Background: Transcriptional control of hCD79b is critical to B cell function and poorly understood.
Results: A fully sufficient and novel secondary promoter was identified within intron 1 of hCD79b.
Conclusion: hCD79b has a complex transcriptional structure comprising two competing promoters.
Significance: The secondary promoter within hCD79b predicts novel pathways of B cell receptor expression in normal and pathologic settings.
Keywords: Gene Expression, Immunology, Transcription Enhancers, Transcription Promoter, Transgenic Mice, B Cell, DNase I Hypersensitive Site, Secondary Promoter, hCD79b
Abstract
The human B cell-specific protein, CD79b (also known as Igβ and B29) constitutes an essential signal transduction component of the B cell receptor. Although its function is central to the triggering of B cell terminal differentiation in response to antigen stimulation, the transcriptional determinants that control CD79b gene expression remain poorly defined. In the present study, we explored these determinants using a series of hCD79b transgenic mouse models. Remarkably, we observed that the previously described hCD79b promoter along with its associated enhancer elements and first exon could be deleted without appreciable loss of hCD79b transcriptional activity or tissue specificity. In this deletion setting, a secondary promoter located within exon 2 maintained full levels and specificity of hCD79b transcription. Of note, this secondary promoter was also active, albeit at lower levels, in the wild-type hCD79b locus. The activity of the secondary promoter was dependent on the action(s) of a conserved sequence element mapping to a chromatin DNase I hypersensitive site located within intron 1. mRNA generated from this secondary promoter is predicted to encode an Igβ protein lacking a signal sequence and thus unable to serve normal B cell receptor function. Although the physiologic role of the hCD79b secondary promoter and its encoded protein remain unclear, the current data suggest that it has the capacity to play a role in normal as well as pathologic states in B cell proliferation and function.
Introduction
CD79b (Igβ, B29) together with CD79α (Igα, mb-1) constitute the heterodimeric signaling component of the B cell receptor (BCR)2 complex. The activities of these two proteins are essential for the initiation of signal transduction following BCR engagement with an antigen molecule (1). The engaged BCR triggers phosphorylation of these subunits with the subsequent triggering of clonal B cell expansion and production of secreted immunoglobulin (2). The expression of the BCR is repressed during the terminal stages of B cell differentiation, and the CD79b/CD79a encoded proteins are undetectable in terminally differentiated plasma cells (3, 4). Abnormal expression of CD79b has been associated with a variety of B cell malignancies (5, 6), including chronic lymphocytic leukemia (7, 8). In addition, naturally occurring null mutations in the human CD79b locus block plasma cell differentiation and production of secreted immunoglobulin, resulting in severe immune deficiency syndromes (7).
Surprisingly little information is available on transcription regulation of human CD79b. Previous studies showed that the gene has a GC-rich and TATA-less promoter. Consistent with this promoter structure, the gene has the expected heterogeneity of transcription start sites that cluster within 100–200 nucleotides 5′ to the translation initiation codon (9, 10). In vitro binding studies along with analyses of transiently transfected CD79b genes in cell culture further revealed associated transcription factor binding sites in the promoter region that could mediate enhancement and/or repression of transcription (11). On the basis of these early studies, it was concluded that this promoter region, along with its linked enhancer/repressor elements, constituted the core determinants of hCD79b transcriptional control (12). However, subsequent in vivo analyses of hCD79b gene expression using transgenic mouse models have brought into question whether these elements have essential roles in the expression of the hCD79b locus in its native genome environment (13).
Nucleosome packaging of DNA into chromatin fibers presents a generally repressive environment that restricts the recruitment of most transcription factors (14). The detection of nucleosome-free regions by DNase I mapping is an effective tool to detect critical regulatory regions that modulate chromatin environments and promote transcriptional activity of a linked gene (15–17). Genome-wide mapping of DNase I hypersensitive sites (HSs) in yeast and humans has revealed a prevalence of DNase I HSs that is much higher than previously thought (18). In yeast chromatin, DNase I HSs co-map with cis-regulatory elements in the promoters of most genes and are generally excluded from the coding regions (19, 20). In human chromatin, DNase I HSs are also enriched in gene regulatory regions, such as promoters, CpG islands, and transcription termination sites. However, in contrast to yeast, half of the HSs in mammalian genomes are located within introns (18, 21–25). The genome-wide analysis of DNase I HS patterns in mouse and human genomes further supports the relevancy of HSs to tissue-specific gene regulation, implying that the corresponding transcription factor complexes associating with these sites, often intragenic, are involved in developmental and dynamic transcriptional controls (18, 26).
The human CD79b gene is located on human chromosome 17 between the human growth hormone gene cluster and the muscle-specific sodium channel gene (SCN4A). This complex locus contains multiple DNase I HSs with distinct tissue and developmental specificities (13, 27–29). The HSs in this region that form in pituitary somatotropes, placental syncytiotrophoblasts, skin fibroblasts, and B lymphocytes have been extensively explored and identified by the combined analyses of chromatin from corresponding cell lines, primary human tissues, and human loci carried in defined transgenic mouse lines (13). These patterns of HSs point to a complex, tissue-specific set of cis-acting elements that are alternatively used to drive mutually exclusive patterns of gene expression in these distinct tissues. Defining the respective functions of these HSs has been instrumental in outlining gene regulatory pathways responsible for selective transcriptional activation of the genes within the growth hormone gene cluster (hGH) in either pituitary or placenta. A similar exploration of the HS involved in hCD79b expression is likely to shed light on controls over the expression of the hCD79b gene in the B cell.
In the present study, we confirm the presence of a set of HS in primary B cells that are linked to the hCD79b gene and explore their contribution to transcriptional controls. The data revealed an unexpected complexity in transcriptional initiation from within the hCD79b locus and its relation to a defined HS. These observations predict novel pathways of hCD79b expression and function.
EXPERIMENTAL PROCEDURES
Cell Lines and Purification of Lymphocytes from Mouse Spleen
Human lymphoblastoid cell line CRL-1484 was maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. Spleen cells were harvested, suspended in phosphate-buffered saline (PBS), underlain with Ficoll-Plaque Plus (Amersham Biosciences), and centrifuged at 650 × g for 30 min without braking. Lymphocytes enriched at the Ficoll/aqueous interface were collected with a pipette and washed twice with PBS prior to analysis.
BAC Transgene Modification by Homologous Recombination
The construction of BAC transgenes (CDΔ0.7/hGH BAC, CDΔ1.6/hGH BAC, CDΔ340/hGH BAC, and CDΔ20/hGH BAC) was based on published BAC recombineering methodology (30). The shuttle vector, pLD53.SCA-E-B, was digested with AscI/PacI, and a 6-kb fragment was purified. The 600-bp “right arm” and “left arm” fragments used for the homologous recombination were amplified by PCR using primer sets (supplemental Table 1); digested with AscI/EcoRI and PacI/EcoRI, respectively; and cloned into the shuttle vector in PIR2 competent cells (Invitrogen). BAC host cells were streaked on an agar plate supplemented with chloramphenicol (Chlr) (20 μg/ml) and incubated overnight at 37 °C. A single colony was picked, inoculated with 5 ml of Luria broth (LB) supplemented with Chlr (20 μg/ml), and grown to A600 of 0.5. The cells were harvested by centrifugation at 3,000 rpm for 10 min at 4 °C. The pellet was suspended with 5 ml of ice-cold 50 mm CaCl2, placed on ice for 5 min, and harvested as above. The pellet was suspended with 300 μl of ice-cold solution of 20% glycerol. 1 μg of cloned shuttle vector was electroporated into 40 μl of BAC DNA competent cells using the Gene PulserTM system (Bio-Rad) at 25 microfarads. Shuttle vector-integrated BAC DNA clones were selected by growth on Chlr (20 μg/ml) and ampicillin (50 μg/μl), and the selected clones were inoculated again in LB with Chlr for 1 h. 100 μl and 10 μl of culture were spread on the TE-agar plate (1% peptone, 0.5% yeast extract, 1.2% agar) supplemented with Chlr (20 μg/ml) and 4.5% of sucrose and incubated at 37 °C overnight. Colonies were picked from the plate and characterized by PCR, DNA sequencing, and Southern blotting. Modified BAC DNA was prepared using BIG DNA kit (Princeton Separations). The 123-kb human DNA insert released by NotI digestion was separated by pulse-field electrophoresis overnight using a low melting point agarose gel. A gel slice containing the 123-kb DNA band was cut from the gel and purified by treating with agarase (New England Biolabs) followed by phenol/chloroform extraction.
Generation of Transgenic Mouse Lines
Purified DNAs were microinjected into fertilized mouse oocytes (C57BL/6×SJL) to generate the transgenic lines according to standard techniques (Transgenic and Chimeric Mouse Facility, University of Pennsylvania). Founders and transgene copy numbers were determined by Southern blot analysis of tail DNA. The CD/hGH BAC and CDΔ1.6/hGH BAC transgenic lines have been described previously (13, 31).
Co-RT-PCR Endonuclease Cleavage Assay for Quantification of hCD79b Transgene Expression
Assays were performed as described previously (9, 13). 0.5 μg of total RNA extracted from tissues or cell lines using RNA-Bee (Tel-Test) was reverse-transcribed with an oligo(dT) primer (Promega) in the presence of avian myeloblastosis virus reverse transcriptase (Promega). Human and mouse CD79b mRNAs were then co-amplified using a primer set corresponding to regions perfectly conserved between hCD79b and mCD79b (supplemental Table 1). The 5′ primer was end-labeled with [γ-32P]ATP and T4 polynucleotide kinase (New England Biolab). This primer set spans intron 5 in order to distinguish cDNA from contaminating genomic DNA. After 32 PCR cycles, products were digested; HinfI specifically cleaves mCD79b cDNA, and SfcI specifically cleaves hCD79b cDNA. Fragments were separated on a 6% polyacrylamide gel, and bands were quantified by PhosphorImager analysis (Storm 840 PhosphorImager, GE Healthcare). The ratio of hCD79b/mCD79b cDNA products was normalized to transgene copy numbers to generate an expression per gene copy value (expression of the endogenous mCD79b was defined as 1.0). All PCR results were determined to be within the linear range of amplification.
