Background: Human dermal fibroblasts (HDFs) from older subjects are known to be more resistant to reprogramming.
Results: Inclusion of SIRT6 can significantly improve the reprogramming efficiency.
Conclusion: Changes in SIRT6 expression and its posttranscriptional regulation may be relevant in aging.
Significance: MiR-766-mediated posttranscriptional regulation of SIRT6 has implications in human aging.
Keywords: Aging, Embryonic Stem Cell, Gene Expression, Induced pluripotent Stem Cells, MicroRNA, Molecular Biology, Transcription Factors
Abstract
Aging is known to be the single most important risk factor for multiple diseases. Sirtuin 6, or SIRT6, has recently been identified as a critical regulator of transcription, genome stability, telomere integrity, DNA repair, and metabolic homeostasis. A knockout mouse model of SIRT6 has displayed dramatic phenotypes of accelerated aging. In keeping with its role in aging, we demonstrated that human dermal fibroblasts (HDFs) from older human subjects were more resistant to reprogramming by classic Yamanaka factors than those from younger human subjects, but the addition of SIRT6 during reprogramming improved such efficiency in older HDFs substantially. Despite the importance of SIRT6, little is known about the molecular mechanism of its regulation. We show, for the first, time posttranscriptional regulation of SIRT6 by miR-766 and inverse correlation in the expression of this microRNA in HDFs from different age groups. Our results suggest that SIRT6 regulates miR-766 transcription via a feedback regulatory loop, which has implications for the modulation of SIRT6 expression in reprogramming of aging cells.
Introduction
Aging is associated with a gradual loss of homeostatic mechanisms that maintain the structure and function of adult tissues. As a result of aging, there is accumulation of mutations that increase the probability of cellular apoptosis, senescence, and malignancy, thus making age the single biggest risk factor for many diseases (1). The emergence of induced pluripotent stem cell (iPSC)3 technology has shown promise for the treatment of many age-related diseases (2). Efforts are underway to better understand how the age of the subjects affects both the process of reprogramming and the nature of the reprogrammed cells. For instance, Tat et al. (3) demonstrated that the age of mouse somatic cells influences reprogramming efficiency, and Baneto et al. (4) observed that cellular senescence acts as a barrier to classic Yamanaka factor reprogramming. Nevertheless, the effect of aging on the reprogramming of human somatic cells has yet to be addressed.
The sirtuin family members are known to be important regulators of silencing at the mating type locus as well as at the telomeres (5). In studies with Saccharomyces cerevisiae (6) and other organisms (7), these proteins were identified as key regulators of life span. They function by catalyzing NAD-dependent lysine deacetylation and a related mono-ADP-ribosylation reaction (8). In mammals, sirtuins (SIRT1–7) have subsequently been shown to play regulatory roles in cellular metabolic pathways (9). Mouse knockout models have been useful for delineating sirtuin functions in mammals. Gene targeting of SIRT1 (10), SIRT3 (11), SIRT4 (11), SIRT5 (11), SIRT6 (12), and SIRT7 (13) has been reported (14). Mice deficient in SIRT6 show an increasingly progeroid phenotype after an initial 4 weeks of apparently normal growth (in a purebred 129 S background) (12). Moreover, it has been shown that overexpression of SIRT1 enhances the reprogramming efficiency of mouse cells through a miR-34a-SIRT1-p53 pathway-dependent mechanism (15).
Biochemically, SIRT6 has been demonstrated to specifically deacetylate lysine 9 on histone H3 (H3K9Ac) (16) and lysine 56 on histone H3 (H3K56Ac) (14). In addition, SIRT6 has been shown to form a macromolecular complex with the DNA double-strand break repair factor DNA-PK (DNA-dependent protein kinase) to promote DNA double-strand break repair (17). It physically interacts with poly[adenosine diphosphate-ribose] polymerase 1 (PARP1) and mono-ADP-ribosylates PARP1 on lysine residue 521, thereby stimulating PARP1 poly-ADP-ribosylase activity and enhancing double-strand break repair under oxidative stress (18). In addition to these functions, SIRT6 is also required for maintenance of the telomere position effect in human cells, in which genes immediately proximal to the capped ends of the chromosome are silenced, indicating that the chromatin environment near the telomere is incompatible with transcription (19). Not surprisingly, SIRT6 knockout mouse cells exhibit DNA damage hypersensitivity and genomic instability (12). These mice have been shown to develop a severe degenerative phenotype quite similar to that of premature aging. Conversely, overexpression of SIRT6 in mice increased the life span of male but not female mice (20). Despite the recent progress in the understanding of SIRT6 biology and its important effects on aging, the molecular regulation of this key molecule remains largely a mystery.
MicroRNAs are a class of non-coding RNAs that have been shown to play a significant role in gene regulation by targeting a variety of transcripts via a short region of imperfect complementarity termed the “seed region.” They modulate numerous biological processes, including aging and age-related pathological diseases (21). In mouse models, the expression of genes targeted by miR-106 and miR-17 is down-regulated in various tissues with age (22). They have been implicated in various pathways related to aging (23), including those for sirtuins. Specifically, numerous microRNAs have been shown to regulate the SIRT1 signaling pathway (24).
In this study, we investigated the influence of age of human subject cells on their reprogramming efficiency and whether the inclusion of SIRT6 in the mix of reprogramming factors could help overcome the observed resistance to reprogramming in somatic cells from older subjects. We showed that human cells reprogrammed with SIRT6 and classical Yamanaka factors were truly pluripotent. Additionally, we investigated the miR-766 mediated posttranscriptional regulation of SIRT6. Our results demonstrate an inverse relationship between the expression of miR-766 and the expression of SIRT6 in human dermal fibroblasts (HDFs) derived from normal subjects from different age groups. Because miR-766 is transcribed as part of a larger transcript that includes the mRNA for the gene SEPT6, we next examined the regulation of the SEPT6 promoter and determined that acetylation levels increase with age at the SEPT6 upstream region, suggesting the presence of a possible feedback loop between SIRT6 and miR-766. We postulate that these components have implications for cellular aging.
