Background: Roles of SoxC genes in the development of retinal ganglion cells (RGCs) are unknown at present.
Results: Targeted deletion of Sox4 and Sox11 in retina results in a complete loss of RGCs.
Conclusion: Sox4 and Sox11 function redundantly to regulate RGC development.
Significance: These findings highlight the essential role of SoxC genes in retinal development.
Keywords: Development, Differentiation, Eye, Gene Knockout, Retina
Abstract
SOX family proteins belong to the high-mobility-group (HMG) domain-containing transcription factors, and function as key players to regulate embryonic development and cell fate determination. The highly related group C Sox genes Sox4 and Sox11 are widely expressed in the development of mouse retina and share a similar expression pattern with each other in this process. Here, to investigate the roles of Sox4 and Sox11 in the retinal development, Sox4, Sox11, and Sox4/Sox11 conditional knock-out (CKO) mice with deletion of Sox4, Sox11, and Sox4/Sox11 in retinas were generated. Our studies demonstrated that targeted disruption of Sox4 or Sox11 in retinas caused a moderate reduction of generation of RGCs. However, a complete loss of RGCs was observed in Sox4/Sox11-null retinas, suggesting the two genes play similar roles in the development of RGCs. Our further analysis confirms that Sox4 and Sox11 function redundantly to regulate the generation of RGCs at early embryonic stages as well as the survival of RGCs at late embryonic stages. In addition, we demonstrated that loss of Math5 impairs the expression of Sox4 and Sox11 in the ganglion cell layer while deletion of Brn3b has no effect on the expression of Sox4 and Sox11. Taken together, these findings elucidate SoxC genes as essential contributors to maintain the survival of RGCs, and imply their intermediate position between Math5 and Brn3b in the genetic hierarchy of RGC development.
Introduction
The well-arranged laminar structure and easy accessibility of the mammalian retina make it an attractive model for studying the development of heterogeneous types of cells in a complex tissue. Mature mammalian retinas consist of six major types of neuronal cells and one type of glial cells, which are organized into three distinct cellular layers photoreceptors (rods and cones) constitute the outer nuclear layer (ONL),2 horizontal, bipolar, amacrine, and Müller glial cells constitute the inner nuclear layer (INL), while ganglion and displaced amacrine cells constitute the ganglion cell layer (GCL). Among all seven major retinal cell types, retinal ganglion cells (RGCs), the only projection neurons in the retina, collect the signals, and transmit them to the target locations within the brain (1, 2). RGCs are the first-born neurons from multipotent retinal progenitor cells (RPCs) during retinogenesis. Formation of RGCs is a stepwise process involving extensive and precise molecular factors. MATH5 (ATOH7-Mouse Genome Informatics) and BRN3B (POU4F2-Mouse Genome Informatics) are two of the most important transcription factors (TFs) involved in regulatory pathway of RGCs (3). MATH5, a basic helix-loop-helix (bHLH) TF, is expressed in post-mitotic RPCs and is required for maintaining the competency of RPCs to become RGCs (4, 5). Loss of Math5 in mice results in nearly complete absence of RGCs. As a POU-homeodomain (POU-HD) factor, BRN3B is one of TFs regulated by Math5 (4, 5). Different from the role of Math5 in early stages of RGC specification, Brn3b functions in terminal differentiation and survival of RGCs by regulating axon outgrowth and apoptosis (6–9). Although it is clear that BRN3B functions downstream of MATH5, it remains unknown what other factors function between MATH5 and BRN3B to regulate RGC differentiation.
The Sry-related high mobility group (HMG) box (SOX) family of transcription factors is characterized by the highly conserved HMG motif and have been reported as critical regulators to control cell fate and differentiation in multiple processes during development (10, 11). In mammals, the Sox family consists of more than 20 members, classified into eight subgroups A-H according to the sequence similarity among the HMG domains (12). Three Sox genes Sox4, Sox11, and Sox12 belong to the mammalian SoxC group. Sox12 knock-out mice are viable and do not exhibit obvious abnormal phenotypes, but the deficiency of either Sox4 or Sox11 is lethal to mice (13–15). The broad expression of SoxC genes has been reported in neural progenitor cells and mesenchymal cells in variant tissues during development (13, 14), and they have been found to regulate cell differentiation, proliferation and survival in multiple organ lineages (16–22). Recent studies on Sox4 and Sox11 imply their roles in retinal cell differentiation (23). However, the regulation of RGC development by the two genes has not been determined. Here we demonstrated that the expression of Sox4 and Sox11 was highly overlapping in the developing retina during embryonic stages. Targeted deletion of either Sox4 or Sox11 in the retina led to a moderate reduction of RGCs, whereas loss of Sox4 and Sox11 resulted in the absence of RGCs. Further analysis on the development of RGCs using three different conditional knock-out (CKO) mice revealed that Sox4 and Sox11 function redundantly to govern the generation and survival of RGCs at early and late embryonic stages, respectively. Furthermore, a Sox4/11-null mutation did not affect the expression of Math5 in retina, but removal of Math5 abolished the expression of Sox4 and Sox11 in the GCL. In addition, we found that the deletion of Brn3b did not alter the expression of Sox4 and Sox11 in retina, and overexpression of Sox4 or Sox11 stimulated BRN3B expression in vitro. These studies strongly suggest that Sox4 and Sox11 function downstream of Math5 while upstream of Brn3b to regulate the development of RGCs.
EXPERIMENTAL PROCEDURES
Animals
Six3-cre and Sox4loxP/loxP mice were described previously (24, 25). To generate Sox11loxP allele, genomic sequences from mouse 129S6 BAC library was isolated using Sox11-coding sequences as a probe. The Sox11loxP targeting construct was created by inserting the Sox11 6.45 kb 5′-flanking arm containing the open reading frame (ORF) and 3.0 kb 3′-flanking arm into the 5′ and 3′ multiple cloning sites of the pKII-2FRT vector, respectively. In this construct, one loxP site was introduced before the ORF of Sox11, and the other loxP site was followed by an frt-flanked Neo-cassette. Thus, the exon of Sox11 could be removed by Cre recombinase. The targeting vector was linearized with NotI and electroporated into 129S6 mouse embryonic stem (ES) cells. Targeted ES cell clones were obtained by G418 positive selection and the negative selection conferred by diphtheria toxin A (DTA) gene. The selected ES cell clones were screened by Southern blotting with a 0.8 kb 5′-probe, which recognizes a 9.3 kb fragment of the wild-type allele and a 7.3 kb fragment of the targeted allele in genomic DNA digested with HindIII. Then the targeted ES cell lines were injected into C57BL/6J blastocysts to generate chimeras. Male chimeric mice were crossed with C57BL/6J female mice to generate F1 heterozygous mice whose genotypes were identified by Southern blotting. The primers for genotyping PCR were designed as follows: 5′-CGTGATTGCAAAGGCAGAGG and 5′-GTACTGAGGTCTAGGCTGTAAGG to detect a 500 bp product of wild-type Sox11 allele and a 550 bp product of Sox11loxP allele; 5′-GAAGGAGGCGGAGAGTAGACGG and 5′-CATAGCTCAACACAAATGCCAACGC to detect a 407 bp product of wild-type Sox4 allele and a 516 bp product of Sox4loxP allele; and 5′-GTGGAATCGCTGAATCTTGAC and 5′-GCCCAAATGTTGCTGGATAGT to detect Six3-cre allele. Embryos were defined as embryonic day (E) 0.5 at noon on the day when vaginal plugs were detected. All procedures regarding animal work in this research were approved by University Committee of Animal Resources (UCAR) at the University of Rochester.
