Background: PerR from Streptococcus pyogenes is critical for bacterial virulence.
Results: We determined the crystal structure of PerR and deduced the molecular mechanism of stress sensing and gene regulation by PerR.
Conclusion: A novel structural motif present at the N terminus of PerR is important for metal binding, oxidative stress sensing, and GAS virulence.
Significance: The structural elements of PerR identified here could potentially be targeted for therapeutic development.
Keywords: Crystal Structure, DNA-binding Protein, Gene Regulation, Oxidative Stress, Pathogenesis
Abstract
Regulation of oxidative stress responses by the peroxide stress regulator (PerR) is critical for the in vivo fitness and virulence of group A Streptococcus. To elucidate the molecular mechanism of DNA binding, peroxide sensing, and gene regulation by PerR, we performed biochemical and structural characterization of PerR. Sequence-specific DNA binding by PerR does not require regulatory metal occupancy. However, metal binding promotes higher affinity PerR-DNA interactions. PerR metallated with iron directly senses peroxide stress and dissociates from operator sequences. The crystal structure revealed that PerR exists as a homodimer with two metal-binding sites per subunit as follows: a structural zinc site and a regulatory metal site that is occupied in the crystals by nickel. The regulatory metal-binding site in PerR involves a previously unobserved HXH motif located in its unique N-terminal extension. Mutational analysis of the regulatory site showed that the PerR metal ligands are involved in regulatory metal binding, and integrity of this site is critical for group A Streptococcus virulence. Interestingly, the metal-binding HXH motif is not present in the structurally characterized members of ferric uptake regulator (Fur) family but is fully conserved among PerR from the genus Streptococcus. Thus, it is likely that the PerR orthologs from streptococci share a common mechanism of metal binding, peroxide sensing, and gene regulation that is different from that of well characterized PerR from Bacillus subtilis. Together, our findings provide key insights into the peroxide sensing and regulation of the oxidative stress-adaptive responses by the streptococcal subfamily of PerR.
Introduction
Bacterial pathogens often encounter highly toxic reactive oxygen species (ROS),2 a key component of the host defense mechanism against invading pathogens (1). The oxidative damage caused by ROS, such as hydroxyl radicals and superoxide anions, is deleterious to cellular proteins, DNA, and membrane lipids and can result in cell death (2–4). To combat the toxic effects of ROS, bacteria have evolved highly sophisticated defense mechanisms that include enzymes to detoxify the free radicals, enzymes to repair the macromolecular damage, mechanisms to protect the macromolecules, and mechanisms to alter the cytosolic metal content (5). In most cases, these mechanisms are components of inducible adaptive responses that are controlled by peroxide-sensing global transcription regulators. With few exceptions, this regulation is mediated by structurally distinct global regulators, OxyR, in Gram-negative bacteria and PerR in Gram-positive bacteria (4, 6).
Group A Streptococcus (GAS, Streptococcus pyogenes) is a human pathogen that causes a variety of clinical manifestations with inflammatory lesions that range from mild pharyngitis to complicated necrotizing fasciitis (7). Given the abundance of ROS at the colonization interface, which is either produced endogenously by GAS (8) or exogenously by the host immune system (1), it is expected that GAS possesses elaborate oxidative stress resistance mechanisms. Consistent with this, GAS senses oxidative stress and mounts inducible responses that confer resistance against oxidative damage (9). However, the adaptive responses induced in GAS differ significantly from the classical oxidative stress defense mechanisms observed in other bacteria. Importantly, GAS does not produce catalase and lacks components of heme biosynthetic pathways, the two major factors that are typically employed in bacterial resistance against oxidative stress (10). In addition, other constituents of an oxidative stress defense mechanism, including alkyl hydroperoxidase (AhpC), glutathione peroxidase (GpoA), superoxide dismutases (SodA), and MrgA, are not part of the GAS peroxide stimulon (9, 11, 12). Thus, the oxidative stress responses of GAS involve novel factors and molecular mechanisms that are distinct from those present in other bacteria.
As in other Gram-positive bacteria, the inducible peroxide stress responses in GAS are predominantly controlled by PerR, a peroxide-sensing regulator (11). PerR influences gene regulation at the level of transcription, and this gene regulation is critical for resistance against ROS-mediated killing by phagocytes and pharyngeal colonization (11–14). When grown in laboratory growth medium, inactivation of perR leads to hyper-resistance against peroxide stress in vitro, and dysregulation of the PerR regulon in an isogenic perR mutant significantly attenuated GAS virulence in several mouse models of infection (9, 15). Depending on the strain M protein serotype, the GAS peroxide regulon varies significantly in composition and number of regulated genes. In the serotype M5 strain, the PerR regulon is composed of only six genes, of which only the transcription of the metal efflux transporter, PmtA, is directly controlled by PerR (11, 14). Conversely, transcriptome studies of a ΔperR mutant in a serotype M3 strain revealed a larger regulon composed of 42 genes (13). Surprisingly, most of the PerR-regulated genes identified were involved in sugar utilization and transport pathways, and only a moderate effect on the expression of peroxidases was observed (13). However, analogous studies in the presence of peroxide stress yielded very different results. First, there is limited overlap in the genes controlled by PerR in the presence and absence of oxidative stress, with most of the regulated genes during oxidative stress being involved in protein and DNA metabolism (12). Second, instead of up-regulating transcription of genes encoding peroxidases and MrgA during oxidative stress, PerR down-regulates the expression of genes involved in protein and DNA metabolism (12). Thus, PerR in GAS controls a novel regulatory circuit in response to oxidative stress. These adaptive responses are critical for GAS survival within the host and pathogenesis.
PerR in GAS is a 155-amino acid protein that belongs to the Fur family of regulators. Members of the Fur family are homodimeric metalloregulators that control the expression of genes involved in metal homeostasis. PerR forms a subfamily of Fur regulators that mediates gene regulation in response to peroxide stress. Although PerR homologs are found in most of the firmicutes and in some Gram-negative bacteria, PerR from Bacillus subtilis (PerRBs) is the best characterized (16–22). Structurally, it exists as a homodimer with two functional domains, an N-terminal DNA-binding domain and a C-terminal dimerization domain (23–25). Each subunit of PerRBs has a structural zinc-binding site in its dimerization domain and a regulatory metal-binding site in the interdomain region (23–25). Under physiological conditions, apoPerRBs (PerR-Zn) binds to either manganese (PerRBs-Zn-Mn) or iron (PerRBs-Zn-Fe). The regulatory metal-bound holorepressor binds to a highly conserved binding motif called a per box in the target promoters to negatively regulate transcription (19). Only the PerRBs-Zn-Fe form, not the PerRBs-Zn-Mn, is responsive to peroxide stress (19, 20). The iron at the regulatory site catalyzes the oxidation of the histidines that coordinate the regulatory metal (20). Conformational changes induced by the oxidation of PerRBs and the resulting loss of regulatory metal lead to its dissociation from DNA and cause derepression of target genes (20, 25). However, the analogous details on the molecular events in GAS PerR that lead to metal binding, DNA binding, peroxide sensing, gene regulation, and the contribution of these events to GAS pathogenesis remain poorly understood.
To understand the mechanistic basis of PerR function, we carried out genetic, biochemical, and structural characterization of serotype M3 GAS PerR. Although the results from this study have parallels with PerRBs, importantly they also reveal several distinct structural and mechanistic differences in the mode of DNA binding, metal binding, and peroxide sensing. The crystal structure of PerR revealed a novel regulatory metal-binding motif that was not previously observed in the structures of PerRBs or any other Fur family regulators. This N-terminal HXH metal-binding motif is unique to PerR from the genus Streptococcus and is critical for regulatory metal binding and GAS virulence.
EXPERIMENTAL PROCEDURES
Ethics Statement
Mouse experiments were performed according to protocols approved by the Methodist Hospital Research Institute Institutional Animal Care and Use Committee. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals, 8th Edition. The protocol was approved by the Institutional Animal Care and Use Committee of The Methodist Hospital Research Institute (OLAW assurance no. A4555-01; United States Department of Agriculture assurance no. 740R-0192). No surgery was performed. All efforts were made to minimize animal suffering.
Bacterial Strains, Plasmids, and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Table 1. Strain MGAS10870 is a previously described invasive serotype M3 isolate whose genome has been fully sequenced (26). MGAS10870 is representative of serotype M3 strains that cause invasive infections and has a wild-type sequence for all major regulatory genes (26). GAS was grown routinely on trypticase soy agar containing 5% (v/v) sheep blood (BSA; BD Biosciences) or in Todd-Hewitt broth containing 0.2% (w/v) yeast extract (THY; Difco). When required, spectinomycin, ampicillin, or chloramphenicol was added to a final concentration of 150, 40, and 5 μg/ml, respectively. All GAS growth experiments were done in triplicate on four separate occasions for a total of 12 replicates. Overnight cultures were inoculated into fresh medium to achieve an initial A600 of 0.03. Growth was monitored by measuring A600. The Escherichia coli strain used for protein overexpression was grown in Luria-Bertani broth (LB broth; Fisher Scientific), and when appropriate ampicillin was added to a final concentration of 40 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Ref. |
|---|---|---|
| Strains | ||
| MGAS10870 | Invasive isolate, serotype M3 | 26 |
| MGAS10870:pDC | MGAS10870 with empty vector, Cm+ | 51 |
| ΔperR | MGAS10870ΔperR::aad9 | This study |
| ΔperR:pDC | MGAS10870ΔperR::aad9, empty vector, Cm+ | This study |
| ΔperR-pDC:perR | MGAS10870ΔperR::aad9, perR+, Cm+ | This study |
| Plasmids | ||
| pSL60-1 | Vector containing aad9 gene encoding spectinomycin resistance | 36 |
| pDC123 | Low copy number plasmid capable of replication in GAS and E. coli, Cm+ | 50 |
| pDC:perR | pDC123 with entire perR gene plus promoter, Cm+ | This study |
Protein Overexpression and Purification
The perR gene of strain MGAS10870 was cloned into plasmid pET-21b, and protein was overexpressed in E. coli strain BL21 (DE3) after induction with 1 mm isopropyl 1-thio-β-d-galactopyranoside at 30 °C. Cell pellets were suspended in 50 ml of buffer A (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 5% (v/v) glycerol, and 1 mm Tris 2-carboxyethyl phosphine hydrochloride (TCEP)) supplemented with one protease inhibitor mixture pellet and DNase I to a final concentration of 5 μg/ml. Cells were lysed by a cell disruptor (Constant Systems), and cell debris was removed by centrifugation at 15,000 rpm for 30 min. PerR was purified by affinity chromatography using a nickel-nitrilotriacetic acid-agarose column, and the protein was eluted using an imidazole gradient. The pooled fractions were concentrated, and buffer was exchanged with buffer B (20 mm Tris-HCl, pH 8.0, 50 mm NaCl, 5% (v/v) glycerol, and 1 mm TCEP). Subsequently, PerR was loaded to a pre-equilibrated Hitrap-heparin (5 ml) column, and bound PerR was eluted using a NaCl gradient of 50–500 mm. Purified PerR fractions were concentrated and subjected to 10 mm EDTA treatment to remove any trace metals bound to the protein. All subsequent steps were carried out with buffer treated with Chelex beads (Sigma). The concentrated PerR was buffer-exchanged into storage buffer (20 mm Tris-HCl, pH 8.0, 200 mm NaCl, 1 mm TCEP) by size exclusion chromatography with a Superdex 200G column. The protein was purified to >95% homogeneity, and purified PerR was concentrated using Amicon YM-10 (10-kDa cutoff) membrane filter to a final concentration of ∼20 mg/ml. The selenomethionine-derivatized PerR (SeMet PerR) was overexpressed using the methionine inhibitory pathway (27) and purified as described for native PerR.