EXPERIMENTAL PROCEDURES
Samples and Cell Culture
Human dermal fibroblasts were punch-biopsied from healthy male individuals with no self-reported disease or disorder. Each experimental (age) group had at least a total of five subjects, except the fetal group, which had two subjects. HDFs were grown in DMEM with high glucose and 10% FBS (Invitrogen). All cells used for reprogramming were within passage five. Derived iPSCs were maintained on Matrigel-coated tissue culture dishes (ES cells qualified, BD Biosciences) with mTESR-1 human ES cell growth medium (StemCell Technologies, Vancouver, Canada). For in vitro analysis of SIRT6 expression and molecular regulation by miR-766, individuals under the age of 18 years were designated as “young,” whereas those over the age of 50 years were designated as “old.”
Cell Culture and Maintenance of Human Pluripotent Stem Cells
Human iPSCs were derived as described in the supplemental Methods. Human iPSCs were cultured on Matrigel-coated plates (ES cells qualified, BD Biosciences) using human ES cell mTeSR-1 cell culture medium (StemCell Technologies) under conditions of 37 °C, 95% air, and 5% CO2 in a humidified incubator as described previously (25). Cells were passaged via disassociation with collagenase IV (Invitrogen) every 4–6 days.
Antibodies and Plasmids
Antibodies specific for SIRT6 (2590) were obtained from Cell Signaling Technology, and antibodies for H3K9Ac (H9286) and β-actin (A5441) were purchased from Sigma. Antibodies for histone H3 (catalog no. ab1791) and SP1 (catalog no. ab13370) were purchased from Abcam. The SIRT6 promoter was cloned in the pGL3 Basic vector from Promega, whereas the SIRT6 3′ UTR was cloned in the pMIR-Report vector from Ambion. The miR-766 Zip vector was purchased from System Biosciences. The two microRNA mimics, miR-766 and cel-miR-67, were purchased from Dharmacon.
Real-time PCR
Total RNA was extracted from cells with the RNeasy mini kit (Qiagen). Total RNA (2 μg) was reverse-transcribed with oligo(dT) primers using the SuperScript first-strand synthesis system for RT-PCR (Invitrogen). All PCR primers and TaqMan probes were from Applied Biosystems. Reactions were in triplicates for each sample and were analyzed using the ABI Prism 7300 sequence detection system. Data were normalized to GAPDH levels. For microRNA real-time PCR, total RNA was reverse-transcribed with random hexamers. For detection, the resulting cDNA for each sample was mixed with TaqMan universal PCR master mix and a set of primers and probes from Applied Biosystems. For endogenous normalization, control small nucleolar RNA was used. Reactions were done in duplicates for each sample using the ABI Prism 7300 sequence detection system.
Transfection and Luciferase Assay
HDFs and HeLa cells were transfected with 100 ng of pMIR-REPORT, pMIR-REPORT-SIRT6, or pMIR-REPORT-SEPT6 3′ UTR with or without end-modified microRNA oligonucleotides using Lipofectamine LTX. Reporter gene assays were performed 24 h after transfection using a luciferase assay kit (Promega). The cells were also cotransfected with 10 ng of firefly luciferase vector for normalizing transfection efficiency.
Embryoid Body (EB) Formation
Human SIRT6-OKSM-iPSCs (iPSC lines derived using Sox-2, Klf-4, Oct-4, and c-Myc along with SIRT6) were collected by collagenase IV treatment (1 μg/μl), resuspended in 20% FBS/DMEM medium, and allowed to form EBs in a six-well plate (Costar 3471) for up to 2 weeks. The EBs were then broken down into smaller clumps using a 200-μl pipette tip and allowed to attach onto gelatin-coated plates for an additional 2 days, followed by fixing and immunohistochemistry for the three embryonic germ layers.
Pluripotency Markers and EB Analysis
Human SIRT6-OKSM-iPSC colonies plated on 6-well tissue culture plates (Sigma Aldrich, St. Louis, MO) were fixed in 4% paraformaldehyde at room temperature for 5 min and then permeabilized with 1 ml of 0.5% Triton for 10 min. After washing with PBS, cells were incubated with primary antibodies (1:100 in PBS) at room temperature for 1 h. The primary antibodies used for staining were Oct3/4 (catalog no. sc-5297, Santa Cruz Biotechnology), Sox2 (catalog no. 630801, Biolegend), Klf-4 (Chemicon), Tra-1–60 (catalog no. MAB4360, Chemicon), Tra-1–81 (catalog no. MAB4381, Chemicon), and Nanog (catalog no. sc-374103, Santa Cruz Biotechnology). After thorough washing with PBS (three times for 5 min each), Alexa Fluor-conjugated secondary antibodies at a dilution of 1:250 (Invitrogen) were added for 20 min. To highlight the nuclei, DAPI (1:200) was added together with secondary antibodies. After three washes with PBS, immunofluorescent images were taken by fluorescent microscopy.
Chromatin Immunoprecipitation
The ChIP protocol was adapted from the one described in greater detail by Kawahara et al. (26). Briefly, DNA was cross-linked for 10 min with 1% formaldehyde and stopped in 0.125 m glycine. Purified chromatin was sonicated to ∼300 bp using the Bioruptor (Diagenode) and incubated with the indicated antibodies. Following reverse cross-linking, ChIP-associated sequences were detected by quantitative real-time PCR as described above.
Statistical Analysis
Results are given as mean ± S.E. Independent two-tailed Student's t test was performed. Differences were considered statistically significant for p < 0.05.