Northern Blot Analysis
Total RNA was extracted from cultured cells with RNA-Bee (Tel-Test). 20 μg of each RNA sample was separated on a 1.2% agarose gel containing 2.2 m formaldehyde in 1× MOPS, and transferred to a Zetaprobe® membrane (Bio-Rad). The membrane was probed with 32P-labeled hCD79b cDNA at 65 °C overnight in hybridization solution (0.5 m NaHPO4, pH 7.2, 0.1 mm EDTA, 1% BSA, 7% SDS) and then washed (1× SSC, 0.1% SDS, 0.5× SSC, 0.1% SDS, and finally 0.1× SSC, 0.1% SDS) at 65 °C. Signals were detected by exposure to a PhosphorImager screen. The probe was demonstrated to be specific to the human CD79b mRNA by analysis of a non-transgenic mouse B cell sample.
5′-Rapid Amplification of cDNA Ends (RACE) Assay
Total RNAs isolated from transgenic mouse lymphocytes were reverse transcribed with Superscript III reverse transcriptase (Invitrogen) from an antisense hCD79b primer corresponding to a site in exon 6 (supplemental Table 1). A poly(G) tail was added to the 3′-end of the transcribed cDNA using terminal dinucleotidyltransferase (Promega). An initial PCR was run using the oligo(dC) adaptor primer and an hCD79b-specific antisense primer in exon 4. A subsequent nested PCR was run between the adaptor primer and exon 3 antisense primer (supplemental Table 1). The amplification products were cloned into the pGEM-T vector (Promega) and sequenced. DNA sequencing was performed by the University of Pennsylvania DNA Sequencing Facility.
DNase I HS Mapping by Indirect End Labeling on Southern Blots and by PCR
Cells from B cell cultures and from purified mouse lymphocytes were collected by centrifugation at 200 × g and washed with cold PBS. The pellets were resuspended at 108 cells/ml in ice-cold sucrose buffer I (0.32 m sucrose, 3 mm CaCl2, 2 mm magnesium acetate, 10 mm Tris-HCl, pH 8.0, 1 mm DTT, 0.5 mm PMSF, 0.05% Triton X-100) by gentle pipetting and then incubated on ice for 10 min. The cell suspensions were centrifuged at 500 × g for 5 min at 4 °C and washed with RB buffer (100 mm NaCl, 50 mm Tris-HCl, pH 8.0, 3 mm MgCl2, 0.1 mm PMSF, 5 mm sodium butyrate). The nuclei were resuspended again at 108 cells/ml RB buffer, and a base-line (0 s) aliquot was withdrawn. One-half volume of RB buffer containing 3 mm CaCl2 (final CaCl2 concentration of 1 mm) and 25 units of DNase I (Invitrogen) per ml was added, and the sample was incubated at 37 °C. Aliquots were withdrawn at the indicated times, adding EDTA to 50 mm, followed by digestion of the nuclei with 120 μg of proteinase K per ml in 0.8 m NaCl, 0.5% SDS overnight at 55 °C, extraction with phenol/chloroform, and a final ethanol precipitation. DNase I-digested DNA samples were subsequently digested with BamHI/XhoI, separated on 1% agarose gel, and transferred to Zetaprobe® membranes (Bio-Rad) for hybridization. 0.9 kb of hybridization probe was purified from hCD79b by digestion with NcoI and NdeI.
For PCR-based HS mapping, purified nuclei were treated with 5 units/ml DNase I for 4 min, and genomic DNA was purified as mentioned for the DNase I mapping method. 100 ng of genomic DNA was used for PCR amplification with [α-32P]dCTP and each primer set (supplemental Table 1). Genomic DNA without DNase I treatment was used in parallel for normalizing primer efficiency. The PCR cycle was one cycle of 94 °C for 2 min, 54 °C for 1 min, 72 °C for 1 min, and 25 cycles of 94 °C for 30 s, 54 °C for 30 s, 72 °C for 30 s, and 72 °C for 2 min. PCR products were separated on 6% acrylamide gels with 1× TBE, exposed to a PhosphorImager screen, and detected with a Storm 840 PhosphorImager (GE Healthcare). The band density was quantified by ImageQuant (GE Healthcare).
Western Blot Analysis
Whole cell extracts were isolated from CRL-1484 and Ficoll gradient-isolated lymphocytes of CD1 (non-transgenic control), 1255B (CD/hGH BAC), and 1284F (CDΔ0.7/hGH BAC). 10 μg of protein was loaded on each lane. Two monoclonal rabbit anti-CD79b antibodies (ab134103 and ab134147, Abcam) were used to detect CD79b. The ab134103 antibody recognizes the human CD79b N terminus specifically, whereas ab134147 detects the C terminus of CD79b of humans, mice, and rats. The polyclonal rabbit anti-L7a serum was used for an internal control measurement (32). Donkey anti-rabbit Ig conjugated with horseradish peroxidase (HRP) (Amersham Biosciences) was used for a secondary antibody. The signal was detected using the ECL Western blotting detection kit (GE Healthcare).
Electrophoretic Gel Mobility Shift Assays (EMSAs)
The EMSAs were performed using CRL-1484 nuclear extract and 32P-labeled synthetic double-stranded oligonucleotides encompassing the hCD79b hypersensitive region in intron 1. The sequences of EMSA probes are listed in supplemental Table 1. Binding reactions (20 μl) contained 4% glycerol, 20 mm Tris-HCl (pH 8.0), 60 mm KCl, 5 mm MgCl2, 100 μg/ml BSA, 0.1 m DTT, 100 ng of poly(dI-dC), ∼2 × 104 cpm of 32P-labeled probe, and nuclear extract protein. The binding reactions were incubated for 40 min at room temperature and loaded onto a 7.5% polyacrylamide gel with 2.5% glycerol in 1× TG buffer (0.025 m Tris-HCl, 0.19 m glycine, pH 8.3). In competition assays, an unlabeled probe (50- and 200-fold excess) was incubated with the nuclear extracts for 10 min before the addition of labeled probe.
RESULTS
Expression of the hCD79b Gene Is Maintained in the Absence of Its Promoter and 5′ Terminus
Our previous studies revealed that the isolated hCD79b gene extending from 500 bp 5′ to the ATG initiation codon through the poly(A) addition site retains robust, copy number-dependent, and tissue-specific expression in transgenic mouse B cells (13). The aim of the current study was to identify the determinant(s) within this defined region that establish the fully activated hCD79b chromatin environment in vivo. To enhance the relevance of these studies to the native chromatin locus, all studies were carried out within the context of an extensive BAC transgene. A 123-kb human genomic transgene contains the hCD79b gene in its native context, flanked at its 5′-end by the SCN4A muscle sodium channel gene and at its 3′-end by hGH and the testicular cell adhesion molecule 1 (TCAM1) gene (Fig. 1A). This CD/hGH BAC has been previously demonstrated to recapitulate appropriate and full expression of the encompassed genes in the host mouse pituitary, placenta, and B cells (13, 31, 33).
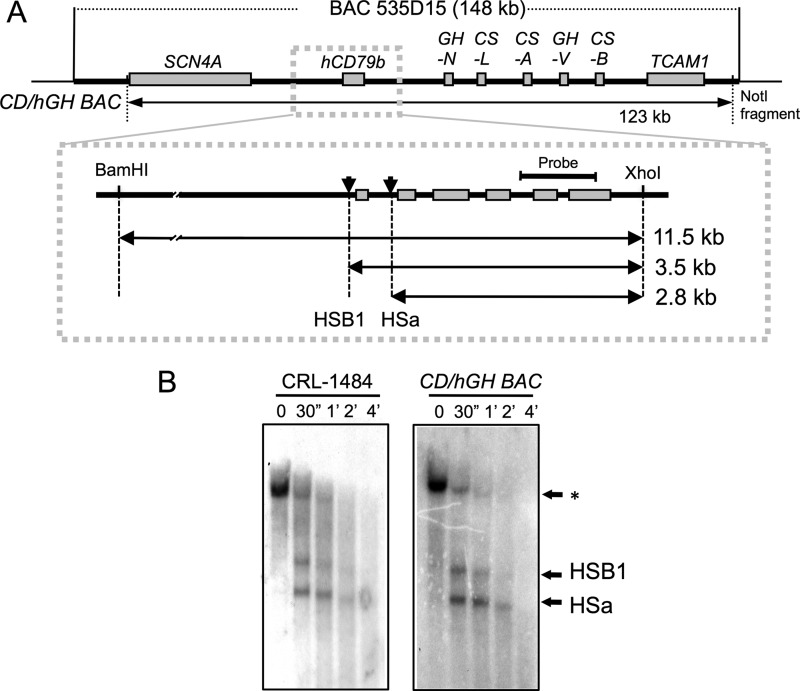
FIGURE 1.
Map of the hCD79b/hGH locus. A, a 123-kb NotI-digested DNA fragment (CD/hGH BAC transgene), isolated from a 148-kb human BAC clone (BAC 535D15), was used to generate a set of transgenic mouse lines. The map of the locus is shown at the top with the respective genes labeled. An expanded view of the hCD79b gene region is shown below this map, with the six exons represented as rectangles. The diagram also summarizes the DNase I HS mapping strategy. After partial digestion of chromatin samples with DNase I, the extracted DNA was digested with BamHI and XhoI. The 3.5- and 2.8-kb sub-bands, released by the partial DNase I-digestion were visualized by Southern blot analysis (B) using the indicated 0.9-kb NdeI/NcoI fragment as a hybridization probe (Probe). B, DNase I mapping and Southern blot analysis. DNase I HS mapping was carried out on nuclei from a human B cell line (CRL-1484) and primary lymphocytes isolated from a CD/hGH BAC transgenic mouse. Purified nuclei were treated with 30 units of DNase I for the indicated times. *, position of the originating 11.5-kb BamHI/XhoI fragment and the two DNase I-generated sub-bands corresponding to cleavage at each of the two DNase I HS sites, HSB1 and HSa. These are indicated and migrated at 3.5 and 2.8 kb, respectively.