Hematoxylin and Eosin (H&E) Staining, Immunochemistry, and in Situ Hybridization
For H&E staining, isolated eyes were fixed by immersion in 2.5% gluteraldehyde and 2% paraformaldehyde at 4 °C for 24 h. Then eyes were dehydrated and embedded in technovit, 2.5 μm sections were cut, and sections cut through the optic disc were stained with hematoxylin and eosin. For immunochemistry and in situ hybridization, staged mouse embryos and enucleated eyes of postnatal mice were dissected followed by immediate fixation in 4% paraformaldehyde in PBS for 1–2 h at 4 °C. After fixation, tissue samples were cryopreserved with 25% sucrose, embedded, and frozen in OCT medium. Horizontal cryosections were prepared at a thickness of 20 μm for in situ hybridization and 16 μm for immunochemistry experiments. BrdU labeling, immunostaining, and in situ hybridization were carried out as previously described (7). The following antibodies and working dilutions used in this study were: mouse anti-BRN3A (1:200; Millipore Bioscience Research Reagents), goat anti-BRN3B (1:200; Santa Cruz Biotechnology), mouse anti-SMI32 (1:1000; Sternberger Monoclonals), mouse anti-PAX6 (1:200; Developmental Studies Hybridoma Bank (DSHB)), sheep anti-CHX10 (1:200; Exalpha), mouse anti-SOX2 (1:200; Santa Cruz Biotechnology), mouse anti-CALBINDIN (1:2000; Sigma), rabbit anti-OPSIN (1:200; Sigma), mouse anti-RHODOPSIN (1:250; Cosmo Bio), mouse anti-BrdU (1:50; DSHB), rabbit anti-activated CASPASE-3 (1:500; R&D Systems), chicken anti-GFP (1:500; Abcam). Alexa-conjugated secondary antibodies (Invitrogen) were used at a dilution of 1:1000. Sox4 and Sox11 probes were generous gifts from Dr. Paul J. Scotting (26). Confocal images were captured with a Zeiss LSM 510 META confocal microscopy. Other pictures were taken with a Nikon Eclipse TE2000-U inverted microscope. Acquired pictures were quantitatively analyzed by manually counting specific marker-stained cells.
Plasmids, in Vitro Electroporation, and Retinal Explant Culture
The ORF of Sox4 and Sox11 were synthesized by Genewiz and cloned into the backbone of the pEGFP-N1 (Clontech) plasmid using SalI and NotI restriction enzymes. pcDNA3 was used as a control plasmid. Retinas at E13.5 from C57BL/6J mice were dissected and transferred to a micro electroporation chamber (Harvard Apparatus #45-0105) containing a DNA solution at the final concentration of 1 μg/μl in 1× PBS. The setting of electroporation was as follows: mode, LV; voltage, 30 V; pulse length, 50 ms; number of pulses, 5; interval, 950 ms; polarity, unipolar. After electroporations, retinas were cultured on Millipore Millicell-CM Low Height Culture Plant Inserts (0.4 μm pore size) in explant culture medium (45% DMEM, 45% HAM F-12 (Bio-Whittaker), 10% fetal bovine serum, 1× pennicilin/streptomycin/l-glutamine (Invitrogen), and 1× insulin (Sigma)) at 37 °C in a humidified 5% incubator. Retinal explant samples were collected after a 48 h culture.
RESULTS
Overlapping Expression of Sox4 and Sox11 during the Development of RGCs
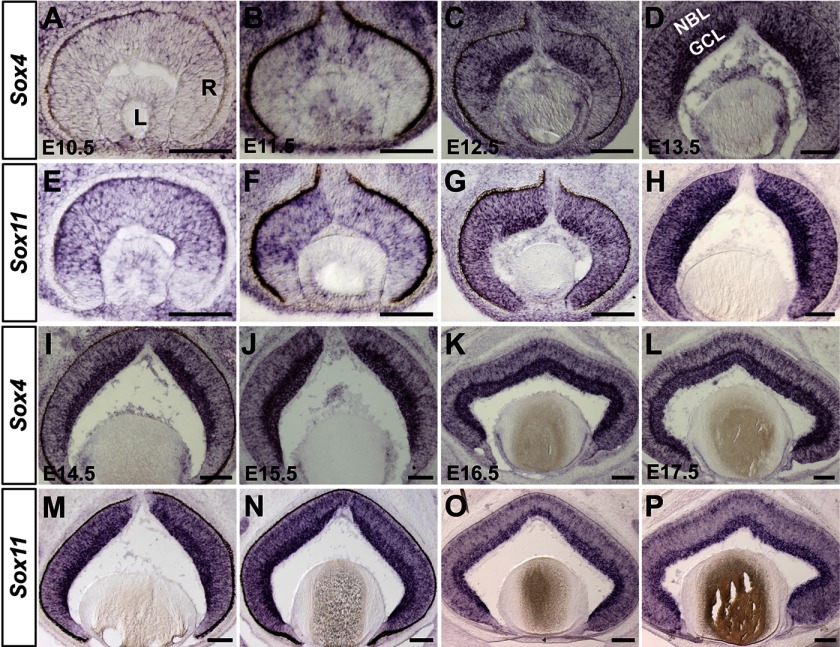
To better elucidate the roles of Sox4 and Sox11 in RGC development, we compared their spatiotemporal expression patterns in embryonic retinas from E10.5 to E18.5. In situ hybridization revealed no detectable Sox4 expression at E10.5 (Fig. 1A), and its expression first appeared in a small patch of cells in the central, dorsal optic cup at E11.5 (Fig. 1B). As retinal development progressed, Sox4 expression rapidly expanded circumferentially toward peripheral retinal regions from E12.5 to E14.5 (Fig. 1, C, D, I). Sox4-expressing cells were distributed in two layers with different levels: dispersive expression in the neuroblast layer (NBL) and a constitutively higher level in the GCL. Besides the GCL, robust expression of Sox4 was also found in the inner most parts of the NBL at E15.5 (Fig. 1J). From E16.5 to E18.5, a slightly intensive expression of Sox4 was detected at the outer boundary of the NBL as well (Fig. 1, K, L; data not shown).