RESULTS
SIRT6 Improves the Reprogramming Efficiency of HDFs from Older Subjects
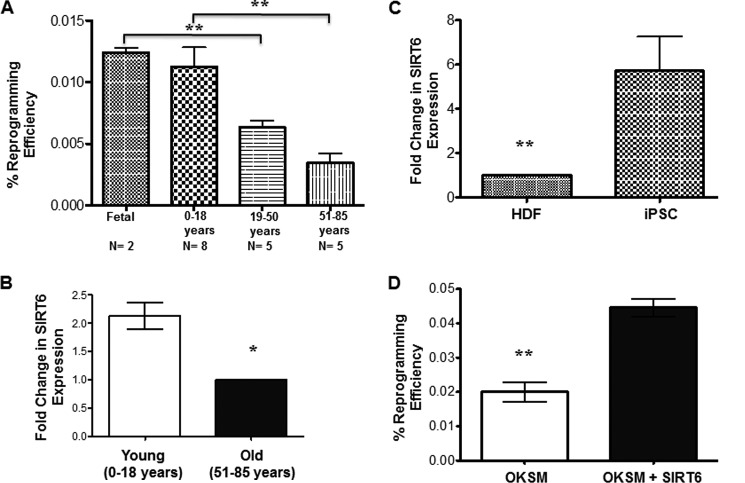
The influence of chronological age on reprogramming efficiency has been of interest in recent years. Consistent with earlier findings, we observed that HDFs from subjects over age 50 were indeed significantly more resistant to reprogramming. HDFs derived from subjects of different age groups were reprogrammed using classic Yamanaka factors (OCT-4, KLF-4, SOX-2, and c-MYC (OKSM)). Colonies were stained for Tra-1–60 on day 16 and counted to calculate reprogramming efficiency. The results in Fig. 1A show significantly lower reprogramming efficiency in the samples from human subjects aged 50–85 years (n = 5) compared with those from fetal samples (n = 2) or subjects aged 0–18 years (n = 8). However, there was no significant difference in reprogramming efficiency between the 20–49 and 50–85 age groups (p = 0.14).
FIGURE 1.
Reprogramming efficiency is influenced by the age of the human subject. A, HDFs from different age groups were transfected with OKSM, and emerging colonies were stained for Tra-1–60 on day 16. The colonies that were positive for Tra-1–60 were counted, and reprogramming efficiency was calculated on the basis of the total number of HDFs seeded. Bars represent the number of Tra-1–60+ colonies/40,000 cells plated initially. B, relative endogenous expression of SIRT6 in HDFs from different age groups. C, relative endogenous expression of SIRT6 in iPSCs (compared with HDFs). Data are represented as the mean ± S.E. *, p < 0.05 and **, p < 0.001. D, HDFs from the old group were infected with OKSM with or without SIRT6 or blank vector, and emerging colonies were stained for Tra-1–60 on day 16. Reprogramming efficiency was calculated as in A. Bars represent the number of Tra-1–60+ colonies/40,000 cells plated initially.
Because SIRT6 has been shown to modulate aging-related signaling pathways, we hypothesized that changes in the expression of SIRT6 may be responsible for the increased barrier to reprogramming cells derived from older subjects. To verify this, we first performed real-time PCR in both HDFs derived from subjects of different age groups and iPSCs derived from those HDFs. Individuals under the age of 18 years were designated as young, whereas those over the age of 50 were designated as old. Our results indicate significantly down-regulated expression of SIRT6 in HDFs from older subjects (Fig. 1B). When testing endogenous SIRT6 expression in iPSCs derived from those lines, we found significantly higher levels of SIRT6 in the pluripotent cells regardless of age (Fig. 1C). These results indicate a possible role of SIRT6 in the reprogramming process. Hence, we cloned human SIRT6 cDNA in pCDH_CMV-MSC_EF1-RFP (supplemental Fig. S1) and then reprogrammed HDFs from older subjects with the four classic Yamanaka factors (OKSM) with or without SIRT6 or a blank vector. When emerging iPSC colonies were stained for Tra-1–60 on day 16 of reprogramming, increased colony numbers were observed upon inclusion of SIRT6 (Fig. 1D). We observed that inclusion of SIRT6 along with classic Yamanaka factors resulted in significant improvements in the reprogramming efficiency of HDFs from older subjects, similar to the reprogramming efficiency from younger HDFs. As proliferation rate of the cells may influence the reprogramming efficiency, a BrdU incorporation assay was performed to confirm any significant difference in proliferation of young and old HDFs. Our results indicate a slight but statistically significant difference in the proliferation rate (supplemental Fig. 2G). Interestingly, inclusion of SIRT6 along with OKSM did not improve the reprogramming efficiency in young HDFs further. In fact, it reduced the number of iPSC colonies (supplemental Fig. S3). A potential explanation may be that SIRT6 acts as a corepressor of c-MYC mediated transcriptional activity at least in cancer cells (27). As the reprogramming efficiency is strongly influenced by c-MYC expression, most likely through enhancing cell proliferation (28), it can be speculated that a certain level of SIRT6 expression is required for the induction of pluripotency and that an elevated SIRT6 protein level impairs the reprogramming.