Crystallization, Data Collection, and Structure Determination
Crystallization was performed using the vapor diffusion method with the crystallization solution containing 1–2 m NaCl and 0.1 m Tris-HCl, pH 8.0. The native and SeMet-PerR crystals were flash-frozen using 30% (v/v) glycerol as a cryoprotectant and diffracted to 2.0 Å resolution. The multiple anomalous dispersion diffraction data were collected at the absorption edges of both selenium and zinc using SeMet-PerR crystals and native crystals, respectively, under cryogenic conditions at Advance Light Source beam line 8.3.1 (Berkeley, CA). Data were processed with MOSFLM (28, 29) and SCALA (30). The crystals belong to space group P 63 2 2 with unit cell dimensions of a = 119 Å, b = 119 Å, and c = 118.6 Å. Seven (six methionines per monomer) out of a possible 12 selenium sites were located using SOLVE (31), and density modification and initial automated model building were carried out using RESOLVE (32). After iterated rounds of model building using “COOT” (33) and refinement using CNS (34), the final refined model has an Rfree of 26.4% and an Rwork of 24.2%. The final model contains residues 2–147 of subunit A, residues 2–148 of subunit B, two zinc ions, two nickel ions, and 153 water molecules. Selected data collection, phasing, and refinement statistics are given in Table 2. All structure-related figures were generated using PyMOL (33). The coordinates and structure factors have been deposited in the Protein Data Bank with accession code 4I7H.
TABLE 2.
Selected crystallographic data and statistics
| Data collection and phasing | ||||
|---|---|---|---|---|
| Dataset | SeMet | Native | ||
| Wavelength | 0.9796 | 0.957 | 0.9798 | 1.116 |
| Resolution | 50.0 to 2.9 Å | 50.0 to 2.0 Å | ||
| Rsyma | 0.1 (0.22) b | 0.099 (0.5) | 0.096 (0.51) | 0.072 (0.54) |
| I/s(I) | 30.2 (8.0) | 30.8 (8.3) | 31.5 (8.5) | 22.7 (5.0) |
| Total reflections | 338,971 | 336,954 | 337,956 | 600,731 |
| Unique reflections | 11,662 | 11,596 | 11,642 | 33,859 |
| Completeness | 100% (100%) | 100% (100%) | 100% (100%) | 99.7% (99.8%) |
| Selenium sites(identified/total no of sites) | 7/12 | |||
| Overall figure of meritc | 0.59 | |||
| Refinement statistics | ||||
| Resolution range | 50.0 to 2.0 Å | |||
| Rwork/Rfreed | 24.1/26.2% | |||
| Atoms | ||||
| Protein | 2369 | |||
| Zinc ions | 2 | |||
| Nickel ions | 2 | |||
| Solvent | 153 | |||
| B factors | 42.3 Å2 | |||
| r.m.s.d. | ||||
| Bond lengths | 0.006 Å | |||
| Bond angles | 1.26° | |||
| Ramachandran analysis | ||||
| Most favored | 97.94% | |||
| Additionally allowed | 2.06% | |||
| Generally allowed | 0.0% | |||
| Disallowed | 0.0% | |||
a Rsym = ΣΣ|Ihkl − Ihkl(j)|/Σ Ihkl, where Ihkl(j) is the observed intensity, and Ihkl is the final average intensity value.
b Values in parentheses are for the highest resolution shell.
c Figure of merit = 〈|SP(a)ei/SP(a)|〉, where a is the phase, and P(a) is the phase probability distribution.
d Rwork = S‖Fobs| − |Fcalc‖/S|Fobs| and Rfree = S‖Fobs| − |Fcalc‖/S|Fobs|; where all reflections belong to a test set of 5% randomly selected reflections.
Site-directed Mutagenesis of PerR
Plasmid pET21b-perR or pDC-perR containing the wild-type perR-coding region was used as template for the site-directed mutagenesis. QuikChange site-directed mutagenesis kit (Stratagene) was used to introduce single amino acid substitutions within the perR coding region, and mutations were verified by DNA sequencing.
Preparation of PerR-Zn and PerR-Zn-Mn Forms of PerR and Its Mutants
Protein overexpression and purification were performed as described previously with the following modifications. PerR eluted by nickel affinity chromatography was subjected to two rounds of overnight dialysis against buffer containing 10 mm EDTA and one more round of dialysis against buffer containing 100 mm EDTA. Dialyzed protein was further purified by size exclusion chromatography using chelexed buffers containing 1 mm EDTA and stored at −80 °C. To reconstitute the PerR-Zn-Mn forms of wild-type PerR and PerR mutant proteins, the apoPerR-Zn was dialyzed overnight against chelexed buffer containing MnCl2 to a final concentration of 10 μm. The metallated states of PerR were verified by ICP-MS.
Metal Content Analysis by ICP-MS
The metal content of purified protein preparations was determined using the ICP-MS DRCII system (PerkinElmer Life Sciences) with gallium as an internal standard as described previously (35). All the measurements were recorded in triplicates using 2% (v/v) ultra-pure nitric acid diluted samples from a 10 μm purified protein stock solution. The final metal content with standard deviation is reported.
Electrophoretic Mobility Shift Assay
Oligonucleotides containing the putative per box from the pmtA promoter (top strand 5′-TTTATTTAGAATTATTATAATTAATA) were annealed by heating an equimolar mixture of top and bottom strand oligonucleotides at 95 °C for 5 min followed by slow cooling to room temperature. Binding reactions were carried out in a 20-μl volume of binding buffer (20 mm Tris, pH 8.0, 200 mm NaCl, 12% (v/v) glycerol and 1 mm TCEP) containing 200 nm oligoduplex, 1 μg of poly(dI/dC), and 4.4 μm of PerR. After 20 min of incubation at room temperature, the reaction mixtures were resolved on a 10% (v/v) native polyacrylamide gel supplemented with 5% (v/v) glycerol (native PAGE) for 45 min at 150 V at 4 °C in Tris borate EDTA (TBE) buffer with 5% (v/v) glycerol. The gels were stained with ethidium bromide and analyzed on a Bio-Rad gel electrophoresis systems.
Fluorescent Polarization Assay
Fluorescence polarization-based assays were performed with a Synergy H1 hybrid microplate reader (Biotek) utilizing the fluorescence of fluorescein-labeled DNA. To determine the effects of metals and chelating agents on PerR-DNA interactions, 10 nm 5′-fluoresceinated oligodeoxynucleotide duplexes containing per box sequences in binding buffer (20 mm Tris-HCl, pH 7.5, 100 mm NaCl, 2.5% (v/v) glycerol, and 1 mm TCEP) were titrated against increasing concentrations of purified PerR. The chelexed binding buffer was supplemented with either 20 μm manganese or 50 μm EDTA. Samples were excited at 485 nm, and emission was measured at 520 nm. The DNA binding data for the metallated form of PerR were fitted to the quadratic equation Y = B + (A/2) (1/P) ((Kd + P + D) − ((Kd + P + D)2 − (4PD))1/2), where B denotes the intercept, A the maximum change in polarization, P the protein concentration, and D the DNA concentration. Binding data for the apoPerR were fitted with the equation P = {(Pbound − Pfree)[protein]/(KD + [protein])} + Pfree, where P is the polarization measured at a given protein concentration; Pfree is the initial polarization of the free ligand; Pbound is the maximum polarization of specifically bound ligand, and [protein] is the protein concentration. Nonlinear least squares analysis was used to determine Pbound, and Kd. The binding constant reported is the average value from three independent experimental measurements.
Construction of perR-inactivated Mutant Strain
Insertional inactivation of the perR gene in wild-type strain MGAS10870 was performed by methods described previously (36, 37). Briefly, a PCR fragment containing a spectinomycin resistance (spc) cassette with the fragment of gene to be deleted on either side was generated in a three-step PCR process. Subsequently, the plasmid with the spc gene disruption cassette was introduced into the parent strain by electroporation, and the gene was disrupted through homologous recombination. The isogenic mutant strains were selected by growth on spectinomycin-containing medium. Inactivation of the gene was confirmed by DNA sequencing. Primers used for the construction of strain 10870 ΔperR are listed in supplemental Table S1.
Construction of perR Trans-complementation Plasmid
To complement isogenic mutant strain ΔperR, the coding sequence of the full-length perR gene together with the perR promoter region were cloned into the E. coli-GAS shuttle vector pDC123 (38). Using the primers listed in supplemental Table S1, the respective fragments were amplified by PCR from GAS genomic DNA, digested with BglII and NdeI, and ligated into digested vector pDC123. The inserts were verified by DNA sequencing and electroporated into the appropriate mutant derivatives of strain MGAS10870.
Transcript Analysis by Quantitative RT-PCR
GAS strains were grown to late exponential phase (A600 ∼1.2) and incubated with 2 volumes of RNAprotect (Qiagen) for 10 min at room temperature. Bacteria were harvested by centrifugation, and the cell pellets were snap-frozen with liquid nitrogen. RNA isolation and purification were performed using an RNeasy kit (Qiagen). Purified RNA was analyzed for quality and concentration with an Agilent 2100 Bioanalyzer. cDNA was synthesized from the purified RNA using Superscript III (Invitrogen), and TaqMan quantitative RT-PCR was performed with an ABI 7500 Fast System (Applied Biosystems). Comparison of transcript levels was done using ΔCT method of analysis using tufA as the endogenous control gene (39). The TaqMan primers and probes used are listed in supplemental Table S1.
Mouse Infection Studies
Virulence of the isogenic mutant GAS strains was tested using an intraperitoneal mouse model of infection. For intraperitoneal infection, 20 female 3–4-week-old CD1 mice (Harlan Laboratories) were used for each GAS strain. Animals were inoculated intraperitoneally with 1 × 107 CFUs, and survival was monitored daily. Data were graphically displayed as a Kaplan-Meier survival curve and analyzed using the log-rank test.