FIGURE 1.
Expression profiles of Sox4 and Sox11 during retinogenesis. Wild type retinas at different embryonic stages (E10.5-E17.5) were collected for in situ hybridization using probes against Sox4 (A–D, I–L) and Sox11 (E–H, M–P). L: lens, R: retina, GCL: ganglion cell layer, NBL: neuroblast layer. Scale bar: 100 μm.
In contrast to Sox4, modest expression of Sox11 had been widely observed in the optic cup at E10.5 (Fig. 1E), which was consistent with the previous report (27). There was a mild increase in expression at E11.5, especially in the central region of the neural retina (Fig. 1F). At E12.5, Sox11-expresssing cells were distributed throughout the retina with the highest expression level in the center (Fig. 1G). From E13.5 to E14.5, the expression of Sox11 was identical to that of Sox4, with a higher level in the GCL and a lower level in the NBL (Fig. 1, H, M). From E15.5 to E18.5, the expression of Sox11 was similar to that of Sox4 (Fig. 1, N–P; data not shown). Scattered expression of Sox11 was detected in the NBL with moderate expression at the outer boundary and strong expression in the GCL. Together, Sox4 and Sox11 share a comparable expression pattern during retinal development, especially in the GCL. Their intensive expression starts from E11.5 in a central-to-peripheral wave and persists in the GCL during the generation of RGCs, indicating Sox4 and Sox11 may play redundant roles in the development of RGCs.
Targeted Disruption of Sox4 and Sox11 in Retina
Conventional Sox11 knock-out mice died immediately at birth due to the congenital cyanosis resulting from heart defects and hypoplasia of the lungs (28). To examine the function of Sox11 in retinal development, we generated a Sox11 CKO allele (Sox11loxP) by flanking the entire open reading frame (ORF) with loxP sites (Fig. 2A). Therefore, the exon of Sox11 could be removed by Cre recombinase. The genotypes of Sox11loxP mice were verified by Southern blotting and PCR (Fig. 2, B and C). To remove Sox11 in the retina, we used Six3-cre mice, which express Cre recombinase in the eye field and ventral forebrain starting at E9 to E9.5 (24, 29). Homozygous Sox11loxP/loxP mice were bred with Six3-cre mice to yield retina-specific Sox11 CKO (Sox11loxP/loxP; Six3-cre) mice. In situ hybridization confirmed the efficient removal of Sox11 in the retinas of Sox11loxP/loxP; Six3-cre mice at E14.5 (Fig. 2D). In the following experiments, Sox11loxP/loxP; Six3-cre mice were defined as Sox11-nulls. Sox11loxP/+ and Sox11loxP/loxP mice were phenotypically indistinguishable and were used as controls hereafter. Sox11-nulls displayed a moderately reduced body size compared with controls at birth. Nevertheless, they reached body weight and size similar to those of the controls after postnatal day (P) 30 and were viable and fertile.
FIGURE 2.
Generation of a Sox11 conditional null allele. A, schematic representation of a portion of the wild type Sox11 allele, the targeting construct and the targeted alleles. Gray-filled box is the open reading frame (ORF) of Sox11. Black-filled boxes are the sequences used to generate the homologous arms in the targeting vector. The Sox11CKO targeting vector is made by inserting the FRT-flanked neomycin gene and one loxP site before the ORF of Sox11. Cre recombinase-mediated deletion of the loxP-flanked Sox11 ORF results in a Sox11-null mutation. Blue-filled box is the 5′ hybridization probe used for Southern blot analysis. Abbreviations: Neo, PGK-neomycin resistance gene; DTA, diphtheria toxin gene for negative selection of embryonic stem cells; loxP, Cre recombinase recognition sequence. B, Southern blot analysis of targeted ES cells. A 5′ probe was used in Southern blot of HindIII-digested genomic DNA to identify the 9.3 kb fragment of the wild-type allele and the 7.3 kb fragment of the targeted allele. C, PCR-based genotyping confirmation of Sox11loxP mice using primers indicated in A distinguishes Sox11loxP and wild type allele. D, Sox11 in situ hybridization of retina sections reveals a complete abolishment of Sox11 expression in the retinas of Sox11loxP/loxP; Six3-Cre mice. Scale bar: 100 μm.
Given the fact that Sox4 and Sox11 share similar structures and expression patterns in the retinal development, Sox4 could compensate for the loss of Sox11. Since conventional Sox4 knock-out mice die at around E14 from the severe malformation of the heart outflow tract (15), we used Sox4loxP/loxP mice (25), and generated the Sox4-null and Sox4/11-null mice using Six3-cre as mentioned above. Sox4-nulls were viable and fertile with no overt discernible physical deficiencies. While Sox4/11-null mice exhibited a smaller body size than the controls from birth to death before postnatal day (P) 14.
Retinal Defects in Sox4/11-null Mice
To examine the possible defects of RGCs in Sox4-, Sox11-, and Sox4/11-null retinas, we analyzed the retinal sections at P30 from Sox4-null and Sox11-null mice, and at P14 from Sox4/11-null mice. H&E staining revealed that deletion of either Sox4 or Sox11 did not alter the retinal structure. The gross organization of Sox4-null or Sox11-null retinas resembled that of the control retinas, while the thickness of the GCL and the INL was reduced in either Sox4-null or Sox11-null retinas compared with those of the control retinas (Fig. 3, A–A′, F–F′). In adult mice retinas, BRN3A and BRN3B label about 70% RGCs separately in a partly overlapping pattern (30). Immunostaining with BRN3A showed fewer RGCs in either Sox4- or Sox11-null retinas in comparison with that of the control retinas in the absence of Sox4 or Sox11 (Fig. 3, B–B′, G–G′). Similar to BRN3A labeling, whole-mount immunostaining revealed a reduction of BRN3B+ RGCs in Sox4- or Sox11-nulls (Fig. 3, C–C′, H–H′). Quantification of BRN3B+ exhibited ∼21 and 28% reduction of RGCs in Sox4-nulls and Sox11-nulls, respectively (Fig. 3, P and Q). To confirm the loss of RGCs in mutants, SMI32, which predominantly marks large RGCs and their nerve fibers (31), was used as another parameter of RGC number. Immunostaining of whole mount retinas with SMI32 revealed that not only Sox4- or Sox11-null retinas had a obviously less number of axon bundles but also their axon bundles was less fasciculated (Fig. 3, D–D′, I–I′). Consistently, optic nerves, which are composed of RGC axons, were thinner in Sox4- or Sox11-null retinas compared with that of controls (Fig. 3, E–E′, J–J′). Therefore, deletion of either Sox4 or Sox11 resulted in a moderate reduction of RGCs. However, removal of both Sox4 and Sox11 completely disrupted the overall structure of retinas. Compared with control retinas, the three nuclear layers were reduced to one thin layer, the entire inner plexiform layer and outer plexiform layer were abolished in Sox4/11-null retinas (Fig. 3, K–K′). Furthermore, neither BRN3A+ nor BRN3B+ cells were observed in Sox4 and Sox11 compound null retinas (Fig. 3, L–L′, M–M′, N–N′). The ventral views of mouse brains showed that optic nerves were absent in Sox4/11-null retinas as well (Fig. 3, O–O′). Taken together, these data demonstrate that deletion of both Sox4 and Sox11 in retinas abolishes RGCs, implying the redundant function of Sox4 and Sox11 in RGC development.