Generation of Bona Fide SIRT6-OKSM iPSCs
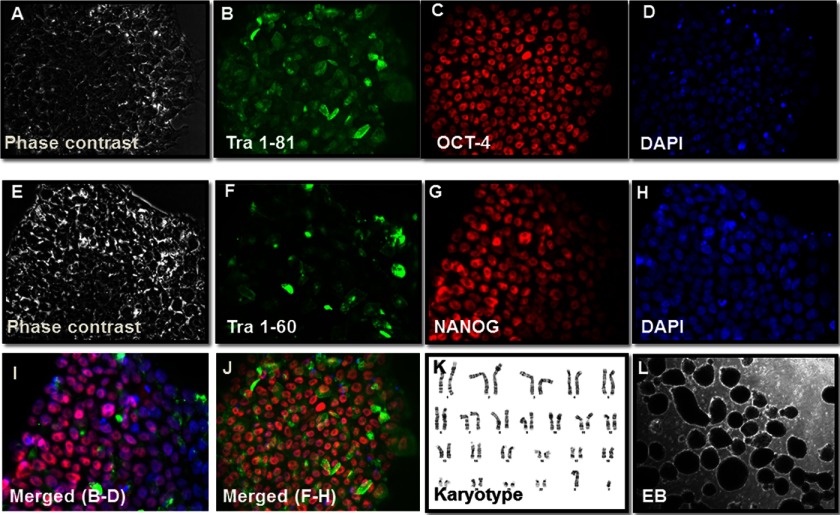
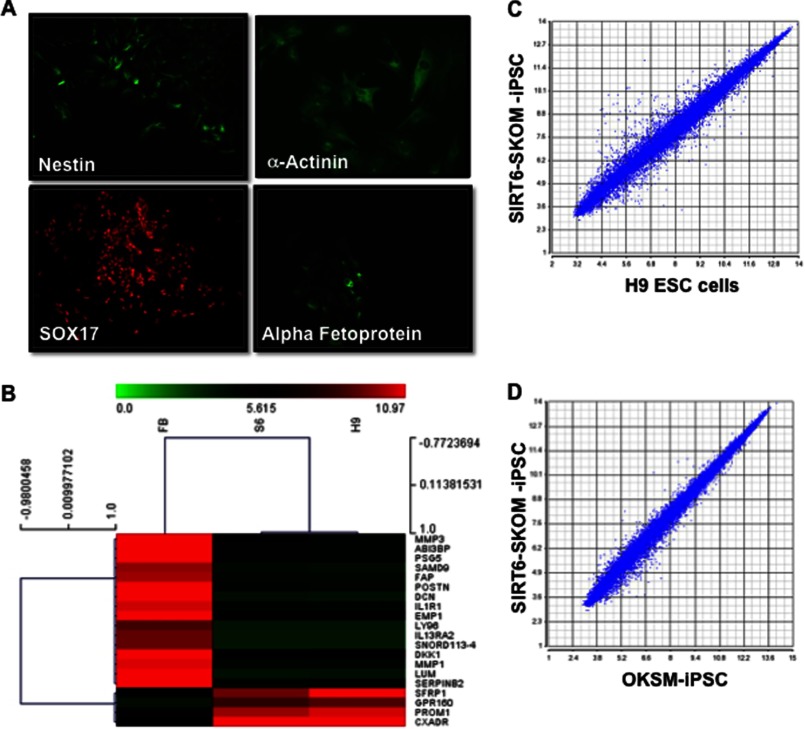
To further investigate the role of SIRT6 in pluripotency, we analyzed the SIRT6-OKSM-iPSC colonies derived from older subjects. These colonies (Fig. 2, A and E) stained positive for the known pluripotency-associated markers Tra-1–81 (Fig. 2B) and OCT4 (Fig. 2C) and nuclear staining by DAPI (Fig. D and H). Colonies also stained positive for Tra-1–60 (Fig. 2F) and NANOG (Fig. 2G). Similar staining for pluripotency markers was also performed for OKSM-iPSC lines (supplemental Fig. S4). To show that these iPSCs can differentiate into other lineages, we confirmed that inclusion of SIRT-6 did not result in karyotypic changes in these cells upon reprogramming (Fig. 2K). SIRT6-OKSM-iPSC lines could be induced to form EBs (Fig. 2L). Spontaneous differentiation of these EBs expressed genes from all three germ layers as assessed by in vitro immunostaining (Fig. 3A). Furthermore, microarray data comparing global gene expression profiles of SIRT6-OKSM-iPSC and human embryonic stem cells (ESCs) to HDFs revealed a high similarity of gene expression between SIRT6-OKSM-iPSC and H9 human ESCs as seen in the heat map (Fig. 3B). Hierarchical clustering analysis by Pearson correlation further demonstrated a tight correlation in gene expression among OKSM-iPSCs, ESCs, and SIRT6-OKSM-iPSCs (Fig. 3, C and D). Taken together, these results indicate that SIRT6-OKSM-iPSCs were indeed pluripotent and highly similar in gene expression to both human ESCs and iPSCs.
FIGURE 2.
Derivation of SIRT6-OKSM iPSC lines. A–H, immunostaining patterns for Tra-1–81, Tra-1–60, Oct4, and Nanog. I, merged images from B, C, and D. J, merged images from F, G, and H. K, the normal karyotype is seen in a representative SIRT6-iPSC colony. L, SIRT6-OKSM-iPSC can form EBs in vitro.
FIGURE 3.
SIRT6-OKSM iPSC lines can spontaneously differentiate in three germ layers. A, expression of markers for neuroectoderm (Nestin), mesoderm (α-actinin), and endoderm SOX17, (α-fetoprotein). B, DNA microarray data comparing global gene expression profiles of human fibroblasts (FB), SIRT6-iPSCs (S6), and human ESCs (H9). Heat map and hierarchical clustering analysis by Pearson correlation show SIRT6-OKSM-iPSCs to be similar to ESCs. C, scatter plots comparing global gene expression patterns between SIRT6-iPSCs and ESCs. D, scatter plots comparing global gene expression patterns between SIRT6-OKSM-iPSCs and OKSM-iPSCs.
Reduced Expression of SIRT6 in Human Dermal Fibroblasts Derived from Older Subjects
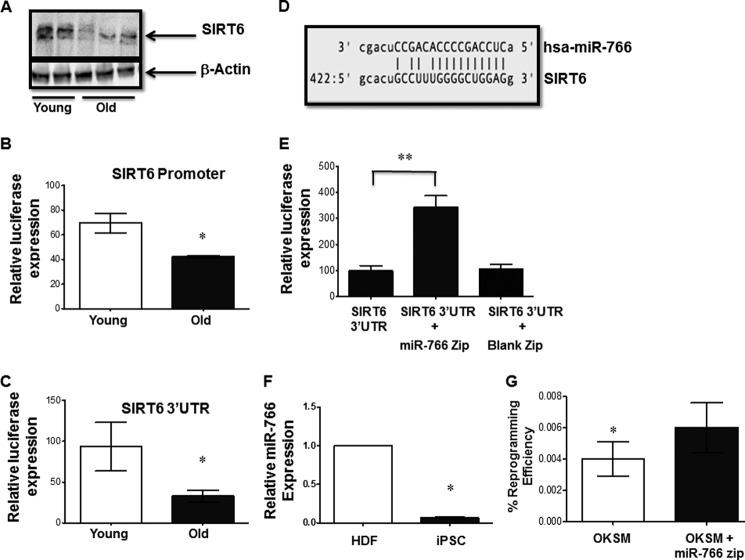
To better understand the role of SIRT6 in aging, we first measured its levels in HDFs derived from young and old subjects. Western blot analyses show higher protein levels in younger subjects compared with older ones (Fig. 4A). This difference was also seen at the mRNA level, in which a 2-fold reduction in expression of the SIRT6 transcript was found among older subjects (Fig. 1B). To elucidate the underlying mechanism(s) by which SIRT6 expression is decreased in cells derived from older subjects, we next cloned the SIRT6 promoter in a pGL3 (Basic) luciferase reporter vector, and its 3′ UTR was cloned in a pMIR-REPORT luciferase vector. Transfection of young and old HDFs with the SIRT6 promoter construct showed lower promoter activity in fibroblasts derived from older subjects (Fig. 4B). Likewise, when the SIRT6 3′ UTR construct was transfected in HDFs, higher luciferase activity was observed in younger fibroblasts compared with older fibroblasts, indicating distinct posttranscriptional regulation in older fibroblasts (Fig. 4C). These results suggest that SIRT6 could also be regulated posttranscriptionally.
