reagents
DR-GFP-U2OS and EJ5-GFP-HEK293 stable cell lines were from Dr. Xingzhi Xu (The Capital Normal University, Beijing, China). The cDNA for wild-type HAT1 was amplified by PCR and ligated into EcoRI/BamHI sites of pcDNA3.1 plasmid containing a FLAG tag. siRNA-resistant pcDNA3.1(−)-FLAG-HAT1 with three synonymous mutations (C510A, T762C, and C1161T) was constructed by site-directed mutagenesis of wild-type plasmid. All clones were confirmed by DNA sequencing. Recombinant baculoviruses containing the coding region of HAT1 and Rbap46 were generated, and the proteins were purified from infected Sf9 cells. Full-length Xenopus histone expression plasmids pET-H2A, pET-H2B, pET-H3, and pET-H4 were from Dr. Karolin Luger (Colorado State University). pBlueScript SK(−) plasmid, Nap1p-plasmid, and Isw1–3×FLAG yeast strain were kindly provided by Dr. Toshio Tsukiyama (University of Washington). Recombinant DnaK ATPase domain (1–384) was from Prospec. Antibodies used were: αH2A, αH2AK5ac, αH3, αH3K14ac, αH3K23ac, αH4, αH4K5ac, αH4K8ac, αH4K12ac, αH4K91ac, and αKu80 from Abcam; αH3.3 from Millipore; αβ-actin, αHIRA, αHIRIP3, and αFLAG from Sigma; goat αHAT1 from Santa Cruz Biotechnology and rabbit αHAT1 from Sigma; rabbit αH2AXp (Ser-139) from Cell Signaling; mouse αH2AXp (Ser-139) from Millipore; αATR, αphospho-ATR (Ser-428) from Cell Signaling; αp53 from MBL. Control siRNA was synthesized by Shanghai GeneChem Inc (Shanghai, China). The sequence for control siRNA was 5′-UUCUCCGAACGUGUCACGU-3′. The siRNAs targeting human HAT1 were purchased from Santa Cruz Biotechnology.
Fluorescence Confocal Microscopy
HeLa, MDA-MB-231, MCF-7, and U2OS cells growing on six-well chamber slides were washed with phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde, permeabilized with 0.1% Triton X-100 in PBS, blocked with 0.8% BSA, and incubated with appropriate primary antibodies followed by staining with TRITC-conjugated secondary antibodies. Cells were washed 4 times. and a final concentration of 0.1 μg/ml 4,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Sigma) was included in the last washing to stain nuclei. Images were visualized and recorded with an Olympus FV1000S confocal microscope.
Cell Cycle Synchronization and Flow Cytometry Analysis
MCF-7 cells were synchronized in the G0/G1 phase by serum starvation, in the G1/S phase by double thymidine block, and in the G2/M phase by thymidine/nocodazole block, and then they were released for various periods of time. In all cases cells were trypsinized, washed with PBS, and fixed in 70% ethanol at 4 °C overnight. After washing with PBS, cells were incubated with RNase A (Sigma) in PBS for 30 min at 37 °C and then staining with 50 mg/ml propidium iodide. Cell cycle data were collected using a FACSCalibur flow cytometer (BD Biosciences) and analyzed with ModFit LT 3.0 (Verity Software House Inc., Topsham, ME).
Cell Viability/Proliferation Assay
For cell proliferation assays, HCT116 or U2OS cells were transfected with HAT1 siRNA and seeded into 96-well plates. Cells were treated or untreated with 0.0001 or 0.0005% methyl methanesulfonate, 2 or 10 mm hydroxyurea, or 100 or 500 nm camptothecin (CPT) for 12 h. On the day of harvesting, the medium was replaced with an equal volume of fresh medium containing 10% Alamar blue. Plates were incubated at 37 °C for 6 h, and cell viability was determined by measuring the absorbance of converted dye at wavelengths 570 and 630 nm (17).
RNA Interference and Western Blotting
Synthesized siRNAs were transfected into cells with the final concentration of 50 nm. For Western blotting, 72 h after transfection total cellular proteins were extracted and resolved on 8 or 15% SDS-PAGE gels and transferred to NC membranes (Millipore). Membranes were incubated with appropriate antibodies for 1 h at room temperature or overnight at 4 °C followed by incubation with a secondary antibody. Immunoreactive bands were visualized using Western blotting luminol reagent (Santa Cruz Biotechnology) according to the manufacturer's recommendation.
ChIP Assay
ChIPs were performed in DR-U2OS cells as described previously (3, 18–21). Briefly, 5 × 107 cells were cross-linked with 1% formaldehyde, sonicated, pre-cleared, and incubated with 5–10 μg of antibody per reaction. Complexes were washed with low and high salt buffers, and the DNA was extracted and precipitated. The enrichment of the DNA template was analyzed by quantitative PCR using primers 500 bp from I-SceI cutting: forward, 5′-GATGGCACAGTGGTCAAGAGC-3′, and reverse, 5′-GAAGGATGGAAGGGTCAGGAG-3′.
Micrococcal Nuclease Sensitivity Assay
siRNA-treated U2OS cells were washed with cold PBS, resuspended in ice-cold Nonidet P-40 cell lysis buffer (10 mm Tris-HCl, pH 7.4, 10 mm NaCl, 3 mm MgCl2, 0.5% Nonidet P-40, 0.15 mm spermine, and 0.5 mm spermidine) and incubated on ice for 5 min. Permeabilized nuclei were washed with micrococcal nuclease digestion buffer (10 mm Tris-HCl, pH 7.4, 15 mm NaCl, 60 mm KCl, 0.15 mm spermine, and 0.5 mm spermidine) and digested at 37 °C for 10 min with 2.5 or 5 units/ml micrococcal nuclease in the digestion buffer containing 1 mm CaCl2. The reaction was stopped by adding 10 mm EDTA and 0.5% SDS, and DNA was then purified after incubation with proteinase K and RNase A and was electrophoresed on 2% agarose gels.
Assembly of Nucleosome Arrays
Assembly of nucleosomal arrays on an immobilized DNA was performed as previously described (22, 23). pBlueScript SK(−) plasmid was used as a template. Dynabeads M-280 (Dynal) were used for coupling with the plasmid DNA linearized by EcoRI and ClaI as instructed. In a nucleosome assembly reaction, Dynabeads coupled with 1.5 μg of DNA were incubated with 1.5 μg of recombinant Xenopus histone octamer, 0.91 pmol of ISW1 complex, 6 μg of purified recombinant Nap1p, and 3 mm ATP in 10 mm HEPES-KOH, pH 7.6, 0.5 mm EGTA, 5 mm MgCl2, 10% glycerol, 1 mm DTT, 0.1 mg/ml BSA, 70 mm KCl in a reaction volume of 180 μl for 4 h at room temperature with constant mixing. The resulting nucleosomal arrays were washed 4 times with 1 ml of buffer H (25 mm HEPES-KOH, pH 7.6, 0.5 mm EGTA, 0.1 mm EDTA, 5 mm MgCl2, 0.02% Nonidet P-40, 10% glycerol, 1 mm DTT, 0.1 mg/ml BSA) containing 0.6 m KCl to remove ATP, Nap1p, ISW1 complex, and non-nucleosomal histones before starting the transfer reaction.
Histone Transfer Assay
In a standard histone transfer assay, immobilized nucleosomal arrays (150 ng of DNA equivalents) were incubated in a 100-μl reaction volume with 6 pmol of free calf thymus histone, titrated FLAG-HAT1 complex, or recombinant HAT1 (reHAT1)/reRbAp46 with or without 1 mm ATP in exchange buffer (25 mm HEPES-KOH, pH 7.6, 0.37 mm EDTA, 0.35 mm EGTA, 5 mm MgCl2, 10% glycerol, 0.02% Nonidet P-40, 1 mm DTT, 0.1 mg/ml BSA, 70 mm KCl) for 60 min at room temperature with constant mixing. Beads were concentrated on a magnetic particle concentrator (Dynal), and the supernatants were saved. Beads were then washed 3 times, each time with 100 μl of buffer H containing 70 mm KCl, and the bound proteins were eluted with SDS-PAGE sample buffer. The wash fractions, and the supernatants were combined, precipitated with TCA, and dissolved in SDS-PAGE sample buffer for analysis of total free (unbound) proteins. Bound and free proteins were analyzed by Western blotting.