FIGURE 3.
Abolishment of RGCs in Sox4/11-null retinas. A–D, A′–D′, F–I, F′–I′, plastic sections of retinas isolated from 30-day-old control, Sox4-null and Sox11-null mice were analyzed by H&E staining (A, A′, F, and F′), immunostaining of cryo-sections with anti-BRN3A (red) (B, B′, G, and G′), immunostaining of whole-mount retina with anti-BRN3B (green) (C, C′, H, and H′) and SMI32 (green) (D, D′, I, and I′). K–N, K′–N″, retinas of 14-day-old control mice and Sox4/11-null mice were analyzed with H&E staining (K and K′), immunostaining of cryo-sections with anti-BRN3A (red) (L and L′) and BRN3B (green) (M and M′). Immunostained sections with anti-BRN3A (red) and anti-BRN3B (green) were merged with DAPI staining pictures. E–E′, J–J′, O–O′, ventral views of brains show thinner optic nerves (arrowheads) in Sox4-null and Sox11-null mice, and no optic nerves in Sox4/11-null mice. P and Q, quantification of BRN3B+ cells in the central region of the whole mounts shows a significant reduction of RGCs in Sox4-null or Sox11-null mice. All experiments were repeated at least three times. Error bars represent S.D. *, p < 0.01. GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer; ON, optical nerve. Scale bar: A–D, A′–D′, F–I, F′–I′, K–N, K′–N′) 50 μm; (E–E′, J–J′, O–O′) 1 mm.
That retinal inactivation of Sox4 or Sox11 leads to a reduced thickness of GCL and INL suggests the potential absence or degeneration of other retinal type cells in addition to RGCs. To test this possibility, immunostaining using distinct cell type-specific markers were performed in retinal sections from mice at P30. Immunostaining with PAX6, a pan-amacrine cell marker, revealed about a 21% reduction of PAX6+ cells in the INL and a 43% reduction of PAX6+ cells in the GCL of Sox4-null retinas, respectively (supplemental Fig. S1, A–A′, S). Immunostaining with CHX10 for pan-bipolar cells revealed a 26% reduction of bipolar cells in Sox4-null retinas (supplemental Fig. S1, B–B′, S). In Sox11 mutant retina, anti-PAX6 labeling showed that amacrine cells were decreased ∼28% in the INL and ∼37% in the GCL (supplemental Fig. S1, G–G′, T), and anti-CHX10 labeling revealed ∼33% reduction of bipolar cells (supplemental Fig. S1, H–H′, T). Whereas, no significant differences in horizontal (CALBINDIN+), Müller glial (SOX2+ cells in the middle parts of INL), cone (OPSIN+) and rod (RHODOPSIN+) cells were detected between Sox4-null or Sox11-null and control retinas (supplemental Fig. S1, C–F, C′–F′, I–L, I′–L′, S, T). Although deletion of Sox4 and Sox11 results in one thin layer of retina, immunostaining with different cell type-specific markers revealed that all retinal cell types mentioned above were present but in much fewer numbers in Sox4/11-null retinas (supplemental Fig. S1, M–R, M′–R′).
Requirement for Sox4 and Sox11 in the Development of RGCs
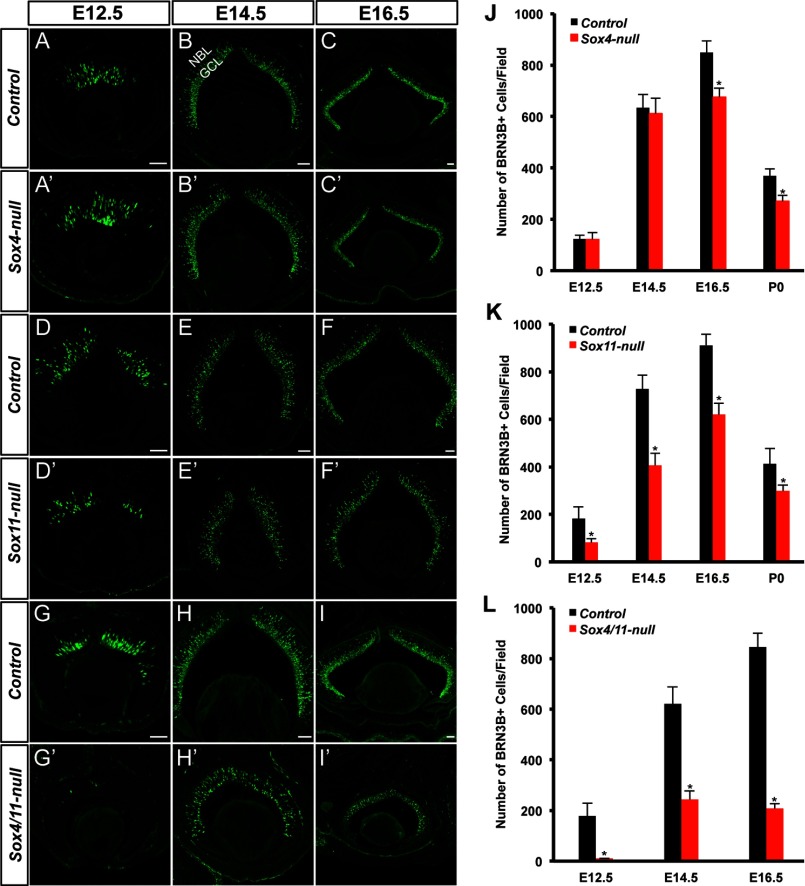
In mice, RGCs are generated from E11.5 to E18.5 (32). BRN3B, whose expression initiates at E11.5, is one of the earliest RGC markers (33, 34). The onset of robust expression of Sox4 and Sox11 in the retina coincides with the differentiation of RGCs in mice, implying their essential roles in RGC differentiation. Thus, we examined the expression of BRN3B in Sox4-, Sox11-, and Sox4/11-null retinas at different embryonic stages. Immunostaining with BRN3B showed no substantial change in number and distribution of RGCs at E12.5 and E14.5 in Sox4-null retinas in comparison to that of control retinas (Fig. 4, A–A′, B–B′, J), but at E16.5 the Sox4 mutant retinas revealed a mild but significant reduction of BRN3B+ RGCs by ∼20% (Fig. 4, C–C′, J). Different from Sox4-null retinas, Sox11-null retinas exhibited similar labeling pattern but reduced number of RGCs, particularly in the GCL, from E12.5 to E16.5 compared with that of control retinas (Fig. 4, D–D′, E–E′, F–F′). Quantification analysis further confirmed that the declines were significant in Sox11 mutants (Fig. 4K). Additionally, the number of BRN3B+ RGCs at P0 in Sox4- or Sox11-null mutants decreased by 27 or 28%, respectively (Fig. 4, J and K), which is comparable to the reduction in adult retinas (Fig. 3, P and Q). However, the combined absence of Sox4 and Sox11 leads to a severe hypoplasia of the developing RGCs. At E12.5, only few BRN3B+ cells were detected in the mutant (Fig. 4, G–G′). In contrast to control retinas, the GCL with BRN3B+ cells was much thinner at E14.5 and eventually disappeared at E16.5. At both E14.5 and E16.5, BRN3B+ cells mainly resided in the NBL (Fig. 4, H–H′, I–I′). By quantifying the number of BRN3B+ cells at E14.5 and E16.5, we found that the number of RGCs in double mutants remained unchanged, which was distinguished from the increasing trend in control retinas (Fig. 4L). In addition, we observed that deletion of Sox11 led to a mild reduction in size of retinas, while deletion of both genes resulted in evident change in retinas at early embryonic stages, indicating that they possibly have impacts on the RPCs. Indeed, immunostaining with progenitor markers, CHX10 and SOX2, confirmed that the two genes affect the RPCs at early embryonic stages (data not shown).