FIGURE 4.
SIRT6 expression is regulated by miR-766. A, Western blot of endogenous SIRT6 levels in HDFs of individuals from different age groups. Equal loading was confirmed by β-actin. The result shown is representative of three individual experiments. B, the SIRT6 promoter construct or the pGL3 basic vector was transfected in HDFs derived from young and old subject cells, and relative luciferase activity was plotted. Results are presented as mean ± S.E. from three independent experiments. *, p < 0.05. C, the SIRT6 3′ UTR construct or the pMIR-Report vector was transfected in HDFs derived from young and old subject cells, and relative luciferase activity was plotted. Results are presented as mean ± S.E. from three independent experiments. *, p < 0.05. D, alignment report showing the predicted binding site for miR-766 in the 3′ UTR of SIRT6. E, HDFs from older subjects were transfected with the SIRT6 3′ UTR construct with or without miR-766 Zip or a blank vector (Blank Zip) construct. Luciferase expression was measured 48 h later, and the relative fold change in luciferase activity for each group was plotted with respect to SIRT6 3′ UTR construct alone. The results are plotted as the relative change in mean luciferase activity from three independent experiments and presented as mean ± S.E. At least four independent experiments were performed. **, p < 0.01. F, relative endogenous expression of miR-766 in iPSCs (compared with HDFs). Data are represented as mean ± S.E. *, p < 0.05. G, HDFs from the old group were infected with OKSM with or without miR-766 Zip or a blank vector, and emerging colonies were stained for Tra-1–60 on day 16. Reprogramming efficiency was calculated as in Fig. 1A. Bars represent the number of Tra-1–60+ colonies/40,000 cells plated initially. *, p < 0.05.
MicroRNA miR-766 Regulates SIRT6 Expression Posttranscriptionally
Thus far, little is known about the molecular mechanisms involved in regulation of SIRT6, and even less is known about its posttranscriptional regulation. To address this posttranscriptional mechanism, we hypothesized that more microRNA(s) may be involved in the regulation of SIRT6. First, we performed an in silico analysis using the miRGen 2.0 database (29), which yielded a single, high-scoring microRNA candidate: miR-766 (10 in the context of a nominal threshold of 7.5 with a precision of 0.65). A pictorial representation of the alignment of miR-766 at the SIRT6 3′ UTR shows the seed sequence match (Fig. 4D). To validate this finding further, we cotransfected both SIRT6–3′ UTR construct along with a miR-766-Zip vector that overexpresses an antisense sequence, which would quench cellular miR-766 and prevent it from interacting with its biological targets within the HDFs of older subjects. For control, we also cotransfected the SIRT6 3′ UTR construct with an “empty” vector that lacks the antisense sequence. Our results indicate that in HDFs, blocking miR-766 function results in a significant increase in luciferase activity via the SIRT6 3′ UTR (Fig. 4E). The effect of miR-766 on SIRT6 expression was also validated in HeLa cells by a series of transfection experiments (supplemental Fig. S5). Taken together, our study demonstrated a novel microRNA-mediated posttranscriptional regulation of SIRT6 by miR-766. In this context, it is interesting to note that miR-766 expression was lower in iPSCs compared with donor HDFs (Fig. 4F). To further validate the involvement of miR-766-mediated regulation of SIRT6 in reprogramming, older HDFs were reprogrammed with OKSM with or without miR-766 Zip. Our results indicate that blocking miR-766 results in a significant improvement in reprogramming efficiency (Fig. 4G).
Expression of miR-766 Is Higher in Older Dermal Fibroblasts
To further investigate the role that miR-766 plays in the physiological regulation of SIRT6 expression and the mechanism by which it modulates cellular phenotypes (especially in the context of aging), real-time PCR was performed for the microRNA in HDFs derived from young and old subjects. Results indicate higher levels of miR-766 in older subjects (Fig. 5A), which, as expected, was inversely correlated with SIRT6 protein levels (Fig. 4A).
FIGURE 5.
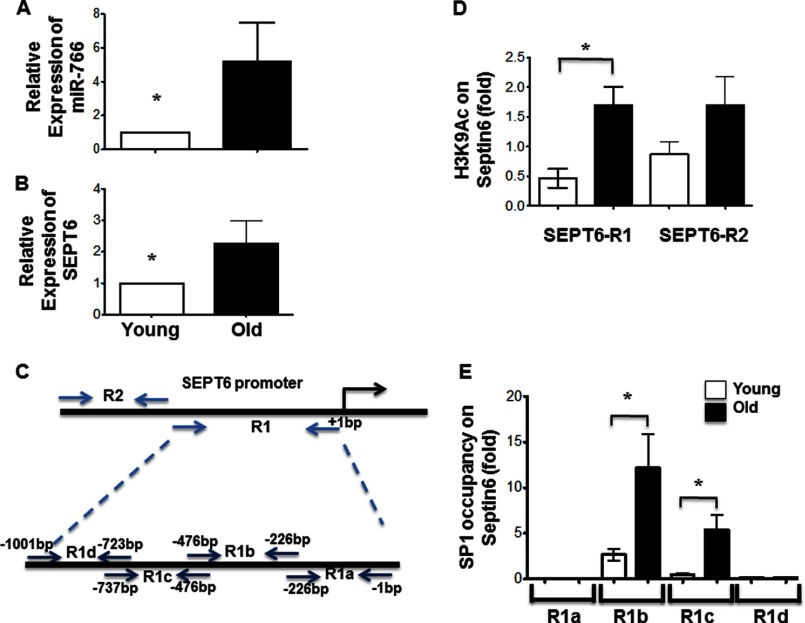
Expression of miR-766 was higher in older subjects. A, relative endogenous expression of miR-766 was determined in HDFs from young versus old age groups by real-time PCR with 6 HDFs both the groups. B, relative endogenous expression of SEPT6 was determined in HDFs from two age groups by real-time PCR. C, schematic of SEPT6 promoter subdivided to identify promoter acetylation and binding of transcription factor(s) to subregions as described under “Experimental Procedures.” D, ChIP was performed at the SEPT6/miR-766 promoter using H3K9Ac and H3 antibodies. H3K9 acetylation at the SEPT6/miR-766 promoter is shown relative to untreated control samples and normalized to total H3 levels. E, SP1 occupancy at the SEPT6/miR-766 promoter was measured for HDFs of older subjects versus younger subjects. Results (A, B, and C) are presented as mean ± S.E. At least four independent experiments were performed. *, p < 0.05.