RESULTS
Mammalian HAT1 Is Involved in DNA Damage Repair
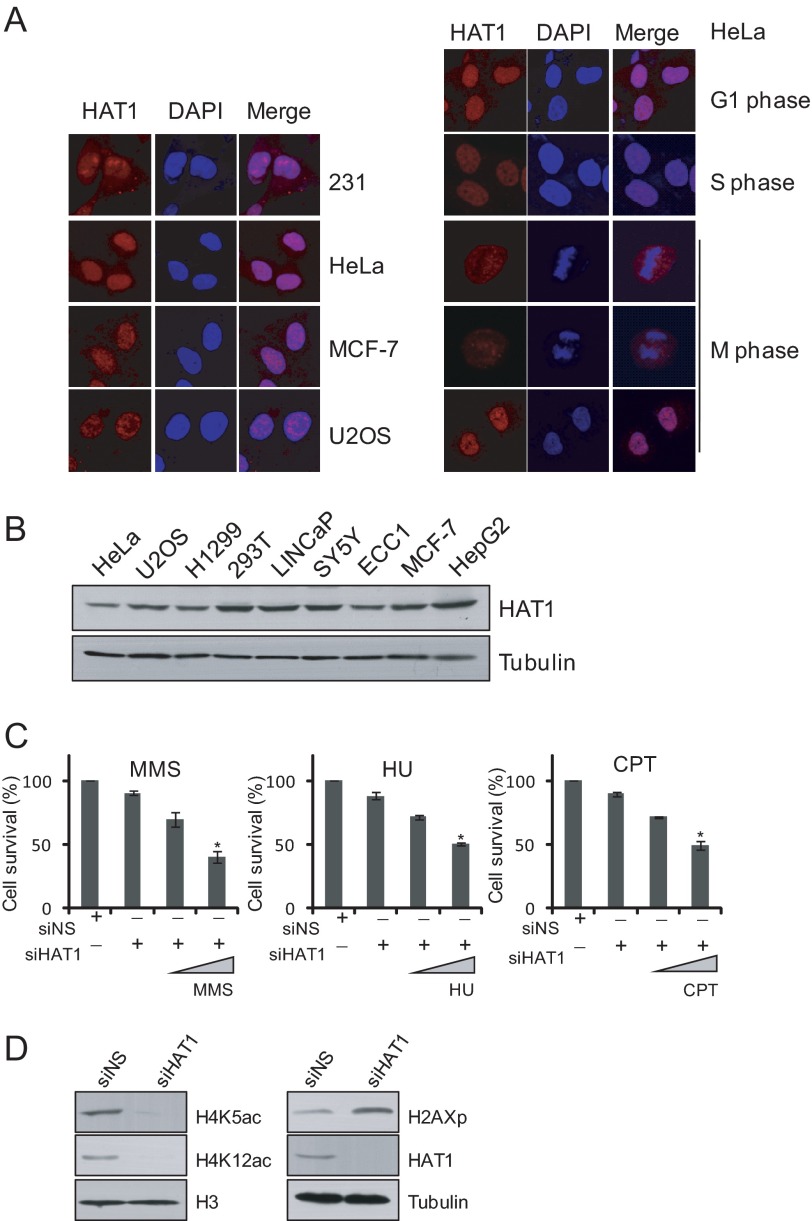
HAT1 belongs to type B histone acetyltransferases and was originally thought to be largely a cytoplasmic enzyme responsible for the acetylation of newly synthesized free histones. However, evidence in yeast and some other eukaryotic cells suggests that HAT1 can also be localized in the nucleus (13). To further explore the biological functions of HAT1 in mammalian cells, we first examined the intracellular localization of endogenous HAT1 in different human cell lines. Exponentially growing MDA-MB-231, HeLa, MCF-7, and U2OS cells were fixed and immunostained with antibodies against HAT1. Immunofluorescent imaging revealed that although HAT1 was detected in the cytoplasm, the majority of this protein was found in the nucleus in all the types of cells examined (Fig. 1A, left panel). We further examined the intracellular localization of HAT1 in different phases of the cell cycle in these cells. The results showed that the nuclear localization of HAT1 is persistent throughout the entire cell cycle (Fig. 1A, right panel). The intracellular localization of HAT1 suggests that, in addition to acetylating newly synthesized free histones in cytoplasm, HAT1 may also function in the nucleus.
FIGURE 1.
Mammalian HAT1 is localized in the nucleus and involved in DNA damage repair. A, shown is immunofluorescent staining of HAT1 protein in different cell lines and in different cell cycle phases of HeLa cells. B, Western blotting analysis of the expression of HAT1 in different cell lines is shown. C, U2OS cells were treated with control and HAT1 siRNA. Thirty-two hours later cells were treated with 0.0001%, 0.0005% methyl methanesulfonate (MMS), 2 or 10 mm hydroxyurea (HU), or 100 nm or 500 nm CPT for 12 h followed by Alamar blue staining to detect their viability. Each bar represents the mean ± S.D. for three independent experiments. Asterisks indicate that the differences in cell viability of those groups are significant compared with the controls (p < 0.01). D, shown is accumulation of γH2AX in the HAT1-depleted cells. U2OS cells were transfected with control and HAT1 siRNA and were harvested after 72 h. Cell lysates were analyzed by immunoblotting. siNS, nonsilencer siRNA.
HAT1 is highly expressed in various human cancer cell lines (Fig. 1B). In fact, HAT1 has been found to be overexpressed in liver and colon cancers compared with their normal counterparts (24, 25). The higher levels of HAT1 seen in tumor cells could reflect its role in chromatin assembly, a process that proliferating cells need to maintain with a high capacity to accompany ongoing DNA replication. Alternatively, as suggested by previous studies in yeast models, HAT1 could play a role in DNA damage repair, which allows tumor cells to fix damaged genome inflicted by various stress conditions. To examine whether HAT1 is involved in the DNA repair process in mammalian cells, HAT1-depleted HCT116 cells were subjected to DNA-damaging sensitivity assays. In these experiments, HCT116 cells treated with control or HAT1 siRNAs were challenged with various DNA-damaging agents including methyl methanesulfonate, hydroxyurea, and CPT and examined for cell viability by Alamar blue staining. The results showed that loss of HAT1 in cells was associated with a significant decrease in the cell viability upon treatment with these genotoxic agents leading to replication stress (26) (Fig. 1C).
To further support a role of HAT1 in DNA-damaging response by the cells, we next investigated the effect of loss-of-function of HAT1 on the level of the phosphorylated H2A.X (γH2AX), an essential mammalian histone variant present at the sites of DSBs (27). U2OS cells were treated with control or HAT1 siRNAs, and whole cell lysates were extracted for Western blotting analysis with antibodies against these proteins. The results indicated that HAT1-deficient cells exhibited increased levels of γH2AX (Fig. 1D). Together, these results support the notion that HAT1 is involved in the process of DNA damage repair.
HAT1 Is Required for HR- but Not NHEJ-mediated DNA Repair
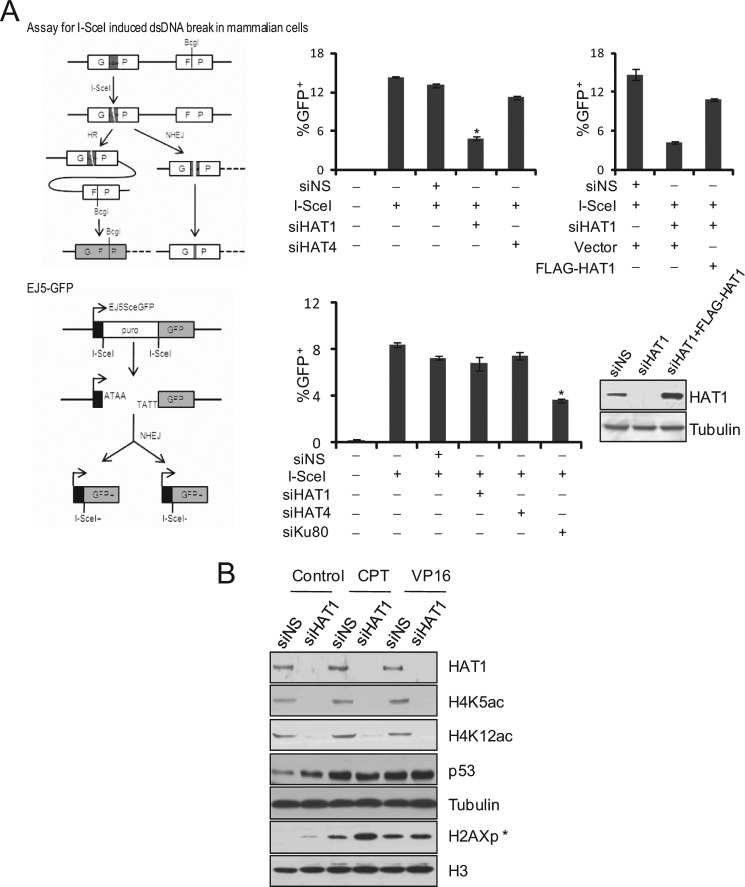
To gain a mechanistic insight into the role of mammalian HAT1 in DNA damage repair, we first investigated the molecular pathway(s) of DNA repair (HR or NHEJ) in which HAT1 might be involved. For this purpose, GFP-based chromosomal reporter assays with two stable cell lines, DR-GFP-U2OS and EJ5-GFP-HEK293, were used to measure HR or NHEJ, respectively (28, 29). The first reporter, DR-GFP, contains a GFP-encoding cDNA that has an endogenous BcgI restriction site replaced by an I-SceI restriction site, thereby rendering its non-functional. A DSB is induced by transfection of cells with a plasmid (pCBASce) that encodes I-SceI enzyme to cut the I-SceI site. An incomplete GFP sequence is located downstream and can serve as a donor for intra-chromosomal homologous recombination. Repair by HR restores a functional GFP cDNA with the original BcgI site, and the resultant GFP+ cells can be detected by FACS (Fig. 2A, upper left panel). The second reporter, EJ5-GFP, detects multiple classes of NHEJ events and thus is considered to be an assay for total NHEJ. Specifically, EJ5-GFP contains a promoter that is separated from a GFP coding cassette by a puro gene flanked by two I-SceI sites in the same orientation. Transfection of cells with I-SceI-encoding constructs leads to the excision of the puro gene, and the restoration of the GFP expression relies on efficient NHEJ repair in these cells (Fig. 2A, lower left panel).
