Background: Fusions of dioxygenase and CBMs have been predicted in cellulolytic microbes.
Results: SACTE_2871 is unique two-domain enzyme that reacts with caffeoyl-CoA and shows preferential binding to synthetic lignins.
Conclusion: SACTE_2871 is an intradiol dioxygenase that is targeted to growing surfaces of lignin.
Significance: SACTE_2871 can destroy precursors needed by the plant for de novo lignin biosynthesis as part of its natural wounding response.
Keywords: Carbohydrate-binding Protein, Dioxygenase, Iron, Lignin Degradation, Protein Structure, SAXS, Caffeoyl-CoA, Lignin
Abstract
Streptomyces sp. SirexAA-E is a highly cellulolytic bacterium isolated from an insect/microbe symbiotic community. When grown on lignin-containing biomass, it secretes SACTE_2871, an aromatic ring dioxygenase domain fused to a family 5/12 carbohydrate-binding module (CBM 5/12). Here we present structural and catalytic studies of this novel fusion enzyme, thus providing insight into its function. The dioxygenase domain has the core β-sandwich fold typical of this enzyme family but lacks a dimerization domain observed in other intradiol dioxygenases. Consequently, the x-ray structure shows that the enzyme is monomeric and the Fe(III)-containing active site is exposed to solvent in a shallow depression on a planar surface. Purified SACTE_2871 catalyzes the O2-dependent intradiol cleavage of catechyl compounds from lignin biosynthetic pathways, but not their methylated derivatives. Binding studies show that SACTE_2871 binds synthetic lignin polymers and chitin through the interactions of the CBM 5/12 domain, representing a new binding specificity for this fold-family. Based on its unique structural features and functional properties, we propose that SACTE_2871 contributes to the invasive nature of the insect/microbial community by destroying precursors needed by the plant for de novo lignin biosynthesis as part of its natural wounding response.
Introduction
Plant biomass represents an abundant source of energy for many organisms. Insects are widely recognized for their capability to use both the cellulosic and hemicellulosic fractions of plant biomass as an abundant source of energy. Termites, leaf cutter ants, pine-boring wood wasps, pine beetles, and other biomass consuming insects (1–4) provide alarming examples of the scale of destruction that can arise from insect attack on plant biomass. In the thoroughly investigated cases, this invasive destruction occurs as a mutually beneficial symbiosis between insects, fungi, and bacteria. Although the role of gut-dwelling microbes in the ability of termites to use cellulose is known (1), other paradigms for symbiotic interactions of insects and microbes are emerging. For example, leaf cutter ants use an elaborate, community-based effort to harvest and shred leaves from specific plant species and to then inoculate the plant matter with a stable fungal/microbial community that carries out the biomass deconstruction (2). The ants subsequently harvest the microbial culture as a food and energy source, and discard the more recalcitrant fraction of biomass.
Pinewood-boring wood wasps also maintain symbiotic communities with fungi and bacteria. One example is the relationship between Sirex noctilio and the white rot fungus Amelyosterum aerolatum (5). S. noctilio is classified by the United States Department of Agriculture as an invasive species (www.invasivespeciesinfo.gov). It has caused massive destruction to pine plantations in South Africa and New Zealand after inadvertent introduction, and has recently been observed in the northeast of North America (6). S. noctilio is currently spreading westward from pine forests in this region, and thus poses a major threat to the multibillion-dollar forest products industry (7).
Recently, the presence of Actinomycetes in the Sirex/Amylosterum community has been established (8). Streptomyces sp. SirexAA-E was isolated from the Sirex ovipositor mycangia, part of a specialized organ that is used to lay eggs into the tree. Consequently, this free-living aerobic bacterium is introduced into the pine tree along with the white rot fungus and wasp eggs. Genomic, transcriptomic, and biochemical characterizations of Streptomyces sp. SirexAA-E have shown that it secretes a full suite of endo- and exocellulases, hemicellulases, pectinases, and polysaccharide monooxygenases when grown on biomass (3). The secreted enzyme mixture has high reactivity with biomass pretreated for conversion to biofuels and thus may provide similar reactivities in infested pine trees.
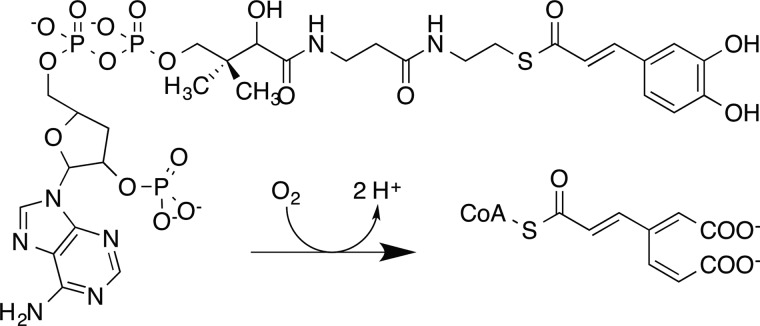
When plants are attacked by insects, the mechanical damage caused by chewing induces a variety of physiological responses, with many of these mediated by hormones derived from the jasmonic acid pathway (9, 10). One of these pathways is lignin biosynthesis, which proceeds by the deamination of phenylalanine and involves successive hydroxylation and methylation reactions (11). Several esterified or CoA-bound compounds as well as the corresponding free acids, aldedehydes, and alcohols are needed for lignin biosynthesis (12). Among these, the p-hydroxyphenyl-, catechyl-, guaiacyl-, 5-OH-guaiacyl, and syringyl compounds are key intermediates in the formation of the three most prevalent monolignols, which are the primary soluble building blocks of lignin. Successful up-regulation of lignin biosynthesis by the plant wound response generates a formidable covalent barrier to further attack. In opposition, the ability to interfere with lignin formation would be potentially advantageous for microbes seeking to carry out an invasive attack on living biomass. For example, enzymatic destruction of key intermediates in lignin biosynthetic pathways might be a promising strategy to short-circuit the plant wound response. In this regard, irreversible O2-dependent dioxygenation of the caffeoyl- and 5-OH-feruoyl (or 5-OH coniferyl) intermediates to form a non-polymerizable substituted cis-muconic acid would potentially impact lignin biosynthesis (Scheme 1). Wood and colleagues (13) demonstrated one example of this type of enzymatic activity, caffeate dioxygenase, in cell extracts obtained from Pseudomonas fluorescens in 1969. Since then, no further work on this enzyme has been reported.
SCHEME 1.
During our genome-enabled studies of Streptomyces sp. SirexAA-E, SACTE_28712 emerged as an intriguing protein because it was a member of the set of proteins and enzymes that were secreted when the organism was grown on either biomass or pure xylan (3). It was not detected among the secreted proteins when the organism was grown on glucose, pure cellulose, or chitin. Moreover, bioinformatics indicated an unusual combination of an intradiol dioxygenase domain with a carbohydrate-binding module assigned to the CBM 5/12 family. Typically, aromatic ring dioxygenases catalyze O2-dependent cleavage of catechyl aromatic rings (14), whereas CBMs are fused to glycoside hydrolases (15). Consequently, the potential function of this unusual hybrid was of interest.
In this work, we combine x-ray crystallography, small angle x-ray scattering, and biochemical analyses to show that SACTE_2871 is an intradiol dioxygenase that also exhibits preferential binding to synthetic lignins mediated by the presence of the CBM 5/12 domain. We propose that this enzyme may disrupt the capacity of the plant to protect itself from invasive attack by destroying one or more key early intermediates in the lignin biosynthetic pathway. In this manner, an enzyme secreted by a bacterium may potentiate the invasive nature of the Sirex symbiotic community.
EXPERIMENTAL PROCEDURES
Bioinformatics
The GenBankTM accession number for the Streptomyces sp. SirexAA-E genome is CP002993. SACTE_2871 is encoded by a gene with UniProt reference number G2NLE6 (16). The primary sequence of SACTE_2871 minus the signal peptide was used to search for homologous enzymes in UniProt. Multiple sequence alignments were done using ClustalW (17). A phylogenetic tree was constructed with MegaAlignTM (DNASTAR, Madison, WI) and displayed using FigTree (tree.bio.ed.ac.uk/software/figtree/).
Cloning and Purification
The mature form of SACTE_2871 (residues 46–291) was generated by removing the twin-arginine translocation signal peptide at the site predicted by the SignalP 4.0 server (18). An additional construct, SACTE_2871cc, which only contained the dioxygenase domain (residues 77–230) was also generated. Both SACTE_2871 and SACTE_2871cc were generated by PCR amplification from Streptomyces sp. SirexAA-E genomic DNA using the following forward and reverse primers: 5′-AACCTGTACTTCCAGTCCGTCCCCCTGGTCGCGGGC and 5′-GCTCGAATTCGTTTAAACTACCCGCGTTCCCACAACGCG for full-length SACTE_2871 and 5′-AACCTGTACTTCCAGTCCGACCCGACCCCCGACCAG and 5′-GCTCGAATTCGTTTAAACTAGGCCACGTCGAGGACGAAG for SACTE_2871cc. The amplified SACTE_2871 constructs were ligated into pVP68K (Center for Eukaryotic Structural Genomics, Madison, WI) containing a tobacco etch virus protease-cleavable His8-maltose-binding protein tag as previously described (19). pVP68K can be obtained from the National Institutes of Health Protein Structure Initiative Materials Repository (http://psimr.asu.edu). The His8-MPB-SACTE_2871 fusion enzymes were expressed in Eschericha coli B834(pRARE2) using autoinduction medium as described previously (20). Cell pellets were resuspended in 25 mm MOPS, pH 7.0, containing 50 mm NaCl and 2% (v/v) glycerol and lysed by sonication. The supernatant was applied to a 2-cm diameter × 20-cm bed height DEAE column equilibrated in the same buffer. The bound protein was eluted by running a 1.6-liter linear gradient from 50 to 500 mm NaCl. Fractions containing the dioxygenase were identified by visual inspection of SDS-PAGE gels. Tobacco etch virus protease (21) was used to remove the affinity/solubility tag, which was subsequently captured by subtractive nickel affinity chromatography using a 1.6-cm diameter × 2.5-cm bed height HisTrap HP column (GE Healthcare). The tag-free target protein was obtained in the flow-through and concentrated in preparation for gel filtration. Gel filtration was carried out in a 1.6-cm diameter × 60-cm bed height HiPrep SephacrylTM S-100 column (GE Healthcare) equilibrated in 25 mm MOPS, pH 7.0, containing 50 mm NaCl, and 2% (v/v) glycerol. The most pure fractions from gel filtration were pooled based on visual inspection of SDS-PAGE gels. The pooled fractions of SACTE_2871 or SACTE_2871cc were concentrated to ∼10 and ∼20 mg ml−1, respectively, dialyzed against 10 mm MOPS, pH 7.0, containing 50 mm NaCl, and drop frozen in liquid N2.