To identify critical transcriptional determinants of hCD79b expression, we carried out a series of DNase I mapping studies on the nuclei of primary B cells isolated from the spleens of the CD/hGH BAC transgenic mice. Two DNase I HS sites were identified within the previously determined limits of the fully functional hCD79b domain. These sites are HSB1 at 200 bp 5′ upstream to the ATG initiation codon and HSa located within intron 1 (Fig. 1, A and B). A 0.7-kb deletion extending from −0.5 kb of the ATG to a site just 3′ to the exon 1/intron I junction was then introduced in the CD/hGH BAC transgene. This deletion encompassed HSB1, along with the hCD79b promoter and its proximal enhancer elements (CDΔ0.7/hGH BAC; Fig. 2A). It was assumed, based on the transient transfection studies by others (10, 12), that this extensive deletion would fully ablate hCD79b transcription and serve as a starting point for more limited alterations.
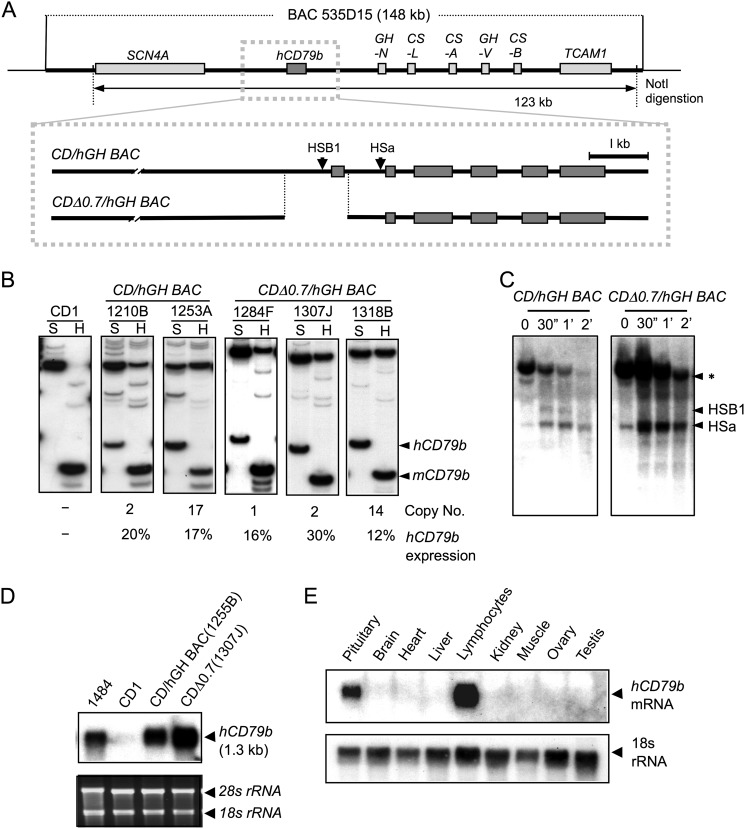
FIGURE 2.
Deletion of the hCD79b promoter and adjacent exon 1 failed to alter levels of hCD79b mRNA expression. A, map of the unmodified wild-type CD/hGH BAC transgene (as in Fig. 1A) and of the derivative transgene lacking a 0.7-kb 5′ segment encompassing the hCD79b promoter and exon 1 (CDΔ0.7/hGH BAC). The 122-kb CDΔ0.7/hGH BAC fragment was released by NotI digestion and used to generate a set of transgenic mouse lines (1284F, 1307J, and 1318B). B, quantification of mouse and human CD79b mRNAs. Human and mouse CD79b mRNA from the B cells of the indicated lines were co-amplified by RT-PCR, and cDNAs were distinguished by restriction enzyme digestions. SfcI (S) digestion of the 32P-5′-end-labeled hCD79b cDNA generated a 95-bp product corresponding to the hCD79b cDNA. Digestion with HinfI (H) exclusively generates a 63-bp mCD79b cDNA product. The transgene copy numbers indicated below each panel were determined by Southern blot analysis. hCD79b expression per transgene copy was normalized to endogenous mCD79b expression, and values were indicated as percentages below each respective lane. The mean expression level was minimally changed by the deletion. C, DNase I hypersensitive site mapping confirms the deletion of HSB1. HS mapping was carried out as described (Fig. 1 and “Experimental Procedures”). The wild-type CD/hGH BAC transgene locus has both HSs intact, whereas the CDΔ0.7/hGH BAC transgene locus assembles HSa but lacks HSB1. D, Northern blot analysis of B cell mRNA from the wild-type CD/hGH BAC and the CDΔ0.7/hGH BAC transgenes revealed no change in hCD79b mRNA size despite the deletion. Total RNAs were extracted from lymphocytes of each sample and separated on a denaturing agarose gel and hybridized with a probe specific to human CD79b mRNA. A human plasma cell line (CRL-1484) was used as a positive control and B cells from a non-transgenic CD1 mouse served as a negative (species specificity) control. Ethidium bromide-stained 28 and 18 S rRNAs were used for loading controls. E, tissue-specific expression of hCD79b was maintained in the absence of the promoter and exon I. Northern blots of RNAs isolated from the tissues of a CDΔ0.7/hGH BAC mouse were analyzed for hCD79b expression. Expression was detected only in lymphocytes and pituitary as observed with the non-deleted transgene (9). 18 S rRNA was used as a loading control.
Three CDΔ0.7/hGH BAC lines were generated with transgene copy numbers of 1, 2, and 14 (lines 1284F, 1307J, 1318B, respectively). The impact of the 5′ deletion on hCD79b expression was assessed by a co-RT-PCR assay that directly compares hCD79b mRNA with the endogenous mCD79b mRNA. The ratio of the human and mouse CD79b mRNA levels in each of the three lines was normalized to the respective gene copy numbers. Quite unexpectedly, we observed that hCD79b mRNA expression per gene copy was unaffected by the extensive 5′ deletion; hCD79b mRNA expression of the three CDΔ0.7/hGH BAC transgenic lines was equivalent to that from the unmodified CD/hGH BAC (Fig. 2B). These data led us to conclude that the 5′-truncated hCD79b locus, lacking the promoter and HSB1, retained determinants that were sufficient to establish an autonomous and active chromatin domain.
DNase I HS mapping of nuclei isolated from CDΔ0.7/hGH BAC splenic B cells confirmed the expected loss of HSBI (Fig. 2C). Of note, however, was the continued formation of HSa. This continued formation of HSa, located within intron 1, indicated that it could assemble independently of HSB1. This observation suggested that HSa might have a unique function, a function that might relate to the continued expression of the promoter-deleted hCD79b transgene.
hCD79b Has a Secondary Promoter within the Gene
The observation that hCD79b transcription is maintained in the absence of its 5′-flanking region, promoter, and first exon was followed by characterization of the encoded mRNA. Northern blot analysis of lymphocyte RNA revealed that the hCD79b mRNA encoded by the CDΔ0.7/hGH BAC was indistinguishable in size and abundance from that of the wild type hCD79b mRNA (Fig. 2D). A tissue survey of expression from the CDΔ0.7/hGH BAC transgene further revealed the same restriction to the lymphocyte and pituitary cell populations as occurs from the intact locus (Fig. 2E). (Note that the hCD79b mRNA present in the pituitary is non-coding and is linked to the activation of the hGH-N gene (9, 31, 34).)
The robust expression, normal size, and conserved tissue distribution of the hCD79b mRNA from the CDΔ0.7/hGH BAC inferred the existence of a secondary promoter site internal to the hCD79b locus. To identify this secondary promoter, we mapped the hCD79b transcription start sites (TSSs) in CD/hGH BAC and CDΔ0.7/hGH BAC B cells by 5′-RACE. 5′-RACE analysis of the intact CD/hGH BAC locus confirmed the expected set of TSS within a 200-bp window 5′ to the ATG initiation codon (Fig. 3A). The heterogeneity of these 5′ termini agreed with prior studies (9) and was consistent with the GC-rich and TATA-less structure of the promoter region (35). Of note, ∼10% of the 5′ termini in this 5′-RACE assay mapped to a site within exon 2 (Fig. 3A). This short form of the hCD79b mRNA (5′-capped) was also detected in an analysis of normal (primary) human B cells (data not shown). The 5′-RACE analysis of the hCD79b mRNA from the CDΔ0.7/hGH BAC mice mapped the 5′ termini to this same position in exon 2 (Fig. 3A). This positioning of 5′ TSS predicts that the 5′ deletion in the CDΔ0.7/hGH BAC, although changing the site of transcription initiation, would have a minimal impact on overall mRNA size, considering the small size of exon 1 (∼70 nt) compared with the overall size of the hCD79b mRNA (∼1,300 nt). This minimal difference was consistent with the apparent lack of a change in the size of the hCD79b mRNA in the Northern comparison (Fig. 2D). The 5′-RACE data thus pointed out the existence of a secondary promoter within the intact hCD79b gene. The quantitative expression studies, along with the 5′-RACE results, further demonstrated that the activity of the secondary promoter was substantially enhanced when the primary promoter and associated 5′-flanking elements were removed from the locus.
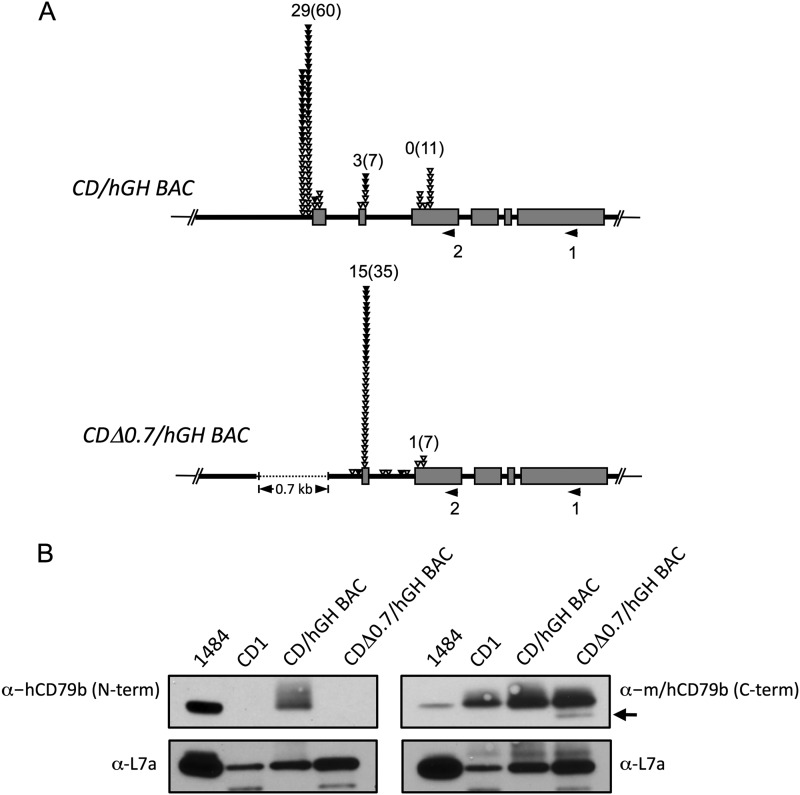
FIGURE 3.