Because miR-766 is an intronic microRNA and is encoded in the introns of the SEPT6 gene, we used real-time PCR to demonstrate the increased expression of SEPT6 in HDFs from older subjects, which is correlated with miR-766 expression (Fig. 5B). These results suggest that the higher levels of miR-766 in HDFs of older subjects are due to increased expression of its parental transcript from the SEPT6 source gene. We then proceeded to investigate this interplay between miR-766 and SIRT6 expression and whether SIRT6 itself regulates miR-766. As SIRT6 is known to modulate gene expression by deacetylating histone residues in the promoter region of target genes, H3K9 acetylation levels at different regions of the SEPT6 promoter were assessed after ChIP, followed by real-time PCR using primers (as indicated in Fig. 5C) that encompassed various fragments of the promoter. Our results indicate a significantly higher acetylation of H3K9 in region R1 in HDFs from older subjects (Fig. 5D). Therefore, SIRT6 could play an important role in miR-766 expression by modulating acetylation levels at its promoter.
We next performed transcription factor activation profiling assay (Signosis) (supplemental Fig. S6). Our preliminary analysis indicated increased binding of SP1 (and possibly p53 at SEPT6 promoter) (supplemental Fig. S7). However, we performed a similar ChIP for transcription factor SP1, which indicated a higher occupancy by SP1 in HDFs from older subjects (Fig. 5E). These results implicate SP1 in the modulation of miR-766 and SEPT6 expression in HDFs.
DISCUSSION
The iPSC technology has emerged as a potential source of therapeutic cells for treating various conditions, such as cardiovascular disease, degenerative cartilage disorders, and neurological diseases that disproportionately affect older patients. However, because earlier studies in mice have shown that older and senescent cells are more resistant to reprogramming (4), efforts are underway to understand the biology of reprogramming in the context of chronological and replicative age of cells. We demonstrate that with increasing age of the subjects, human dermal fibroblasts become increasingly resistant to reprogramming using classical Yamanaka factors.
Numerous studies have previously shown the critical role SIRT6 plays in modulating global gene expression changes and various pathways known for their relevance in aging. In yeast and in mice, SIRT6 (along with other members of its family) have been shown to regulate epigenetic gene silencing and DNA repair. We examined the effects of modulating SIRT6 expression during reprogramming. Our results indicate that inclusion of SIRT6 along with classical Yamanaka factors resulted in significant improvements in the reprogramming efficiency of HDFs from older subjects and that including SIRT6 during reprogramming has no negative effect on the pluripotency of these iPSCs (as demonstrated by expression of stem cell-related genes and in vitro differentiation). The similarity in global gene expression among SIRT6-OKSM-iPSCs, OKSM-iPSCs, and human ESCs as shown by hierarchical clustering of microarray data also indicated that SIRT6-OKSM-iPSCs are indeed pluripotent.
Despite recent investigations into SIRT6 biology, little is known regarding its molecular regulation. The decrease in SIRT6 expression in older HDFs (as shown by real-time PCR and Western blot analysis) may be relevant because several age-related genes that are silenced because of a higher activity of SIRT6 in young cells have been found to be transcriptionally up-regulated by NF-kB with increasing age, perhaps because of higher acetylation of histone residues near the promoters of these gene (30). This prompted us to investigate the possibility that SIRT6 expression is subject to posttranscriptional control via miR-766. Interestingly, another microRNA (miR-33a/b) known to regulate a host of genes involved in fatty acid metabolism and insulin signaling pathways (31) was also found to regulate SIRT6 (31), highlighting the vital role microRNAs in general play in regulating SIRT6 and cellular aging-related signaling pathways.
The influence of microRNAs on aging by virtue of their ability to modulate longevity genes has been proposed before (32). In one such study, microRNAs were shown to tightly regulate longevity in Caenorhabditis elegans (33), with some promoting longevity and others antagonizing it. Our results indicate a higher expression of miR-766 in HDFs of older subjects. Yu et al. (34) have reported similar increases in miR-766 expression (among others) in mesenchymal stem cells from old rhesus macaque bone marrow, which is interesting because aging is known to affect tissue-resident mesenchymal cell properties (35). Whether the miR-766/SIRT6 regulatory axis also plays a role in this context remains to be investigated.
The microRNA miR-766 has not yet been annotated in mice. However, it has been predicted in chimpanzees and orangutans and has been shown to be present in humans and monkeys, suggesting that the evolution of this microRNA may be relatively recent (miRBase, mIR-766 (Accession Number MI0003836)). Hence, it would be worthwhile to investigate the expression of miR-766 with increasing age in mice. It is also important to identify other miR-766 targets to shed more light on the hypothesis that the evolution of miR-766 has added complexity to gene regulation in higher mammals.