Enzyme Assays
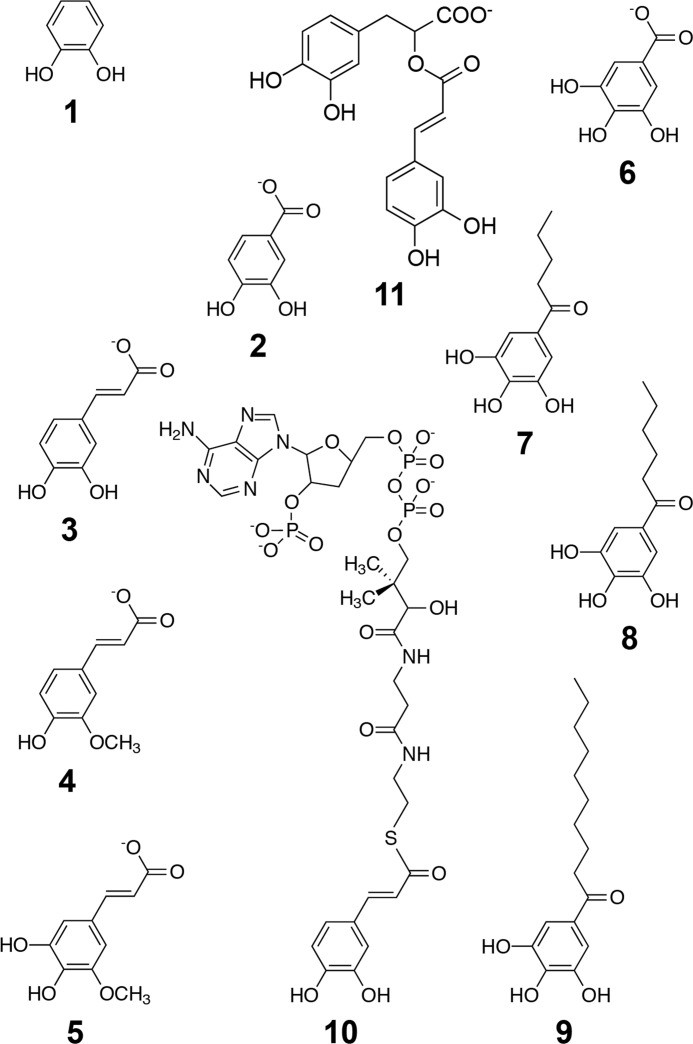
SACTE_2871-catalyzed reactions were studied by monitoring the substrate-dependent consumption of O2 (22) by polarography (Hansatech Instruments, Norfolk, England). The electrode was calibrated to a maximal response in the presence of air-saturated water and to a minimal response by the addition of excess sodium dithionite to the reaction chamber. All activity assays were performed in 500 μl of air-saturated 100 mm phosphate, pH 7.0, at 25 °C. The following aromatic substrates were added to the reaction chamber from saturated solutions: 1,2-dihydroxybenzene (catechol, 1); 3,4-dihydroxybenzoic acid (protocatechuate, 2); 3,4-dihydroxycinammic acid (caffeic acid, 3); 3-methoxy-4-hydroxycinnamic acid (ferulic acid, 4); 5-OH-ferulic acid (5); 3,4,5-trihydroxybenzoic acid (gallic acid, 6); propyl gallate (7); butyl gallate (8); octyl gallate (9); and rosmarinic acid (11). 5 and 11, as well as synthetic lignin compounds (G-DHP and G/S-DHP), were a gift from Prof. John Ralph's group (Great Lakes Bioenergy Research Center); all other compounds were purchased from Sigma-Aldrich. Caffeoyl-CoA (10 mg, 10) was synthesized using recombinant Nt4CL1 as described previously (23) and quantified using optical spectroscopy (molar absorptivity at 346 nm in 50:50 methanol:water (24)) and mass spectroscopy (m/z = 463.56). The optical spectrum of caffeoyl-CoA (10) shown in Fig. 2 was collected in 100 mm phosphate, pH 7.0.
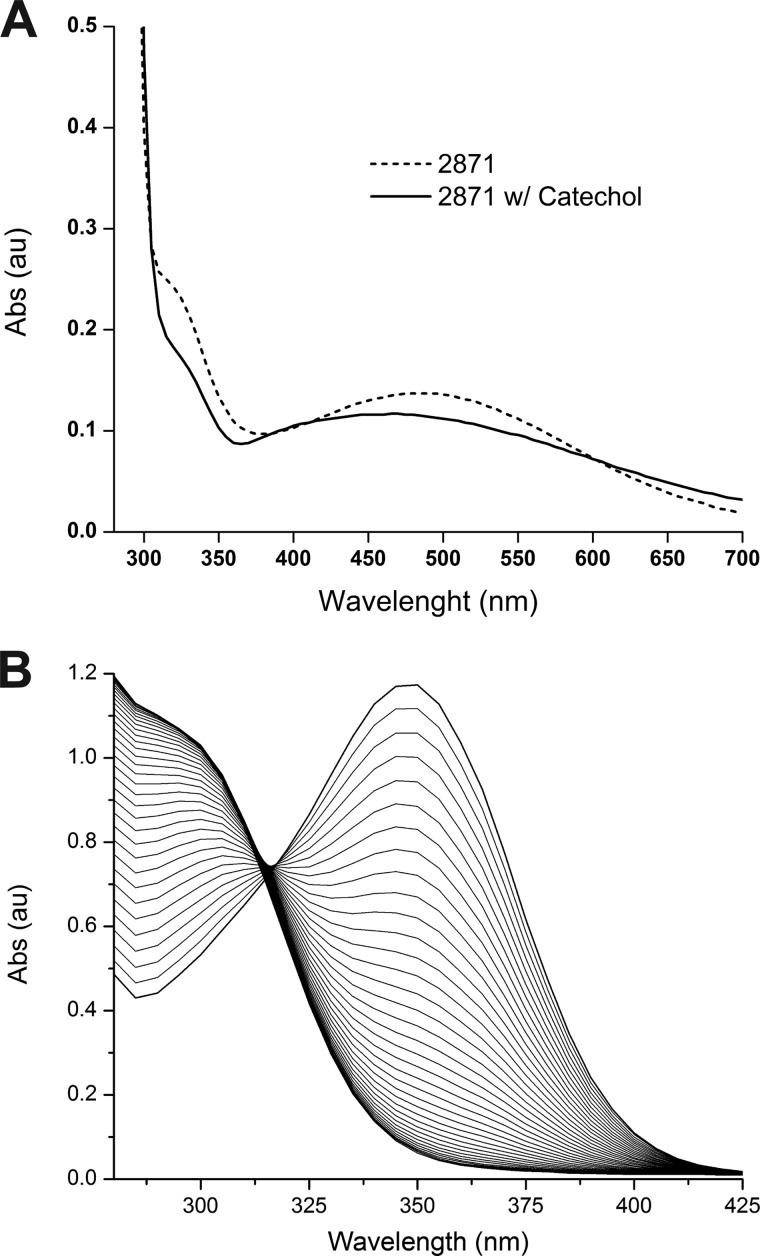
FIGURE 2.
A, absorption spectrum of full-length SACTE_2871 (dotted line). After addition of the model substrate catechol there is a distinct shift in the absorbance spectra (solid line). B, time dependent disappearance of the visible absorption band of caffeoyl-CoA (10) in the presence of SACTE_2871.
For polarography assays, buffer and substrate were placed into the reaction chamber and a stable baseline was established. Reactions were initiated by the addition of SACTE_2871 to a final concentration of 5 μm in the reaction chamber. Initial reaction velocities were determined by monitoring the decrease in O2 concentration in the early, linear portion of the reaction at 0.1-s intervals. Kinetic constants were calculated by nonlinear least squares fitting of the experimental results to the Michaels-Menten equation using Origin9 (OriginLab, Northampton, MA).
CBM Binding Studies
Binding of SACTE_2871 to insoluble polysaccharides was studied by a pull-down approach (25) using Sigmacell-20 (Sigma), birchwood xylan (Sigma), shrimp shell chitin (Sigma), 1–4β-d-mannan (Megazyme, Wicklow, Ireland), and two synthetic lignin compounds (G-DHP and G/S-DHP). The synthetic lignins were generated by in vitro peroxidase-catalyzed polymerization of monolignols as previously described (26). For the binding studies, 25 μg of SACTE_2871 was incubated with 1 mg of substrate in 50 mm phosphate buffer, pH 6.0, for 1 h at 4 °C. After incubation, the samples were centrifuged at 12,000 × g for 5 min at 4 °C. The unbound fraction (supernatant) and the substrate-bound fraction (pellet) were separated and run on 4–20% gradient SDS-PAGE gels. Binding experiments with SACTE_2871cc and with no substrate were performed as controls for binding of the catalytic domain in the absence of the CBM 5/12 domain and the presence of insoluble protein, respectively.
Crystallization
Crystals of SACTE_2871cc were grown in a hanging drop vapor diffusion set-up by mixing 2 μl of protein solution described above with an equal volume of 24% polyethylene glycol 3350, 5 mm CoCl2, 5 mm NiCl2, 5 mm CdCl2, 5 mm MnCl2, and 100 mm HEPES, pH 7.0, at 277 K. Hanging drop vapor diffusion crystallization trials were also conducted on full-length SACTE_2871 by mixing 2 μl of protein solution described above with an equal volume of 16% polyethylene glycol 3350, 200 mm sodium malonate, and 100 mm BisTris, pH 5.5, at 277 K. Crystals from both SACTE_2871cc and SACTE_2871 were cryoprotected by the addition of 15% 1,2-ethanediol to the final well solutions described above and then frozen directly in liquid N2.
Structure Determination
Diffraction data were collected at the Life Sciences Collaborative Access Team 21-ID-G and 21-ID-F beamlines at the Advanced Photon Source, Argonne National Laboratory (Argonne, IL). Collected diffraction images from both constructs were indexed, integrated, and scaled using HKL2000 (27). After processing in Chainsaw (28), catechol 1,2-dioxygenase from Rhodococcus opacus 1CP (PDB code 3HGI, (29)) was used as the initial model for molecular replacement with Phenix AutoMR (30). The refined SACTE_2871cc structure was used as the molecular replacement model in subsequent work. The SACTE_2871 structures were completed with alternating cycles of manual model building in Coot and refinement in Phenix (31). TLS was used during the final rounds of refinement for SACTE_2871 (32). All refinement steps for both structures were monitored using an Rfree value based on selection of 5.0% of the independent reflections. Model quality was assessed using MolProbity (33). Figures with structure representations were generated using PyMOL (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC) with the exception of Fig. 7D, which was generated using Chimera (34).