A secondary promoter for the hCD79b is markedly enhanced subsequent to deletion of the primary promoter, and its mRNA encodes an N-terminally deleted CD79b protein. A, 5′-RACE assay analyses of B cell mRNA from the wild-type CD/hGH BAC revealed two sites of polymerase II transcription initiation. Each RNA sample was reverse transcribed using hCD79b-specific exon 6 primer (1), and the 3′-ends of these cDNAs were extended with poly(G). Poly(G)-tailed cDNAs were then amplified with exon 3 primer (2) and poly(C) adaptor primer and cloned into pGEM-T vector for sequencing. Each triangle indicates a transcription start site from each sequenced clone in the 50-bp window. The total numbers of cDNAs containing an additional non-templated terminal C (corresponding to the 5′-cap structure) are shown, and the total numbers of cDNAs mapping to the indicated position (with and without the additional C) are included in parentheses. The position of the 0.7-kb deletion that removes the primary promoter and exon 1 is indicated by the dotted line. B, hCD79b proteins were detected by human-specific rabbit monoclonal antibody that recognizes the N terminus of the protein (left; ab134103) and a rabbit monoclonal antibody that detects the C terminus of both the mouse and human CD79b (right; ab134147). The inability to detect an hCD79b protein in lymphocytes from the CDΔ0.7/hGH BAC transgene with the N terminus-specific antibody (left) was consistent with the exon 1-truncated structure of the mRNA initiated from the secondary promoter. The antibody recognizing the C terminus of both mouse and human CD79b revealed a smaller hCD79b (∼30 kDa) in the protein extract from the CDΔ0.7/hGH BAC lymphocytes (arrow), supporting the presence of a protein encoded by the mRNA originating from the secondary promoter. Ribosomal protein, L7a (30 kDa), detected using rabbit polyclonal antibody, served as an internal loading control.
Western blot analysis was performed to examine whether the mRNA initiated at the secondary promoter was translated to a protein product in vivo. The first AUG encountered in this mRNA is located in exon 3 and remains in-frame with the full-length hCD79b mRNA. Consistent with the prediction that the protein encoded by this 5′-shortened mRNA lacks the signal peptide (encoded in exon 1), the fluorescence-activated cell sorting (FACS) failed to detect a hCD79b protein on the surface of lymphocytes from the CDΔ0.7/hGH BAC line (data not shown), and Western analysis with an antibody the 5′ terminus of the hCD79b protein failed to reveal a protein. Of note, however, an antibody to the 3′ terminus detected an hCD79b protein in CDΔ0.7/hGH BAC lymphocytes that migrated slightly faster than the major protein band and was not detected in lymphocytes isolated from the wild type CD/hGH/BAC transgene (Fig. 3B). The size of this protein was consistent with that predicted from the sequence of the mRNA originating from the secondary promoter.
Activity of the Secondary Promoter Is Dependent on the Segment of Intron 1 Encompassing HSa
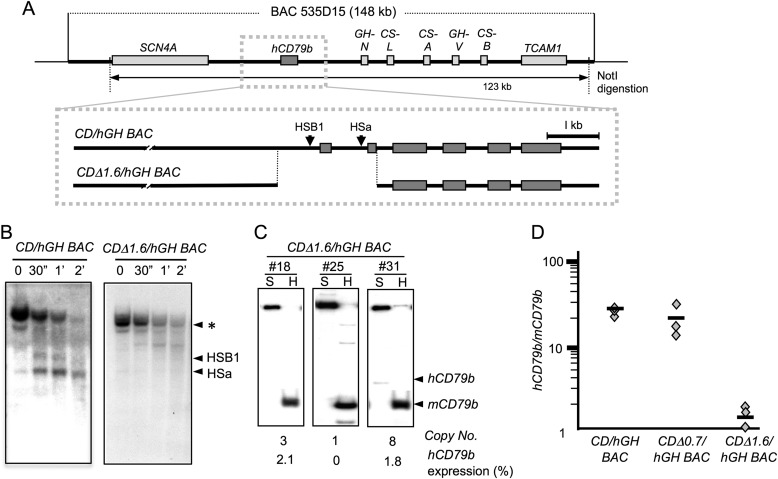
The CDΔ0.7/hGH BAC transgene study confirms that HSa can form independently of the determinants surrounding the primary promoter (Fig. 2B). The initiation of transcription from the secondary promoter of hCD79b, within exon 2, would eliminate exon 1 from the mRNA. This exon encodes the signal peptide of the trans-membrane CD79b protein. Thus, usage of this secondary promoter would have a major impact on function of the hCD79b protein product. To explore this secondary promoter activity in more depth, we first attempted to rule out the possibility that the secondary TSS reflected a general promiscuity of transcriptional control in this genomic region. This was accomplished by extending the 5′ deletion of the hCD79b further 3′ to include intron 1 and exon 2 and terminate in intron 2 (CDΔ1.6/hGH BAC; Fig. 4A). This deleted region would be predicted to remove HSa as well as the secondary promoter site defined by the 5′-RACE (Fig. 3). Three lines carrying this CDΔ1.6/hGH BAC transgene were established, and DNase I HS mapping of chromatin from B cells of these mice confirmed the loss of HSa as well as HSB1 (Fig. 4B). Expression analysis revealed that this extension of the 5′ deletion essentially ablates hCD79b expression in the splenic B cells (Fig. 4, C and D). An analysis of nascent nuclear transcripts confirmed that the loss of mRNA expression from the CDΔ1.6/hGH BAC was due to a dramatic loss of transcriptional activity rather than to instability of the encoded mRNA as might be triggered by nonsense mRNA-mediated decay (data not shown). These data support the presence of specific determinants capable of triggering high level transcription from the secondary promoter within hCD79b when the primary promoter and associated elements are deleted from the locus. In particular, the retention of full levels of expression of hCD79b mRNA in CDΔ0.7/hGH BAC and its quantitative loss in the CDΔ1.6/hGH BAC are consistent with a role for HSa in this process.
FIGURE 4.
A deletion of the hCD79b gene that encompasses the primary and secondary promoters ablates gene expression. A, map of the CDΔ1.6/hGH BAC transgene. A 1.6-kb segment of the hCD79b gene was removed from the CD/hGH BAC transgene by homologous recombination. This deleted region extended the 5′-flanking region through exon 2. The modified 121-kb transgene, released by NotI digestion, was purified and used to generate a set of transgenic mouse lines (#18, #25, and #31). B, DNase I HS mapping of chromatin from CDΔ1.6/hGH BAC lymphocytes confirmed elimination of HSB1 and HSa. The origins of the chromatin for the two assays are indicated above the Southern blot images, and the migration of HS fragments is shown to the right. C, expression of hCD79b mRNA was dramatically diminished by loss of the primary and secondary transcription start sites in the CDΔ1.6/hGH BAC lymphocytes. hCD79b mRNA levels were normalized to endogenous mCD79b expression and to transgene copy number. These normalized values are indicated as percentages of mCD79b mRNA expression below each respective lane. Three independent lines with unique transgene insertion sites were analyzed. The transgene copy numbers were determined by Southern blot analysis. D, summary of hCD79b mRNA expression levels, from intact and deleted constructs, in B cells of the transgenic lines. Each symbol represents the normalized level of hCD79b mRNA in a separate transgenic mouse line. The loss of the primary promoter (Δ0.7) had no significant effect on mRNA levels, whereas extension of the deletion to encompass the secondary promoter (Δ1.6) resulted in a dramatic loss of gene expression.
Structural and Functional Characterization of HSa
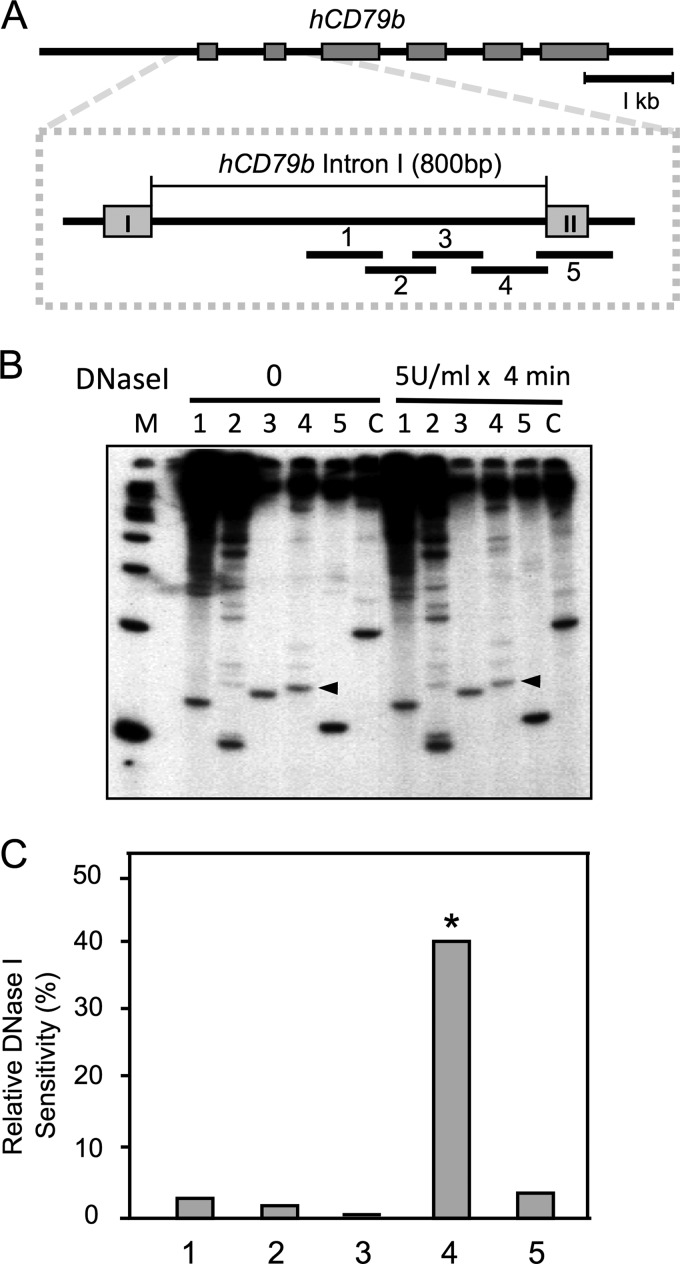
The position of HSa was confirmed and further resolved using a PCR-based assay. Five sets of primer pairs were designed to span the 3′ region of intron 1 (Fig. 5A). This region encompasses the site of HSa as predicted by DNase I HS Southern blot mapping. PCR amplification was performed with [α-32P]dCTP at low cycle numbers and separated on polyacrylamide gel. Each band was normalized to the PCR signal from the input genomic DNA (non-DNase I-treated). The data revealed a single amplimer product that was selectively reduced in intensity compared with the band from non DNase I-treated genomic DNA (Fig. 5B). This region, detected by primer pair 4 (129 bp), was concluded to encompass HSa.