FIGURE 2.
HAT1 is required for HR but not for NHEJ. A, left panel, shown is a schematic illustration of I-SceI-mediated DSB induction and repair using the DR-GFP and EJ5-GFP transgenes. Right panels, the efficiency of HR in DR-GFP transgenic U2OS cells and of NHEJ in EJ5-GFP transgenic 293 cells were analyzed after HAT1 knockdown. DR-U2OS cells were treated with 50 μm Nonsilencer, HAT1 siRNA, or HAT4 siRNA 6 h before transfection with pCBASce or pCAGGS-Ds-red (transfection indicator), whereas EJ5-HEK293 cells were treated with comparable amounts of Nonsilencer, HAT1 siRNA, HAT4 siRNA, or Ku80 siRNA. HR or NHEJ repair efficiency was measured 48 h later by measuring the fraction of GFP+ cells. Ds-red was used to determine transfection efficiency. The ratio of GFP+ to Ds-red+ cells is graphed for untreated and siRNA-treated cells. Each bar represents the mean ± S.D. for three independent experiments. Asterisks denote the significance of the difference in the repair efficiency between HAT1- or Ku80-depleted cells and control (or HAT4-depleted) cells (p < 0.01). RNAi-resistant FLAG-HAT1 construct could rescue the HR-deficient effect of HAT1 knockdown. siRNA-treated DR-U2OS cells were transfected with I-SceI plus empty vector, I-SceI plus RNAi resistant pcDNA3.1(−)-FLAG-HAT1, or pCAGGS-Ds-red. 48 h later the ratio of GFP+ to Ds-red+ cells was graphed (upper right panel). Each bar represents the mean ± S.D. for three independent experiments. Western analysis of FLAG-HAT1 resisting HAT1 RNAi is shown (lower right panel). B, accumulation of γH2AX in the HAT1-depleted cells after CTP stimulation is shown. U2OS cells were transfected with control and HAT1 siRNA and then treated with 100 nm CPT or 40 μm VP16 for 8 h before harvesting. Cell lysates were analyzed by immunoblotting. siNS, nonsilencer siRNA.
DR-U2OS and EJ5-HEK293 cells were plated in 6-well plates and transfected with control siRNAs or siRNAs for HAT1 or HAT4 (another B-type histone acetyltransferase that we recently identified) (30). The cells were then transfected with the pCBASce plasmid or pCAGGS-Ds-red (transfection indicator). After 48 h, the percentage of GFP+ cells per well was determined by FACS. The ratio of GFP+ to Ds-red+ cells is graphed for untreated and siRNA-treated cells. We observed that the percentage of GFP+ cells was significantly reduced in HAT1-depleted DR-U2OS cells compared with cells treated with control siRNA or HAT4 siRNA (Fig. 2A, upper middle panel). In contrast, the level of total-NHEJ had no obvious change in HAT1-deficient EJ5-HEK293 cells compared with cells transfected with control siRNAs (Fig. 2A, lower middle panel), whereas knockdown of the expression of Ku80, a factor that is known to be involved in NHEJ, led to a significant decrease of total-NHEJ events in EJ5-HEK293 cells, validating the sensitivity of this assay. Furthermore, the negative effect of HAT1 depletion on HR efficiency of DR-U2OS cells could be rescued by overexpression of siRNA-resistant HAT1 plasmid, which had three synonymous mutations (C510A, T762C, and C1161T) (Fig. 2A, upper right panel).
Next, U2OS cells were transfected with HAT1 siRNAs and treated with DNA damaging agent CPT or VP16. CPT activates S or G2-M arrest and the HR repair pathway in tumor cells, whereas VP16 damages DNA without affecting a particular cell cycle phase; thus, NHEJ is the major repair pathway responsible for VP16 induced DSBs (31, 32). Compared with the cells treated with control siRNAs, HAT1-depleted cells exhibited an elevated level of γH2AX but only under the treatment of CPT (Fig. 2B). When HAT1-depleted cells were treated with VP16, no evident increase in the level of γH2AX was detected. These results favor the idea that HAT1 depletion impairs HR but not NHEJ repair of DSBs.
HAT1 Is Physically and Functionally Associated with HR Repair Proteins
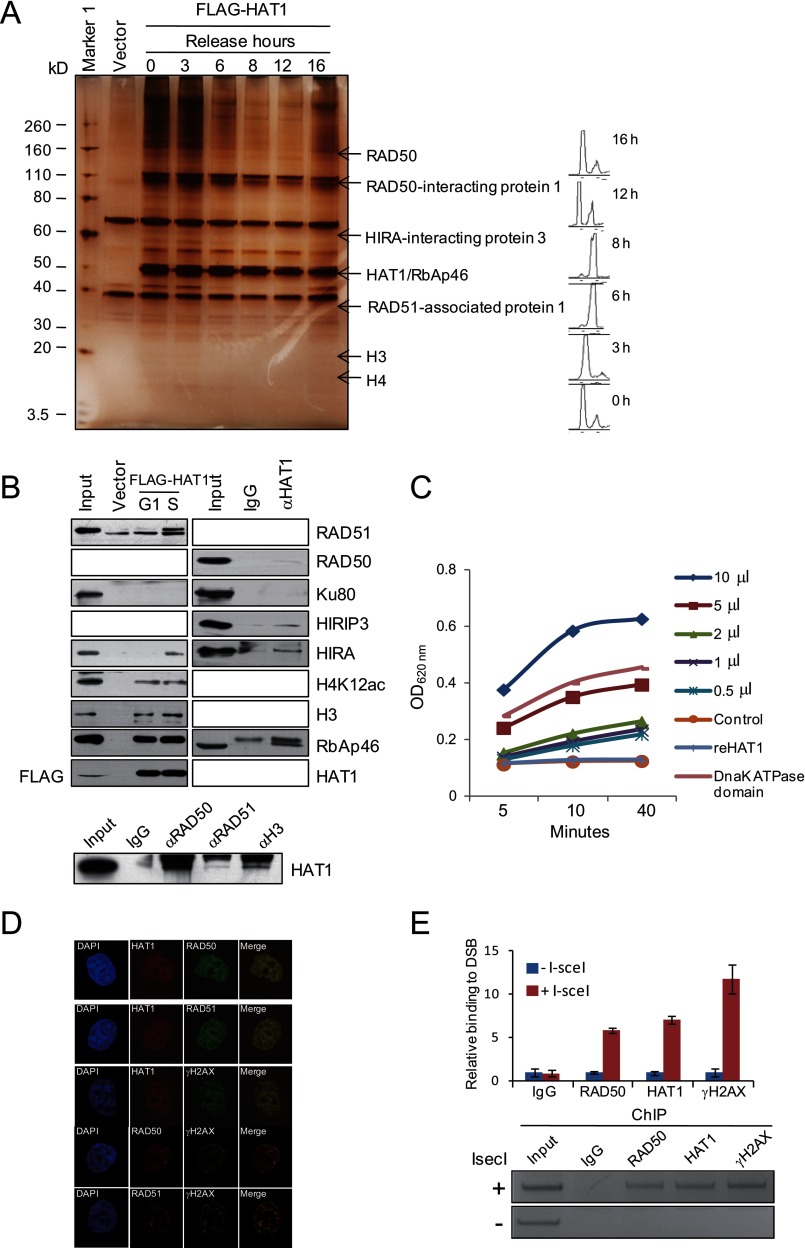
In an effort to better understand the mechanistic roles of HAT1 in the HR repair process, we employed affinity purification and mass spectrometry to identify cellular proteins that are associated with HAT1. Although NHEJ is active throughout the cell cycle and is favored in G1 phase, HR-DSB is more prevalent after DNA replication, mainly because an identical sister chromatid is available as a template for repair at this stage (33). Therefore, we performed affinity purification with the cell lysates collected from different cell cycle stages. In these experiments, FLAG-tagged HAT1 (FLAG-HAT1) was stably expressed in HeLa cells. Cells were synchronized in G1/S transition by double thymidine block and released for different hours to allow cells to enter S, G2/M, and G1 phases. Cellular extracts were prepared from cells in different cell cycle phases and subjected to affinity purification using an anti-FLAG affinity gel. After extensive washing, the bound protein complex was eluted with excess FLAG peptides. Bound proteins were resolved by SDS-PAGE and visualized by silver staining. The protein bands on the gel were retrieved and analyzed by mass spectrometry. Although we did not observe obvious differences in the staining profiles with the extracts from different phases of cell cycle, the total amount of HAT1 interacting proteins increased in S (represented by 0, 3, and 16 h after release) and G2 (6 h after release), 2 phases when HR repair is predominant (Fig. 3A). The detailed results of the mass spectrometric analysis are provided in supplemental File 1.