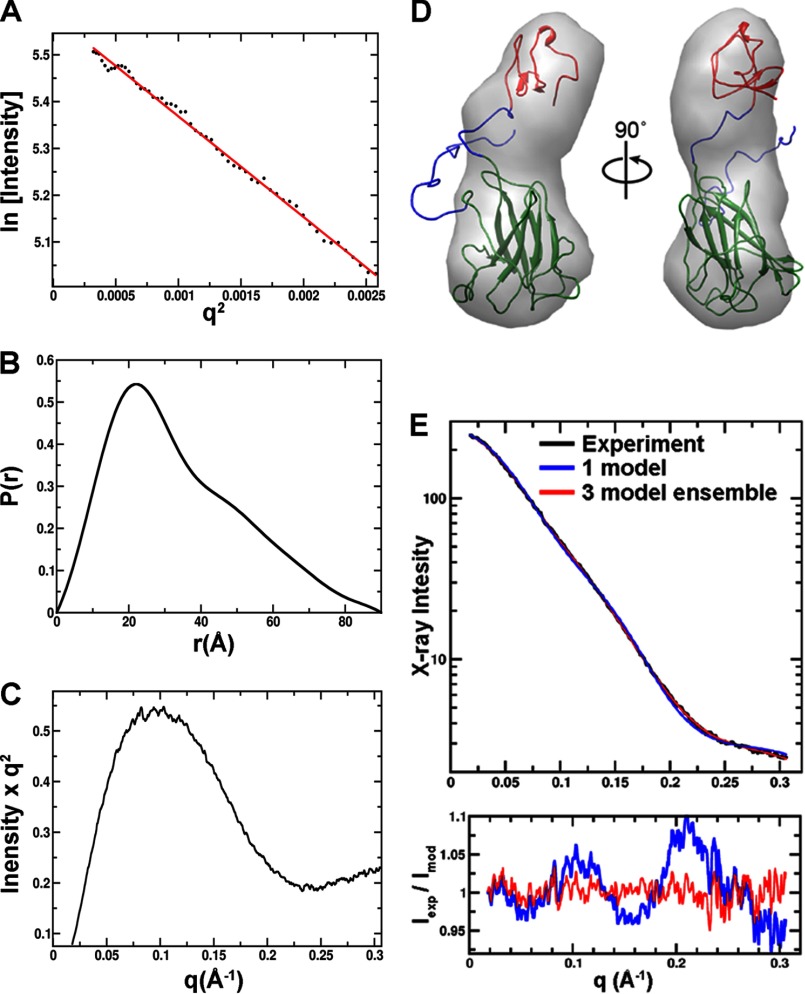
FIGURE 7.
SAXS data and analysis from full-length SACTE_2871. A, Guineir plot; B, pair distribution function; C, Kratky plot of SAXS data. D, an envelope generated from the SAXS data with a best fitting atomic resolution model. There is sufficient space to accommodate the dioxygenase domain (green schematic) at one end of the envelope. At the opposite end, the shape narrowed and only had sufficient volume to accommodate a homology model of SACTE_2871CBM (schematic). E, the experimental SAXS data (black) as fit by the single best model (blue) and a 3 model ensemble (red). The experimental signal divided by the models is shown below the SAXS data.
Mass Spectrometry
The molecular weight of protein present in SACTE_2871 crystals was determined by the Mass Spectrometry Facility at the University of Wisconsin-Madison Biotechnology Center. Crystals were washed five times with 16% polyethylene glycol 3350, 200 mm sodium malonate, and 100 mm BisTris, pH 5.5, to remove any protein that was not part of the crystal. The washed SACTE_2871 crystals were precipitated in 60% acetone, washed once in ice-cold methanol, solubilized in neat formic acid, and diluted 10-fold in 50:50 methanol:water for analysis. Analyses of acid-solubilized protein were carried out on an LC/MSD-TOF mass spectrometer (Agilent, Palo Alto, CA) using an auto-syringe delivery system (Harvard Apparatus, Holliston, MA). The following instrumental parameters were used to generate the optimal protonated ions [M + H+] in positive mode: capillary voltage 3500 V; drying gas 6.0 liter/min; nebulizer 20 psig; gas temperature 325 °C; Oct DC1 39.5 V; fragmentor 180 V; Oct RF 250 V; skimmer 60 V. Acquired data were processed using Analyst QS 1.1 build:9865 software (Agilent, Palo Alto, CA) to monitor masses observed in the range from 100 to 3200 atomic mass units. Intact protein species deconvolution was carried out using the ProteinApp module within the Agilent BioConfirm Software version A.02.00.
SAXS Data Collection
Small angle x-ray scattering from full-length SACTE_2871 were collected at the SIBYLS beamline at the Advanced Light Source (Berkeley, CA) as described previously (35). SACTE_2871 was prepared in three concentrations (10, 6.6, and 3.3 mg/ml) in 10 mm MOPS, pH 7.0, containing 50 mm NaCl. A buffer blank was collected both before and after the concentration series. The subtraction of either buffer from each sample yielded identical results to within experimental error (∼1% of signal). The sample was exposed for four different time durations (0.5, 1, 2, and 4 s) and no radiation damage was observed. A small concentration dependence was corrected for using standard procedures (36). The mass was calculated by using glucose isomerase as a standard at 1 mg/ml. The samples were placed 1.5 m from a MAR165 CCD detector arranged co-axial with the 12 keV monochromatic beam; 1012 photons/s were impingent on the sample. The spot size at the sample was 4 × 1-mm convergent to a 100-μm spot at the detector. Buffer subtraction and raw image data were integrated by beamline software specific for this arrangement.
SAXS Data Processing
Initial processing of SAXS data were conducted utilizing the ATSAS package (37). Utilizing the SAXS curve alone, 10 three-dimensional structural envelopes were generated and averaged by GASBOR (38). To combine the SAXS results with the crystallographic results an atomic model of the full-length SACTE_2871 was created by combining the structure of the dioxygenase domain reported here (PDB 4ILT) with a homology model of CBM 5/12. For the homology model, residues 245–291 from SACTE_2871 were modeled using the chitin-binding CBM 5/12 domain of chitinase A1 from Bacillus circulans WL-12 (PDB ID 1ED7 (39)). In addition, 30 residues on the N terminus of the dioxygenase domain (SVPLVAGGGAALARDTGAGAVPLAPTPACD) and a 14-residue linker between the dioxygenase and CBM5/12 domains (PQQPDPTDPPTDPG) were added using MODELLER. The built-in residues allowed flexibility in subsequent analysis by BILBOMD (40) and minimal ensemble search. BILBOMD generates a large ensemble of conformations by carrying out a molecular dynamics simulation imposing only forces required for a self-avoiding chain. The minimal ensemble search algorithm calculates a scattering profile from each conformation and identifies the minimal ensemble of SAXS curves required to fit the experimental data. Molecular graphics analyses were performed with UCSF Chimera.
RESULTS
SACTE_2871 Domain Structure
SACTE_2871 is a two-domain enzyme that consists of a twin-arginine translocation signal peptide (residues 1–45), an intradiol dioxygenase domain (residues 77–232), and a CBM 5/12 carbohydrate-binding module (residues 245–291). Interestingly, the twin-arginine translocation signal motif is frequently associated with secreted proteins in Streptomyces sp. SirexAA-E (3). All SACTE_2871 homologs contain four strictly conserved residues (2 Tyr and 2 His) that provide ligands to the active site Fe(III) (41). The CBM of SACTE_2871, designated here as SACTE_2871CBM, is attached to the dioxygenase domain via a 14-residue proline/threonine-rich flexible linker. To our knowledge, this study is the first structural and biochemical characterization of this combination of protein domains.
SACTE_2871 Sequence Homologs
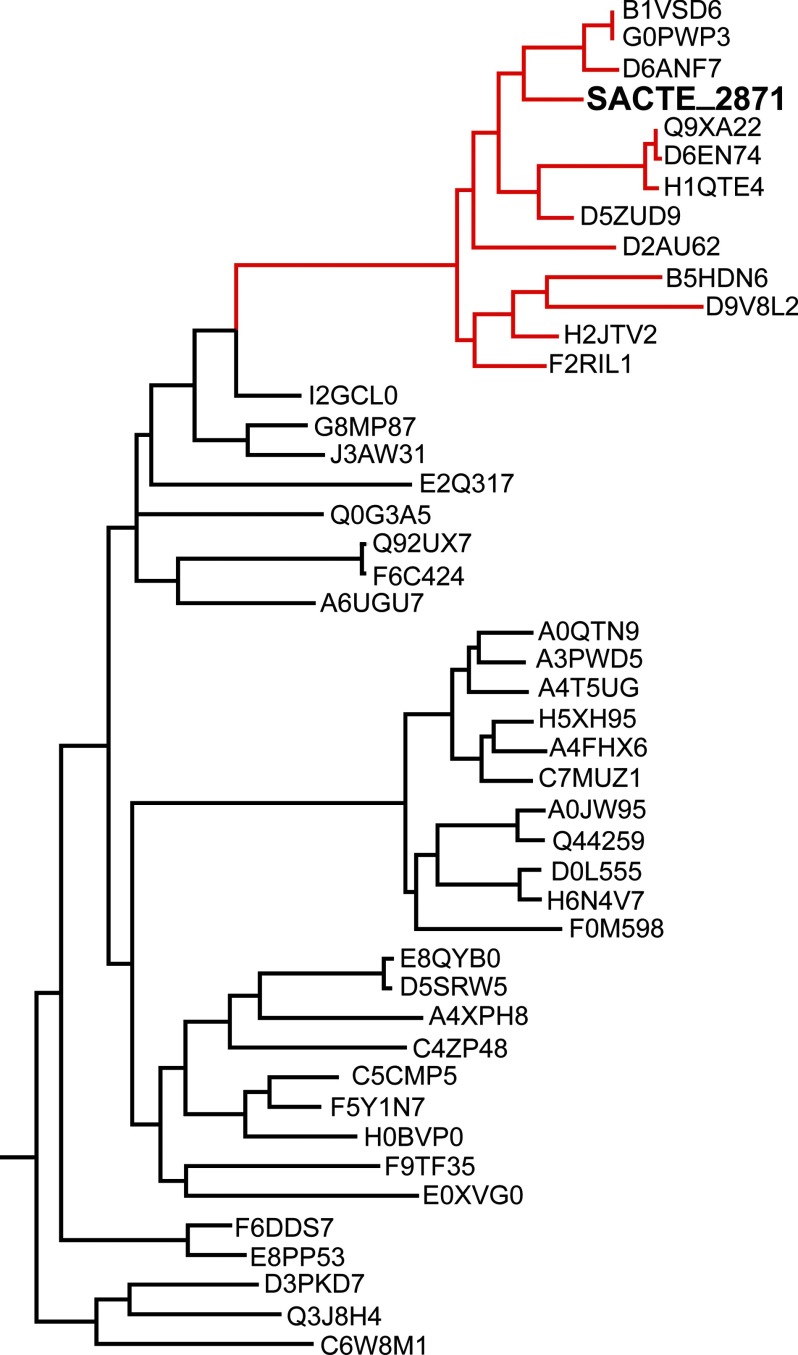
A BLAST search using the full-length SACTE_2871 sequence revealed numerous homologs from a wide range of bacterial and fungal species. However, the vast majority of these homologs only showed a low level of sequence identity to the dioxygenase domain and lacked any annotated CBM domain. Interestingly, some cellulolytic fungi have homologous dioxygenase domains that are also predicted to be secreted, but these are not fused to a CBM domain. Sequence alignments including the 45 closest homologs to SACTE_2871 show that the pairing of a dioxygenase domain with a CBM constitutes a single clade that is almost exclusively composed of proteins from various Streptomyces sp. (Fig. 1). Many of these organisms are known to be either cellulolytic or hemicellulolytic.