RESULTS AND DISCUSSION
Purified Recombinant PerR Exists in Apo-PerR-Zn Form
To investigate the metal binding, metal dependence for DNA binding, and metal selectivity for peroxide sensing by PerRGAS, we constructed a C-terminal hexahistidine-tagged PerRGAS. For the purpose of clarity, the PerRGAS will be referred to hereafter as PerR. Because the DNA binding and oxidation of PerRBs is influenced by contaminating trace metals (19, 20), we purified the recombinant PerR under metal-free conditions using chelating agent EDTA in the buffer conditions. The protein was dialyzed extensively against buffer containing 100 mm EDTA and stored in chelexed metal-free buffer. To ensure that the purified protein exists in the apoPerR-Zn form, we assayed the metal content of the purified PerR by ICP-MS. The results indicate that PerR has only one zinc per subunit (Table 3). Because structural zinc binding is critical for the stability and dimerization of PerR and most of the Fur family regulators (12, 21), we conclude that zinc in PerR is likely bound to its structural site, with its regulatory site unoccupied.
TABLE 3.
Metal content analysis by ICP-MS of crystallized and EDTA treated PerR
| Protein | EDTA treatment | Moles of zinc/mol of PerRa | Moles of nickel/mol of PerRa |
|---|---|---|---|
| Crystallized wild type | No | 1.14 ± 0.01 | 0.95 ± 0.01 |
| Wild type | Yes | 0.97 ± 0.01 | Not detectedb |
| H6A | Yes | 1.08 ± 0.02 | Not detectedb |
| H19Ac | Yes | 1.21 ± 0.05 | Not detectedb |
| H44A | Yes | 0.9 ± 0.02 | 0.5 ± 0.00 |
| H97A | Yes | 1.16 ± 0.02 | Not detectedb |
| H99A | Yes | 1.04 ± 0.02 | Not detectedb |
a Measurements were reported as average from triplicate studies with standard deviation.
b Metal content less than 10% of the protein molar ratio is not reported.
c This has a copper content of >10% of the protein molar ratio.
PerR Does Not Require Regulatory Metal to Bind Per Box Sequences
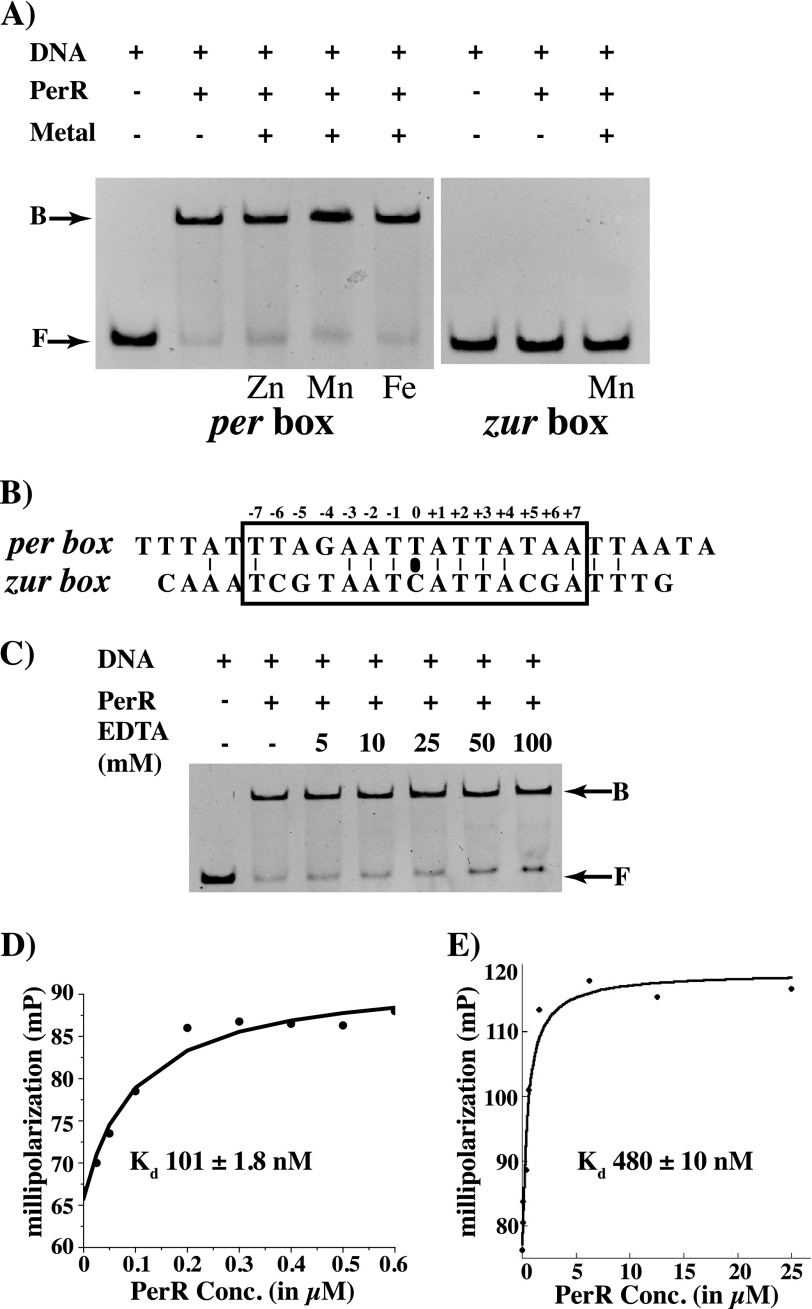
The paradigm of repression by Fur family members holds that metal occupancy at the regulatory site is required for the repressor to interact with DNA (19, 40, 41). We tested the metal dependence of PerR for its DNA binding in metal-free conditions by gel mobility shift assay. When PerR was incubated with oligoduplexes containing the putative per boxes from the pmtA promoter, PerR bound to the per box sequences in the absence of metals, and the metal ions did not alter PerR-per box interactions, indicating that PerR does not require regulatory metal to bind cognate DNA (Fig. 1A). To ensure that the observed interactions are sequence-specific, we performed similar experiments with oligoduplexes containing the binding sites for zinc uptake regulator (zur box) from B. subtilis (18). Comparison of the nucleotide sequences between the two binding sites indicated that they are identical at 9 out of 14 bases within the pseudo-palindromes and symmetrical differences occur at positions 5 and 6 (Fig. 1B). However, when PerR-zur box interactions were tested, PerR did not bind to the zur box, even in the presence of metal (Fig. 1A). To rule out the possibility that trace metals present in the reaction conditions might influence PerR-DNA interactions, we tested the PerR-DNA interactions in the presence of increasing concentrations of chelating agent EDTA. Consistent with our previous observation that regulatory metal is not required for DNA binding, DNA binding by PerR is not affected by EDTA even at concentrations as high as 100 mm (Fig. 1C). Based on these results, we conclude that PerR interacts directly with per box sequences in a sequence-specific fashion, and these interactions do not require the occupancy of the regulatory metal-binding site.
FIGURE 1.
Metal-independent DNA binding by PerR. A, PerR-DNA interactions were probed by gel mobility shift assay. Oligoduplexes (0.2 μm) containing the sequences corresponding to either the putative per box from the pmtA promoter (left panel) or zur box from B. subtilis (right panel) were incubated with purified PerR (4.4 μm) in the presence and absence of 7.5 μm of the indicated metals. B, alignment of the nucleotide sequence corresponding to per box from pmtA promoter and zur box from B. subtilis. The positions of each nucleotide within the palindrome are numbered on top, and the characteristic 7-1-7 motif identified in the B. subtilis PerR-binding site is boxed. The black oval indicates the center of the inverted repeat. C, effect of chelating agent EDTA on PerR-DNA interactions probed by gel mobility shift assay. PerR-DNA interactions were titrated against indicated concentrations of EDTA, and the reaction mixture was resolved on a 10% (v/v) native-PAGE. The positions of free probe (F) and protein-bound probe (B) are indicated by arrows. Binding isotherms were determined by fluorescent polarization assay for PerR binding to fluoresceinated oligoduplex containing per box sequences (10 nm) in the presence of 20 μm MnCl2 (D) or 100 μm EDTA (E). The change in polarization (mP) was plotted against PerR concentration (in micromolar), and the resulting curve was fitted to a quadratic equation (D) and simple bimolecular model by nonlinear least square regression analysis (E). The apparent binding constants with standard deviations are shown in each plot.
To better understand the effect of metals on PerR-DNA interactions, fluorescence polarization assays were carried out with oligoduplexes containing per box sequences in the presence of either 20 μm MnCl2 or 100 μm EDTA. In the presence of metal, PerR binds to operator sequences with a dissociation constant (Kd) of 100 nm (Fig. 1D). However, when the similar experiment was performed with apoPerR under metal-free conditions, a 5-fold reduction (Kd = 480 nm) in the affinity of PerR for per box sequences was observed (Fig. 1E). Together, these data demonstrate that apoPerR retained the ability to bind DNA in a sequence-specific fashion, and metallation at the regulatory site of PerR promotes tighter PerR-DNA interactions.
Peroxide Sensing by PerR Requires Regulatory Metal
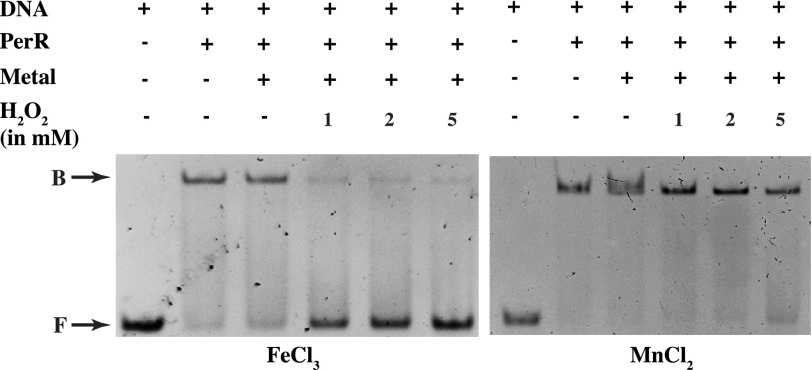
Given its role as a key regulator of the GAS-adaptive responses to oxidative stress, we next investigated whether PerR directly senses peroxide stress by metal-catalyzed oxidation (MCO) and responds by dissociating from the operator sequences. Using gel mobility shift assay, we tested increasing concentrations of hydrogen peroxide for its ability to dissociate the active PerR-Zn-Fe form from the cognate DNA-binding site. Purified PerR-Zn was incubated with ferric chloride to reconstitute the PerR-Zn-Fe form. Results clearly demonstrate that exposure to peroxide in the presence of iron resulted in complete loss of DNA binding by PerR (Fig. 2, left panel). Because iron is a key mediator of the generation of toxic hydroxyl radicals and MCO of PerRBs (19), we next investigated the metal specificity for peroxide stress sensing and inactivation of PerR. We performed a similar experiment but substituted iron with manganese and assessed peroxide-mediated inactivation of PerR. Consistent with the hypothesis that iron is required for peroxide sensing by PerR, PerR-Zn-Mn is insensitive to the peroxide stress, and no loss of DNA binding was observed (Fig. 2, right panel). Together, these data indicate that PerR directly senses peroxide stress through iron-dependent MCO and relieves the repression of the target genes by dissociating from its binding sites in the target promoters.