FIGURE 5.
The specific location of HSa was mapped with a tiled amplimer array. A, chromatin from CD/hGH BAC lymphocytes was subjected to partial DNase I digestion followed by amplification of the purified DNA with five partially overlapping amplimer sets. The positioning of the amplimer sets is shown in the expanded view of the 5′-end of the hCD79b gene. B, amplicons were incorporated with [32P] α-dCTP and separated on a 6% polyacrylamide gel. Correctly sized bands (amplimer 1, 120 bp; amplimer 2, 100 bp; amplimer 3, 127 bp; amplimer 4, 129 bp; amplimer 5, 113 bp; mGAPDH, 236 bp) were quantified. The arrowheads indicate the amplicons (4) that show the most sensitivity to DNase I treatment. C, the products of each amplimer set from DNase I-digested samples were normalized to each signal from non-digested control sample (see “Experimental Procedures”). The decrease in the amplicon signal due to DNase I digestion was calculated and plotted as a percentage of relative sensitivity. *, amplimer with the greatest level of DNase I sensitivity and thus the location of HSa. mGAPDH was used for the normalization of PCR amplifications.
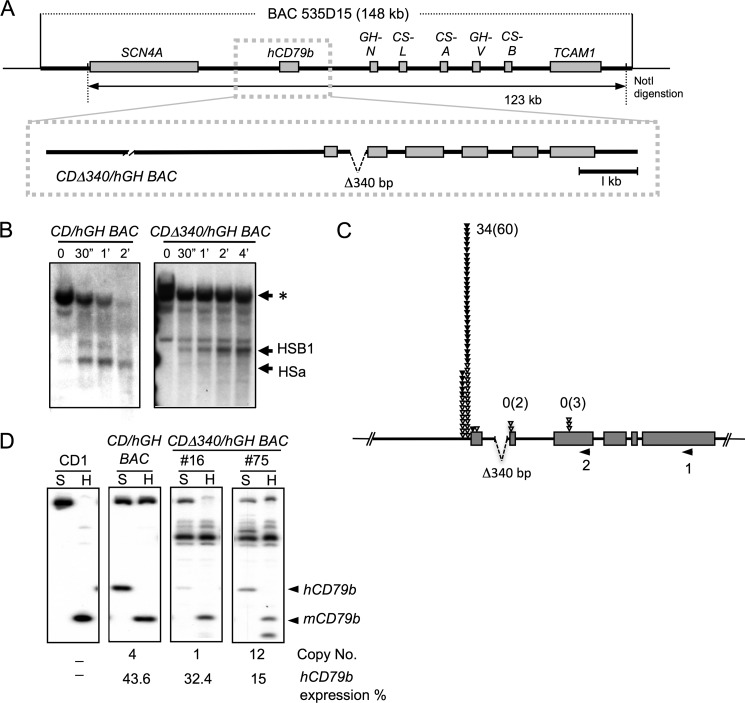
A new transgene was generated to define the role of HSa in hCD79b expression. This CDΔ340/hGH BAC-derived transgene contained a deletion that encompasses the HSa segment. Because the determinant responsible for the formation of DNase I HS might be adjacent to the HSa rather than coincident with it (36), a 340-bp segment from intron 1 that encompassed the 129-bp putative HSa region along with limited extents of additional 5′- and 3′-flanking sequences was deleted. This deletion left the last 27 nucleotides of the intron intact in order to preserve the splice acceptor site. Two CDΔ340/hGH BAC lines were generated (Fig. 6A). HS mapping of B cell nuclei from these lines confirmed that this deletion eliminated HSa. Of note, HSB1 remained intact (Fig. 6B). This result, along with the HS mapping of the 5′ deletion transgene (Fig. 2C), demonstrated that both HSB1 and HSa are formed independently of each other and thus may support independent functions and independent sets of TSSs.
FIGURE 6.
Deletion of HSa results in a selective loss of transcription initiation from the hCD79b secondary promoter. A, deletion of HSa from the CD/hGH BAC transgene. A 340-bp segment of intron 1 encompassing HSa was deleted from the CD/hGH locus, and the modified human gene was released from its BAC 535D15 vector. After purification, the fragment was injected into mouse embryos to generate a set of CDΔ340/hGH BAC transgenic mice (#16 and #75). B, DNase I HS mapping of CDΔ340/hGH BAC lymphocyte chromatin confirmed selective elimination of HSa. Nuclei from the indicated wild type and 340 bp-deleted human transgenic mouse lymphocytes were analyzed as in Fig. 1B. HSB1 remained intact despite the loss of the HSa determinant. C, hCD79b transcription initiation from exon 2 was selectively repressed in the HSa-deleted transgene. hCD79b TSSs in the CDΔ340/hGH BAC were mapped and quantitiated by 5′-RACE. Each inverted triangle indicates a transcription start site from each sequenced clone in a 50-bp window. The filled triangles indicate individual cDNA clones generated by the 5′-RACE that contain an additional non-templated C corresponding to the 5′-7meG cap. The numbers of C-containing clones are shown for each site, and the total numbers of clones are noted in parentheses. D, the 340-bp deletion encompassing HSa fails to alter the net expression from the hCD79b locus. Co-RT-PCR was performed to examine hCD79b expression in CDΔ340/hGH BAC. The origin of each lymphocyte RNA sample is indicated above the respective panel; CD1, nontransgenic mouse; BAC17, CD/hGH BAC; #16 and #75, CDΔ340/hGH BAC.
Of note, the deletion of HSa in the CDΔ340/hGH BAC transgene was accompanied by a selective loss of transcriptional activity corresponding to the secondary promoter region (Fig. 6C). In addition, mRNA analysis of the CDΔ340/hGH BAC lines revealed that the overall levels of hCD79b mRNA in the absence of HSa were maintained in the range of wild type hCD79b transgene expression (∼10–53%) (Fig. 6D). These results support the conclusion that HSa plays a critical and specific role in the activation of PolII transcription from the secondary promoter within the hCD79b gene.
In Vitro Analysis of the HSa Region for trans-Factor Binding
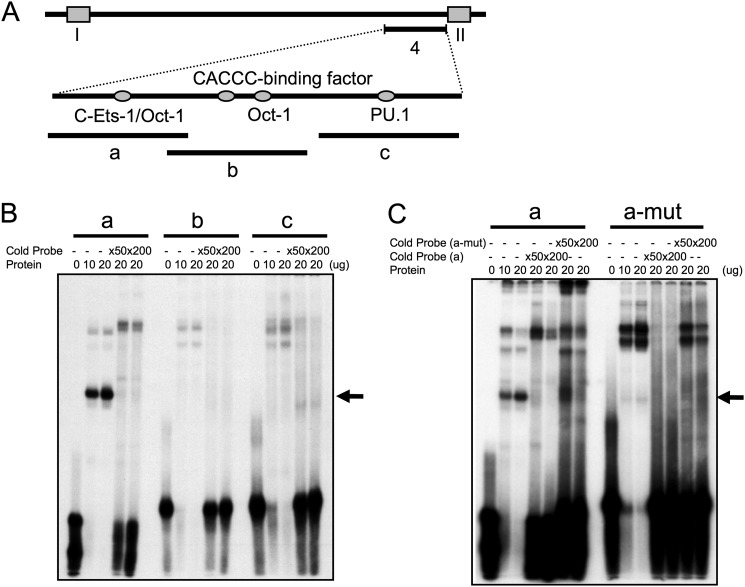
The sequence encompassing the HSa region was scanned for trans-factor binding sites (see the TESS (Transcription Element Search System) Web site). Strong predictions included the sites for c-Ets-1/PU.1, Oct-1, and CACCC-binding protein (Fig. 7A). Gel shift assays were carried out to determine whether any of these predicted proteins bind to HSa. Each of three 40-bp probes corresponding to the DNase I-hypersensitive region in hCD79b intron 1 were designed and used for narrowing the search for binding proteins (Fig. 7A). After incubation of each probe with human B cell nuclear extract, DNA-protein complexes were separated on a polyacrylamide gel (Fig. 7B). DNA probe 1 generated a strong single DNA shift. Probe 1 contains a predicted binding site for an Ets protein (37). The c-Ets-1 was of particular interest because it had been identified in a number of studies as a key factor in lymphocyte activation. Of additional importance was the observation that this c-Ets-1 binding sequence, GAAGTA, is highly conserved at this site from mice to humans. Remarkably, a two-nucleotide substitution (GAAGTA → AGAGTA) at this site eliminated the band shift (Fig. 7C). These data suggest that this Ets-1 binding site is a determinant of HSa and/or is involved in the activity of the secondary promoter within the hCD79b gene.