FIGURE 1.
SACTE_2871 sequence homologs. A phylogenic tree constructed from 45 homologous sequences (Uniprot IDs). Enzymes that share the same domain structure as SACTE_2871 are clustered into the same clade, highlighted in red. The majority of these enzymes are from various Streptomyces sp.
A BLAST search was also completed using the SACTE_2871CBM sequence (residues 245–291) as the search model. The search results show that the closest homologs to SACTE_2871CBM comprise one domain of a multidomain protein. Although there was some variation in the composition of the attached domains, the majority of the closest SACTE_2871CBM homologs were attached to a putative dioxygenase domain, yielding a full-length protein similar to SACTE_2871. The second most frequently observed fusion of a SACTE_2871CBM homolog was to a chitinase-like domain.
Protein Expression and Purification
SACTE_2871 was expressed in B834(pRARE2) as a fusion to His8-maltose-binding protein, and purified by a combination of ion exchange, subtractive immobilized metal affinity, and gel filtration chromatographies. After the ion exchange step, fractions containing the fusion protein had a distinct purple color comparable with the ligand to metal absorption of Fe(III) ligated by tyrosine observed in other intradiol dioxygenases (42). The optical spectrum of the purified protein obtained after subtractive immobilized metal affinity chromatography and gel filtration is shown in Fig. 2 (dotted line). Upon introduction of catechol to an anoxic sample of SACTE_2871 (solid line), the optical spectrum underwent a spectral shift consistent with the binding of catechol to iron (43). Polarographic studies also showed that the enzyme consumed O2 in the presence of catechol and other related compounds (see “Catalytic Studies,” below). These experiments provided the first evidence for function of the two-domain enzyme.
Structure Determination
Data collection, refinement, and model statistics are summarized in Table 1. Crystallization screens were set up with SACTE_2871 (residues 46–291, full-length protein) and SACTE_2871cc (residues 77–230, dioxygenase catalytic domain only), and crystals were obtained from both preparations. The SACTE_2871cc crystals belonged to the P21 space group and contained four monomers per asymmetric unit. The SACTE_2871cc structure was solved at 2.56 Å and well defined electron density was observed for residues 77–230. An additional serine residue, which derives from the cloning method, was observed at the N terminus in two of the four monomers.
TABLE 1.
Summary of crystal parameters, data collection, and refinement statistics
Values in parentheses are for the highest resolution shell.
| SACTE_2871 (residues 58–232) | SACTE_2871cc (residues 77–230) | |
|---|---|---|
| Crystal parameters | ||
| Space group | P43212 | P21 |
| Unit-cell parameters (Å) | a = 62.46, b = 62.46, c = 196 | a = 49.19, b = 85.13, c = 70.26, β = 95.53° |
| Data collection statistics | ||
| Wavelength (Å) | 0.97857 | 0.97872 |
| Resolution range (Å) | 32.81 − 2.059 (2.13 − 2.06) | 42.56 − 2.56 (2.69 − 2.56) |
| No. of reflections (measured / unique) | 24,644/2,345 | 18,534/1,589 |
| Completeness (%) | 98.53 (96.66) | 98.53 (85.34) |
| Rmergea | 0.076 (0.736) | 0.155 (0.651) |
| Redundancy | 9.5 (8.2) | 3.7 (3.6) |
| Mean I /σ (I) | 13.5 (2.7) | 7.2 (1.7) |
| Refinement and model statistics | ||
| Resolution range (Å) | 32.81 − 2.06 | 42.56 − 2.56 |
| No. of reflections (work / test) | 23,376 / 1232 | 17,589 / 930 |
| Rcrystb | 0.183 (0.233) | 0.203 (0.256) |
| Rfreec | 0.237 (0.299) | 0.268 (0.34) |
| Root mean square deviation bonds (Å) | 0.015 | 0.010 |
| Root mean square deviation angles (°) | 1.09 | 1.30 |
| Average B-factor (Å2) | 39.50 | 36.72 |
| No. of protein atoms | 2,388 | 4,695 |
| No. of waters | 211 | 220 |
| No. of auxiliary molecules | 3 1,2-Ethanediol | 2 Chloride |
| Ramachandran plot (%) | ||
| Favorable | 99 | 98.3 |
| Allowed | 1 | 1.97 |
| Disallowed | 0.0 | 0.0 |
| PDB code | 4ILV | 4ILT |
a Rmerge = Σh Σi|Ii (h) − 〈I(h)〉|/ΣhΣiIi(h), where Ii(h) is the intensity of an individual measurement of the reflection and 〈I(h)〉 is the mean intensity of the reflection.
b Rcryst = Σh‖Fobs| − |Fcalc‖/Σh|Fobs|, where Fobs and Fcalc are the observed and calculated structure-factor amplitudes, respectively.
c Rfree was calculated as Rcryst using ∼5% of randomly selected unique reflections that were omitted from the structure refinement.
Unlike crystals of SACTE_2871cc, which grew to a reasonable size in a week, crystals of full-length SACTE_2871 took 3–4 months to grow. The SACTE_2871 crystals belonged to the P43212 space group and contained two monomers per asymmetric unit. The structure was solved at 2.06 Å resolution, and gave interpretable electron density for residues 77–231 in the A monomer and 74–230 in the B monomer. Even though both the dioxygenase and CBM 5/12 domains were present in the protein used for the crystallization screening, only the dioxygenase domain was observed in the electron density. Mass spectrometry performed on SACTE_2871 crystals taken from the same well that yielded the crystal used to solve the structure revealed that the crystallized SACTE_2871 consisted of a mixture of polypeptides with masses of 18,165 and 18,574 Da. These two fragments correspond to polypeptides Ala63–Gln232 and Asp58–Gln232, respectively. Thus, SACTE_2871 was degraded from both the N and C termini during the long time required for crystallization, yielded a shorter form that eventually crystallized and yielded a structure.
Dioxygenase Domain
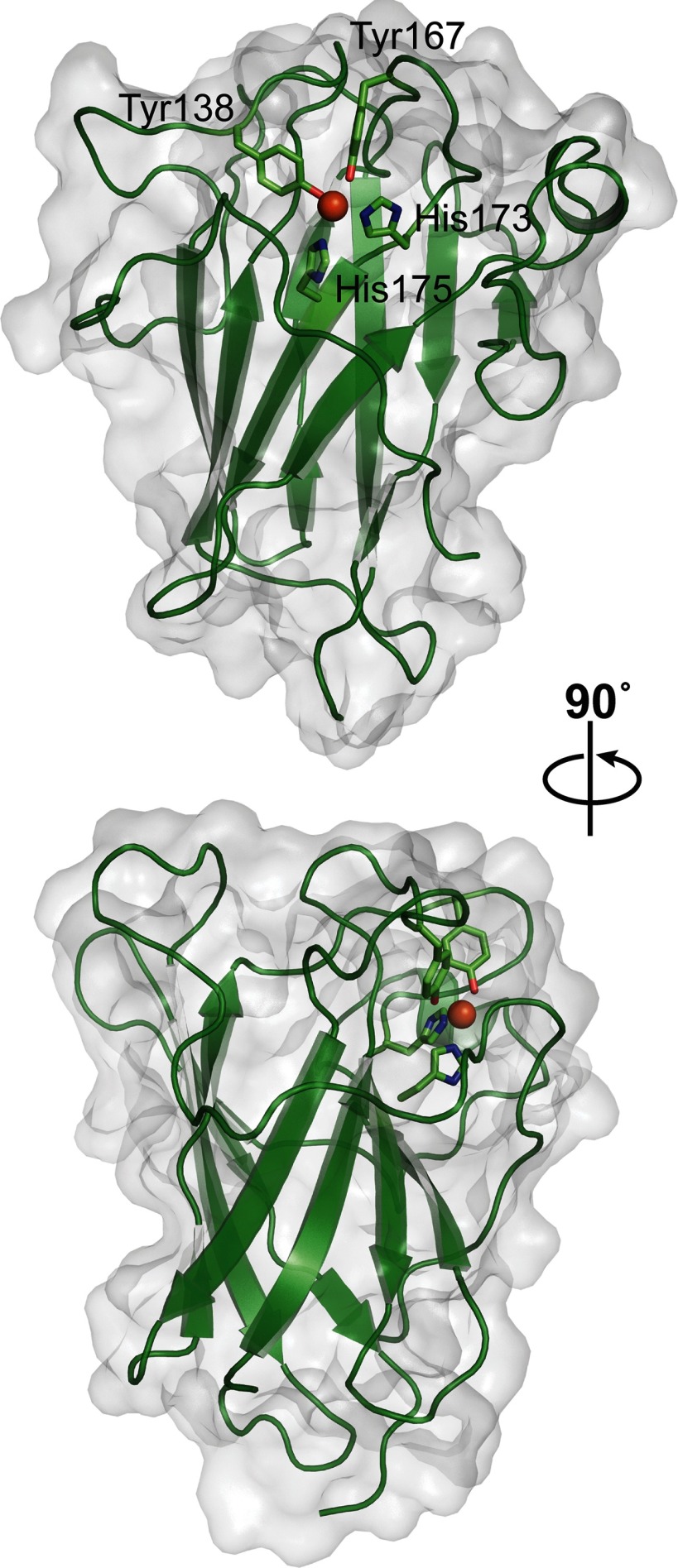
The core of the SACTE_2871 dioxygenase domain consists of two four-stranded β-sheets that interact to form a β-sandwich (Fig. 3). This core is similar to other intradiol dioxygenases (29, 44–48). The remainder of the SACTE_2871 dioxygenase domain is composed of a single α-helix and several extended loops that connect the β-sheets. Residues (Tyr138, Tyr167, His173, and His175) that form the active site and coordinate the active site iron are located on these loops.
FIGURE 3.
Overall structure of the dioxygenase domain of SACTE_2871. Schematic representation of the dioxygenase domain of SACTE_2871 with active site iron shown as an orange sphere and the iron coordinating residues shown as sticks. The bottom panel is a 90° rotation from the top panel. The solvent accessible surface is shown as a transparent surface.