FIGURE 3.
HAT1 is physically and functionally associated with HR repair proteins. A, shown is immunoaffinity purification of HAT1-containing protein complexes. HeLa cells stably expressing FLAG-HAT1 were synchronized by double thymidine block, and cellular extracts were immunopurified with anti-FLAG affinity columns and eluted with FLAG peptide at specific cell cycle phases. The eluates were resolved by SDS-PAGE and visualized by silver-staining. The proteins bands were retrieved and analyzed by mass spectrometry. Detailed results from the mass spectrometric analysis are provided supplemental File 1. B, left panels, shown is Western blotting analysis of the identified proteins in the purified fractions as in A. Right panels, endogenous co-immunoprecipitation using whole cell lysates from S phase U2OS cells with antibodies against the indicated proteins. C, ATPase assays in 96-well plates are shown. 0.5, 1, 2, 5, or 10 μl of FLAG-HAT1 complex purified from mammalian cells or 2 μg of reHAT1 were used for ATPase assays. 2 μg of recombinant Dnak ATPase domain was used as a positive control validating the sensitivity of the assay. Absorbance of A620 nm represents the concentration of free propidium iodide. D, colocalization of HAT1 and HR proteins is shown. U2OS cells were treated with 100 nm CPT for 4 h and immunostained with HAT1, RAD50, RAD51, and γH2AX antibodies. E, DR-U2OS cells were transfected with control or pCBASce plasmids. Forty hours later ChIP assays and quantitative PCR were performed. The y axis represents the relative enrichment of the indicated proteins compared with the IgG control (after normalization with a PCR internal control to a locus other than the DSB). Each bar represents the mean ± S.D. for three independent experiments. Thr right panel shows the ethidium bromide staining of ChIP samples analyzed by semiquantitative PCR.
Among the proteins that were co-purified with HAT1, RAD50 is a component of the MRN complex (composed by RAD50, MRE11, and NBS1), which binds to DNA ends and functions as a nuclease enzyme. RAD51-associated protein 1 is a protein interacting with RAD51 (34, 35), which is a key factor in homologous pairing and strand transfer of DNA in HR repair in mammalian cells (36, 37). HIRIP3 (HIRA-interacting protein 3) is a protein that is closely related to HIRA (38), a histone chaperone that preferentially places the variant histone H3.3 in nucleosomes (39). As expected, various histone species were also detected in HAT1 co-eluted complexes. Based on the roles of these proteins in DNA repair, their association with HAT1 further supports a role for HAT1 in HR of DNA repair.
We confirmed the interaction of HAT1 with RAD50, RAD51, HIRA, HIRIP3, and histones by co-immunoprecipitation assays with specific antibodies against these proteins, whereas no interaction was detected between HAT1 and other repair proteins such as Ku80, which is responsible for NHEJ repair. Notably, HAT1 interacted with these proteins more efficiently in S phase of the cell cycle (Fig. 3B), suggesting that interaction of HAT1/Rbap46 with these proteins is cell cycle-regulated, a notion that is consistent with their roles in HR repair and chromatin assembly.
Based on previous reports that RAD50 and RAD51 exhibit ATPase activity (40, 41), we next examined whether the HAT1-purified complex was indeed associated with an ATPase activity. To this end, BioAssay Systems QuantiChromTM ATPase/GTPase assay kit was used in a microplate format. We set up the experiments with a series dilution of FLAG-HAT1 complex purified from HeLa cells or in vitro purified reHAT1 as well as a negative control with no enzyme and a positive control with recombinant Dnak ATPase domain in separate wells. The reactions were incubated for 30 min in room temperature before the addition of the malachite green reagent, which forms a dark green color with liberated phosphate after desired incubation time. The color change was measured at 620 nm on a plate reader. As expected, we observed that purified FLAG-HAT1 complex, but not reHAT1, possessed a strong and dose-dependent ATPase activity (Fig. 3C), supporting our observation that RAD50 and RAD51 are associated with HAT1 in vivo.
The interaction of HAT1 with HR proteins was further demonstrated by immunofluorescent analysis. The colocalization of HAT1 and RAD50/RAD51 were shown at the damaged DNA, which was represented by the accumulation of γH2AX (Fig. 3D).
Because it is well established that RAD50 and RAD51 are recruited to DSB lesions (37, 42), the interaction of HAT1 with RAD50 and RAD51 could mean that HAT1 is also present at the sites of DSBs. To verify this, chromatin immunoprecipitation (ChIP) assays were carried out in DR-U2OS cells with antibodies against human HAT1. PCR primers that amplify a region surrounding the I-SceI restriction site were used, and samples were taken at 14 h after I-SceI cutting. The results indicated that HAT1 was clearly recruited to the site of DNA damage (Fig. 3E).
HAT1 Complex Possesses an ATP-independent and Histone Chaperone-dependent Histone Turnover Activity
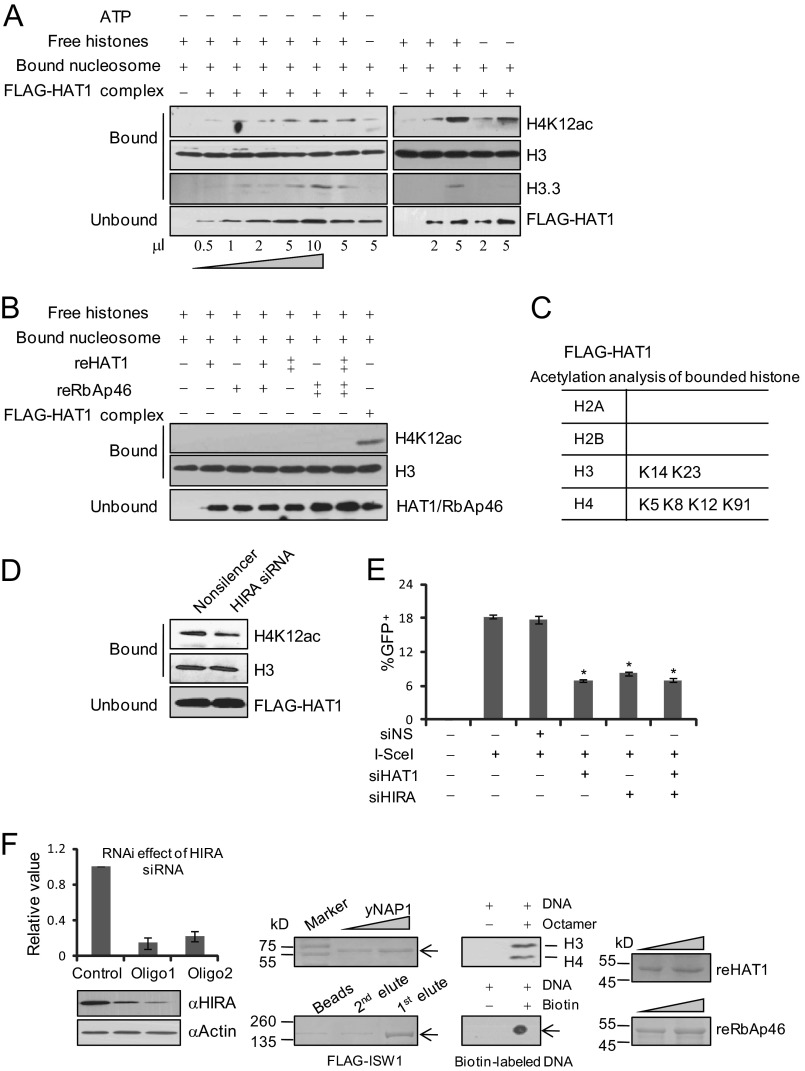
To further explore the molecular mechanisms underlying the role of HAT1 in HR of DNA DSB repair, we next performed in vitro nucleosome assembly assays. In these experiments, immobilized nucleosome arrays were prepared with recombinant Xenopus histones and DNAs that were bound to magnetic beads (22, 23). Because these histones were purified from bacteria Escherichia coli, such preassembled nucleosomes did not have any posttranslational modifications. HAT1 complex was then purified from HeLa cells, and reHAT1 and RbAp46 (reRbAp46) proteins were purified from baculovirus system (21, 43). Incubation of the immobilized nucleosomes with HAT1 complex or calf thymus-free histones led to marked increases in ATP-independent transfer of acetylated histone H4 to nucleosomes (Fig. 4A). In addition to histones associated with HAT1, free calf thymus histones that were added in the system can be used as substrates and transferred by the HAT1 complex, as presenting calf thymus histones in the reaction resulted in a modest increase in the amount of histones being transferred (Fig. 4A). However, recombinant HAT1 or/and recombinant RbAp46 could not transfer histones to the immobilized nucleosomes (Fig. 4B), suggesting that an additional factor(s) in the HAT1 complex is required for its proper histone turnover activity.