FIGURE 7.
Ets family protein binds to the DNase I hypersensitive region of hCD79b. A, diagram of the HSa region in hCD79b intron 1 (see also Fig. 5). Three probes (a–c) encompassing the HSa (region 4; see Fig. 5) were used for gel shift analyses. Transcription factor binding sites predicted by informatics analyses of the corresponding sequence are indicated. B, band shift analysis. Nuclear extract from the human plasma cell line CRL-1484 was incubated with each 32P-labeled probe (a–c) and resolved on a native 7.5% polyacrylamide gel. Non-labeled probes were added at the indicated excess to the labeled probe (×50 and ×200) for self-competition studies. The arrow to the right indicates the position of the putative Ets protein-DNA complex. C, band shift analysis with probe “a” containing a mutation at the Ets binding site (a-mut) (GAAGTA → AGAGTA). The mutant probe was used to confirm the specific binding of Ets family proteins. The arrow indicates the specific band shift with Ets protein using probe “a” and its selective loss when the Ets site is mutated (probe “a-mut”).
The c-Ets-1 Binding Site at the 3′-End of Intron I Selectively Enhances Secondary Promoter Activity
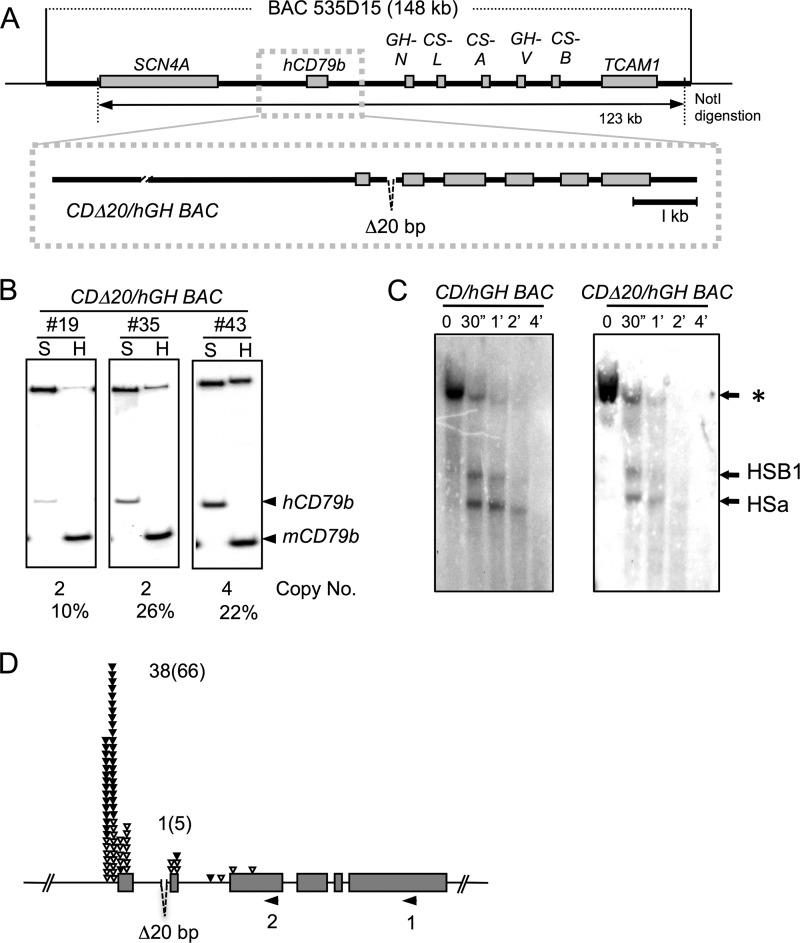
DNase I HSs are assumed to represent binding sites for protein complexes involved in genome organization and/or transcriptional control (15, 18). Gel shift assays revealed that the Ets protein binds to a site within the hCD79b intron 1 that is situated in close proximity to HSa. To examine if this Ets family protein binding was critical to HSa formation and/or to the support of hCD79b transcription, the putative c-Ets-1 binding site was deleted form the CD/hGH BAC transgene (Fig. 8A). This was accomplished by a site-directed deletion of a 20-bp segment by homologous recombination. Three transgenic lines carrying the 123-kb CDΔ20/hGH BAC were generated and assessed for levels of hCD79b expression in B cells. Co-RT-PCR results indicated that the expression of hCD79b mRNA (Fig. 8B) and the formation of HSa (Fig. 8C) were both maintained in the absence of the c-Ets-1 binding site. In contrast, the 5′-RACE analysis revealed a selective diminution in TSSs at the secondary promoter (Fig. 8D) (also, see “Discussion”). These results indicate that the c-Ets-1 binding site, although not essential to HSa formation, may play a role in the activation of transcription from the secondary promoter.
FIGURE 8.
Deletion of the Ets protein binding site in hCD79b intron 1 selectively represses use of the secondary promoter. A, a 20-bp segment of intron I encompassing the c-Ets-1 binding site (GAAGTA) was deleted from the CD/hGH BAC human transgene. The resulting NotI digestion released a 123-kb insert that was used to establish a set of CDΔ20/hGH BAC transgenic mice (#19, #35, and #43). B, hCD79b mRNA expression in mouse lymphocytes of transgenic mouse lines. Human and mouse CD79b mRNA were co-amplified by RT-PCR, and cDNAs were distinguished by restriction enzyme digestion. Expression per copy number is indicated below each gel segment and is relatively unchanged. C, deletion of the c-Ets-1 site in intron I fails to inhibit formation of HSa. Shown is DNase I HS mapping of purified lymphocytes isolated from the wild-type CD/hGH BAC and the derived transgene with deletion of the c-Ets-1 site in intron I (CDΔ20/hGH BAC). *, position of the originating 11.5-kb BamHI/XhoI fragment and the two DNase I-generated sub-bands corresponding to cleavage at each of the two DNase I HS sites, HSB1 and HSa. These are indicated and migrated at 3.5 and 2.8 kb, respectively. D, 5′-RACE analysis of CDΔ20/hGH BAC transcription initiation. The analysis reveals a ratio of 38:1 for verified (presence of a non-templated 3′-terminal C) TSSs in the hCD79b transcription start sites in the CDΔ20/hGH BAC. All explanations and designations used in this figure are same as in Fig. 3A and Fig. 6C.
DISCUSSION
Human CD79b is essential for B cell function. Its encoded protein is a trans-membrane component of the B cell receptor that is necessary for B cell proliferation and activation subsequent to antigen engagement (38). Mutation of this gene has been identified in a number of hematologic malignancies, most prominently in chronic lymphocytic leukemia (7). Although of critical importance to normal B cell function and linked to malignant transformation, the basis for regulation of hCD79b gene expression remains poorly understood. Initial in vitro studies identified a 200-bp region encompassing a set of transcription factor binding sites and a GC-rich promoter that were predicted to be essential to hCD79b activation (10, 12). An additional enhancer element was identified by other studies in the 3′-flanking region of the rat CD79b (39). To assess hCD79b in a more physiologic context, we reassessed these findings with a set of transgenic mouse lines in a native B cell setting. Using this approach, we demonstrated that transcription of a delimited hCD79b transgene with as little as a 0.5-kb 5′-flanking region (promoter) and 0.8 kb of 3′-flanking sequences was sufficient to support full levels of hCD79b expression in the mouse B cell (13). These studies thus argued against a functional role for enhancer elements 3′ of the gene or in the remote 5′-flanking region in vivo. Two DNase I HSs were identified within this defined hCD79b critical region in B cell chromatin: HSB1, located approximately −0.2 kb from the initiation ATG, and HSa, located within intron 1. Based on these studies, we had concluded that these two HSs were probably critical to and possibly sufficient for the establishment of an autonomous chromatin domain that supports full levels of hCD79b transcription in the B cell.
Remarkably, in the current study, we observed that deletion of the previously defined promoter region of hCD79b, including the linked HSB1, failed to alter the level or tissue specificity of hCD79b transcription (Fig. 2). Extensive studies confirmed that the mRNA expressed from the 5′-deleted locus were processed normally and generated a discrete mRNA product of approximately the same size and abundance as that originating from the intact wild-type locus. 5′-RACE analyses mapped transcription initiation from the 5′-deleted hCD79b locus to a cluster of sites within exon 2. Importantly, HSa, located directly 5′ to the TSSs remained intact. These data demonstrated that HSa can form independently of HSB1 and that robust transcription of hCD79b can be maintained independently of the native promoter region. These findings support a model in which the hCD79b gene contains a fully functional alternative promoter internal to the gene.
The relevance of HSa to hCD79b expression was next assessed by two additional transgenes. In the first, the 5′ deletion of hCD79b was extended to exon 2. As with all of the transgenes in this study, this was done in the context of an extended and native locus configuration in the hGH/CD BAC transgene. As predicted from our HS mapping study, this extended deletion effectively removed HSa as well as the secondary promoter region within intron 1 and resulted in a dramatic reduction in hCD79b mRNA levels (Fig. 4). A more refined deletion was then introduced to remove HSa based on mapping of the HS site using a tiled array of amplimers (Fig. 5). This 340-bp deletion within intron 1 effectively removed HSa and ablated TSS from the secondary promoter. Analysis of the locus revealed the normal assembly of HSB1 and full levels of gene expression (Fig. 6). These results suggest that these two HSs, HSB1 and HSa, are formed independently of each other. The data further suggest that either of these HS is individually sufficient to establish an hCD79b chromatin locus that supports full levels of transcription in the B cell.
Detailed analysis of HSa, based on structural mapping, informatic analysis, and gel shift studies (Figs. 5 and 7) pointed to an important binding site for an Ets family protein. Ets family proteins, including c-Ets-1, c-Ets-2, and PU.1, have been reported to be expressed in the hematopoietic tissues and play a pivotal role in hematopoietic cell development. Functional analysis was then undertaken by site-specific deletion of this Ets family protein binding site from 123-kb human CD/hGH BAC. Remarkably, this mutation failed to eliminate HSa (Fig. 8C) and failed to decrease overall expression from the hCD79b gene (Fig. 8B). It did, however, selectively decrease transcriptional initiation from the secondary promoter. These results suggested that Ets factor binding in the HSa region, although not necessary for HSa formation, is involved in the support of local transcriptional activation and initiation from the secondary promoter.