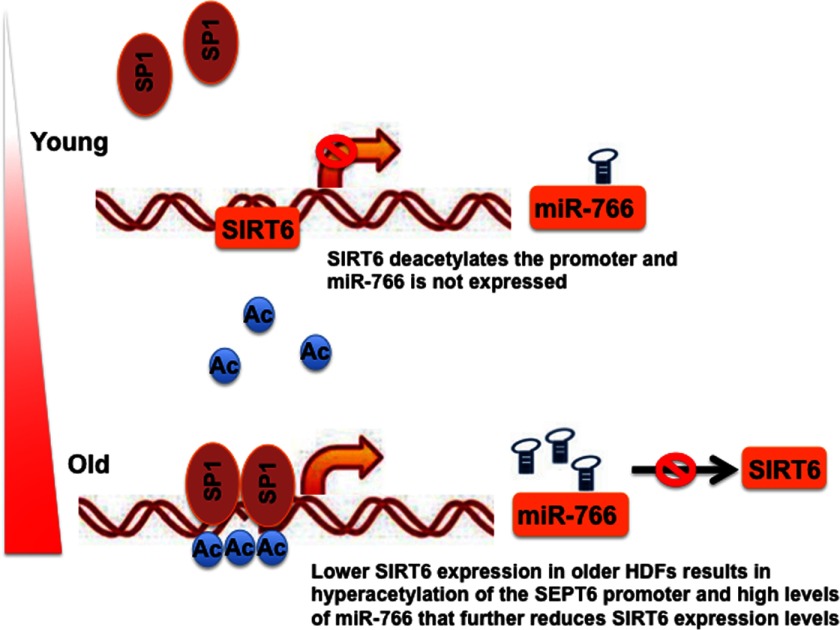
The interaction between microRNAs and target genes is complex. Negative and positive feedback loops between the two have been demonstrated before (36). Thus, we investigated such an interaction between miR-766 and SIRT6. A series of ChIP analyses on miR-766 and its source gene SEPT6 indicated the involvement of SP1 in its regulation. The ubiquitous transcription factor SP1 has been implicated previously in regulating several genes, including the age-related gene WNR (37) and other age-related genes (38). In younger subjects, high levels of SIRT6 activity ensure deacetylation at H3K9 at various promoters, including the SEPT6/miR-766 promoter, which results in low expression of the microRNA. As proposed in Fig. 6, older subjects may have a slight decrease in expression or activity of SIRT6 that may result in higher acetylation levels of H3K9 and elevated levels of miR-766, which further decreases the expression of SIRT6. These results imply a potential feedback loop between SIRT6 and miR-766.
FIGURE 6.
A schematic indicating proposed feedback regulation between miR-766 and SIRT6 expression. In younger subjects, high levels of SIRT6 activity ensure deacetylation at H3K9 at various promoters, including the SEPT6/miR-766 promoter, which results in a lower expression of the microRNA. However, in older subjects, slight changes in expression or activity of SIRT6 may result in higher acetylation (Ac) levels of H3K9 and elevated levels of miR-766, which further reduce the expression of SIRT6.
In summary, we have shown that age-related down-regulation of SIRT6 can contribute to increased resistance to reprogramming of HDFs from older subjects and that inclusion of SIRT6 may improve reprogramming efficiency significantly. Furthermore, we demonstrated, for the first time, that SIRT6 expression may be regulated by miR-766 in a feedback manner, which may be relevant in age-related changes in SIRT6 expression.
Supplementary Material
Acknowledgments
We thank the Stanford Functional Genomics Facility for assistance with the microarray experiment. We also thank Joseph Gold for critical reading of the manuscript and Paul Burridge for help with cardiomyocyte differentiation.
This work was supported, in whole or in part, by National Institutes of Health Grants DP2 OD004437, RC1 HL099117, R01 HL113006, P01 GM099130, UO1 HL099776, and CIRM RB3–05129 (to J. C. W.). This work was also supported by DFG (German Research Foundation) (to S. D.).

This article contains supplemental Figs. S1–S7, Video S1, Methods, and References.
- iPSC
- induced pluripotent stem cell
- HDF
- human dermal fibroblast
- EB
- embryoid body
- ESC
- embryonic stem cell.
REFERENCES
- 1. Harman D. (1991) The aging process. Major risk factor for disease and death. Proc. Natl. Acad. Sci. U.S.A. 88, 5360–5363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yoshida Y., Yamanaka S. (2010) Recent stem cell advances. Induced pluripotent stem cells for disease modeling and stem cell-based regeneration. Circulation 122, 80–87 [DOI] [PubMed] [Google Scholar]
- 3. Tat P. A., Sumer H., Pralong D., Verma P. J. (2011) The efficiency of cell fusion-based reprogramming is affected by the somatic cell type and the in vitro age of somatic cells. Cell Reprogram 13, 331–344 [DOI] [PubMed] [Google Scholar]
- 4. Banito A., Rashid S. T., Acosta J. C., Li S., Pereira C. F., Geti I., Pinho S., Silva J. C., Azuara V., Walsh M., Vallier L., Gil J. (2009) Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 23, 2134–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Finkel T., Deng C. X., Mostoslavsky R. (2009) Recent progress in the biology and physiology of sirtuins. Nature 460, 587–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kaeberlein M., McVey M., Guarente L. (1999) The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13, 2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tissenbaum H. A., Guarente L. (2001) Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410, 227–230 [DOI] [PubMed] [Google Scholar]
- 8. Liszt G., Ford E., Kurtev M., Guarente L. (2005) Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J. Biol. Chem. 280, 21313–21320 [DOI] [PubMed] [Google Scholar]
- 9. Donmez G., Guarente L. (2010) Aging and disease. Connections to sirtuins. Aging Cell 9, 285–290 [DOI] [PubMed] [Google Scholar]
- 10. Cheng H. L., Mostoslavsky R., Saito S., Manis J. P., Gu Y., Patel P., Bronson R., Appella E., Alt F. W., Chua K. F. (2003) Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 100, 10794–10799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lombard D. B., Alt F. W., Cheng H. L., Bunkenborg J., Streeper R. S., Mostoslavsky R., Kim J., Yancopoulos G., Valenzuela D., Murphy A., Yang Y., Chen Y., Hirschey M. D., Bronson R. T., Haigis M., Guarente L. P., Farese R. V., Jr., Weissman S., Verdin E., Schwer B. (2007) Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol. Cell Biol. 27, 8807–8814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mostoslavsky R., Chua K. F., Lombard D. B., Pang W. W., Fischer M. R., Gellon L., Liu P., Mostoslavsky G., Franco S., Murphy M. M., Mills K. D., Patel P., Hsu J. T., Hong A. L., Ford E., Cheng H. L., Kennedy C., Nunez N., Bronson R., Frendewey D., Auerbach W., Valenzuela D., Karow M., Hottiger M. O., Hursting S., Barrett J. C., Guarente L., Mulligan R., Demple B., Yancopoulos G. D., Alt F. W. (2006) Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124, 315–329 [DOI] [PubMed] [Google Scholar]
- 13. Vakhrusheva O., Smolka C., Gajawada P., Kostin S., Boettger T., Kubin T., Braun T., Bober E. (2008) Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ. Res. 102, 703–710 [DOI] [PubMed] [Google Scholar]