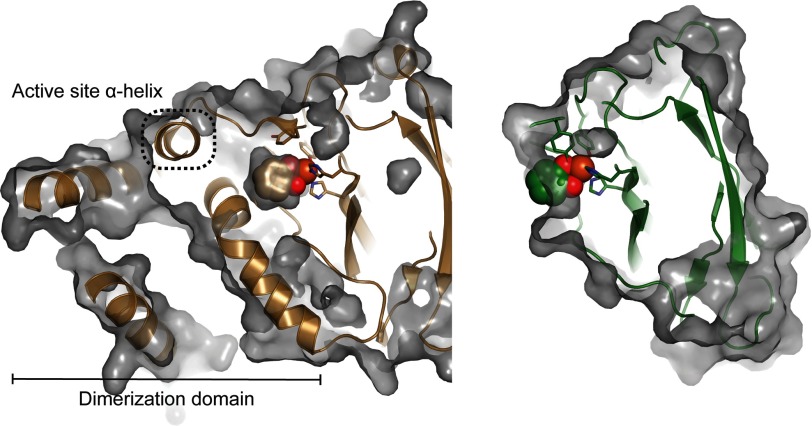
There are several notable differences in the overall structure of the SACTE_2871 dioxygenase domain when compared with other dioxygenases. In SACTE_2871, the most significant departure from the typical dioxygenase-fold is the absence of the extensive N-terminal dimerization domain observed in the closely related dioxygenases (Fig. 4). In these related structures, the dimerization domain can include up to ∼100 residues and also contains a hydrophobic pocket that binds a phospholipid (44). In both SACTE_2871 structures, the interactions between monomers in the asymmetric unit are sufficient to allow crystal formation, but are unlikely to be sufficient to form a stable dimer or higher oligomer in solution. Thus, unlike most known intradiol dioxygenases, which form a dimeric (44) or higher multimeric quaternary structure (41), SACTE_2871 appears to be monomeric.
FIGURE 4.
Comparison of the active sites of catechol 1,2-dioxygenase from R. opacus 1CP (PDB code 3HHY in brown) and SACTE_2871. The extensive N-terminal dimerization domain and α-helix (highlighted with a dashed box) that defines part of the active site pocket in catechol 1,2-dioxygenase are absent in SACTE_2871. The iron center of SACTE_2871 is exposed to solvent and lacks the defined active site pocket that is observed in other dioxygenases. The active site iron is exposed to the solvent along a flat surface.
In addition to the loss of the N-terminal dimerization domain, SACTE_2871 also lacks an α-helix (Fig. 4) that provides residue-specific contacts with bound substrate in other structurally related dioxygenases. Indeed, mutation to residues provided by this helix in the catechol 1,2-dioxygenase IsoB from Acinetobacter radioresistens LMG S13 alter substrate specificity (48). A structural alignment of SACTE_2871 with catechol 1,2-dioxygenase from R. opacus 1CP, the closest sequence homologue, (PDB code 3HHY, Z-score 21, root mean square deviation 1.8 Å) calculated using Dali (49) illustrates the location and potential consequences of the missing dimerization domain and α-helix.
The absence of the dimerization domain and α-helix places the catalytic iron center at the surface of a solvent-exposed depression instead of being deeply buried in an active site pocket observed in all other dioxygenases (Figs. 3 and 4). To illustrate the accessibility of the SACTE_2871 active site, catechol was docked to the iron center based on the structural alignment of SACTE_2871 with catechol 1,2-dioxygenase bound to catechol (PDB code 3HHY). Compared with the bound catechol in the R. opacus structure, which is enclosed by the dimerization domain and an α-helix, the catechol docked into the SACTE_2871 active site is exposed to solvent, which is consistent with the ability of SACTE_2871 to react with 3,4-dihydroxyphenyl compounds such as caffeoyl-CoA (Fig. 4).
SACTE_2871 Iron Center
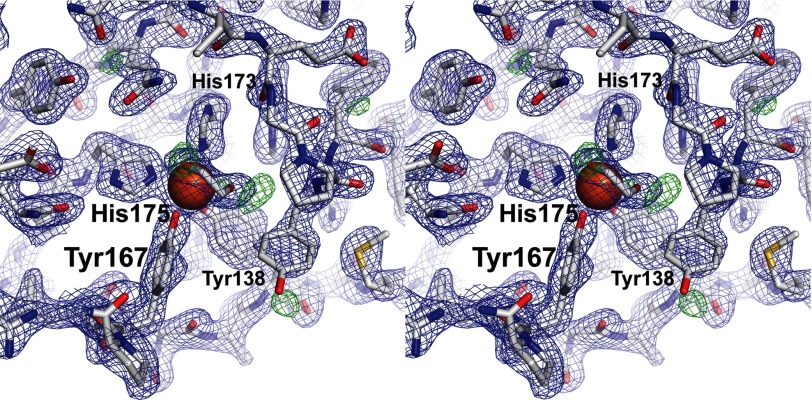
Intradiol dioxygenases coordinate mononuclear ferric ions using four conserved residues (41). In SACTE_2871, these are Tyr138, Tyr167, His173, and His175. The two structures of the dioxygenase domain of SACTE_2871 determined in this work were obtained from crystals grown with PEG3350 as the precipitant, but at different pH values. The only differences in the two structures are observed in residues near the iron center. In the SACTE_2871 structure (determined using crystals grown at pH 5.5), Tyr138, Tyr167, His173, and His175 coordinate the iron atom (Figs. 3 and 5). The bond distances and coordination geometry are indistinguishable from catechol 1,2-dioxygenase from R. opacus (PDB code 3HHY). Furthermore, in the SACTE_2871 structure, both monomers in the asymmetric unit have a cryoprotectant-derived 1,2-ethanediol bound to the iron in a bidentate geometry (Fig. 5).
FIGURE 5.
Detailed view of the iron center in SACTE_2871. Stereo view of the SACTE_2871 2Fo − Fc map contoured at 1.5 σ (blue) and the Fo − Fc map is contoured at 3.0 σ (green). A 1,2-ethanediol molecule interacts with the bound iron in a bidentate fashion. The iron coordinating residues are labeled.
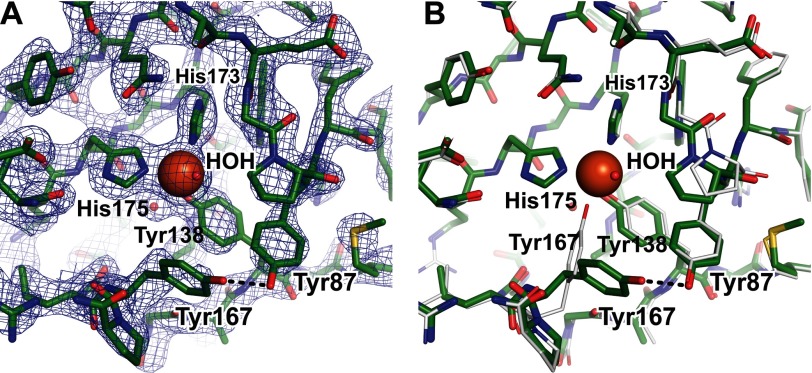
In the SACTE_2871cc structure, the iron was coordinated by only Tyr138, His173, and His175 (Fig. 6A), whereas Tyr167 had rotated to place the tyrosyl OH into hydrogen bonding distance with Tyr87. Although an identical amount of 1,2-ethanediol was used as a cryoprotectant for the crystals of SACTE_2871cc (grown at pH 7) and SACTE_2871 (grown at pH 5.5), there was no electron density corresponding to an endogenous molecule bound to Fe(III) in SACTE_2871cc. The displacement of an active site tyrosine is coordinated with substrate binding during the course of the intradiol dioxygenase reaction (50). Residual difference electron density in the active site suggested that Tyr167 might also be present in the iron-bound conformation, but with an occupancy too low to model with confidence. In both SACTE_2871 structures, Tyr167 had elevated B-factors when compared with Tyr138, His173, and His175, giving further evidence for the conformational flexibility of Tyr167.
FIGURE 6.
Differences between the two SACTE_2871 structures. A, 2Fo − Fc map contoured at 1.5σ of the SACTE_2871cc iron center. Unlike the SACTE_2871 structure, a water molecule is bound to the iron center and Tyr167 no longer coordinates the iron but instead hydrogen bonds with Tyr87. B, an alignment of the SACTE_2871 (gray) and SACTE_2871cc (green) structures is shown in the same orientation as in panel A and Fig. 4. The movement of Tyr167 is the only significant difference in the active site.
A single water was also bound to the iron in three of the four monomers of the SACTE_2871cc structure approximately opposite to the position where Tyr167 was bound to iron in the SACTE_2871 structure (Fig. 6B). The Fe-O distances were 1.9, 2.4, and 2.1 Å for the A, B, and C monomers, respectively. There was no density assignable to a bound water in the D monomer.
Solution Structure of SACTE_2871
The open access to the active site observed in SACTE_2871 is different from most dioxygenases. Consequently, we considered whether the CBM 5/12 domain might provide an oligomerization interface or otherwise restrict access of small molecules to the active site. Small angle x-ray scattering (SAXS) was used to investigate these possibilities, and the results are summarized in Table 2 and Fig. 7.
TABLE 2.
SAXS summary
| Wavelength (Å) | 1.03 |
| Signal/noise | ∼99% |
| Molecular mass (kDa) | 20 |
| Rg (Å) | 26 |
| Rg (from P(r)) | 26.6 |
| Dmax (Å) | 85 − 100 |
The extracted global experimental parameters from the SAXS analysis indicated a molecular mass of 20 kDa (26 kDa expected mass of a monomer), a radius of gyration (Rg) of 26 Å, and a maximum molecular dimension (Dmax) between 85 < Dmax < 100 Å. For comparison, the sum of Dmax from the dioxygenase and CBM domains was 66 Å. A Kratky plot of the data shows evidence of flexibility (Fig. 7, A–C). The larger Dmax and indicators of flexibility suggests that the dioxygenase and CBM domains behave as separate domains connected by a flexible linker rather than a compact globular complex.
The shape generated from the SAXS curve represent an average of all conformations in solution (Fig. 7D). At the widest end, the shape has sufficient volume to accommodate the dioxygenase domain lengthwise (green schematic). At the opposite end, the shape narrowed and only had sufficient volume to accommodate a homology model of SACTE_2871CBM (red schematic) but not the dioxygenase domain.