FIGURE 4.
HAT1 complex possesses an ATP-independent and histone chaperone-dependent histone turnover activity. A and B, histone turnover activity of FLAG-HAT1 complex and reHAT1 or/and reRbAp46 was analyzed. pBluescript SK-DNA (150 ng; 0.08 pmol) immobilized on beads was incubated (100-μl reaction volume) for 1 h with 0.5, 1, 2, 5, or 10 μl of FLAG-HAT1 eluate (A) or 0.5 or 1 μg of reHAT1 and/or reRbAp46 (B) in the presence or absence of free histones (6 pmol) with or without 1 mm ATP as indicated. Beads were concentrated and washed 3 times with buffer containing 70 mm KCl. The supernatant and wash fractions were combined, and proteins were precipitated by TCA. Bound proteins were eluted with SDS-PAGE sample buffer. H4K12 acetylation and H3.3 levels were analyzed by Western blotting. C, mass spectrometry analysis of immobilized nucleosome after the exchange assay is shown. D, histone turnover activity analysis of FLAG-HAT1 complex purified from control or HIRA depleted HeLa cells is shown. E, efficiency of HR in DR-U2OS cells was analyzed after knockdown of HAT1, HIRA, or both. DR-U2OS cells were treated with 50 μm Nonsilencer, HAT1 siRNA, or/and HIRA siRNA for 6 h before being transfected with PCBASce or pCAGGS-Ds-red. Cells were collected to measure GFP+ cells after 48 h. The ratio of GFP+ to Ds-red+ cells is graphed for untreated and siRNA-treated cells. Each bar represents the mean ± S.D. for three independent experiments. Asterisks indicate that the differences in repair efficiency of those groups are significant compared with the control (p < 0.01). F, SDS-PAGE electrophoresis combined with Coomassie Blue staining or Western blotting was performed to monitor the protein purification or assembly of immobilized nucleosome arrays. Verification of HIRA knockdown in U2OS and MCF-7 cells was done by real-time RT PCR and Western blotting. Each bar represents the mean ± S.D. for three independent experiments.
Because histones co-purified with HAT1 exhibit acetylations more than just H4K5/K12ac that is directly added by HAT1 (30, 44, 45), histone turnover activity could provide a flexible way of affecting the stability of chromatin marks, which results in changes of a wide range of acetylation sites on the exchanged nucleosomes (45). Mass spectrometry analysis of the acetylated lysines in the bound nucleosomes after the exchange assays revealed that in addition to H4K5 and K12 acetylation, acetylations of H4K8, H4K91, H3K14, and H3K23 could also be detected on the bound nucleosomes (Fig. 4C).
As stated above, our results showed that HAT1 complex purified from cells, but not recombinant HAT1, had a histone turnover activity in vitro, suggesting that an additional factor(s) is required. In this regard, it is interesting to note that biochemical analysis revealed that HAT1 interacts with histone chaperone HIRA, which promotes histone variant H3.3 deposition independent of DNA synthesis (46). To determine if the histone turnover activity of HAT1 is HIRA-dependent, we examined incorporation of histone variant H3.3 in the immobilized nucleosomes by Western blotting after the exchange assays. We found that the deposition profile of H3.3 was similar to that of H4K12 acetylation (Fig. 4A). However, knockdown of HIRA in HeLa cells impaired the histone turnover activity of the purified HAT1 complex (Fig. 4D), indicating that HIRA is required for histone turnover activity of HAT1.
To gain further support of the notion that HIRA is required for histone turnover activity of HAT1, we compared HR efficiency of the DR-U2OS cells treated with HAT1 siRNA or/and HIRA siRNA. We reasoned that if the repair efficiency of HAT1/HIRA co-depletion cells is lower than that of cells with HAT1 or HIRA individually depleted, it could reflect a possibility that HAT1 and HIRA affect different aspects of HR; conversely, a similar HR efficiency would suggest that HAT1 and HIRA affect the same DNA repair pathway. The results of our experiments favor an argument that HIRA and HAT1 function in the same pathway in HR repair (Fig. 4E).
Regulation of Chromatin Dynamics by HAT1 during DNA Repair
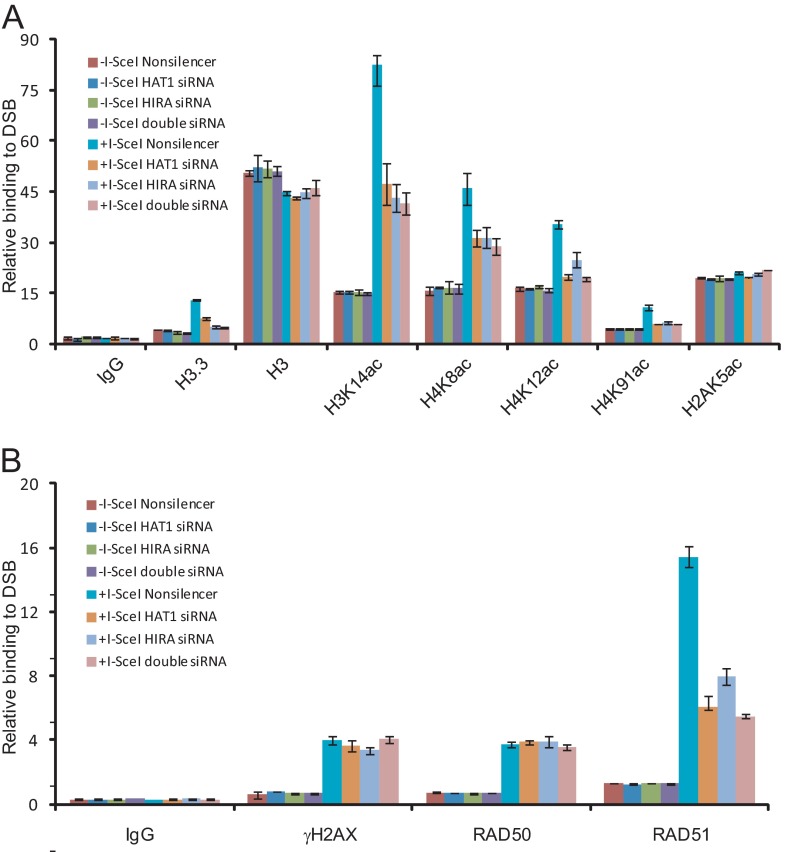
To further investigate the roles of HAT1 in histone dynamics during DNA repair, DR-U2OS cells were transfected with HAT1 siRNA or/and HIRA siRNA for 36 h before transfection with pCBASce for an additional 24 h. ChIP experiments with specific antibodies against various histone modifications indicated that the acetylation levels of histone H4K8, K12, K91, and H3K14 and the incorporation of H3.3 surrounding the DSB sites increased significantly after I-SceI cutting (Fig. 5A). Knockdown of either HAT1 or HIRA led to significant decreases of acetylations of H4K8, K12, K91, H3K14, or H3.3 at the DSB sites. No changes were observed on the levels of H2AK5 acetylation, consisting with the previous prediction that HAT1-induced histone exchange is specific for the substitution of histone H3/H4 tetramers (45) (Fig. 5A). Double knockdown of HAT1 and HIRA did not result in further changes of these modifications, supporting the notion that HAT1 and HIRA function in the same pathway in HR repair (Fig. 5A). The histone turnover activity of HAT1 is further confirmed by the observation that the level of H3.3 enrichment at DSB sites is also correlated with that of HAT1 (Fig. 5A). In addition, knockdown of HAT1 and/or HIRA was associated with a significant decrease in the recruitment of HR key factor RAD51 to the DSB sites, whereas the accumulation of RAD50, which is known to be recruited to DSBs at the beginning step of either HR or NHEJ, was not affected (Fig. 5B). The data suggest that HAT1 acts upstream of RAD51. Together, these results indicate that HAT1 contributes to the histone dynamics at the sites of DSBs through its HIRA-dependent histone turnover activity.
FIGURE 5.
Regulation of chromatin dynamics by HAT1 in DNA repair. A and B, shown is quantitative PCR analysis of ChIP samples from U2OS DR-GFP cells transfected with indicated siRNA or/and PCBASce. The y axis represents the relative enrichment of the indicated proteins compared with the IgG control. Each bar represents the mean ± S.D. for three independent experiments.