FIGURE 2.
Peroxide sensing by PerR requires iron. PerR-per box interactions were challenged by increasing concentrations of hydrogen peroxide (H2O2) in the presence of either ferric chloride (left panel) or manganese chloride (right panel). The reaction mixture was resolved on 10% (v/v) native-PAGE. The reaction conditions for this assay were devoid of chelating agent EDTA. Bands corresponding to unbound (F) and PerR-bound probe (B) are indicated by arrows.
Crystal Structure of PerR
To better understand the molecular mechanism of metal binding, operator recognition, peroxide sensing, and gene regulation by PerR, we determined the crystal structure of PerR to 2.9 Å resolution by multiple anomalous dispersion phasing method using selenomethionine-derivatized protein. Subsequently, the high resolution native data set was used for structure refinement. The final model was refined to 2.0 Å resolution with a Rfree and Rwork of 26.4 and 24.2, respectively. The resulting model has excellent stereochemistry, with 98% of the residues in the most allowed region of Ramachandran plot and none in the disallowed region (Table 2). The data collection, phasing, and structure refinement statistics are shown in Table 2.
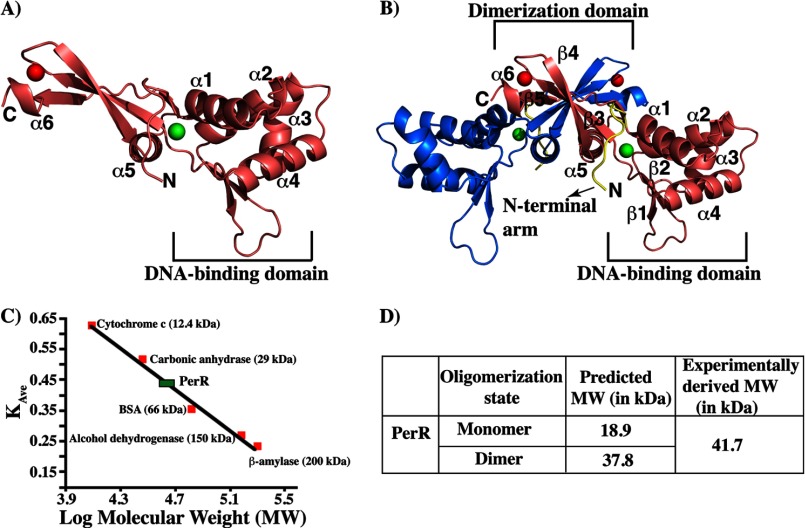
The overall topology of the secondary structure elements of PerR dimer is similar to the structures of Fur family regulators (Fig. 3A). However, PerR has an 11-amino acid N-terminal arm that is not present in the structures of other Fur family regulators. This extended N-terminal region has a significant role in metal binding and peroxide sensing (Fig. 3B) (discussed below). Analysis of purified PerR in solution by size exclusion chromatography indicates that PerR exists as a dimer (Fig. 3, C and D). Consistent with this observation, each asymmetric unit has one biologically relevant PerR dimer (Fig. 3, B–D). As observed in the structures of other members of the Fur family of regulators, PerR has a two-domain architecture as follows: an N-terminal DNA-binding domain (2–94) and a C-terminal dimerization domain (98–148) that are connected to each other by a hinge region (95–97) (Fig. 3, A and B). The three helices (α2-α4) of the N-terminal domain along with the two β-strands (β1-β2) form the characteristic winged helix-turn-helix motif (wHTH), in which helix 4 (α4) forms the “recognition helix.” The recognition helix is predicted to make sequence-specific contacts within the major groove of its cognate DNA sequences, whereas the wing motif interacts with the bases in the adjacent minor grooves (42). The C-terminal dimerization domain has the typical α/β-fold with most of the inter-subunit interactions occurring between helix α5 and strands β5 and β6 and their symmetry related partners (helix α5′ and strands β5′ and β6′, ′ indicates the second subunit) (Fig. 3B). This dimerization interface is extensive and buries ∼1500 Å2 surface area within the C-terminal domain of the molecule.
FIGURE 3.
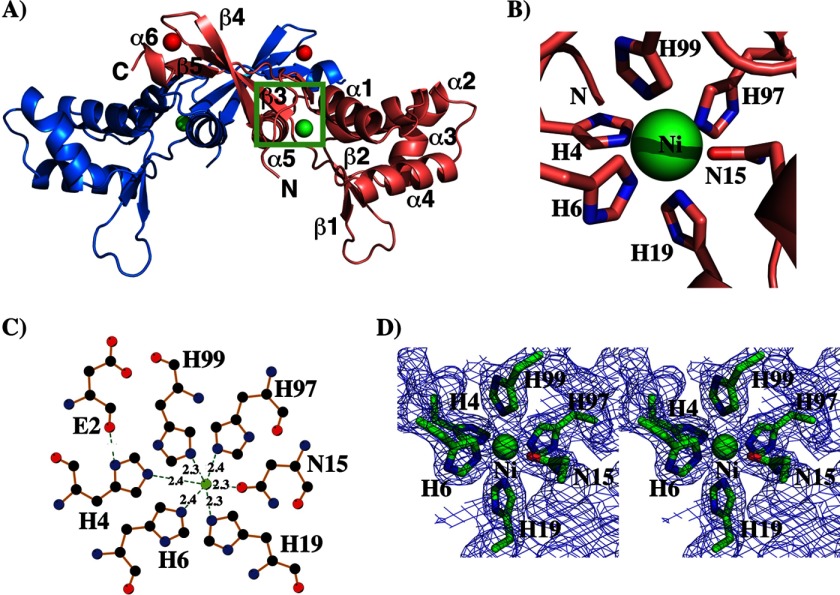
Structural and biochemical characterization of PerR dimer. Ribbon representation of the crystal structure of an individual subunit (A) and dimeric form of PerR (B). The individual subunits of a PerR dimer are color-coded. The two metals bound to each PerR subunit are shown as spheres, and the structural and regulatory metals are color-coded in red and green, respectively. The secondary structural elements of one subunit are labeled, and the N and C termini of the molecule are labeled as N and C, respectively. The N-terminal DNA-binding domain and the C-terminal dimerization domain in PerR structure are marked and labeled. The N-terminal extension of both subunits of PerR is colored in yellow. C, assessment of the oligomerization state of PerR in solution by size exclusion chromatography. The linear fit of the elution volumes (KAve) of five protein molecular weight standards, cytochrome c (12.4 kDa), carbonic anhydrase (29 kDa), bovine serum albumin (BSA, 66 kDa), alcohol dehydrogenase (150 kDa), and β-amylase (200 kDa), to their log molecular weight is shown as a black line on the graph. The elution volume (KAve) of the purified PerR was plotted on the graph (green rectangle). D, PerR exists as a dimer in solution. The theoretical molecular weights of different oligomeric states of PerR were calculated based on their amino acid sequence, and the experimentally calculated molecular weights of PerR based on the elution profile in a Superdex-200 size exclusion chromatography are shown.
PerR Has Two Metal-binding Sites
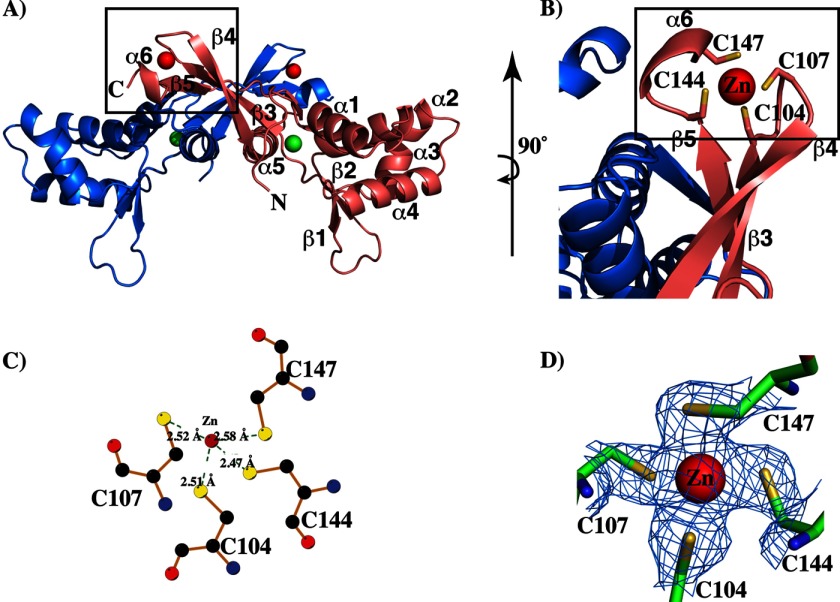
Each subunit of PerR has two metal-binding sites. Site 1 is located within the dimerization domain, and site 2 is located at the interdomain region (Fig. 3, A and B). To confirm the identity and the position of the metals, we used the anomalous scattering data collected at the absorption edge of zinc, and the data indicated that each PerR subunit has one zinc located at site 1. The zinc at this site is tetra-coordinated by four highly conserved cysteines from the C-terminal domain, Cys-104, Cys-107, Cys-144, and Cys-147 (Fig. 4, A–D). With few exceptions, the CXXC motifs and the zinc coordination at this site are highly conserved among Fur regulators (23, 41, 43, 44). Metal occupancy at this site is known to be critical for protein stability and dimerization, and it is likely that zinc binding at site 1 contributes to the structural integrity and dimerization of PerR (21). Consistent with this hypothesis, results from previous studies showed that substitutions at one of the site 1 metal ligand, C104S, compromises the stability of PerR (12). Together, these results suggest that site 1 of PerR is a structural zinc site.
FIGURE 4.
Structural zinc-binding site within the dimerization domain of PerR. A, ribbon representation of the PerR dimer with the structural metal-binding site (boxed). The individual subunits of PerR dimer are color-coded, and the secondary structure elements of one subunit is labeled. The structural and regulatory metals in PerR structure are shown as spheres and color-coded in red and green, respectively. B, magnified view of the structural zinc-binding site. The structural elements in the proximity of the zinc-binding site are shown, and the zinc-coordinating side chains are labeled. C, diagram showing the interactions between structural zinc and the metal ligands from PerR as depicted by the program LIGPLOT (39). The bonds in the protein are shown in brown, and the carbon, nitrogen, oxygen, and sulfur atoms in each residue are colored in black, blue, red, and yellow, respectively. The zinc (at the center) is shown as a red sphere and labeled. The green broken lines indicate the bonds between the cysteines and the zinc, and the length of each is given. D, view of the final 2Fo − Fc electron density map contoured at 1σ showing the zinc site. Zinc and the side chains of the zinc-coordinating cysteines are shown as a sphere and sticks, respectively.