To better understand the ensemble of conformations possible in solution, we constructed a full-length model for SACTE_2871 from a flexible N-terminal sequence, the dioxygenase domain (x-ray coordinates from PDB 4ILT), another flexible linker sequence, and a homology model for the CBM domain produced using PDB code 1ED7 as the template. This molecular construct was used to calculate a large ensemble of possible conformations that matched the SAXS shape. The individual conformation that best matched the SAXS data had a χ of 1.82 (Fig. 7D). A further improvement in fit was attained by including 3 conformations (χ of 0.6) with a fractional contribution of 44, 29, and 27% and the center to center distance between domains were 53, 39, and 45 Å, respectively (Fig. 7E). The average distance between domains was thus 46 Å, and is in agreement with the position of a shoulder in the P(r) function. Inclusion of additional conformations to this minimal ensemble did not substantially improve the χ value. The improvement in fit obtained by including an ensemble supports the conclusion that the two domains of SACTE_2871 are flexibly linked in solution and so can adopt slightly different configurations.
Catalytic Studies
Given the unique features of the dioxygenase domain revealed by the crystal structure, and the likelihood that the enzyme has an exposed active site when the enzyme is in solution, we examined catalytic reactions with a series of aromatic compounds that are either observed during lignin biosynthesis or are structurally related (Scheme 2). The steady-state kinetics results from assays utilizing SACTE_2871 are presented in Table 3. In summary, SACTE_2871 reacted with all of the simple catechyl substrates listed, including catechol (1), 3,4-dihydroxybenzoate (2), caffeate (3), and 5-OH-ferulate (5). Dioxygenase activity was confirmed by mass spectrometry by comparison of 10 (m/z = 463.56) and the substituted cis-muconic acid product (m/z = 479.56) (Scheme 1). Ferulate (4) was not a dioxygenase substrate, which is consistent with the presence of a blocking O-methylation on the catechyl group. SACTE_2871 reacted with gallic acid (6) as well as its propyl (7), butyl (8), and octyl (9) esters. The gallic acid esters demonstrated the ability of SACTE_2871 to react with aromatic catechols with extended ring substituents including 3,4,5-dihydroxyphenyl units. These ester compounds also lack the charged carboxylic acid group that is important for stabilizing the bound substrate in protocatechuate 3,4-dioxygenase (i.e. 3,4-dihydroxybenzoate). The kcat/Km values for 1-4, 6, and 7 were similar, with values in the range of 0.14 to 0.63 min μm−1. Due to the low solubility of 8 and 9, further steady-state kinetics analyses were not performed with these compounds.
SCHEME 2.
TABLE 3.
Kinetic parameters
| Compound | Name | Km | Vmax | Vmax | kcat | kcat/Km |
|---|---|---|---|---|---|---|
| μm | μm min−1 | % | min−1 | min−1 μm−1 | ||
| 1 | Catechol | 197 | 185 | 100 | 37 | 0.19 |
| 2 | Protochatechuate | 24 | 23 | 12 | 4.6 | 0.19 |
| 3 | Caffeate | 130 | 215 | 116 | 43 | 0.33 |
| 4 | Ferulate | NRa | NR | NR | NR | NR |
| 5 | 5-OH-ferulate | 41 | 45 | 24 | 8.9 | 0.22 |
| 6 | Gallic acid | 148 | 102 | 55 | 20 | 0.14 |
| 7 | Propyl gallate | 33 | 104 | 56 | 21 | 0.63 |
| 10 | Caffeoyl-CoA | 0.005 | 24.5 | 14 | 4.9 | 980 |
| 11 | Rosmarinic acid | 0.04 | 52.7 | 28 | 10.5 | 263 |
a NR, no reaction.
Caffeoyl-CoA (10) was enzymatically synthesized using Nt4CL1 and also found to be a substrate for SACTE_2871. Caffeoyl-CoA is yellow-colored, with an absorption maximum at 346 nm in a 50:50 methanol:water solution (24) and exhibits a shift to ∼350 nm when dissolved in 100 mm phosphate buffer, pH 7.0. The ∼350 nm absorption band disappeared with an isosbestic point at ∼320 nm after SACTE_2871 was added (Fig. 2B). Caffeoyl-CoA had kcat/Km = 980 min μm−1, the highest value among the substrates tested. These results establish the capacity of SACTE_2871 to dioxygenate, and thus destroy, several key early intermediates needed for lignin biosynthesis. Rosmarinic acid (11), a natural product from plants in the family Lamiaceae such as rosemary, sage, basil and other aromatic plants was also a substrate with kcat/Km = 263 min μm−1.
SACTE_2871CBM
CBMs are commonly associated with cellulases or other carbohydrate-active enzymes (15), so the potential role(s) of SACTE_2871CBM in the function of a dioxygenase domain was of interest, particularly as steady-state catalysis with caffeoyl-CoA indicated no contribution for binding the kinetically preferred substrate (data not shown). SACTE_2871CBM is annotated as a member of the CBM 5/12 family, which has been divided into three subgroups with binding to cellulose or chitin experimentally established for some representatives (51). Among the structurally characterized CBM family 5/12 family members, SACTE_2871CBM has the highest identity and similarity (58 and 70%, respectively) with the chitin-binding domain of chitinase A1 from B. circulans WL-12, designated here as ChBDChiA1 (PDB ID 1ED7 (39)).
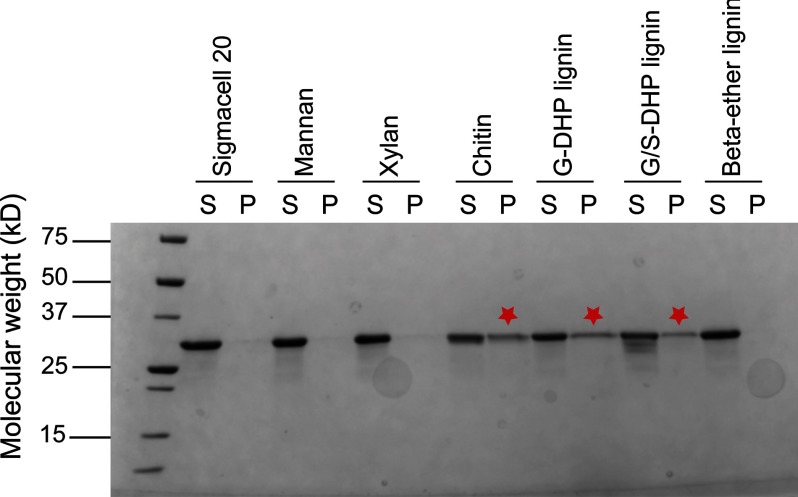
The binding affinity of SACTE_2871CBM was determined with insoluble substrates using a pull-down assay format. For these studies, comparisons of the binding of SACTE_2871 (including the CBM 5/12) and SACTE_2871cc (lacking the CBM 5/12) were made. SACTE_2871 was bound to synthetic G-DHP and G/S-DHP lignin (26) and to chitin. In contrast, SACTE_2871 showed no appreciable binding to cellulose, mannan, or xylan, or to a synthetic lignin that contained only β-ether linkages between guaiacyl groups (Fig. 8). Furthermore, SACTE_2871cc showed no binding to any of the insoluble materials tested (data not shown). These results suggest that SACTE_2871CBM may have a role in targeting the dioxygenase domain to lignin surfaces.
FIGURE 8.
Binding affinity of CBM 5/12 from SACTE_2871 with insoluble polysaccharides. The binding affinity of SACTE_2871CBM was tested on several substrates including Sigmacell 20, mannan, xylan, chitin, G-DHP lignin, G/S-DHP lignin, and β-ether-linked lignin model. SACTE_2871CBM was observed to bind to G-DHP lignin, chitin, and to a lesser extent to G/S-DHP lignin. No binding was observed with cellulose, xylan, mannan, or β-ether linked lignin.
DISCUSSION
The combination of a glycoside hydrolase domain with a CBM is frequently observed in cellulases, hemicellulases, and chitinases (15), and in some copper-containing polysaccharide monooxygenases (52–54). In these enzymes, the presence of the CBM helps to localize the catalytic domain to the surface of the insoluble substrate and so promotes catalysis. Here we provide the first characterization of SACTE_2871, a secreted intradiol dioxygenase that binds synthetic lignin polymers. BLAST searching indicates that homologs containing both a dioxygenase domain and a CBM are present in many Streptomyces, including some considered to be cellulolytic. Furthermore, although some distantly related SACTE_2871 homologs are found in cellulolytic fungi such as Phanerochaete carnosa and Aspergillus niger, these homologs lack the attached CBM domain observed in SACTE_2871. Thus SACTE_2871 may represent a new class of multidomain dioxygenases that facilitate invasive microbial attack on living plant biomass.
Structure of the Dioxygenase Domain
The SACTE_2871 dioxygenase domain adopts a β-sandwich fold that is similar to other known dioxygenases (Fig. 3). The iron-coordinating residues are also conserved and Fe(III) is found in the active site (Figs. 3 and 4). However, SACTE_2871 lacks the N-terminal dimerization domain and an active site α-helix observed in other dioxygenases, which gives rise to a unique monomeric configuration with a solvent-exposed active site pocket (Fig. 4). As a consequence of this open active site architecture, SACTE_2871 lacks distal residues that could provide stabilizing hydrogen bonding or hydrophobic packing interactions with para substituents on the aromatic ring of substrates. For example, protocatechuate 3,4-dioxygenase has a stabilizing electrostatic interaction between Arg457 and the carboxylate group of 3,4-dihydroxybenzoate (2), the highly preferred substrate (55). Moreover, the active site pocket of catechol 1,2-dioxygenase is similar in size to that of protocatechuate 3,4-dioxygenase but is more hydrophobic as it is composed of mostly Leu, Ile, Pro, and Ala residues (44). These types of specific interactions with substrates are not possible in SACTE_2871, allowing reaction with substrates containing large substituents extending away from the aromatic ring, such as caffeoyl-CoA.
Due to the inherent flexibility of the linker between catalytic domains and CBMs and the confounding effect of disorder on the production of diffraction-quality protein crystals, the majority of known cellulase structures consist solely of the catalytic domain. With SACTE_2871, the dynamic linker connecting the dioxygenase domain to the CBM and the unstructured N-terminal region apparently prevented crystallization. However, when flexible linker regions of the enzyme were cut by a slow proteolytic event during the crystallization trials, the truncated SACTE_2871 formed diffraction quality crystals. Likewise, the truncated SACTE_2871cc produced by molecular cloning gave diffracting crystals albeit under different crystallization conditions, yielding two structures of the dioxygenase domain differing only in the active site.