The Effect of HAT1 on Chromatin Integrity, Cell Proliferation, and Survival
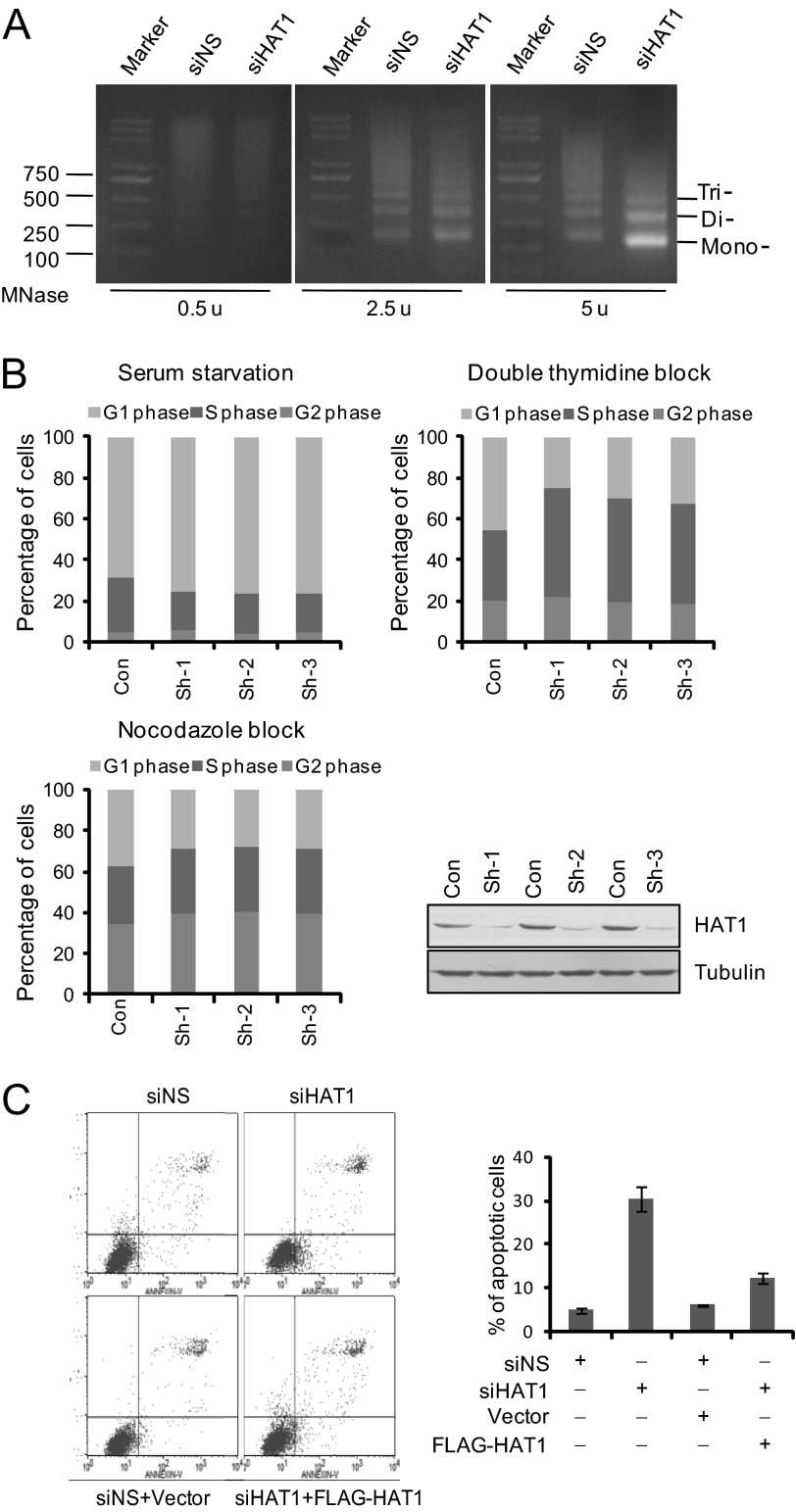
Based on our observations that HAT1 functions in HR repair and replication-coupled chromatin assembly, we investigated the effect of HAT1 on global chromatin structure in mammalian cells by micrococcal nuclease sensitivity assays. In these experiments, equal amounts of nuclei from control or HAT1 siRNA-treated cells were digested with micrococcal nuclease, and extracted chromosomal DNAs were resolved on agarose gels and stained with ethidium bromide. Our data indicated that HAT1 depletion resulted in an increased sensitivity of the cellular DNAs to micrococcal nuclease digestion (Fig. 6A), supporting a role for HAT1 in the maintenance of genome integrity.
FIGURE 6.
The effect of HAT1 on chromatin integrity, cell proliferation, and survival. A, U2OS cells were treated with control or HAT1 siRNA. Nuclei were prepared and treated with 2.5 or 5 Worthington units of micrococcal nuclease (MNase)/ml as indicated. Genomic DNA was purified and fractionated by agarose gel electrophoresis. Mononucleosome, dinucleosome, and trinucleosome are indicated. u, units. B, MCF-7 cells were transfected with pGCsi-Nonsilencer, pGCsi-HAT1 shRNA1#, pGCSi-HAT1 shRNA2#, and pGCSi-HAT1 shRNA3# and then synchronized by serum starvation, double thymidine block, or thymidine nocodazole block at G0/G1, G1/S, or the G2/M boundary, respectively. Cell cycle progression after releasing was analyzed by flow cytometry. RNAi efficiency of HAT1 by these three HAT1 shRNA plasmids was monitored by Western blotting. C, HAT1 knockdown promotes cell apoptosis. siRNA-treated or siRNA and vector/FLAG-HAT1(RNAi-resistant) co-transfected U2OS cells were double-stained with annexin V and propidium iodide. Cell apoptosis was determined by flow cytometry. Each bar represents the mean ± S.D. for three independent experiments. siNS, nonsilencer siRNA.
Defects in genome stability will inevitably affect the growth and homeostasis of cells (47). To investigate whether the involvement of HAT1 in histone turnover in HR repair can be extended to a physiologically relevant cell response, we determined the effect of loss-of-function of HAT1 on the growth and proliferation of MCF-7 cells. The cells were transfected with three different HAT1 shRNA plasmids and were subsequently synchronized in the G0/G1 phase by serum starvation, at the G1/S boundary by double thymidine blocking, and at the M phase with nocodazole blocking. Cell cycle profiling by flow cytometry indicated that, after release from nocodazole or thymidine blocking, HAT1-depleted cells proceeded through the cell cycle with a much lower efficiency compared with control cells (Fig. 6B). Moreover, consistent with a role of HAT1 in maintaining genomic stability, flow cytometry analysis revealed that knockdown of HAT1 resulted in an ∼30% increase in the number of U2OS cells that underwent apoptosis, which could be rescued by RNAi-resistant HAT1 construct (Fig. 6C). These experiments indicate that HAT1 is required for the proliferation and survival of cells.
DISCUSSION
Through their effects on chromatin configurations, chromatin remodeling complexes and histone modification enzymes function at multiple stages during the DNA repair process (48, 49). Due to the ability of HAT1 to acetylate newly synthesized histones rather than nucleosomal histone substrates, it was thought that HAT1 most likely plays a role in repackaging of DNA and free histones into chromatin after the damaged DNA has been repaired. However, a previous study found that the HAT1 yeast homolog Hat1p could still be recruited to chromatin in strains that cannot repair an endonuclease-induced DSB (45), suggesting that HAT1 may be recruited at an early stage.
We showed in this study that HAT1-depleted cells display an increased sensitivity to DNA-damaging reagents. We demonstrated that knockdown of HAT1 in cells compromises HR, but not NHEJ, repair of DSBs. HAT1 interacts with HR-associated proteins RAD50 and RAD51 and is recruited to the sites of DNA DSBs. We propose that HAT1 is directly involved in the histone turnover during HR repair, analogous to the histone exchange activity seen in the replication-independent histone variant assembly in transcriptionally active chromatin (6, 50–54).
Coupling of histone acetylation with histone exchange at the sites of DNA damage has been observed with the Drosophila Tip60 complex (11), indicating that histone exchange factors play important roles during DNA repair. In addition, increase of the levels of H4K12 acetylation, which is mediated by HAT1, is thought to be an early event at DSB sites (55–57). It is also interesting to note that HAT1 was found in both replication-dependent histone H3.1 and replication-independent histone H3.3 nucleosome assembly complexes (46). In our system, although knockdown of either HAT1 or HIRA leads to changes of acetylation at multiple sites of H3 and H4, H2AK5 acetylation and H2A phosphorylation levels remained unaltered. HAT1-induced histone exchange is likely to be specific for the substitution of histone H3/H4 tetramers, as also suggested by a previous report (45).