FIGURE 4.
Targeted disruption of Sox4 and Sox11 impairs the development of RGCs. A–I, A′–I′, cryo-sections of retinas from the control and mutant mice (Sox4-null, Sox11-null, and Sox4/11-null) at E12.5, E14.5, and E16.5 were immunolabeled with anti-BRN3B (green). J–L, quantification of RGCs in control and mutant mice at different embryonic stages. All experiments were repeated at least three times and error bars represent S.D. *, p < 0.01. GCL: ganglion cell layer, NBL: neuroblast layer. Scale bar: 50 μm.
It is possible that the gradual loss of BRN3B+ RGCs was caused by increased apoptosis in mutant retinas. Therefore, we examined the number of CASP3+ cells in control and mutant retinas. Previous research has shown that apoptosis is rare throughout embryogenesis (35, 36). We found no significant change of the number of apoptotic cells at E12.5 in both control and mutant retinas (data not shown). However, starting from E14.5, the expression of CASP3 was up-regulated in all three mutants compared with that in the controls (Fig. 5, A–F, A′–F′). Quantification analysis revealed that Sox4- or Sox11-null mutants had an ∼2-fold increase in the number of apoptotic cells, while ∼6-fold and more than 10-fold increase were found in Sox4/11-null mutant at E14.5 and E16.5, respectively (Fig. 5, G–I). Moreover, compared with Sox4- or Sox11-null retina, more cells undergoing apoptosis were detected in the GCL of Sox4/11-null retinas at E14.5 (Fig. 5, A′, C′, E′), the time consistent with the extensive loss of BRN3B+ RGCs.
FIGURE 5.
Increased number of apoptotic cells in Sox4-null and Sox11-null mice. A–F, A′–F′, immunostaining of cryo-sections of retinas from control and mutant (Sox4-null, Sox11-null, Sox4/11-null) mice at different embryonic stages with anti-CASPASE3 (green). G–H, quantification of number of apoptotic cells in control and mutant (Sox4-null, Sox4-null, and Sox4/11-null) retinas. All experiments were repeated three times, and error bars are S.D. *, p < 0.01 Scale bar: 50 μm.
Taken together, deletion of either Sox4 or Sox11 has no or a minor influence on the formation of most RGCs, suggesting that either of them is dispensable for the development of major RGCs. However, removal of both genes results in a severe hypoplasia of RGCs at early stages and progressive loss of RGCs at late stages, indicating they play similar roles in different time windows during the development of RGCs: Sox4 and Sox11 are essentially required for the generation of primary RGCs at early stages and maintaining RGC survival during late stages.
Roles of Sox4 and Sox11 in the Genetic Regulatory Network of RGCs
Previous studies have identified an essential Math5→Brn3b pathway in the development of RGCs (4, 5). To determine the relationship between SoxC genes and Math5 or Brn3b in the RGC genetic hierarchy, we examined the expression of Sox4 and Sox11 in Math5-null or Brn3b-null retinas, separately. Compared with control retina, both Sox4 and Sox11 expression was reduced in the NBL and was abolished in the GCL of Math5-null retina at E13.5 (Fig. 6, A–D), indicating that Math5 acts upstream of Sox4 and Sox11 during RGC development. In contrast, targeted deletion of Brn3b had no effect on the expression of SoxC group genes in retinas at E14.5 (Fig. 6, E–H). Our findings suggest that Sox4 and Sox11 likely function between Math5 and Brn3b to regulate RGC development. Next, we tested whether the deletion of SoxC could influence the expression of Math5. As shown by in situ hybridization, similar to controls, normal onset of Math5 expression was detected in all three mutants including the Sox4/11-null mutant (Fig. 6, I–K, I′–K′), further confirming that Sox4 and Sox11 function downstream of Math5.
FIGURE 6.
Functional mechanisms of Sox4 and Sox11 in the development of RGCs. A–H, expression of Sox4 and Sox11 in Math5−/− and Brn3b−/− retinas. In situ hybridization analysis of Sox4 and Sox11 expression in retinas of control and Math5-null mice at E13.5 (A–D). In situ hybridization analysis of Sox4 and Sox11 expression in retinas of control and Brn3b-null mice at E14.5 (E–H). I–K, I′–K′, expression of Math5 in Sox4-null, Sox11-null, and Sox4/11-null retinas. In situ hybridization analysis of Math5 expression in retinas of control and mutant mice at E12.5. L–N, L′–N′, cryo-sections of in vitro cultured Sox4 overexpressing retinal explants were immunostained with anti-GFP (green) and anti-BRN3B (red) as well as counterstained with DAPI (blue). O–Q, O′–Q′, cryo-sections of in vitro cultured Sox11 overexpressing retinal explants were immunostained with anti-GFP (green) and anti-BRN3B (red) as well as counterstained with DAPI (blue). R, quantification of RGCs in control and Sox4 or Sox11-overexpressing retina explants. S, schematic model of RGC development by Sox4 and Sox11 regulation. Solid lines, direct regulation of identified; broken lines, indirect or proposed regulation. All experiments were repeated at least three times, and error bars are S.D. *, p < 0.01. Scale bar: (A–K, I′–K′) 100 μm; (L–Q, L′–Q′) 50 μm.