Dioxygenase Active Site
Examination of the two structures revealed different positions for Tyr167, a conserved iron ligand. This residue is important in the dioxygenase mechanism, as it is displaced from the iron upon substrate binding and is rebound to iron upon product release (50, 56). The displacement is proposed to assist in deprotonation of the incoming catechyl OH by the leaving tyrosyl O−, thus maintaining active site charge balance as well as promoting binding of the substrate to iron. In the SACTE_2871cc structure, Tyr167 is predominantly present in the unbound configuration, even though the enzyme is in a substrate-free state. The unbound configuration is stabilized by the formation of a hydrogen bond between the OH atoms of Tyr167 and Tyr87. A similar hydrogen bonding interaction stabilizing the unbound configuration is observed in the substrate-bound forms of other dioxygenases (29, 56), but this residue pairing is not strictly conserved. The iron atom in SACTE_2871cc is also bound by a water molecule. Given that the crystallization buffer was pH 7 and the likely pKa of water bound to Fe(III) is ∼5.5, it is likely that stronger bonding provided by hydroxide could weaken the binding of Tyr167 and thus increase its fraction in the unbound configuration.
Structure in Solution
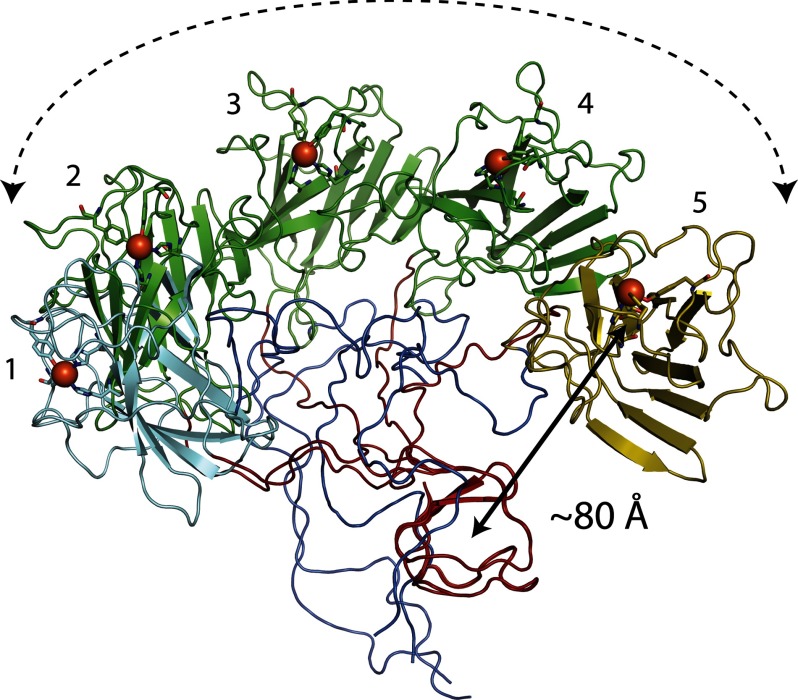
The monomeric state indicated by the crystal structure was confirmed in solution by use of SAXS. These studies revealed that SACTE_2871 preferentially assumed an extended configuration, further supporting the conclusion that SACTE_2871CBM does not directly interact with the dioxygenase domain. Consequently, the dioxygenase active site is solvent-exposed in solution. These findings suggest the possibility that the CBM 5/12 domain serves to localize the dioxygenase domain to a surface, as is observed with fusions of CBM and cellulase domains. Fig. 9 shows the potential spatial range of the three major conformers used to fit the SAXS scattering profile assuming a fixed position of the CBM domain and inherent flexibility of the linker region. Assuming these configurations extend from a single point on a surface, a single molecule of SACTE_2871cc could potentially sample a hemisphere of ∼106 Å3 (0.1 picoliter) as its effective volume for catalysis.
FIGURE 9.
Visualization of the ability of surface-bound SACTE_2871 to sweep a hemispherical volume with radius of ∼80 Å. The CBM 5/12 domains (red) from five configurations are overlaid. The 27% SAXS conformer is show as a cyan-colored dioxygenase domain (position 1), whereas the 29% SAXS conformer is shown as a gold-colored dioxygenase domain (position 5). The green-colored dioxygenase domains represent ∼30° transformations of the dioxygenase from position 1 to 2 (44% SAXS conformer), and then to positions 3–5.
Catalytic Function
Catalytic assays showed that SACTE_2871 reacts with many different catechyl substrates including lignin biosynthetic precursors with side-chain substituents meta-para to the phenolic hydroxyls on the aromatic ring. Thus the solvent-exposed active site is capable of hydrolyzing all compounds listed in Table 3 and Scheme 2 except 4, which is blocked from dioxygenation by the presence of the methoxy group. When compared with catechol (1), protocatechuate (2), and gallate (6), the other substrates shown are likely too large to fit in the confined active sites of most intradiol dioxygenases (Fig. 4). The alteration in active site architecture allows SACTE_2871 to react with a broad spectrum of substituted catechols, including aromatic acyl-CoAs and possibly other esters (shikimate or quinate) that are generated during the biosynthesis of lignin. Dioxygenation of 5-OH ferulate (5) is likely supported by the presence of an ∼6 Å deep spherical cavity lined by residues Met83, Glu84, Gly85, Arg170, Gln189, and Leu203, with surface-exposed Met83 and Leu203 particularly well positioned to accommodate the presence of the methoxy group. There is also a small cavity with a depth of ∼5 Å adjacent to the iron center that would provide a convenient position for O2 binding prior to interaction with substrate. This cavity is lined by residues Glu84, Pro86, Tyr87, Trp130, and His175. NE2 of Trp130 provides the surface of this cavity directly opposite of the iron atom.
One consequence of the open active site of SACTE_2871 is that no residues are available to interact with para substitutions on the bound substrate. The lack of additional coordinating residues may account for the similarity in Kcat/Km observed for most of the compounds tested (Table 3, Scheme 2) as only the catechol functional group productively interacts with the enzyme. In this regard, it is noted that SACTE_2871 has a turnover number for 1 that is only ∼5% of that observed for catechol dioxygenase reacting with its preferred substrate (29). However, the two more complex substrates, caffeoyl-CoA (10) and rosmarinic acid (11), have kcat/Km parameters that are much closer to those typically observed for intradiol dioxygenases. These parameters also provide strong support for the assignment of SACTE_2871 as a caffeoyl-CoA dioxygenase.
Significant research is now underway to introduce hydrolyzable ester linkages into lignin as a replacement for covalent β-ether linkages (57–59). This change makes lignin easier to remove from plant biomass by chemical pretreatments (59). Research on ester-linked lignin has been inspired by the observation that monolignol conjugates (esters) such as monolignol p-coumarates and monolignol p-hydroxybenzoates are already incorporated into the lignins of some plants. Other phenolic monomers, including various catechols, are also compatible with lignification suggesting that there may be viable approaches to modifying the lignin other than via monolignol ferulates (58). Rosmarinic acid (11) is one such promising “alternative lignin monomer” (Scheme 2, ester of 3,4-dihydroxyphenyl-lactate and caffeate). It is found naturally in Lamiaceae such as rosemary, basil, sage, and other aromatic plants but, as far as is known, it is not used as a monomer for lignification in these plants. Recent research has shown that 11 can, however, be incorporated into in vitro synthesized plant cell walls as evidenced by the diagnostic appearance of benzodioxane substructures in the lignin (26). The in vitro lignified cell walls also exhibit enhanced properties for removal of lignin and saccharification of the remaining polysaccharides, suggesting lignification incorporating rosmarinate may have utility in improving biomass processing to biofuels and many other useful materials (26). However, Table 2 shows that 11 is also an effective substrate for SACTE_2871, raising the question of whether naturally occurring microbes might already be well equipped to attack transgenic biomass crops enhanced for biosynthesis of 11 and other catechyl monomers.
SACTE_2871CBM, a Lignin Binding Module?
Insoluble substrate pull-down assays showed that SACTE_2871CBM could bind to the synthetic G-DHP and G/S-DHP lignin compounds (and also to chitin), but not to other polysaccharides (Fig. 8). Given that the SACTE_2871 dioxygenase domain can destroy caffeate, 5-OH-ferulate, and caffeoyl-CoA, three key lignin precursors, we propose that SACTE_2871CBM localizes the enzyme to lignin surfaces, thus helping to position the enzyme for interception of potential biosynthetic intermediates that are poised to be added to the nascent lignin polymer. It is also intriguing that the SACTE_2871CBM might provide a specific interaction with G-DHP lignin. G-DHP is a guaiacyl-type polymer representative of the lignin commonly found in gymnosperm plants such as pine trees (11). In contrast, G/S-DHP is a mixed composition guaiacyl/syringyl-type polymer that is more typical of the lignin found in angiosperm plants.
The question of whether SACTE_2871CBM has been evolutionarily specialized to interact with the predominant form of lignin in the pinewood being attacked by the Sirex/microbe symbiotic community warrants further consideration. The CBM 5/12 family has three major subclades, with functional properties of some members from each clade partially elucidated. In many CBM families, solvent-exposed Trp, Tyr, Phe, and His residues adsorb to the repetitive surface present in chitin or other crystalline polysaccharides. For example, the CBM 5/12 domain of endoglucanase Z from Erwinia chrysanthemi (PDB code 1AIW (60)) has a planar, surface-exposed arrangement of 2 Trp and 1 Tyr residues that span ∼25 Å and interacts with cellulose. Although the three synthetic lignins tested here are chemically homogeneous, they adopt an irregular structure distinct from crystalline polysaccharides. This suggests that SACTE_2871CBM may interact with lignin through an alternative binding mode.