We demonstrated that HAT1 complex exhibited an ATP-independent histone turnover activity in vitro. Because histone turnover activity was not detected with recombinant HAT1 or/and recombinant RbAp46, it is reasonable to believe that an additional factor(s) in the HAT1 complex is needed. Among the proteins that are associated with HAT1, we propose that HIRA is the chaperone that facilitates HAT1 to fulfill its histone turnover activity at DSB sites. HIRA is a histone chaperone implicated in H3.3-specific deposition and DNA synthesis-independent nucleosome assembly (39, 58–60). Moreover, HIRA has been shown to localize to laser micro-irradiation-induced DNA lesions (61). These observations are consistent with the role of HIRA in DNA repair and support our argument. Interestingly, knockdown of both HAT1 and HIRA in DR-U2OS cells had similar defects in HR repair compared with individual depletion of either one of the factors, supporting that HAT1 and HIRA function in the same pathway of HR repair.
In a previous report, HAT1 was shown to be recruited to DNA lesions in HR deficient RAD52Δ yeast strains, indicating the recruitment of HAT1 is upstream to that of HR key factor at DSB sites (45). We demonstrated that the recruitment of HAT1 is required for the subsequent deposition of histone variant H3.3 and altered histone modifications. The change of the histone dynamics at DSBs may function to define a chromatin state for the subsequent recruitment of mammalian HR key factor RAD51 to the sites for successful HR repair.
HAT1 is highly conserved from yeast to human. Depletion of HAT1 in yeast and chicken cells does not significantly change the overall cell growth or viability (62, 63). In S. cerevisiae, HAT1 is only required for HR repair in strains with H3K14R and K23R mutation, suggesting that HAT1 may be functionally redundant in these species (21). Our results showed that knockdown of HAT1 causes significant HR repair defects, cell cycle delay, and growth retardation even in the absence of DNA damaging agents, indicating that mammalian cells are more sensitive to HAT1 deficiency due to the absence of a compensatory activity.
HR repair of DSBs in mammalian cells is a complex, multi-step process. Although considerable progress has been made in understanding the role of histone dynamics in HR repair, important questions remain to be answered. For instance, it is not clear how chromatin disassembly/assembly and sequential recruitment of effectors to DSBs are affected by histone dynamics; that is, whether and how histone dynamics-flanking DSBs are regulated during cell cycle. We found that HAT1 promotes acetylation of histone H4 as well as incorporation of histone variant H3.3 at DSBs with its HIRA-dependent histone turnover activity. It is likely that HAT1-induced histone dynamics distinguishes damaged chromatin from the surrounding normal area, facilitating subsequent recruitment of key HR protein RAD51, for the ultimate HR repair. Although our present study cannot exclude the possibility that HAT1 participates in multiple events of HR repair of DSBs, it is clear that HAT1 plays an important role in the maintenance of genomic stability in higher eukaryotic cells.
Supplementary Material
This work was supported by National Natural Science Foundation of China Grants 91219201 and 81130048 (to Y. S.) and by Ministry of Science and Technology of China grants (973 Program: 2011CB504204; to Y. S.).

This article contains supplemental File 1.
- DSB
- DNA double-strand break
- HR
- homologous recombination
- NHEJ
- non-homologous end joining
- HAT1
- histone acetyltransferase 1
- TRITC
- tetramethylrhodamine isothiocyanate
- CPT
- camptothecin
- reHAT1
- recombinant HAT1
- reRbAp46
- recombinant RbAp46.
REFERENCES
- 1. Tsukuda T., Fleming A. B., Nickoloff J. A., Osley M. A. (2005) Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 438, 379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Downs J. A., Lowndes N. F., Jackson S. P. (2000) A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408, 1001–1004 [DOI] [PubMed] [Google Scholar]
- 3. Pei H., Zhang L., Luo K., Qin Y., Chesi M., Fei F., Bergsagel P. L., Wang L., You Z., Lou Z. (2011) MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 470, 124–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang H., Zhai L., Xu J., Joo H. Y., Jackson S., Erdjument-Bromage H., Tempst P., Xiong Y., Zhang Y. (2006) Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell 22, 383–394 [DOI] [PubMed] [Google Scholar]
- 5. Althaus F. R. (1992) Poly ADP-ribosylation. A histone shuttle mechanism in DNA excision repair. J. Cell Sci. 102, 663–670 [DOI] [PubMed] [Google Scholar]
- 6. Verzijlbergen K. F., van Welsem T., Sie D., Lenstra T. L., Turner D. J., Holstege F. C., Kerkhoven R. M., van Leeuwen F. (2011) A barcode screen for epigenetic regulators reveals a role for the NuB4/HAT-B histone acetyltransferase complex in histone turnover. PLoS Genet. 7, e1002284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen D., Dundr M., Wang C., Leung A., Lamond A., Misteli T., Huang S. (2005) Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J. Cell Biol. 168, 41–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kimura H., Cook P. R. (2001) Kinetics of core histones in living human cells. Little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153, 1341–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mizuguchi G., Shen X., Landry J., Wu W. H., Sen S., Wu C. (2004) ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348 [DOI] [PubMed] [Google Scholar]
- 10. Luk E., Ranjan A., Fitzgerald P. C., Mizuguchi G., Huang Y., Wei D., Wu C. (2010) Stepwise histone replacement by SWR1 requires dual activation with histone H2A.Z and canonical nucleosome. Cell 143, 725–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kusch T., Florens L., Macdonald W. H., Swanson S. K., Glaser R. L., Yates J. R., 3rd, Abmayr S. M., Washburn M. P., Workman J. L. (2004) Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306, 2084–2087 [DOI] [PubMed] [Google Scholar]
- 12. Parthun M. R., Widom J., Gottschling D. E. (1996) The major cytoplasmic histone acetyltransferase in yeast. Links to chromatin replication and histone metabolism. Cell 87, 85–94 [DOI] [PubMed] [Google Scholar]
- 13. Ai X., Parthun M. R. (2004) The nuclear Hat1p/Hat2p complex. A molecular link between type B histone acetyltransferases and chromatin assembly. Mol. Cell 14, 195–205 [DOI] [PubMed] [Google Scholar]
- 14. Ge Z., Wang H., Parthun M. R. (2011) Nuclear Hat1p complex (NuB4) components participate in DNA repair-linked chromatin reassembly. J. Biol. Chem. 286, 16790–16799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Verreault A., Kaufman P. D., Kobayashi R., Stillman B. (1998) Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr. Biol. 8, 96–108 [DOI] [PubMed] [Google Scholar]
- 16. Dutnall R. N., Tafrov S. T., Sternglanz R., Ramakrishnan V. (1998) Structure of the histone acetyltransferase Hat1. A paradigm for the GCN5-related N-acetyltransferase superfamily. Cell 94, 427–438 [DOI] [PubMed] [Google Scholar]
- 17. Nociari M. M., Shalev A., Benias P., Russo C. (1998) A novel one-step, highly sensitive fluorometric assay to evaluate cell-mediated cytotoxicity. J. Immunol. Methods 213, 157–167 [DOI] [PubMed] [Google Scholar]
- 18. Zhang H., Yi X., Sun X., Yin N., Shi B., Wu H., Wang D., Wu G., Shang Y. (2004) Differential gene regulation by the SRC family of coactivators. Genes Dev. 18, 1753–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu H., Chen Y., Liang J., Shi B., Wu G., Zhang Y., Wang D., Li R., Yi X., Zhang H., Sun L., Shang Y. (2005) Hypomethylation-linked activation of PAX2 mediates tamoxifen-stimulated endometrial carcinogenesis. Nature 438, 981–987 [DOI] [PubMed] [Google Scholar]
- 20. Li Q., Shi L., Gui B., Yu W., Wang J., Zhang D., Han X., Yao Z., Shang Y. (2011) Binding of the JmjC demethylase JARID1B to LSD1/NuRD suppresses angiogenesis and metastasis in breast cancer cells by repressing chemokine CCL14. Cancer Res. 71, 6899–6908 [DOI] [PubMed] [Google Scholar]
- 21. Wang Y., Zhang H., Chen Y., Sun Y., Yang F., Yu W., Liang J., Sun L., Yang X., Shi L., Li R., Li Y., Zhang Y., Li Q., Yi X., Shang Y. (2009) LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell 138, 660–672 [DOI] [PubMed] [Google Scholar]
- 22. Sandaltzopoulos R., Becker P. B. (1994) Solid phase DNase I footprinting. Quick and versatile. Nucleic Acids Res. 22, 1511–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gelbart M. E., Rechsteiner T., Richmond T. J., Tsukiyama T. (2001) Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays. Analyses using recombinant yeast histones and immobilized templates. Mol. Cell. Biol. 21, 2098–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seiden-Long I. M., Brown K. R., Shih W., Wigle D. A., Radulovich N., Jurisica I., Tsao M. S. (2006) Transcriptional targets of hepatocyte growth factor signaling and Ki-ras oncogene activation in colorectal cancer. Oncogene 25, 91–102 [DOI] [PubMed] [Google Scholar]