- 14. Michishita E., McCord R. A., Boxer L. D., Barber M. F., Hong T., Gozani O., Chua K. F. (2009) Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 8, 2664–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee Y. L., Peng Q., Fong S. W., Chen A. C., Lee K. F., Ng E. H., Nagy A., Yeung W. S. (2012) Sirtuin 1 facilitates generation of induced pluripotent stem cells from mouse embryonic fibroblasts through the miR-34a and p53 pathways. PLoS ONE 7, e45633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Michishita E., McCord R. A., Berber E., Kioi M., Padilla-Nash H., Damian M., Cheung P., Kusumoto R., Kawahara T. L., Barrett J. C., Chang H. Y., Bohr V. A., Ried T., Gozani O., Chua K. F. (2008) SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452, 492–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McCord R. A., Michishita E., Hong T., Berber E., Boxer L. D., Kusumoto R., Guan S., Shi X., Gozani O., Burlingame A. L., Bohr V. A., Chua K. F. (2009) SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging 1, 109–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mao Z., Hine C., Tian X., Van Meter M., Au M., Vaidya A., Seluanov A., Gorbunova V. (2011) SIRT6 promotes DNA repair under stress by activating PARP1. Science 332, 1443–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tennen R. I., Bua D. J., Wright W. E., Chua K. F. (2011) SIRT6 is required for maintenance of telomere position effect in human cells. Nat. Commun. 2, 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kanfi Y., Naiman S., Amir G., Peshti V., Zinman G., Nahum L., Bar-Joseph Z., Cohen H. Y. (2012) The sirtuin SIRT6 regulates lifespan in male mice. Nature 483, 218–221 [DOI] [PubMed] [Google Scholar]
- 21. Mendell J. T., Olson E. N. (2012) MicroRNAs in stress signaling and human disease. Cell 148, 1172–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hackl M., Brunner S., Fortschegger K., Schreiner C., Micutkova L., Mück C., Laschober G. T., Lepperdinger G., Sampson N., Berger P., Herndler-Brandstetter D., Wieser M., Kühnel H., Strasser A., Rinnerthaler M., Breitenbach M., Mildner M., Eckhart L., Tschachler E., Trost A., Bauer J. W., Papak C., Trajanoski Z., Scheideler M., Grillari-Voglauer R., Grubeck-Loebenstein B., Jansen-Dürr P., Grillari J. (2010) miR-17, miR-19b, miR-20a, and miR-106a are down-regulated in human aging. Aging Cell 9, 291–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gorospe M., Abdelmohsen K. (2011) MicroRegulators come of age in senescence. Trends Genet. 27, 233–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yamakuchi M. (2012) MicroRNA Regulation of SIRT1. Front Physiol. 3, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wilson K. D., Sun N., Huang M., Zhang W. Y., Lee A. S., Li Z., Wang S. X., Wu J. C. (2010) Effects of ionizing radiation on self-renewal and pluripotency of human embryonic stem cells. Cancer Res. 70, 5539–5548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kawahara T. L., Michishita E., Adler A. S., Damian M., Berber E., Lin M., McCord R. A., Ongaigui K. C., Boxer L. D., Chang H. Y., Chua K. F. (2009) SIRT6 links histone H3 lysine 9 deacetylation to NF-κB-dependent gene expression and organismal life span. Cell 136, 62–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sebastián C., Zwaans B. M., Silberman D. M., Gymrek M., Goren A., Zhong L., Ram O., Truelove J., Guimaraes A. R., Toiber D., Cosentino C., Greenson J. K., MacDonald A. I., McGlynn L., Maxwell F., Edwards J., Giacosa S., Guccione E., Weissleder R., Bernstein B. E., Regev A., Shiels P. G., Lombard D. B., Mostoslavsky R. (2012) The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 151, 1185–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wernig M., Meissner A., Cassady J. P., Jaenisch R. (2008) c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell 2, 10–12 [DOI] [PubMed] [Google Scholar]
- 29. Alexiou P., Vergoulis T., Gleditzsch M., Prekas G., Dalamagas T., Megraw M., Grosse I., Sellis T., Hatzigeorgiou A. G. (2010) miRGen 2.0. A database of microRNA genomic information and regulation. Nucleic Acids Res. 38, D137–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kawahara T. L., Rapicavoli N. A., Wu A. R., Qu K., Quake S. R., Chang H. Y. (2011) Dynamic chromatin localization of Sirt6 shapes stress- and aging-related transcriptional networks. PLoS Genet. 7, e1002153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fernández-Hernando C., Moore K. J. (2011) MicroRNA modulation of cholesterol homeostasis. Arterioscler. Thromb. Vasc. Biol. 31, 2378–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen L. H., Chiou G. Y., Chen Y. W., Li H. Y., Chiou S. H. (2010) MicroRNA and aging. A novel modulator in regulating the aging network. Ageing Res. Rev. 9, S59–66 [DOI] [PubMed] [Google Scholar]
- 33. de Lencastre A., Pincus Z., Zhou K., Kato M., Lee S. S., Slack F. J. (2010) MicroRNAs both promote and antagonize longevity in C. elegans. Curr. Biol. 20, 2159–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yu J. M., Wu X., Gimble J. M., Guan X., Freitas M. A., Bunnell B. A. (2011) Age-related changes in mesenchymal stem cells derived from rhesus macaque bone marrow. Aging Cell 10, 66–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Alt E. U., Senst C., Murthy S. N., Slakey D. P., Dupin C. L., Chaffin A. E., Kadowitz P. J., Izadpanah R. (2012) Aging alters tissue resident mesenchymal stem cell properties. Stem Cell Res. 8, 215–225 [DOI] [PubMed] [Google Scholar]
- 36. Tsang J., Zhu J., van Oudenaarden A. (2007) MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell 26, 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dunah A. W., Jeong H., Griffin A., Kim Y. M., Standaert D. G., Hersch S. M., Mouradian M. M., Young A. B., Tanese N., Krainc D. (2002) Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science 296, 2238–2243 [DOI] [PubMed] [Google Scholar]
- 38. Wu J., Xue L., Weng M., Sun Y., Zhang Z., Wang W., Tong T. (2007) Sp1 is essential for p16 expression in human diploid fibroblasts during senescence. PLoS ONE 2, e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.