The relative activities of the primary and secondary promoters in the hCD79b gene were established by 5′-RACE analyses. As a technical note, it is important to comment on the validity of these TSS mapping studies. The identification of a TSS by the 5′-RACE is most reliably scored by the generation of an additional C at the 3′ terminus of the primer extension product, corresponding to the 5′-terminal 7-methylguanosine cap nucleotide (40). This non-templated C is present in 50% of the clones mapping to the primary and secondary promoters (Fig. 3). Using this metric, the analysis of the wild-type locus (CD/hGH BAC transgene) reveals that 10% of steady-state hCD79b mRNAs originate from the secondary promoter (3 of 29 TSSs; Fig. 3). The mutation of the Ets-1 binding site adjacent to HSa (CDΔ20/hGH BAC transgene) reduces this percentage to 2.5% (1 of 38 TSSs). These data, along with the complete loss of secondary promoter activity subsequent to HSa deletion (Fig. 6), suggest that transcription initiation from the secondary promoter is dependent on HSa and the adjacent Ets-1 binding site.
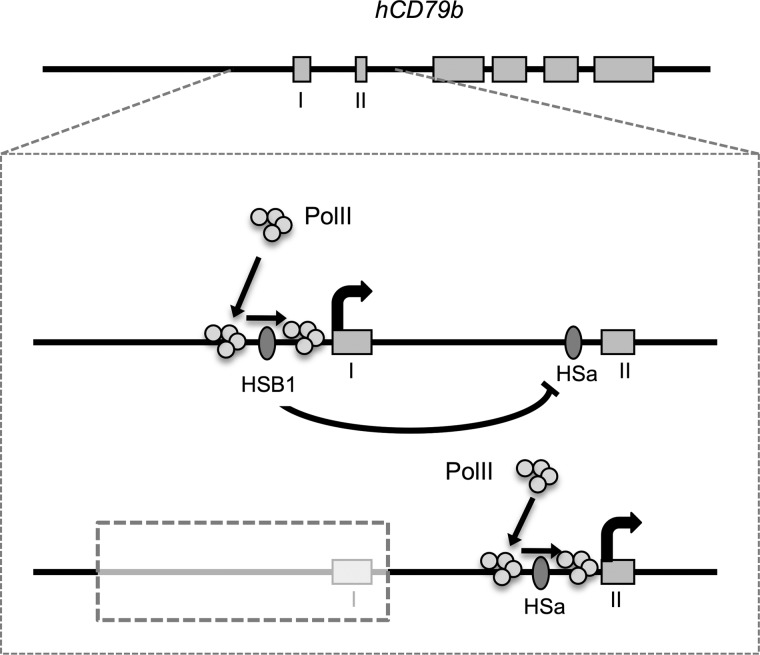
A model that reflects our findings is summarized in Fig. 9. HSB1 and HSa both assemble at the wild-type hCD79b chromatin locus (Fig. 1). These HSs can form independently of each other (Figs. 2C and 6B). However, the activity of the secondary promoter is under the negative influence of the primary promoter and HSa (Figs. 3 and 6C). Under normal circumstances, HSB1 function appears preferential to that of HSa, resulting in a 10:1 ratio of transcription initiation from the promoter sites 5′ of exon 1 and within exon 2, respectively. This ratio may reflect different intrinsic “strengths” of the two sites, or, as shown in the model diagram, this predominance may reflect direct repression of the secondary promoter by the more active primary site. The predominance may also reflect transcriptional interference of the more 5′ promoter on the more 3′ secondary site of polymerase II assembly. Whatever the mechanism, it is clear that deletion of the primary promoter results in dramatic enhancement of the secondary promoter at rates and with tissue specificity comparable with those of the primary site (Fig. 2).
FIGURE 9.
Proposed model of interactions between the primary and secondary promoters at the hCD79b locus. HSB1 recruits chromatin remodeling factors and B cell-specific transcriptional factors to the primary promoter. Transcriptional machineries, including RNA polymerase II, recognize the −0.5 kb to −0.2 kb region and track through open chromatin in the promoter region and start transcription. Activity of the primary promoter inhibits the activity of HSa and the secondary promoter. When the primary promoter is deleted or inactivated, HSa and associated cis-acting determinants are able to mediate full levels of B cell-specific transcription from the adjacent secondary promoter.
What is the functional relevance of the secondary promoter to hCD79b expression? Does it simply represent an evolutionary accident, or does it serve an essential function? Comparison of the human and mouse genomic sequences reveals conservation of sequence over the HSa region. This conservation would suggest a level of functional importance to this region. However, more direct evidence for a critical function for the secondary promoter remains lacking at present.
Promoters generally comprise a complex array of cis-acting regulatory elements required for accurate and efficient initiation of transcription, and in some cases, promoters are also responsible for controlling levels or specificity of transcriptional activity. Simple transcription units encode a single protein product, whereas more complex transcriptional units can produce multiple mature mRNAs that can give rise to distinct protein products. Such complex transcriptional units can generate diversity by a variety of mechanisms. These mechanisms include use of alternative transcript processing pathways for splicing and polyadenylation as well as the use of alternative promoters. Alternative promoters can be regulated in a manner that is specific to tissues or to developmental progression or may be used in a specific setting to generate two discrete mRNAs and encoded protein products (41, 42). In the case of the hCD79b gene, the secondary promoter generates an mRNA that encodes a protein lacking the signal sequence critical to assembly and integration of the wild-type CD79b protein into a functional BCR (Fig. 3B). Thus, activity of the secondary promoter would either shunt the hCD79b gene to a nonproductive pathway or redirect the N-terminally truncated protein to some as yet undefined cytoplasmic function. Whether this altered protein would act as a dominant negative, suppressing BCR assembly and function, or serve an independent role is open to speculation. Further analysis of this hypothetical protein will be necessary to distinguish between these various fates.
Activation of secondary promoters can participate in cell transformation pathways. For example, high levels of c-myb expression can be generated subsequent to deletion of a region encompassing the promoter and first exon human c-myb proto-oncogene. This deletion has been identified in cases of acute lymphocytic leukemia (43, 44) in which high levels of expression initiated from a secondary promoter determinant in intron 1 generate high levels of an N-terminally truncated oncogenic c-myb protein (43). It is apparent from our studies that deletion of the primary promoter of the hCD79b gene would in a similar fashion trigger robust expression from the secondary promoter, with the potential to generate high levels of the truncated protein product. Although we do not at this point have direct evidence that such deletions occur in pathologic settings, the present data support this possibility. In the setting of B cell differentiation, loss of primary promoter function via the native pathway of somatic hypermutation might result in a similar scenario. The somatic hypermutation of hCD79b during B cell maturation was reported previously (45). Thus, the loss of primary hCD79b promoter activity in B cells by a number of potential pathways would radically diminish the synthesis of a functional BCR and generate a novel and potentially detrimental protein product.
In summary, we have identified a secondary promoter of hCD79b. This promoter is located in the 3′ region of intron 1 and is closely linked to and functionally dependent on an internal HS, and its activity is dramatically enhanced when the primary promoter function is abrogated. The presence of a functional secondary promoter in the hCD79b gene sets the stage for complex patterns of gene expression and potential dysregulation of B cell function.
Supplementary Material
Acknowledgments
We thank the University of Pennsylvania Transgenic and Chimeric Mouse Facility (supported by National Institutes of Health P30 Grants DK019525, DK050306, and CA016520) for generation of the transgenic mice.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HD/DK25147 and R01 HD/DK046737 (to N. E. C. and S. A. L.).

This article contains supplemental Table 1.
- BCR
- B cell receptor
- HS
- hypersensitive site
- Chlr
- chloramphenicol
- BAC
- bacterial artificial chromosome
- RACE
- rapid amplification of cDNA ends
- TSS
- transcription start site.