The second metal-binding site is located at the interdomain interface, and metal at this site is coordinated by metal ligands from the N-terminal arm, and helix α1 of the N-terminal domain and hinge region (Figs. 3B and 5A). To identify the metal bound at site 2, we performed metal content analysis of crystallized PerR by ICP-MS. The results revealed that each subunit of PerR has one zinc and one nickel ion (Table 3). Because the anomalous scattering from zinc indicated that site 1 has zinc, we conclude the metal ion at site 2 is nickel, which was likely acquired during protein purification. The nickel is coordinated in an octahedral geometry by the nitrogen atoms from histidines 4 and 6 of the N-terminal arm, His-19 of helix α1, His-97 and His-99 of the hinge region, and the oxygen atom of Asn-15 (Fig. 5, B–D). In addition, the main chain carboxyl atom of Asp-2 also contributes indirectly to metal coordination at site 2. It interacts with the imidazole ring of His-4, which facilitates proper positioning of the side chain of His-4 to participate in nickel coordination (Fig. 5D). In PerRBs, His-37 is the primary site of oxidation, and oxidation-induced loss of regulatory metal triggers the conformational changes to inactivate PerRBs (20, 25). Interestingly, the side chain of His-44 that is analogous to His-37 in PerRBs does not participate in metal binding but is involved in interdomain interactions.
FIGURE 5.
Regulatory metal-binding site in the inter-domain region of PerR dimer. A, ribbon representation of PerR dimer with its regulatory metal-binding site is marked by a green rectangle. The individual subunits of PerR dimer are color-coded, and the secondary structure elements of one subunit are labeled. The structural and regulatory metals in PerR structure are shown as spheres and color-coded in red and green, respectively. B, close-up view of the regulatory metal-binding site. The structural elements in the proximity of the metal-binding site are shown, and the coordinating side chains are labeled. C, diagram showing the interactions between regulatory metal, nickel (Ni), and the metal ligands from PerR as depicted by the program LIGPLOT (39). The bonds in the protein are shown in brown, and the carbon, nitrogen, oxygen, and sulfur atoms in each residue is colored in black, blue, red, and yellow, respectively. The metal (at the center) is shown as a green sphere. The green broken lines indicate the bonds between the metal ligands, and the metal and the length of each bonds is given. D, stereoview of final 2Fo − Fc electron density maps contoured at 1σ, showing the nickel-binding site. The nickel ion and the side chains of coordinating amino acids in PerR are shown as spheres and sticks, respectively.
In the crystal structures of regulatory metal-bound PerRBs (PerR-Zn-Mn) and other Fur family regulators, the regulatory metal-binding site is located in the interdomain region, and the metal ligands are derived from both the N-terminal DNA-binding and C-terminal dimerization domains (23, 44–46). Metal occupancy at this site promotes interdomain interactions and metal-induced allostery in the regulator, which results in a conformation compatible for DNA binding (23). Because site 2 of PerR is located in the interdomain region, and the regulatory metal is coordinated by the metal ligands from both domains, it is likely that metal occupancy at this site of PerR is critical for the allosteric changes in PerR and influences the gene regulation function of PerR. Hence, we designate site 2 as the regulatory site.
Mutations at Site 2 Metal Ligands Affect Metal Binding by PerR
To test the hypothesis that the site 2 metal ligands of PerR are critical for regulatory metal binding, we used site-directed mutagenesis to generate single alanine substitutions at 4 out of 5 histidines of site 2 that are unique for PerR. We also mutated His-44, which is analogous to the N-terminal metal ligand His-37 in PerRBs. Recombinant mutant proteins were overexpressed and purified under metal-free conditions. ICP-MS analysis of the mutant proteins indicated that with the exception of His-44, all mutant proteins exist in apoPerR-Zn form with only the structural site occupied (Table 3). The mutant protein H44A has one structural zinc and 0.5 nickel per subunit, and repeated attempts to remove this metal ion failed. We next investigated the metal-binding properties of mutant PerR proteins by ICP-MS. The apo-forms of purified mutant proteins were metallated by dialysis against chelexed buffer containing 10 μm MnCl2, and the metal content of the dialyzed proteins were assayed. In accordance with our hypothesis that site 2 metal ligands participate in regulatory metal binding, mutant proteins H6A, H19A, and H99A exhibited a drastic reduction in the metal occupancy at site 2. The fold difference in the metal occupancy at site 2 between PerR and its mutant derivatives was determined by calculating the ratio of total regulatory metal content of a mutant protein to that of PerR WT. Compared with wild-type PerR, the metal occupancy of H6A, H19A, and H99A mutant proteins was reduced by 2.8-, 2.0-, and 2.0-fold, respectively (Table 4). The metal occupancy in H97A was only slightly affected. Consistent with our structural observation that the side chain of His-44 is not involved in metal binding at site 2, the H44A mutant protein displayed wild-type-like metal-binding properties with both regulatory and structural sites occupied (Table 4). Although the mutant proteins are defective in metal binding compared with wild type, most of the mutants have their regulatory sites partially occupied. Because of the high reactivity of hydrogen peroxide in the presence of iron, we were unable to test the in vitro peroxide sensing ability of the mutant proteins. Together, these data indicate that the structural integrity of site 2 is critical for regulatory metal binding, and alterations in the form of alanine substitutions significantly affect the regulatory metal-binding properties of PerR.
TABLE 4.
Metal content analysis of metallated PerR-Zn-Mn by ICP-MS
| Protein | Mol of zinc per mol of PerRa | Mol of manganese per mol of PerRa | Mol of nickel per mol of PerRa |
|---|---|---|---|
| Wild type | 0.98 ± 0.08 | 0.85 ± 0.02 | 0.42 ± 0.02 |
| H6A | 0.95 ± 0.04 | 0.46 ± 0.01 | NDb |
| H19A | 0.91 ± 0.05 | 0.67 ± 0.01 | NDb |
| H44A | 1.09 ± 0.09 | 0.97 ± 0.01 | 0.45 ± 0.00 |
| H97Ac | 0.89 ± 0.09 | 1.09 ± 0.01 | NDb |
| H99A | 0.83 ± 0.07 | 0.69 ± 0.01 | NDb |
a Measurements were reported as average from triplicate studies.
b ND, not detected.
c Sample has copper levels of >10% of the protein molar ratio.
To corroborate our earlier finding that PerR does not require metal occupancy at site 2 for DNA binding and to better elucidate the interplay between metal binding and DNA binding by PerR, we tested the ability of the mutant proteins to bind DNA using a gel mobility shift assay. As shown in Fig. 6A, substitutions at the site 2 metal ligands did not affect PerR-per box interactions under metal-free conditions. However, mutant protein H44A exhibited defective DNA binding compared with wild type as it formed weak, smeary protein-DNA complexes (Fig. 6A). Because PerR does not require regulatory metal to bind the per box sequences from the pmtA promoter, the DNA-binding phenotypes of the site 2 metal ligand mutants are not surprising. Contrarily, given that H44A mutant protein displayed wild-type-like dimerization (data not shown) and metal-binding properties, its inability to bind DNA under metal-free conditions is intriguing. Subsequently, we tested whether the metal occupancy at site 2 restores the DNA-binding defect of the H44A mutant protein by assessing its DNA binding activity in the presence of 100 μm MnCl2. The H44A mutant failed to bind DNA under these conditions, in contrast to the other mutant proteins that bound DNA similarly to wild type (Fig. 6B). These findings are consistent with the results from previous in vivo data that demonstrated derepression of pmtA in an H44A mutant of PerR (12).
FIGURE 6.
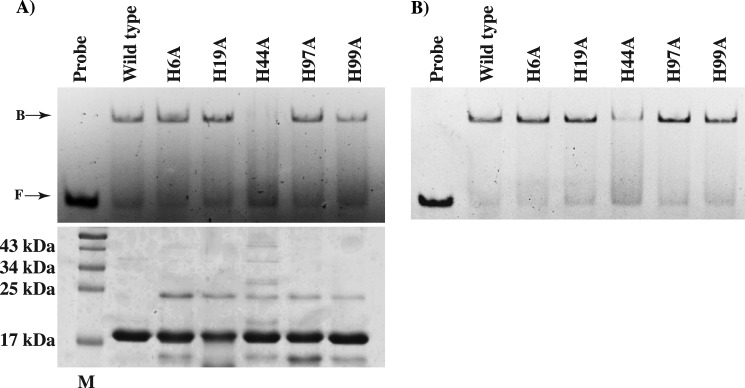
Analysis of DNA binding activity of the mutant proteins of PerR in the absence (A) or presence (B) of 100 μm MnCl2. Purified apo-forms (left) and metallated forms (right) of the wild-type or mutant proteins of PerR with single amino acid substitutions in H6A, H19A, H44A, H97A, and H99A were assayed for binding with per box sequences (0.2 μm) from the pmtA promoter by gel mobility shift assay. Bands corresponding to unbound (F) and PerR-bound probe (B) are labeled and indicated by arrows. A Coomassie-stained SDS-PAGE with equal amounts of wild-type or mutant proteins of PerR is shown (bottom left), and the lane with the molecular mass marker corresponding to 17 kDa is marked M. The lane with the molecular mass marker is marked, and the corresponding masses are indicated (in kDa).