To further test roles of Sox4 and Sox11 in the development of RGCs, gain-of-function experiments in embryonic retinal explant cultures were conducted. C57BL/6J mouse retinas at E13.5 were dissected and electroporated with vectors pCMV-Sox4 or pCMV-Sox11 and the pEGFP-N1 vector in a ratio of 1:1. Retinal explants were harvested and examined after 2 days of culture. Overexpression of either Sox4 or Sox11 in retinas significantly increased the percentage of BRN3B+ cells by ∼30% in comparison to pcDNA3 control vector electroporated retinas (Fig. 6, N–N′, Q–Q′, R), indicating SoxC group genes enhance the generation of RGCs. We also observed robust expression of GFP, which served as the internal control, in both control and experimental retina sections (Fig. 6, L–L′, O-O′), indicating that a high electroporation efficiency was consistently achieved and the possibility that fluctuated electroporation efficiencies affected results was ruled out. In summary, our data indicates that SoxC group genes are essential intermediate factors between Math5 and Brn3b to promote the development of RGCs.
DISCUSSION
In this report, we generated Sox4, Sox11, and Sox4/11 CKO mice with specific deletions in retinas. Using the three mice models, our studies demonstrate that Sox4 and Sox11 are essential factors orchestrating the development of RGCs by promoting the differentiation and survival of RGCs. Previous studies on their potential functions in development have been mainly focused on their roles in maintaining the differentiation ability of progenitor cells and promoting cell proliferation. Overexpression of Sox4 or Sox11 in cultured retinal explants at E17 stimulates the differentiation of progenitor cells into cone cells at the cost of losing rod cells (23). Abrogation of Sox4/11 in vivo suppresses the differentiation of adult neuron stem cells and does not affect the apoptosis (20). In addition, knocking down of Sox4 is reported to damage proliferation of osteoblasts in vitro (37). The proliferation of pancreatic islet cells is also impaired in Sox4-null pancreas cultured in vitro but Sox4 deficiency has no impact on cell survival of insulin producing cells (38). However, it has been reported that Sox4 and Sox11 are up-regulated in several types of tumors (39, 40), indicating their roles in anti-apoptosis or pro-survival. In recent research, attention has been paid to their pro-survival roles in the development of mice. Ablation of both factors greatly reduced the survival of neural and mesenchymal progenitor cells during organogenesis (17), as well as of neuron progenitor cells during the spinal cord development (22). While Sox11 was reported to play an indispensable role in the proliferation of tyrosine hydroxylase expressing cells at early stages, Sox4 is crucial to their survival at late stages (21). Further lines of evidence demonstrate that Sox11 also helps improve the survival of differentiated post-mitotic sensory neurons (19). Our data about the regulation of RGC development, for the first time, provides direct evidence indicating that Sox4 and Sox11 not only share functions in controlling differentiation and proliferation but also work in the same pathway of regulating cell survival of post-mitotic RGCs.
Targeted deletion of either Sox4 or Sox11 in RGCs results in a moderate inhibitory effect on RGC survival. However, loss of both genes results in dramatic increase in the number of CASP3+ RGCs during development in comparison to controls, demonstrating that they play redundant roles in promoting the survival of RGCs. Why do the two genes have redundant functions? The SoxC group comprises 3 genes, Sox4, Sox11, and Sox12. They are structurally similar to one another with a highly conserved HMG box and a less conserved transcriptional activation domain (13). Interesting, very similar to the phenotype of Sox11−/− mice, Sox4+/−; Sox11+/− mice died at birth from heart malformation, suggesting the dosage of the two genes are essential to heart development and they might function redundantly (17). During the development of the spinal cord, deletion of either Sox4 or Sox11 had no significant effect on the cell survival in the spinal cord, but loss of both reveals an up to a 70% decrease of cell number, strongly supporting the hypothesis that Sox4 and Sox11 regulate certain biological processes simultaneously (22). In comparison to Sox4 and Sox11, although the expression pattern of Sox12 in certain developing tissues including lung, gut and pancreas is similar, the biochemical property of weak binding activity to promoter DNA makes Sox12 a less important factor in the regulation of development (13). And indeed, no phenotype of Sox12 knock-out mice was observed (14). In addition, our study shows that abolishment of both Sox4 and Sox11 by Six3-cre recombinase disrupts the formation of RGCs by 80% at E16.5 and leads to the complete loss of RGCs at later stages, suggesting Sox12 either does not participate or plays a limited role in regulating the development of RGCs.
Moreover, in the present study, we delineated the function of Sox4 and Sox11 in the genetic pathway regulating the generation of RGCs. Previous research has established that Math5 and Brn3b are two pivotal TFs in controlling development of RGCs (3). In the mouse retina, the expression of Math5 first appears at E11 and its expression expands circumferentially to the peripheral retina from E11–16.5 (4, 41). Its expression is limited to the RPCs and the nascent, migrating BRN3B+ RGCs in the NBL but is turned off in the post-migration RGCs in the GCL. MATH5 likely promotes the cell cycle exit of the RPCs as Math5-null mutation causes the failure of the progenitors to exit cell cycle (42). Math5 determines RGC competence acquisition but does not specify the fate of RGCs (5). Moreover, Brn3b functions in the terminal differentiation and survival of RGCs (7). The factors responsible for turning RGC-competent progenitors into post-mitotic RGCs are yet to be identified. Based on the spatiotemporal expression pattern of Sox4 and Sox11 in the NBL as well as in the GCL during early retinogenesis, we hypothesized that Sox4 and Sox11 are the genes promoting RGC fate commitment. Expressions of Sox4 and Sox11 have been shown to be down-regulated by ∼30 and 50% in Math5−/− retinas at E14.5 in comparison to control using microarray analysis (29), however, no further study was conducted to confirm the finding. Our analysis using Math5-null mutant provides direct evidence proving that loss of Math5 greatly impairs the expression of both SoxC genes in the GCL. On the contrary, loss of Sox4/11 does not alter the expression of Math5 in retinas at E12.5. Therefore, Sox4 and Sox11 function downstream of Math5 in RGC development. Math5 starts to express in retinas at E11 and our findings show that in the control retinas at E11.5, the number of RPCs is 1.5-fold higher than that in Sox4/11-null retinas whereas interestingly early RGCs generated at E12.5 in the control is ∼18-fold higher than those in Sox4/11-null retinas, strongly favoring that after Math5 endows RPCs with RGC competence, Sox4 and Sox11 are necessary for specification or early differentiation of RGCs. The increased generation of RGCs observed in Sox4 and Sox11 overexpression further supports the role of SoxC genes in RGC differentiation. Whether MATH5 as a TF directly targets the promoter of Sox4 and Sox11 to regulate their expression is still unclear. A further experiment using ChIP assay would help answer this question. In addition, no apparent change of Sox4 or Sox11 expression is observed in the retina lacking Brn3b at E14.5, indicating that Sox4 and Sox11 unlikely acts downstream of Brn3b in RGC development. In contrast, loss of Sox4 and Sox11 alone or together causes a reduction or loss of BRN3B+ RGCs, arguing for a role of Sox4 and Sox11 upstream of Brn3b but downstream of Math5 in the Math5→Brn3b regulatory pathway of RGC development (Fig. 6S). The moderate increase in the number of BRN3B+ cells conferred by Sox4 or Sox11 overexpression further supports the role of Sox4 and Sox11 upstream of Brn3b.