REFERENCES
- 1. Gold M. R., Matsuuchi L., Kelly R. B., DeFranco A. L. (1991) Tyrosine phosphorylation of components of the B-cell antigen receptors following receptor crosslinking. Proc. Natl. Acad. Sci. 88, 3436–3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Benschop R. J., Cambier J. C. (1999) B cell development. Signal transduction by antigen receptors and their surrogates. Curr. Opin. Immunol. 11, 143–151 [DOI] [PubMed] [Google Scholar]
- 3. Calame K. L., Lin K. I., Tunyaplin C. (2003) Regulatory mechanisms that determine the development and function of plasma cells. Annu. Rev. Immunol. 21, 205–230 [DOI] [PubMed] [Google Scholar]
- 4. Huang X., Takata K., Sato Y., Tanaka T., Ichimura K., Tamura M., Oka T., Yoshino T. (2011) Downregulation of the B-cell receptor signaling component CD79b in plasma cell myeloma. A possible post-transcriptional regulation. Pathol. Int. 61, 122–129 [DOI] [PubMed] [Google Scholar]
- 5. Dobbs A. K., Yang T., Farmer D., Kager L., Parolini O., Conley M. E. (2007) Cutting edge. A hypomorphic mutation in Igβ. J. Immunol. 179, 2055–2059 [DOI] [PubMed] [Google Scholar]
- 6. Ferrari S., Lougaris V., Caraffi S., Zuntini R., Yang J., Soresina A., Meini A., Cazzola G., Rossi C., Reth M., Plebani A. (2007) Mutation of the Igβ gene causes agammaglobulinemia in man. J. Exp. Med. 204, 2047–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gordon M. S., Kato R. M., Lansigan F., Thompson A. A., Wall R., Rawlings D. J. (2000) Aberrant B cell receptor signaling from B29 (Igβ, CD79b) gene mutations of chromic lumphocytic leukemia B cells. Proc. Natl. Acad. Sci. 97, 5504–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thompson A. A., Talley J. A., Do H. N., Kagan H. L., Kunkel L., Berenson J., Cooper M. D., Saxon A., Wall R. (1997) Aberrantions of the B-cell receptor B29 (CD79b) gene in chromic lymphocytic leukemia. Blood 90, 1387–1394 [PubMed] [Google Scholar]
- 9. Cajiao I., Zhang A., Yoo E. J., Cooke N. E., Liebhaber S. A. (2004) Bystander gene activation by a locus control region. EMBO J. 23, 3854–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thompson A. A., Wood W. J., Jr., Gilly M. J., Damore M. A., Omori S. A., Wall R. (1996) The promoter and 5′ flanking sequences controlling human B29 gene expression. Blood 87, 666–673 [PubMed] [Google Scholar]
- 11. Malone C. S., Wall R. (2002) Bob1 (OCA-B/OBF-1) differential transactivation of the B cell-specific B29 (Igβ) and mb-1 (Igα) promoters. J. Immunol. 168, 3369–3375 [DOI] [PubMed] [Google Scholar]
- 12. Omori S. A., Wall R. (1993) Multiple motifs regulate the B-cell-specific promoter of the B29 gene. Proc. Natl. Acad. Sci. U.S.A. 90, 11723–11727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yoo E. J., Cajiao I., Kim J. S., Kimura A. P., Zhang A., Cooke N. E., Liebhaber S. A. (2006) Tissue-specific chromatin modifications at a multigene locus generate asymmetric transcriptional interactions. Mol. Cell. Biol. 26, 5569–5579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Paranjape S. M., Kamakaka R. T., Kadonaga J. T. (1994) Role of chromatin structure in the regulation of transcription by RNA polymerase II. Annu. Rev. Biochem. 63, 265–297 [DOI] [PubMed] [Google Scholar]
- 15. Gross D. S., Garrard W. T. (1988) Nuclease hypersensitive sites in the chromatin. Annu. Rev. Biochem. 57, 159–197 [DOI] [PubMed] [Google Scholar]
- 16. Stalder J., Larsen A., Engel J. D., Dolan M., Groudine M., Weintraub H. (1980) Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNase I. Cell 20, 451–460 [DOI] [PubMed] [Google Scholar]
- 17. Wu C. (1980) The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature 286, 854–860 [DOI] [PubMed] [Google Scholar]
- 18. Thurman R. E., Rynes E., Humbert R., Vierstra J., Maurano M. T., Haugen E., Sheffield N. C., Stergachis A. B., Wang H., Vernot B., Garg K., John S., Sandstrom R., Bates D., Boatman L., Canfield T. K., Diegel M., Dunn D., Ebersol A. K., Frum T., Giste E., Johnson A. K., Johnson E. M., Kutyavin T., Lajoie B., Lee B. K., Lee K., London D., Lotakis D., Neph S., Neri F., Nguyen E. D., Qu H., Reynolds A. P., Roach V., Safi A., Sanchez M. E., Sanyal A., Shafer A., Simon J. M., Song L., Vong S., Weaver M., Yan Y., Zhang Z., Zhang Z., Lenhard B., Tewari M., Dorschner M. O., Hansen R. S., Navas P. A., Stamatoyannopoulos G., Iyer V. R., Lieb J. D., Sunyaev S. R., Akey J. M., Sabo P. J., Kaul R., Furey T. S., Dekker J., Crawford G. E., Stamatoyannopoulos J. A. (2012) The accessible chromatin landscape of the human genome. Nature 489, 75–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bernstein B. E., Liu C. L., Humphrey E. L., Perlstein E. O., Schreiber S. L. (2004) Global nucleosome occupancy in yeast. Genome Biol. 5, R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yuan G. C., Liu Y. J., Dion M. F., Slack M. D., Wu L. F., Altschuler S. J., Rando O. J. (2005) Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309, 626–630 [DOI] [PubMed] [Google Scholar]
- 21. Crawford G. E., Davis S., Scacheri P. C., Renaud G., Halawi M. J., Erdos M. R., Green R., Meltzer P. S., Wolfsberg T. G., Collins F. S. (2006) DNase-chip. A high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nat. Methods 3, 503–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Crawford G. E., Holt I. E., Mullikin J. C., Tai D., Blakesley R., Bouffard G., Young A., Masiello C., Green E. D., Wolfsberg T. G., Collins F. S. (2004) Identifying gene regulatory elements by genome-wide recovery of DNase hypersensitive sites. Proc. Natl. Acad. Sci. 101, 992–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Crawford G. E., Holt I. E., Whittle J., Webb B. D., Tai D., Davis S., Margulies E. H., Chen Y., Bernat J. A., Ginsburg D., Zhou D., Luo S., Vasicek T. J., Daly M. J., Wolfsberg T. G., Collins F. S. (2006) Genome-wide mapping of DNase hypersinsitive sites using massively parallel signature sequencing (MPSS). Genome Res. 16, 123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sabo P. J., Humbert R., Hawrylycz M., Wallace J. C., Dorschner M. O., McArthur M., Stamatoyannopoulos J. A. (2004) Genome-wide identification of DNase I-hypersensitive sites using active chromatin sequence libraries. Proc. Natl. Acad. Sci. 101, 4537–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sabo P. J., Kuehn M. S., Thurman R., Johnson B. E., Johnson E. M., Cao H., Yu M., Rosenzweig E., Goldy J., Haydock A., Weaver M., Shafer A., Lee K., Neri F., Humbert R., Singer M. A., Richmond T. A., Dorschner M. O., McArthur M., Hawrylycz M., Green R. D., Navas P. A., Noble W. S., Stamatoyannopoulos J. A. (2006) Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat. Methods 3, 511–518 [DOI] [PubMed] [Google Scholar]
- 26. He H. H., Meyer C. A., Chen M. W., Jordan V. C., Brown M., Liu X. S. (2012) Differential DNase I hypersensitivity reveals factor-dependent chromatin dynamics. Genome Res. 22, 1015–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bennani-Baïti I. M., Cooke N. E., Liebhaber S. A. (1998) Physical linkage of the human growth hormone gene cluster and the CD79b (Igβ/B29) gene. Genomics 48, 258–264 [DOI] [PubMed] [Google Scholar]
- 28. Jones B. K., Monks B. R., Liebhaber S. A., Cooke N. E. (1995) The human growth hormone gene is regulated by a multicomponent locus control region. Mol. Cell. Biol. 15, 7010–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shewchuk B. M., Asa S. L., Cooke N. E., Liebhaber S. A. (1999) Pit-1 binding sites at the somatotrope-specific DNase I hypersensitive sites I, II of the human growth hormone locus control region are essential for in vivo hGH-N gene activation. J. Biol. Chem. 274, 35725–35733 [DOI] [PubMed] [Google Scholar]
- 30. Gong S., Zheng C., Doughty M. L., Losos K., Didkovsky N., Schambra U. B., Nowak N. J., Joyner A., Leblanc G., Hatten M. E., Heintz N. (2003) A gene expression atlas of the central nervous system base on bacterial artificial chromosomes. Nature 425, 917–925 [DOI] [PubMed] [Google Scholar]
- 31. Yoo E. J., Cooke N. E., Liebhaber S. A. (2012) An RNA-independent linkage of noncoding transcription to long-range enhancer function. Mol. Cell. Biol. 32, 2020–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ji X., Kong J., Liebhaber S. A. (2003) In vivo association of the stability control protein αCP with actively translating mRNAs. Mol. Cell. Biol. 23, 899–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kimura A. P., Sizova D., Handwerger S., Cooke N. E., Liebhaber S. A. (2007) Epigenetic activation of the human growth hormone gene cluster during placental cytotrophoblast differentiation. Mol. Cell. Biol. 27, 6555–6568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ho Y., Elefant F., Liebhaber S. A., Cooke N. E. (2006) Locus control region transcription plays an active role in long-range gene activation. Mol. Cell 23, 365–375 [DOI] [PubMed] [Google Scholar]
- 35. Anish R., Hossain M. B., Jacobson R. H., Takada S. (2009) Characterization of transcription from TATA-less promoters. Identification of a new core promoter element XCPE2 and analysis of factor requirements. PloS One 4, e5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shenkar R., Shen M. H., Arnheim N. (1991) DNase I-hypersensitive sites and transcription factor-binding motifs within the mouse E β meiotic recombination hot spot. Mol. Cell. Biol. 11, 1813–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sharrocks A. D. (2001) The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2, 827–837 [DOI] [PubMed] [Google Scholar]
- 38. Reth M. (1992) Antigen receptors on B lymphocytes. Annu. Rev. Immunol. 10, 97–121 [DOI] [PubMed] [Google Scholar]
- 39. Komatsu A., Otsuka A., Ono M. (2002) Novel regulatory regions found downstream of the rat B29/Igβ gene. Eur. J. Biochem. 269, 1227–1236 [DOI] [PubMed] [Google Scholar]
- 40. Hirzmann J., Luo D., Hahnen J., Hobom G. (1993) Determination of messenger RNA 5′-ends by reverse transcription of the cap structure. Nucleic Acids Res. 21, 3597–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ayoubi T. A., Van De Ven W. J. (1996) Regulation of gene expression by alternative promoters. FASEB J. 10, 453–460 [PubMed] [Google Scholar]
- 42. Landry J. R., Mager D. L., Wilhelm B. T. (2003) Complex controls. The role of alternative promoters in mammalian genomes. Trends Genet. 19, 640–648 [DOI] [PubMed] [Google Scholar]
- 43. Jacobs S. M., Gorse K. M., Kennedy S. J., Westin E. H. (1994) Characterization of a rearrangement in the c-MYB promoter in the acute lymphoblastic leukemia cell line CCRF-CEM. Cancer Genet. Cytogenet. 75, 31–39 [DOI] [PubMed] [Google Scholar]
- 44. Jacobs S. M., Gorse K. M., Westin E. H. (1994) Identification of a second promoter in the human c-myb proto-oncogene. Oncogene 9, 227–235 [PubMed] [Google Scholar]
- 45. Gordon M. S., Kanegai C. M., Doerr J. R., Wall R. (2003) Somatic hypermutation of the B cell receptor genes B29 (Igβ, CD79b) and mb1 (Igα, CD79a). Proc. Natl. Acad. Sci. U.S.A. 100, 4126–4131 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.