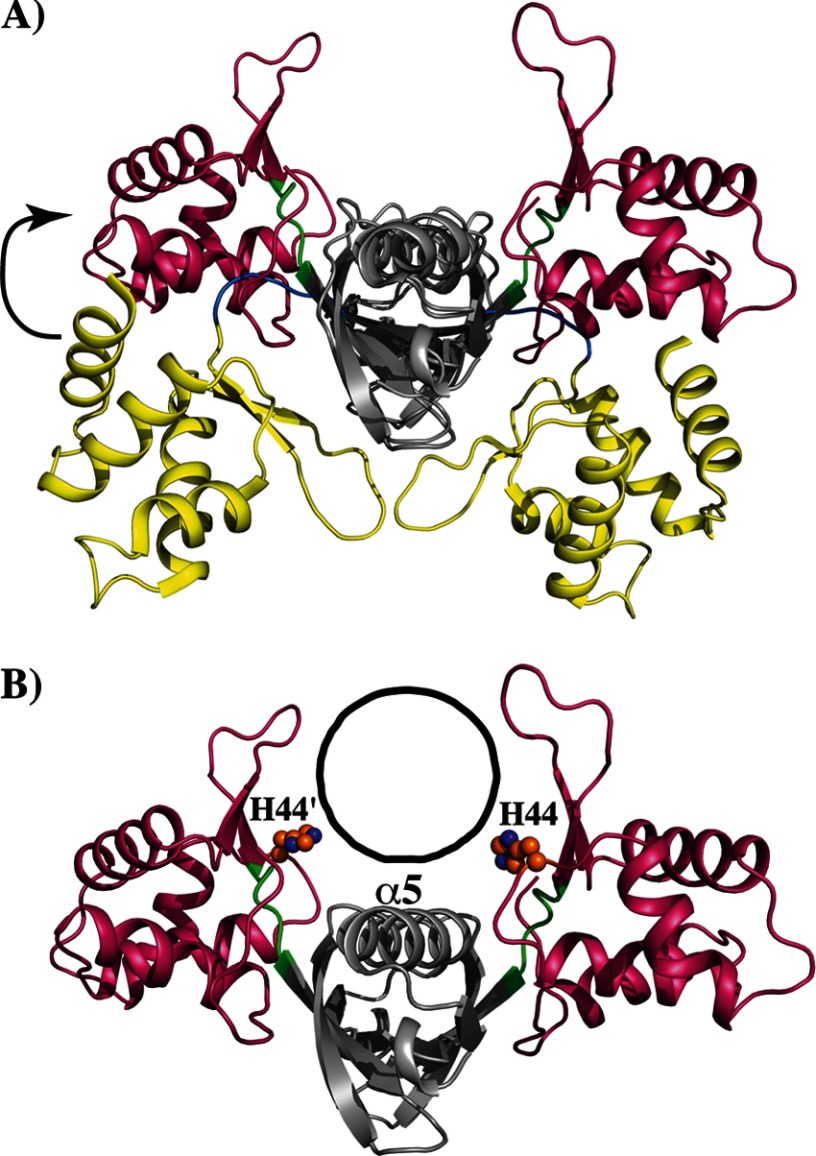
To elucidate the contribution of the side chain of His-44 to PerR-per box interactions, we performed docking studies with a B-DNA docked into the cleft between the DNA-binding domains of PerR dimer (Fig. 7, A and B). Based on these structural analyses, we propose two possible roles for His-44 in DNA binding. First, when the DNA was oriented in the region between the two wHTH motifs of PerR, the side chain of His-44 is pointing toward the cleft and positioned in close proximity to interact with either the bases or the phosphate backbone of DNA (Fig. 7B). Alternatively, a detailed look at the immediate vicinity of His-44 also revealed an indirect role for His-44 in the proper positioning of the DNA-binding domains of PerR to interact with DNA. Specifically, the side chain of His-44 engaged in interactions with amino acids from the wHTH motif of the N-terminal domain and α5 of the C-terminal domain The side chain of His-44 is 2.8 Å away from the carboxyl group of Asp-93 from the wHTH motif and involved in electrostatic interactions. However, it is also positioned to engage in water-mediated interactions with the charged residues Lys-123 and Asp-120, of α5. It is possible that these interactions might play a role in proper positioning of the DNA-binding structural elements of PerR to interact with DNA. Disruption of these interactions in H44A might result in the displacement of the DNA-binding domains of PerR, thus inhibiting PerR-DNA interactions. This unexpected role of His-44 in PerR is contrary to its role in PerRBs. In PerRBs, the side chain of His-37 is involved in the coordination of regulatory metal and is the primary site for the MCO (20, 25). Although the oxidation of His-44 in PerR cannot be ruled out, its relatively distant location from the regulatory metal-binding site of PerR makes this scenario unlikely. Given that the side chain of His-44 is conserved between PerR and PerRBs, it remains a possibility that His-44 might be involved in additional metal-binding events in PerR. Further biochemical and structural characterization of metal binding and MCO in PerR will be required to fully understand the role of His-44 in PerR. Collectively, results from the mutational analysis of site 2 metal ligands of PerR are consistent with our structural data that these amino acids are important for regulatory binding. Additionally, these data indicate a role for His-44 that is different from its counterpart in PerRBs.
FIGURE 7.
Structural comparisons of PerR and PerRBs-Zn-Mn. A, superposition of the main chains of PerR and PerRBs-Zn-Mn dimers. The PerRBs-Zn-Mn was used as a reference molecule, and the superposition of PerR was optimized for the dimerization domain using LSQKAB (47). The aligned dimerization domains are shown in gray, and the DNA-binding domains of PerR (red) and PerRBs-Zn-Mn (yellow) are color-coded. The flexible hinge region of PerR and PerRBs-Zn-Mn that connects the two functional domains are shaded in green and blue, respectively. The upward movement of PerR DNA-binding domain is indicated by the arrow on the left. B, novel mode of DNA binding by PerR. The structure of PerR dimer is oriented and color-coded as in A. The DNA-binding cleft between the two DNA-binding wHTH motifs is indicated as a black circle. The side chains of His-44 from each subunit is shown as ball and sticks and labeled (′ indicates His-44 from the second subunit of the PerR dimer). Similarly, the position of helix α5 is indicated and labeled.
Metal Binding at Site 2 Is Critical for GAS Virulence
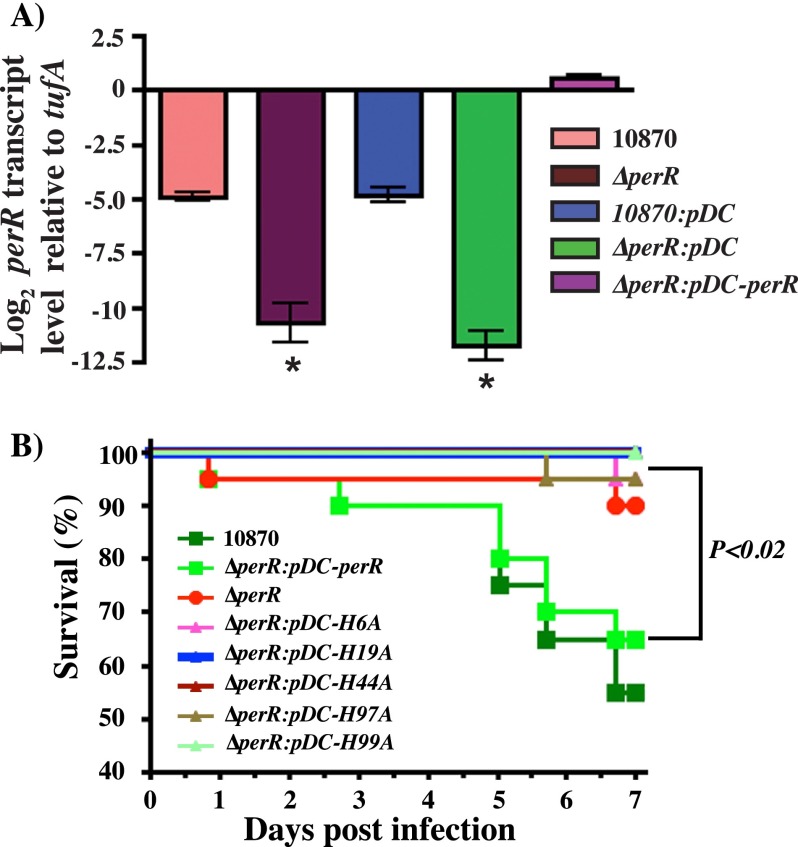
To test the hypothesis that an intact regulatory site of PerR is critical for in vivo peroxide sensing, gene regulation, and GAS virulence, we carried out animal infection studies using an intraperitoneal mouse model of infection. The parental strain MGAS 10870 trans-complemented with empty vector pDC123 was used as wild type, and the insertionally inactivated isogenic perR mutant trans-complemented with empty vector pDC123 was used as a negative control. To trans-complement the isogenic mutant ΔperR strain, we constructed plasmid pDC-perR containing the coding region of perR and its putative promoter region. To ensure that the pDC-perR construct contains a functional transcriptional unit, we compared the perR transcript levels in the wild-type (10870:pDC) and trans-complemented (ΔperR:pDC-perR) strains by TaqMan qRT-PCR. Results indicate that perR is transcribed from the pDC-perR and consistent with the high copy number of the plasmid pDC123, and we found an overall increase (∼32-fold) in the perR transcript levels (Fig. 8A). Using this construct as a template, we generated single alanine substitutions at four (H6A, H19A, H97A, and H99A) out of the total five histidines of the metal-binding site 2 of PerR and His-44 that is not involved in metal binding by Quik-Change site-directed mutagenesis. When grown in standard medium, the wild-type strain, isogenic ΔperR mutant strain, and mutant strain trans-complemented with either the wild-type perR or one of the mutant strains did not differ significantly in growth characteristics, indicating that the single amino acid substitutions introduced in PerR do not affect GAS growth (data not shown).
FIGURE 8.
Single amino acid substitutions at the metal ligands of PerR regulatory site significantly attenuate GAS virulence. A, transcript levels of perR in the indicated strains as measured by TaqMan qRT-PCR. Samples were collected at mid-exponential (A600 −1.0) phase of growth for transcript level analysis. Duplicate biological replicates were grown on two different occasions and analyzed in duplicate. Data graphed are mean ± S.D. *, transcript levels were not detectable. B, 20 CD-1 mice were inoculated with each indicated strain intraperitoneally, and the near mortality was assessed. Results are graphically represented as a Kaplan-Meier survival curve with p values derived by log rank test.
To determine the effects of PerR mutants on GAS virulence, mice were infected intraperitoneally and monitored for near mortality for 7 days post-infection. Based on the virulence phenotype, the strains can be categorized into two groups as follows: the wild-type control (10870:pDC) and isogenic perR mutant trans-complemented with perR (ΔperR:pDC-perR) showed increased virulence compared with either the isogenic perR mutant strain (ΔperR:pDC) or any of the mutant strains (p < 0.02 when comparing any two strains in different groups). There was no significant difference in mortality among strains within either of the two groups (Fig. 8B). Together, these data indicate that substitutions at the regulatory site metal ligands of PerR significantly attenuate GAS virulence, likely due to the defective metal binding, peroxide sensing, and the consequent misregulation of the PerR regulon.