- 25. Parthun M. R. (2007) Hat1. The emerging cellular roles of a type B histone acetyltransferase. Oncogene 26, 5319–5328 [DOI] [PubMed] [Google Scholar]
- 26. Kleiman N. J., Wang R. R., Spector A. (1990) Ultraviolet light induced DNA damage and repair in bovine lens epithelial cells. Curr. Eye Res. 9, 1185–1193 [DOI] [PubMed] [Google Scholar]
- 27. Morrison A. J., Highland J., Krogan N. J., Arbel-Eden A., Greenblatt J. F., Haber J. E., Shen X. (2004) INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119, 767–775 [DOI] [PubMed] [Google Scholar]
- 28. Bennardo N., Cheng A., Huang N., Stark J. M. (2008) Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 4, e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oberdoerffer P., Michan S., McVay M., Mostoslavsky R., Vann J., Park S. K., Hartlerode A., Stegmuller J., Hafner A., Loerch P., Wright S. M., Mills K. D., Bonni A., Yankner B. A., Scully R., Prolla T. A., Alt F. W., Sinclair D. A. (2008) SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell 135, 907–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang X., Yu W., Shi L., Sun L., Liang J., Yi X., Li Q., Zhang Y., Yang F., Han X., Zhang D., Yang J., Yao Z., Shang Y. (2011) HAT4, a Golgi apparatus-anchored B-type histone acetyltransferase, acetylates free histone H4 and facilitates chromatin assembly. Mol. Cell 44, 39–50 [DOI] [PubMed] [Google Scholar]
- 31. Adachi N., Suzuki H., Iiizumi S., Koyama H. (2003) Hypersensitivity of nonhomologous DNA end-joining mutants to VP-16 and ICRF-193. Implications for the repair of topoisomerase II-mediated DNA damage. J. Biol. Chem. 278, 35897–35902 [DOI] [PubMed] [Google Scholar]
- 32. Horwitz S. B., Horwitz M. S. (1973) Effects of camptothecin on the breakage and repair of DNA during the cell cycle. Cancer Res. 33, 2834–2836 [PubMed] [Google Scholar]
- 33. Chapman J. R., Taylor M. R., Boulton S. J. (2012) Playing the end game. DNA double-strand break repair pathway choice. Mol. Cell 47, 497–510 [DOI] [PubMed] [Google Scholar]
- 34. Dunlop M. H., Dray E., Zhao W., Tsai M. S., Wiese C., Schild D., Sung P. (2011) RAD51-associated protein 1 (RAD51AP1) interacts with the meiotic recombinase DMC1 through a conserved motif. J. Biol. Chem. 286, 37328–37334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xiao J., Liu C. C., Chen P. L., Lee W. H. (2001) RINT-1, a novel Rad50-interacting protein, participates in radiation-induced G2/M checkpoint control. J. Biol. Chem. 276, 6105–6111 [DOI] [PubMed] [Google Scholar]
- 36. Bai Y., Symington L. S. (1996) A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 10, 2025–2037 [DOI] [PubMed] [Google Scholar]
- 37. Sugawara N., Wang X., Haber J. E. (2003) In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell 12, 209–219 [DOI] [PubMed] [Google Scholar]
- 38. Lorain S., Quivy J. P., Monier-Gavelle F., Scamps C., Lécluse Y., Almouzni G., Lipinski M. (1998) Core histones and HIRIP3, a novel histone-binding protein, directly interact with WD repeat protein HIRA. Mol. Cell. Biol. 18, 5546–5556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ray-Gallet D., Quivy J. P., Scamps C., Martini E. M., Lipinski M., Almouzni G. (2002) HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9, 1091–1100 [DOI] [PubMed] [Google Scholar]
- 40. Hopfner K. P., Karcher A., Shin D. S., Craig L., Arthur L. M., Carney J. P., Tainer J. A. (2000) Structural biology of Rad50 ATPase. ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101, 789–800 [DOI] [PubMed] [Google Scholar]
- 41. Sung P. (1994) Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 265, 1241–1243 [DOI] [PubMed] [Google Scholar]
- 42. He J., Shi L. Z., Truong L. N., Lu C. S., Razavian N., Li Y., Negrete A., Shiloach J., Berns M. W., Wu X. (2012) Rad50 zinc hook is important for the Mre11 complex to bind chromosomal DNA double-stranded breaks and initiate various DNA damage responses. J. Biol. Chem. 287, 31747–31756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang Y., Yang X., Gui B., Xie G., Zhang D., Shang Y., Liang J. (2011) Corepressor protein CDYL functions as a molecular bridge between polycomb repressor complex 2 and repressive chromatin mark trimethylated histone lysine 27. J. Biol. Chem. 286, 42414–42425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Verreault A., Kaufman P. D., Kobayashi R., Stillman B. (1996) Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87, 95–104 [DOI] [PubMed] [Google Scholar]
- 45. Qin S., Parthun M. R. (2006) Recruitment of the type B histone acetyltransferase Hat1p to chromatin is linked to DNA double-strand breaks. Mol. Cell. Biol. 26, 3649–3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y. (2004) Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61 [DOI] [PubMed] [Google Scholar]
- 47. Li Q., Zhou H., Wurtele H., Davies B., Horazdovsky B., Verreault A., Zhang Z. (2008) Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 134, 244–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vidanes G. M., Bonilla C. Y., Toczyski D. P. (2005) Complicated tails. Histone modifications and the DNA damage response. Cell 121, 973–976 [DOI] [PubMed] [Google Scholar]
- 49. Thiriet C., Hayes J. J. (2005) Chromatin in need of a fix. Phosphorylation of H2AX connects chromatin to DNA repair. Mol. Cell 18, 617–622 [DOI] [PubMed] [Google Scholar]
- 50. Ahmad K., Henikoff S. (2002) Histone H3 variants specify modes of chromatin assembly. Proc. Natl. Acad. Sci. U.S.A. 99, 16477–16484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ahmad K., Henikoff S. (2002) The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9, 1191–1200 [DOI] [PubMed] [Google Scholar]
- 52. McKittrick E., Gafken P. R., Ahmad K., Henikoff S. (2004) Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. U.S.A. 101, 1525–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schwartz B. E., Ahmad K. (2005) Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 19, 804–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Thiriet C., Hayes J. J. (2005) Replication-independent core histone dynamics at transcriptionally active loci in vivo. Genes Dev. 19, 677–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Unnikrishnan A., Gafken P. R., Tsukiyama T. (2010) Dynamic changes in histone acetylation regulate origins of DNA replication. Nat. Struct. Mol. Biol. 17, 430–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tamburini B. A., Tyler J. K. (2005) Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol. Cell. Biol. 25, 4903–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Qin S., Parthun M. R. (2002) Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol. Cell. Biol. 22, 8353–8365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lamour V., Lécluse Y., Desmaze C., Spector M., Bodescot M., Aurias A., Osley M. A., Lipinski M. (1995) A human homolog of the S. cerevisiae HIR1 and HIR2 transcriptional repressors cloned from the DiGeorge syndrome critical region. Hum. Mol. Genet. 4, 791–799 [DOI] [PubMed] [Google Scholar]
- 59. Ray-Gallet D., Woolfe A., Vassias I., Pellentz C., Lacoste N., Puri A., Schultz D. C., Pchelintsev N. A., Adams P. D., Jansen L. E., Almouzni G. (2011) Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol. Cell 44, 928–941 [DOI] [PubMed] [Google Scholar]
- 60. Goldberg A. D., Banaszynski L. A., Noh K. M., Lewis P. W., Elsaesser S. J., Stadler S., Dewell S., Law M., Guo X., Li X., Wen D., Chapgier A., DeKelver R. C., Miller J. C., Lee Y. L., Boydston E. A., Holmes M. C., Gregory P. D., Greally J. M., Rafii S., Yang C., Scambler P. J., Garrick D., Gibbons R. J., Higgs D. R., Cristea I. M., Urnov F. D., Zheng D., Allis C. D. (2010) Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Adamson B., Smogorzewska A., Sigoillot F. D., King R. W., Elledge S. J. (2012) A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat. Cell Biol. 14, 318–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Barman H. K., Takami Y., Ono T., Nishijima H., Sanematsu F., Shibahara K., Nakayama T. (2006) Histone acetyltransferase 1 is dispensable for replication-coupled chromatin assembly but contributes to recover DNA damages created following replication blockage in vertebrate cells. Biochem. Biophys. Res. Commun. 345, 1547–1557 [DOI] [PubMed] [Google Scholar]
- 63. Benson L. J., Phillips J. A., Gu Y., Parthun M. R., Hoffman C. S., Annunziato A. T. (2007) Properties of the type B histone acetyltransferase Hat1. H4 tail interaction, site preference, and involvement in DNA repair. J. Biol. Chem. 282, 836–842 [DOI] [PubMed] [Google Scholar]
- 64. Kleff S., Andrulis E. D., Anderson C. W., Sternglanz R. (1995) Identification of a gene encoding a yeast histone H4 acetyltransferase. J. Biol. Chem. 270, 24674–24677 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.