In Sox4/11-null retinas, BRN3B+ RGCs die from apoptosis beginning at E14.5, indicating that Sox4/11 promotes RGC survival by either directly regulating apoptosis or mediating the regulation of apoptosis by Brn3b. Deletion of Brn3b starts to have a negative impact on RGC survival as late as E16.5 (33, 43) while loss of Sox4/11 starts to suppress the survival at the stage of E14.5, suggesting that Sox4/11 attains a pro-survival function independently of Brn3b. It is still plausible that Sox4/11 may mediate the regulation of RGC survival by Brn3b.
Compared with the Math5-null or Brn3b/Isl1-null retinas with a loss of nearly all RGCs (4, 9), the defect observed in Sox4/11-null retinas is more severe with a more profound reduction in other retinal cell types in addition to RGCs, suggesting a possible, direct involvement of Sox4 and Sox11 in the differentiation or survival or both of other retinal cell types. Consistent with this, the expression of Sox4 and Sox11 continues after the peak of RGC genesis in the GCL and NBL (Fig. 1) and persists until at least P11 in the developing INL (data not shown). Future experiments using specific Cre deleter mouse lines to remove Sox4 and Sox11 after RGC development will help define their role in the development of other retinal cell types.
Supplementary Material
Acknowledgments
We thank Amy Kiernan, Patricia White, and members of the Gan laboratory for their insightful discussions and technical assistance.
This work was supported by National Natural Science Foundation of China Grant No. 81271006, and Hangzhou City Health Science Foundation Grant No. 20120633B01, and by a Research to Prevent Blindness Challenge Grant to the Department of Ophthalmology at the University of Rochester.

This article contains supplemental Fig. S1.
- ONL
- outer nuclear layer
- GCL
- ganglion cell layer
- RGC
- retinal ganglion cell
- RPC
- retinal progenitor cell
- bHLH
- basic helix-loop-helix
- HMG
- high mobility group
- NBL
- neuroblast layer
- TF
- transcription factor
- INL
- inner nuclear layer
- H&E
- hematoxylin and eosin.
REFERENCES
- 1. Wässle H., Boycott B. B. (1991) Functional architecture of the mammalian retina. Physiol. Rev. 71, 447–480 [DOI] [PubMed] [Google Scholar]
- 2. Vaney D. I. (2002) Retinal neurons: cell types and coupled networks. Progress Brain Res. 136, 239–254 [DOI] [PubMed] [Google Scholar]
- 3. Mu X., Klein W. H. (2004) A gene regulatory hierarchy for retinal ganglion cell specification and differentiation. Semin. Cell Dev. Biol. 15, 115–123 [DOI] [PubMed] [Google Scholar]
- 4. Wang S. W., Kim B. S., Ding K., Wang H., Sun D., Johnson R. L., Klein W. H., Gan L. (2001) Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 15, 24–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang Z., Ding K., Pan L., Deng M., Gan L. (2003) Math5 determines the competence state of retinal ganglion cell progenitors. Dev. Biol. 264, 240–254 [DOI] [PubMed] [Google Scholar]
- 6. Erkman L., Yates P. A., McLaughlin T., McEvilly R. J., Whisenhunt T., O'Connell S. M., Krones A. I., Kirby M. A., Rapaport D. H., Bermingham J. R., O'Leary D. D., Rosenfeld M. G. (2000) A POU domain transcription factor-dependent program regulates axon pathfinding in the vertebrate visual system. Neuron 28, 779–792 [DOI] [PubMed] [Google Scholar]
- 7. Pan L., Yang Z., Feng L., Gan L. (2005) Functional equivalence of Brn3 POU-domain transcription factors in mouse retinal neurogenesis. Development 132, 703–712 [DOI] [PubMed] [Google Scholar]
- 8. Gan L., Wang S. W., Huang Z., Klein W. H. (1999) POU domain factor Brn-3b is essential for retinal ganglion cell differentiation and survival but not for initial cell fate specification. Dev. Biol. 210, 469–480 [DOI] [PubMed] [Google Scholar]
- 9. Pan L., Deng M., Xie X., Gan L. (2008) ISL1 and BRN3B co-regulate the differentiation of murine retinal ganglion cells. Development 135, 1981–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schepers G. E., Teasdale R. D., Koopman P. (2002) Twenty pairs of sox: extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev. Cell 3, 167–170 [DOI] [PubMed] [Google Scholar]
- 11. Lefebvre V., Dumitriu B., Penzo-Méndez A., Han Y., Pallavi B. (2007) Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int. J. Biochem. Cell Biol. 39, 2195–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bowles J., Schepers G., Koopman P. (2000) Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 227, 239–255 [DOI] [PubMed] [Google Scholar]
- 13. Dy P., Penzo-Méndez A., Wang H., Pedraza C. E., Macklin W. B., Lefebvre V. (2008) The three SoxC proteins–Sox4, Sox11 and Sox12–exhibit overlapping expression patterns and molecular properties. Nucleic acids research 36, 3101–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoser M., Potzner M. R., Koch J. M., Bösl M. R., Wegner M., Sock E. (2008) Sox12 deletion in the mouse reveals nonreciprocal redundancy with the related Sox4 and Sox11 transcription factors. Mol. Cell Biol. 28, 4675–4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schilham M. W., Oosterwegel M. A., Moerer P., Ya J., de Boer P. A., van de Wetering M., Verbeek S., Lamers W. H., Kruisbeek A. M., Cumano A., Clevers H. (1996) Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature 380, 711–714 [DOI] [PubMed] [Google Scholar]
- 16. Bergsland M., Werme M., Malewicz M., Perlmann T., Muhr J. (2006) The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 20, 3475–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bhattaram P., Penzo-Méndez A., Sock E., Colmenares C., Kaneko K. J., Vassilev A., Depamphilis M. L., Wegner M., Lefebvre V. (2010) Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors. Nature Commun. 1, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Y., Wang J., Zheng Y., Zhao Y., Guo M., Li Y., Bao Q., Zhang Y., Yang L., Li Q. (2012) Sox11 modulates neocortical development by regulating the proliferation and neuronal differentiation of cortical intermediate precursors. Acta Biochim. Biophys. Sinica 44, 660–668 [DOI] [PubMed] [Google Scholar]