N-terminal HXH Metal-binding Motif Is Unique in PerR Structure and Is Conserved among Genus Streptococcus
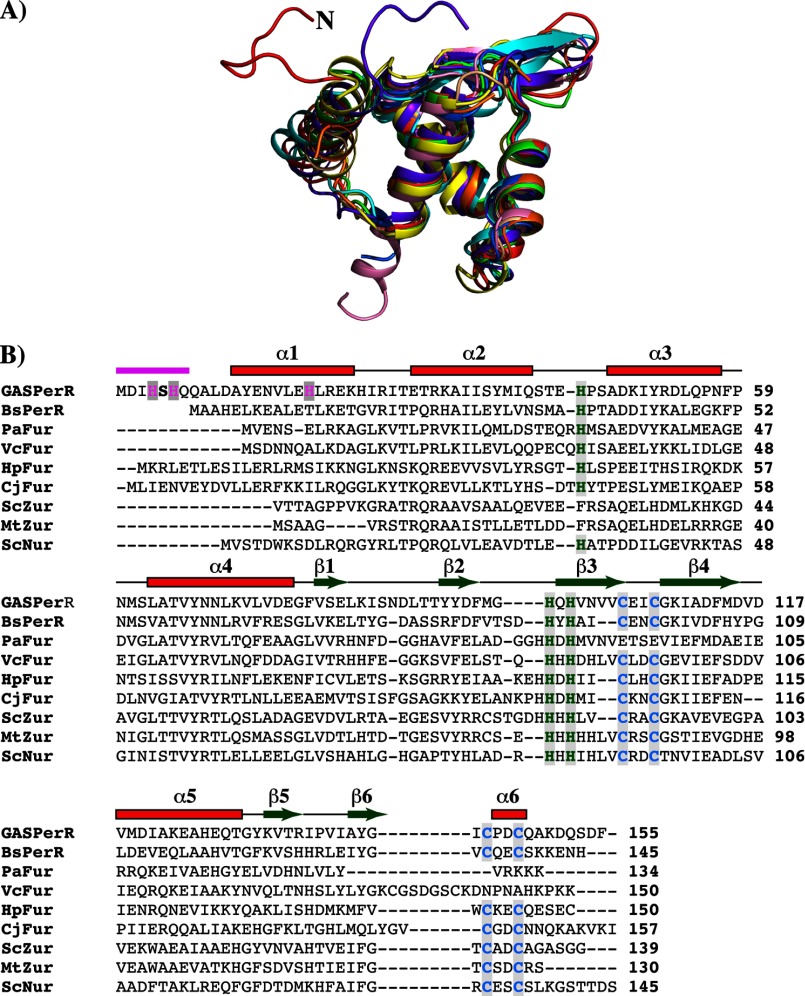
Structural alignment of the N-terminal DNA-binding domain of PerR with that of other structurally characterized members of the Fur family of regulators revealed a high degree of structural homology. However, major difference occurs at the N terminus of PerR (Fig. 9A). The N-terminal extension of PerR that has the site 2 metal-binding ligands, His-4 and His-6, is not present in the structures of the majority of the Fur regulators (Fig. 9A). In addition to PerR, only Fur from Helicobacter pylori (FurHp) has an extended N terminus, but the FurHp extension does not have the characteristic HXH motif within this region (46). Furthermore, the crystal structure of FurHp showed that the N-terminal extension does not participate in metal binding but instead is involved in hydrogen bonding interactions with the hinge region and the loop between α2 and α3 that has a potential role in apoFurHp-operator interactions (46). The presence of the novel HXH metal-binding motif in PerR is more evident with the structure-based amino acid sequence alignment of all the structurally characterized Fur regulators (Fig. 9B). Although the structural zinc binding CXXC motifs and the C-terminal regulatory metal ligands are highly conserved, the HXH motif is present only in the N terminus of PerR (Fig. 9B). Interestingly, this distinction is extended to other N-terminal metal ligands of PerR, as Asn-15 and His-19 of α1 are unique to PerR and not conserved among the structurally characterized Fur regulators. Together, these data indicate that the GAS PerR uses a previously unknown novel structural motif for regulatory metal binding and peroxide sensing.
FIGURE 9.
N-terminal extension with the HXH metal-binding motif is unique in the structure of PerR. A, superposition of residues 2–94 of the N-terminal domain of PerR (red) with the analogous residues from the structures of PerRBs (green, PDB code 2FE3) (23) and other Fur regulators (Fur from Pseudomonas aeruginosa (orange, PDB code 1MZB), C. jejuni (yellow, PDB code 4ETS), H. pylori (purple, PDB code 2XIG), and Vibrio cholerae (light brown, PDB code 2W57); Zur from Streptomyces coelicolor (blue, PDB code 3MWM) and Mycobacterium tuberculosis (pink, PDB code 2O03); and Nur from Streptomyces coelicolor (cyan, PDB code 3EYY)) (43). The N-terminal extension of PerR is colored red and labeled (N). Structural superposition was performed with “LSQKAB”(47). B, amino acid sequence alignment of PerR with other structurally characterized Fur family regulators (B. subtilis PerR-BsPerR, Fur from P. aeruginosa, V. cholerae, H. pylori, and C. jejuni, PaFur, VcFur, HpFur, and CjFur, respectively; Zur from M. tuberculosis and S. coelicolor, MtZur and ScZur, respectively; and S. coelicolor Nur-ScNur). The multiple sequence alignment was performed with ClustalW (49). The secondary structure elements are labeled and marked above the alignment; α-helices are shown as rectangles and β-strands as arrows. The region corresponding to the N-terminal extension is shown in magenta. The highly conserved CXXC structural zinc motif is colored in blue, and the C-terminal metal-binding ligands are colored in green. The N-terminal HXH motif unique for PerR is highlighted in magenta.
Because distinctive structural motifs are likely conserved among closely related organisms, we investigated whether this metal-binding motif is conserved in PerR and PerR-like proteins from other members of the genus Streptococcus. Remarkably, when the amino acid sequences of the streptococcal PerR proteins were aligned, all the identified PerR orthologs from streptococci contain the N-terminal extension with the metal-binding HXH motif (Fig. 10). Furthermore, the metal ligand from helix α2, His-19, is also conserved among the members of this subgroup (Fig. 10). This indicates that despite the core structural features shared between PerR and other Fur regulators, PerR from streptococci possess distinctive structural and amino acid features that have important functional implications for their ability to sense peroxide stress and mediate gene regulation. Given the high degree of conservation of this structural motif among these proteins, it is likely that these related groups of proteins share a common mechanism of peroxide sensing and gene regulation. In this regard, the structure of PerR from GAS reported here provides the framework for understanding the mechanistic basis for the regulatory functions of PerR from streptococci.
FIGURE 10.
Amino acid sequence alignment of PerR and PerR-like proteins from the genus Streptococcus. The alignment was carried out with ClustalW (49). The secondary structure elements derived from the structure of PerR are indicated above the alignment. The color coding of the key residues involved in metal binding is the same as Fig. 9.
Functional Implications from the Structure
The PerR dimer is symmetrical as the individual subunits can be superimposed onto each other with a root square mean deviation (r.m.s.d.) of ∼1 Å for 147 corresponding residues. The two-domain modular architecture of PerR is similar to the structures of other Fur family regulators. With the exception of Fur from Campylobacter jejuni (FurCj), the individual domains of PerR can be structurally aligned with the respective domains of other Fur regulators with r.m.s.d. of <2 Å for corresponding Cα atoms. Because the crystal structure of apoFurCj has a unique arrangement of the DNA-binding elements compared with other Fur regulators (43), it was not included in the structural analysis. Comparison of the dimeric structures of PerR to the structures of apoPerRBs-Zn and active PerRBs:Zn-Mn form using LSQKAB (47) revealed significant misalignment of the positions of the individual domains within the dimer (r.m.s.d. of >17 Å). To understand the mode of DNA binding by PerR, we overlaid the dimerization domains of PerR and PerRBs-Zn-Mn and analyzed the position of the DNA-binding domains. Interestingly, the DNA-binding domain of PerR is flipped to the opposite face of the molecule compared with that of PerRBs and adopts a unique conformation (Fig. 7A). The impetus for this conformation stems from the orientation of the flexible hinge region that connects the two domains. The hinge region of PerR is elongated and extends upwards in the opposite direction relative to that of manganese-bound PerRBs (Fig. 7A). This conformation places the wHTH motif facing the DNA-binding cleft, a position suitable for PerR-DNA interactions (Fig. 7B). However, the recognition helices of each HTH motif are misaligned to interact with the major grooves of DNA, and additional conformational changes might be required to bring them closer. Interestingly, in this arrangement, the helix α5 is located on top of the DNA-binding cleft between the two DNA-binding domains and positioned to interact with DNA (Fig. 7B). Despite these drastic structural rearrangements of the DNA-binding domains in PerR, it still maintains several interdomain contacts, including the metal ligands of the regulatory metal-binding site. This is contrary to the lack of interdomain interactions observed in the structure of apoPerRBs (24). Given the lack of DNA-bound structures for the members of Fur family regulators, we propose that the crystal structure of PerR might represent an alternative conformation that is capable of DNA binding. It is possible that PerR has two modes of DNA binding, and this dual mode of DNA binding is consistent with the transcriptome studies that showed that PerR controls two different regulons in the presence and absence of oxidative stress (12, 13). Thus, it is likely that PerR might mediate gene regulation in its apo- and metal-bound form and the DNA-binding properties of PerR diverge depending on the metal occupancy at site 2. Similar gene regulation properties were observed in the metalloregulation by Fur from H. pylori and C. jejuni (43, 48).
Conclusion
We demonstrated that PerR from GAS contains a unique structural element and employs a mechanism of peroxide sensing that is distinct from any of the structurally characterized Fur regulators, thus highlighting the divergence in the repertoire of metal-binding motifs and coordination geometry used by the Fur family of regulators.
Supplementary Material
Acknowledgments
We thank the beamline scientists and staff at Advanced Light Source beamline 8.3.1 for their great help with data collection. We also acknowledge the Advanced Light Source supported by the Director, Office of Science, Office of Basic Energy Sciences, Material Sciences Division, of the United States Department of Energy at Lawrence Berkeley National Laboratory.
This work was supported in part by an American Heart Association Scientist Development grant (to M. K.).

This article contains supplemental Table S1.
The atomic coordinates and structure factors (code 4I7H) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- ROS
- reactive oxygen species
- GAS
- group A Streptococcus
- PerR
- peroxide stress regulator
- PDB
- Protein Data Bank
- Fur
- ferric uptake regulator
- TCEP
- tris(2-carboxyethyl)phosphine
- ICP-MS
- inductively coupled plasma mass spectrometry
- SeMet
- selenomethionine
- r.m.s.d.
- root mean square deviation
- wHTH
- winged helix-turn-helix
- MCO
- metal-catalyzed oxidation.
REFERENCES
- 1. Robinson J. M. (2009) Phagocytic leukocytes and reactive oxygen species. Histochem. Cell Biol. 131, 465–469 [DOI] [PubMed] [Google Scholar]
- 2. Nunoshiba T., Obata F., Boss A. C., Oikawa S., Mori T., Kawanishi S., Yamamoto K. (1999) Role of iron and superoxide for generation of hydroxyl radical, oxidative DNA lesions, and mutagenesis in Escherichia coli. J. Biol. Chem. 274, 34832–34837 [DOI] [PubMed] [Google Scholar]
- 3. Storz G., Imlay J. A. (1999) Oxidative stress. Curr. Opin Microbiol. 2, 188–194 [DOI] [PubMed] [Google Scholar]
- 4. Imlay J. A. (2008) Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77, 755–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Faulkner M. J., Helmann J. D. (2011) Peroxide stress elicits adaptive changes in bacterial metal ion homeostasis. Antioxid. Redox Signal. 15, 175–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dubbs J. M., Mongkolsuk S. (2012) Peroxide-sensing transcriptional regulators in bacteria. J. Bacteriol. 194, 5495–5503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Olsen R. J., Shelburne S. A., Musser J. M. (2009) Molecular mechanisms underlying group A streptococcal pathogenesis. Cell. Microbiol. 11, 1–12 [DOI] [PubMed] [Google Scholar]
- 8. Gibson C. M., Mallett T. C., Claiborne A., Caparon M. G. (2000) Contribution of NADH oxidase to aerobic metabolism of Streptococcus pyogenes. J. Bacteriol. 182, 448–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. King K. Y., Horenstein J. A., Caparon M. G. (2000) Aerotolerance and peroxide resistance in peroxidase and PerR mutants of Streptococcus pyogenes. J. Bacteriol. 182, 5290–5299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Condon S. (1987) Responses of lactic acid bacteria to oxygen. FEMS Microbiol. Lett. 46, 269–280 [Google Scholar]
- 11. Brenot A., King K. Y., Caparon M. G. (2005) The PerR regulon in peroxide resistance and virulence of Streptococcus pyogenes. Mol. Microbiol. 55, 221–234 [DOI] [PubMed] [Google Scholar]
- 12. Grifantini R., Toukoki C., Colaprico A., Gryllos I. (2011) Peroxide stimulon and role of PerR in group A Streptococcus. J. Bacteriol. 193, 6539–6551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gryllos I., Grifantini R., Colaprico A., Cary M. E., Hakansson A., Carey D. W., Suarez-Chavez M., Kalish L. A., Mitchell P. D., White G. L., Wessels M. R. (2008) PerR confers phagocytic killing resistance and allows pharyngeal colonization by group A Streptococcus. PLoS Pathog. 4, e1000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brenot A., Weston B. F., Caparon M. G. (2007) A PerR-regulated metal transporter (PmtA) is an interface between oxidative stress and metal homeostasis in Streptococcus pyogenes. Mol. Microbiol. 63, 1185–1196 [DOI] [PubMed] [Google Scholar]
- 15. Ricci S., Janulczyk R., Björck L. (2002) The regulator PerR is involved in oxidative stress response and iron homeostasis and is necessary for full virulence of Streptococcus pyogenes. Infect. Immun. 70, 4968–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hahn J.-S., Oh S.-Y., Chater K. F., Cho Y.-H., Roe J.-H. (2000) H2O2-sensitive Fur-like repressor CatR regulating the major catalase gene in Streptomyces coelicolor. J. Biol. Chem. 275, 38254–38260 [DOI] [PubMed] [Google Scholar]
- 17. van Vliet A. H., Baillon M.-L., Penn C. W., Ketley J. M. (1999) Campylobacter jejuni contains two Fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J. Bacteriol. 181, 6371–6376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fuangthong M., Helmann J. D. (2003) Recognition of DNA by three ferric uptake regulator (Fur) homologs in Bacillus subtilis. J. Bacteriol. 185, 6348–6357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Herbig A. F., Helmann J. D. (2001) Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol. Microbiol. 41, 849–859 [DOI] [PubMed] [Google Scholar]
- 20. Lee J.-W., Helmann J. D. (2006) The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440, 363–367 [DOI] [PubMed] [Google Scholar]
- 21. Lee J.-W., Helmann J. D. (2006) Biochemical characterization of the structural Zn2+ site in the Bacillus subtilis peroxide sensor PerR. J. Biol. Chem. 281, 23567–23578 [DOI] [PubMed] [Google Scholar]
- 22. Ortiz de Orué Lucana D., Schrempf H. (2000) The DNA-binding characteristics of the Streptomyces reticuli regulator FurS depend on the redox state of its cysteine residues. Mol. Gen. Genet. 264, 341–353 [DOI] [PubMed] [Google Scholar]
- 23. Jacquamet L., Traoré D. A., Ferrer J. L., Proux O., Testemale D., Hazemann J. L., Nazarenko E., El Ghazouani A., Caux-Thang C., Duarte V., Latour J. M. (2009) Structural characterization of the active form of PerR: insights into the metal-induced activation of PerR and Fur proteins for DNA binding. Mol. Microbiol. 73, 20–31 [DOI] [PubMed] [Google Scholar]
- 24. Traoré D. A., El Ghazouani A., Ilango S., Dupuy J., Jacquamet L., Ferrer J.-L., Caux-Thang C., Duarte V., Latour J.-M. (2006) Crystal structure of the apo-PerR-Zn protein from Bacillus subtilis. Mol. Microbiol. 61, 1211–1219 [DOI] [PubMed] [Google Scholar]
- 25. Traoré D. A., Ghazouani A. E., Jacquamet L., Borel F., Ferrer J.-L., Lascoux D., Ravanat J.-L., Jaquinod M., Blondin G., Caux-Thang C., Duarte V., Latour J.-M. (2009) Structural and functional characterization of 2-oxo-histidine in oxidized PerR protein Nat. Chem. Biol. 5, 53–59 [DOI] [PubMed] [Google Scholar]
- 26. Beres S. B., Carroll R. K., Shea P. R., Sitkiewicz I., Martinez-Gutierrez J. C., Low D. E., McGeer A., Willey B. M., Green K., Tyrrell G. J. (2010) Molecular complexity of successive bacterial epidemics deconvoluted by comparative pathogenomics. Proc. Natl. Acad. Sci. U.S.A. 107, 4371–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Doubliè S. (1997) Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276, 523–530 [PubMed] [Google Scholar]
- 28. Leslie A. (1992) Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography, No. 26, 27–33 [Google Scholar]
- 29. Leslie A. (2006) The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 30. Collaborative Computational Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 31. Terwilliger T. C., Berendzen J. (1999) Discrimination of solvent from protein regions in native Fouriers as a means of evaluating heavy-atom solutions in the MIR and MAD methods. Acta Crystallogr. D Biol. Crystallogr. 55, 501–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Terwilliger T. (1999) Reciprocal-space solvent flattening. Acta Crystallogr. D Biol. Crystallogr. 55, 1863–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. DeLano W. L. (2002) The PyMOL Molecular Graphics System. DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
- 34. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J.-S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR System: A New software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 35. Faulkner M. J., Ma Z., Fuangthong M., Helmann J. D. (2012) Derepression of the Bacillus subtilis PerR peroxide stress response leads to iron deficiency. J. Bacteriol. 194, 1226–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lukomski S., Hoe N. P., Abdi I., Rurangirwa J., Kordari P., Liu M., Dou S.-J., Adams G. G., Musser J. M. (2000) Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect. Immun. 68, 535–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kuwayama H., Obara S., Morio T., Katoh M., Urushihara H., Tanaka Y. (2002) PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 30, E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chaffin D. O., Rubens C. E. (1998) Blue/white screening of recombinant plasmids in Gram-positive bacteria by interruption of alkaline phosphatase gene (phoZ) expression. Gene 219, 91–99 [DOI] [PubMed] [Google Scholar]
- 39. Virtaneva K., Porcella S. F., Graham M. R., Ireland R. M., Johnson C. A., Ricklefs S. M., Babar I., Parkins L. D., Romero R. A., Corn G. J., Gardner D. J., Bailey J. R., Parnell M. J., Musser J. M. (2005) Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc. Natl. Acad. Sci. U.S.A. 102, 9014–9019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ahn B.-E., Cha J., Lee E.-J., Han A.-R., Thompson C. J., Roe J.-H. (2006) Nur, a nickel-responsive regulator of the Fur family, regulates superoxide dismutases and nickel transport in Streptomyces coelicolor. Mol. Microbiol. 59, 1848–1858 [DOI] [PubMed] [Google Scholar]
- 41. Lucarelli D., Russo S., Garman E., Milano A., Meyer-Klaucke W., Pohl E. (2007) Crystal structure and function of the zinc uptake regulator FurB from Mycobacterium tuberculosis. J. Biol. Chem. 282, 9914–9922 [DOI] [PubMed] [Google Scholar]
- 42. Huffman J. L., Brennan R. G. (2002) Prokaryotic transcription regulators: more than just the helix-turn-helix motif. Curr. Opin Struct. Biol. 12, 98–106 [DOI] [PubMed] [Google Scholar]
- 43. Butcher J., Sarvan S., Brunzelle J. S., Couture J.-F., Stintzi A. (2012) Structure and regulon of Campylobacter jejuni ferric uptake regulator Fur define apo-Fur regulation. Proc. Natl. Acad. Sci. U.S.A. 109, 10047–10052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shin J.-H., Jung H. J., An Y. J., Cho Y.-B., Cha S.-S., Roe J.-H. (2011) Graded expression of zinc-responsive genes through two regulatory zinc-binding sites in Zur. Proc. Natl. Acad. Sci. U.S.A. 108, 5045–5050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sheikh M. A., Taylor G. L. (2009) Crystal structure of the Vibrio cholerae ferric uptake regulator (Fur) reveals insights into metal co-ordination. Mol. Microbiol. 72, 1208–1220 [DOI] [PubMed] [Google Scholar]
- 46. Dian C., Vitale S., Leonard G. A., Bahlawane C., Fauquant C., Leduc D., Muller C., de Reuse H., Michaud-Soret I., Terradot L. (2011) The structure of the Helicobacter pylori ferric uptake regulator Fur reveals three functional metal-binding sites. Mol. Microbiol. 79, 1260–1275 [DOI] [PubMed] [Google Scholar]
- 47. Kabsch W. (1976) A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. Sect. A 32, 922–923 [Google Scholar]
- 48. Delany I., Spohn G., Rappuoli R., Scarlato V. (2001) The Fur repressor controls transcription of iron-activated and -repressed genes in Helicobacter pylori. Mol. Microbiol. 42, 1297–1309 [DOI] [PubMed] [Google Scholar]
- 49. Thompson J. D., Higgins D. G., Gibson T. J. (1994) Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li J., Kasper D. L., Ausubel F. M., Rosner B., Michel J. L. (1997) Inactivation of the α C protein antigen gene, bca, by a novel shuttle/suicide vector results in attenuation of virulence and immunity in group B Streptococcus. Proc. Natl. Acad. Sci. U.S.A. 94, 13251–13256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shelburne S. A., 3rd, Olsen R. J., Makthal N., Brown N. G., Sahasrabhojane P., Watkins E. M., Palzkill T., Musser J. M., Kumaraswami M. (2011) An amino-terminal signal peptide of Vfr protein negatively influences RopB-dependent SpeB expression and attenuates virulence in Streptococcus pyogenes. Mol. Microbiol. 82, 1481–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.