- 19. Lin L., Lee V. M., Wang Y., Lin J. S., Sock E., Wegner M., Lei L. (2011) Sox11 regulates survival and axonal growth of embryonic sensory neurons. Dev. Dyn. 240, 52–64 [DOI] [PubMed] [Google Scholar]
- 20. Mu L., Berti L., Masserdotti G., Covic M., Michaelidis T. M., Doberauer K., Merz K., Rehfeld F., Haslinger A., Wegner M., Sock E., Lefebvre V., Couillard-Despres S., Aigner L., Berninger B., Lie D. C. (2012) SoxC transcription factors are required for neuronal differentiation in adult hippocampal neurogenesis. J. Neurosci. 32, 3067–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Potzner M. R., Tsarovina K., Binder E., Penzo-Méndez A., Lefebvre V., Rohrer H., Wegner M., Sock E. (2010) Sequential requirement of Sox4 and Sox11 during development of the sympathetic nervous system. Development 137, 775–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thein D. C., Thalhammer J. M., Hartwig A. C., Crenshaw E. B., 3rd, Lefebvre V., Wegner M., Sock E. (2010) The closely related transcription factors Sox4 and Sox11 function as survival factors during spinal cord development. J. Neurochem. 115, 131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Usui A., Mochizuki Y., Iida A., Miyauchi E., Satoh S., Sock E., Nakauchi H., Aburatani H., Murakami A., Wegner M., Watanabe S. (2013) The early retinal progenitor-expressed gene Sox11 regulates the timing of the differentiation of retinal cells. Development 140, 740–750 [DOI] [PubMed] [Google Scholar]
- 24. Furuta Y., Lagutin O., Hogan B. L., Oliver G. C. (2000) Retina- and ventral forebrain-specific Cre recombinase activity in transgenic mice. Genesis 26, 130–132 [PubMed] [Google Scholar]
- 25. Penzo-Méndez A., Dy P., Pallavi B., Lefebvre V. (2007) Generation of mice harboring a Sox4 conditional null allele. Genesis 45, 776–780 [DOI] [PubMed] [Google Scholar]
- 26. Cheung M., Abu-Elmagd M., Clevers H., Scotting P. J. (2000) Roles of Sox4 in central nervous system development. Brain Res. Mol. Brain Res. 79, 180–191 [DOI] [PubMed] [Google Scholar]
- 27. Wurm A., Sock E., Fuchshofer R., Wegner M., Tamm E. R. (2008) Anterior segment dysgenesis in the eyes of mice deficient for the high-mobility-group transcription factor Sox11. Exp. Eye Res. 86, 895–907 [DOI] [PubMed] [Google Scholar]
- 28. Sock E., Rettig S. D., Enderich J., Bösl M. R., Tamm E. R., Wegner M. (2004) Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol. Cell Biol. 24, 6635–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mu X., Fu X., Sun H., Beremand P. D., Thomas T. L., Klein W. H. (2005) A gene network downstream of transcription factor Math5 regulates retinal progenitor cell competence and ganglion cell fate. Dev. Biol. 280, 467–481 [DOI] [PubMed] [Google Scholar]
- 30. Xiang M., Zhou L., Macke J. P., Yoshioka T., Hendry S. H., Eddy R. L., Shows T. B., Nathans J. (1995) The Brn-3 family of POU-domain factors: primary structure, binding specificity, and expression in subsets of retinal ganglion cells and somatosensory neurons. J. Neurosci. 15, 4762–4785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nixon R. A., Lewis S. E., Dahl D., Marotta C. A., Drager U. C. (1989) Early posttranslational modifications of the three neurofilament subunits in mouse retinal ganglion cells: neuronal sites and time course in relation to subunit polymerization and axonal transport. Brain Res. Mol. Brain Res. 5, 93–108 [DOI] [PubMed] [Google Scholar]
- 32. Young R. W. (1985) Cell differentiation in the retina of the mouse. Anat. Rec. 212, 199–205 [DOI] [PubMed] [Google Scholar]
- 33. Gan L., Xiang M., Zhou L., Wagner D. S., Klein W. H., Nathans J. (1996) POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc. Natl. Acad. Sci. U.S.A. 93, 3920–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xiang M. (1998) Requirement for Brn-3b in early differentiation of postmitotic retinal ganglion cell precursors. Dev. Biol. 197, 155–169 [DOI] [PubMed] [Google Scholar]
- 35. Farah M. H., Easter S. S., Jr. (2005) Cell birth and death in the mouse retinal ganglion cell layer. J. Comp. Neurol. 489, 120–134 [DOI] [PubMed] [Google Scholar]
- 36. Perry V. H., Henderson Z., Linden R. (1983) Postnatal changes in retinal ganglion cell and optic axon populations in the pigmented rat. J. Comp. Neurol. 219, 356–368 [DOI] [PubMed] [Google Scholar]
- 37. Nissen-Meyer L. S., Jemtland R., Gautvik V. T., Pedersen M. E., Paro R., Fortunati D., Pierroz D. D., Stadelmann V. A., Reppe S., Reinholt F. P., Del Fattore A., Rucci N., Teti A., Ferrari S., Gautvik K. M. (2007) Osteopenia, decreased bone formation and impaired osteoblast development in Sox4 heterozygous mice. J. Cell Sci. 120, 2785–2795 [DOI] [PubMed] [Google Scholar]
- 38. Wilson M. E., Yang K. Y., Kalousova A., Lau J., Kosaka Y., Lynn F. C., Wang J., Mrejen C., Episkopou V., Clevers H. C., German M. S. (2005) The HMG box transcription factor Sox4 contributes to the development of the endocrine pancreas. Diabetes 54, 3402–3409 [DOI] [PubMed] [Google Scholar]
- 39. Castillo S. D., Matheu A., Mariani N., Carretero J., Lopez-Rios F., Lovell-Badge R., Sanchez-Cespedes M. (2012) Novel transcriptional targets of the SRY-HMG box transcription factor SOX4 link its expression to the development of small cell lung cancer. Cancer Res. 72, 176–186 [DOI] [PubMed] [Google Scholar]
- 40. Sander B. (2011) Mantle cell lymphoma: recent insights into pathogenesis, clinical variability, and new diagnostic markers. Sem. Diagnostic Pathol. 28, 245–255 [DOI] [PubMed] [Google Scholar]
- 41. Brown N. L., Kanekar S., Vetter M. L., Tucker P. K., Gemza D. L., Glaser T. (1998) Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development 125, 4821–4833 [DOI] [PubMed] [Google Scholar]
- 42. Feng L., Xie Z. H., Ding Q., Xie X., Libby R. T., Gan L. (2010) MATH5 controls the acquisition of multiple retinal cell fates. Mol. Brain 3, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Erkman L., McEvilly R. J., Luo L., Ryan A. K., Hooshmand F., O'Connell S. M., Keithley E. M., Rapaport D. H., Ryan A. F., Rosenfeld M. G. (1996) Role of transcription factors Brn-3.1 and Brn-3.2 in auditory and visual system development. Nature 381, 603–606 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.