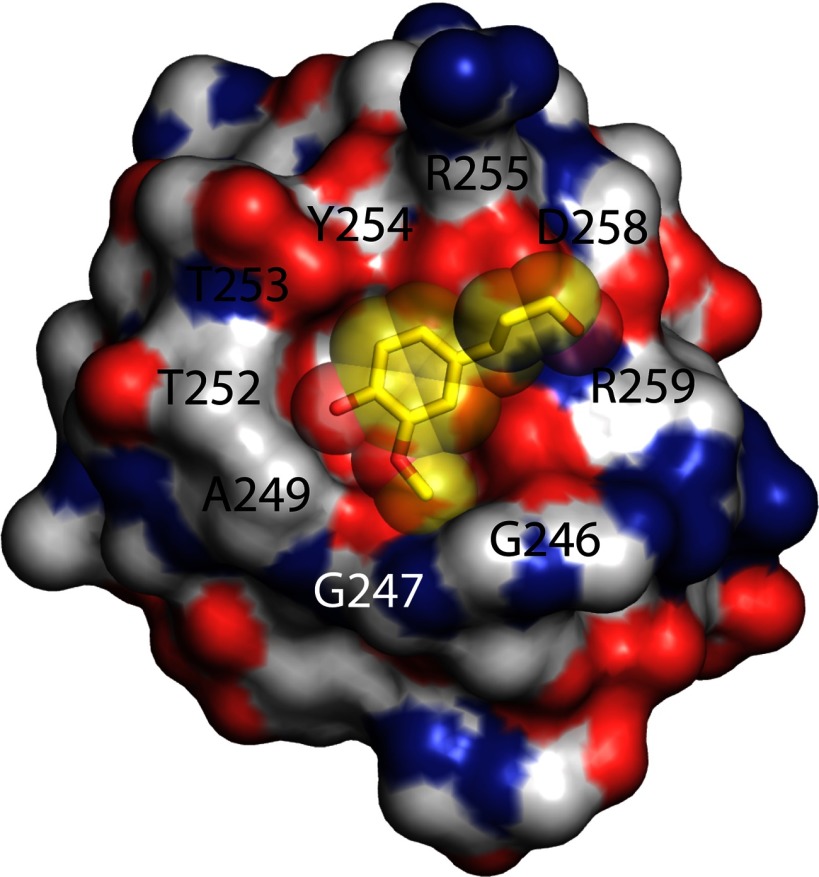
The possibilities for alternative binding modes in the CBM 5/12 family are supported by the structure of another CBM 5/12, ChBDChiA1, where all aromatic residues are buried within the protein and not available for binding to surfaces (39). Consequently, Ikegami et al. (39) suggested that non-aromatic surface residues might provide hydrophobic contacts for binding ChBDChiA1 to crystalline chitin. Alignment of SACTE_2871CBM and ChBDChiA1 shows a high level of sequence identity and further shows that the aromatic residues are conserved in identity and position. Thus it is unlikely that SACTE_2871CBM will have solvent-exposed aromatic residues. However, ChBDChiA1 and SACTE_2871CBM have a considerably different complement of residues that form a ring around one surface of the protein that surrounds a conserved, buried Tyr residue (Fig. 10). Thus, residues 655AWQVNTA-Tyr662-TAGQL667 forms a distinct surface in the chitin-binding enzyme relative to 246GWAAGTT-Tyr254-RAGDR259 in SACTE_2871. Fig. 10 also shows that the surface given by these residues in SACTE_2871 is predicted to form a pocket that is overall highly complementary to the shape of coniferyl alcohol.
FIGURE 10.
A unique surface feature predicted for SACTE_2871CBM by homology modeling with ChBDChiA. The surface ring defined by residues Gly246-Gly247-W-A249AGTT-Tyr254-RAGD-Arg259 surrounding buried Tyr254 that may provide pocket for interaction with lignin constituents. Docked coniferyl alcohol is shown as yellow sticks and a transparent surface.
Conclusion
This study provides biochemical and structural characterization of SACTE_2871, an enzyme from the highly cellulolytic Streptomyces sp. SirexAA-E. The results show that the enzyme is a novel hybrid of CBM and dioxygenase domains with capacity to bind to lignin and dioxygenate caffeoyl-CoA, which is an important early substrate in the lignin biosynthetic pathway. By secreting an enzyme with this unique domain structure and reactivity, SirexAA-E may contribute to the invasive nature of the S. noctilio microbial community symbiosis by interfering with the ability of the plant to protect itself by de novo lignin biosynthesis.
Acknowledgments
We thank the Dr. Craig A. Bingman (University of Wisconsin Center for Eukaryotic Structural Genomics) for access to crystallization robotics, Grzegory Sabat (Biotechnology Center, University of Wisconsin-Madison) for assistance with mass spectrometry, Dr. John Ralph and Dr. Yuki Tobimatsu (Great Lakes Bioenergy Research Center, University of Wisconsin) for gifts of synthetic lignins and 5-OH-ferulate, and Dr. Curtis Wilkerson and Saunia Withers (Great Lakes Bioenergy Research Center, Michigan State University) for the gift of the caffeoyl-CoA synthesis enzyme Nt4CL1. We also thank Dr. Ralph for many stimulating discussions on the complexities of lignin. Use of the Advanced Photon Source was supported by the United States Department of Energy, Basic Energy Sciences, Office of Science, under contract number W-31-109-ENG-38. Use of the Life Science Collaborative Access Team at the Advanced Photon Source was supported by the College of Agricultural and Life Sciences, Department of Biochemistry, the Graduate School of the University of Wisconsin, the Michigan Economic Development Corporation, and Michigan Technology Tri-Corridor Grant 085P1000817). X-ray scattering studies at the SIBYLS was supported by DOE program Integrated Diffraction Analysis Technologies (IDAT-DE-AC02-05CH11231).
This work was supported by the Department of Energy Great Lakes Bioenergy Research Center Office of Science Grant DE-FC02-07ER64494.
The atomic coordinates and structure factors (codes 4ILT and 4ILV) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- SACTE_2871
- two domain enzyme from Streptomyces sp. SirexAA-E encoded by the SACTE_2871 gene
- SACTE_2871cc
- dioxygenase domain of SACTE-2871 consisting of residues 77–230
- SACTE_2871CBM
- CBM 5/12 domain of SACTE_2871 consisting of residues 245–291
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PDB
- Protein Data Bank
- SAXS
- small angle x-ray scattering
- CBM
- carbohydrate binding module
- G-DHP
- guiaiacyl synthetic ligin
- G/S-DHP
- guiaiacyl/Syringyl synthetic ligin.
REFERENCES
- 1. Warnecke F., Luginbühl P., Ivanova N., Ghassemian M., Richardson T. H., Stege J. T., Cayouette M., McHardy A. C., Djordjevic G., Aboushadi N., Sorek R., Tringe S. G., Podar M., Martin H. G., Kunin V., Dalevi D., Madejska J., Kirton E., Platt D., Szeto E., Salamov A., Barry K., Mikhailova N., Kyrpides N. C., Matson E. G., Ottesen E. A., Zhang X., Hernández M., Murillo C., Acosta L. G., Rigoutsos I., Tamayo G., Green B. D., Chang C., Rubin E. M., Mathur E. J., Robertson D. E., Hugenholtz P., Leadbetter J. R. (2007) Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450, 560–565 [DOI] [PubMed] [Google Scholar]
- 2. Suen G., Scott J. J., Aylward F. O., Adams S. M., Tringe S. G., Pinto-Tomás A. A., Foster C. E., Pauly M., Weimer P. J., Barry K. W., Goodwin L. A., Bouffard P., Li L., Osterberger J., Harkins T. T., Slater S. C., Donohue T. J., Currie C. R. (2010) An insect herbivore microbiome with high plant biomass-degrading capacity. PLoS Genet. 6, e1001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Takasuka T. E., Book A. J., Lewin G. R., Currie C. R., Fox B. G. (2013) Aerobic deconstruction of cellulosic biomass by an insect-associated Streptomyces. Sci. Rep. 3, 1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scott J. J., Oh D. C., Yuceer M. C., Klepzig K. D., Clardy J., Currie C. R. (2008) Bacterial protection of beetle-fungus mutualism. Science 322, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kukor J. J., Martin M. M. (1983) Acquisition of digestive enzymes by siricid woodwasps from their fungal symbiont. Science 220, 1161–1163 [DOI] [PubMed] [Google Scholar]
- 6. Hoebeke E. R., Haugen D. A., Haack R. A. (2005) Sirex noctilio. Discovery of a Palearctic siricid woodwasp in New York, Newsletter of the Michigan Entomological Society, pp. 24–25, Michigan Entomological Society [Google Scholar]
- 7. Carnegie A. J., Matsuki M., Haugen D. A., Hurley B. P., Ahumada R., Klasmer P., Sun J. H., Iede E. T. (2006) Predicting the potential distribution of Sirex noctilio (Hymenoptera: Siricidae), a significant exotic pest of Pinus plantations. Ann. For. Sci. 63, 119–128 [Google Scholar]
- 8. Adams A. S., Jordan M. S., Adams S. M., Suen G., Goodwin L. A., Davenport K. W., Currie C. R., Raffa K. F. (2011) Cellulose-degrading bacteria associated with the invasive woodwasp Sirex noctilio. ISME J. 5, 1323–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Denness L., McKenna J. F., Segonzac C., Wormit A., Madhou P., Bennett M., Mansfield J., Zipfel C., Hamann T. (2011) Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species- and jasmonic acid-dependent process in Arabidopsis. Plant Physiol. 156, 1364–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gonzales-Vigil E., Bianchetti C. M., Phillips G. N., Jr., Howe G. A. (2011) Adaptive evolution of threonine deaminase in plant defense against insect herbivores. Proc. Natl. Acad. Sci. U.S.A. 108, 5897–5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boerjan W., Ralph J., Baucher M. (2003) Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546 [DOI] [PubMed] [Google Scholar]
- 12. Vanholme R., Demedts B., Morreel K., Ralph J., Boerjan W. (2010) Lignin biosynthesis and structure. Plant Physiol. 153, 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Seidman M. M., Toms A., Wood J. M. (1969) Influence of side-chain substituents on the position of cleavage of the benzene ring by Pseudomonas fluorescens. J. Bacteriol. 97, 1192–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dagley S. (1971) Catabolism of aromatic compounds by micro-organisms. Adv. Microb. Physiol. 6, 1–46 [DOI] [PubMed] [Google Scholar]
- 15. Ohmiya K., Sakka K., Karita S., Kimura T. (1997) Structure of cellulases and their applications. Biotechnol. Genet. Eng. Rev. 14, 365–414 [DOI] [PubMed] [Google Scholar]
- 16. UniProt (2012) Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res. 40, D71–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thompson J. D., Higgins D. G., Gibson T. J. (1994) CLUSTAL W. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011) SignalP 4.0. Discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 19. Blommel P. G., Martin P. A., Wrobel R. L., Steffen E., Fox B. G. (2006) High efficiency single step production of expression plasmids from cDNA clones using the Flexi Vector cloning system. Protein Expr. Purif. 47, 562–570 [DOI] [PubMed] [Google Scholar]
- 20. Blommel P. G., Becker K. J., Duvnjak P., Fox B. G. (2007) Enhanced bacterial protein expression during auto-induction obtained by alteration of lac repressor dosage and medium composition. Biotechnol. Prog. 23, 585–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blommel P. G., Fox B. G. (2007) A combined approach to improving large-scale production of tobacco etch virus protease. Protein. Expr. Purif. 55, 53–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Whittaker J. W., Orville A. M., Lipscomb J. D. (1990) Protocatechuate 3,4-dioxygenase from Brevibacterium fuscum. Methods Enzymol. 188, 82–88 [DOI] [PubMed] [Google Scholar]
- 23. Beuerle T., Pichersky E. (2002) Enzymatic synthesis and purification of aromatic coenzyme a esters. Anal. Biochem. 302, 305–312 [DOI] [PubMed] [Google Scholar]
- 24. Stockigt J., Zenk M. H. (1975) Chemical syntheses and properties of hydroxycinnamoyl coenzyme A derivatives. Zeitschrift Fur Naturforschung C, A J. Biosci. 30, 352–358 [DOI] [PubMed] [Google Scholar]
- 25. Gilkes N. R., Jervis E., Henrissat B., Tekant B., Miller R. C., Jr., Warren R. A., Kilburn D. G. (1992) The adsorption of a bacterial cellulase and its two isolated domains to crystalline cellulose. J. Biol. Chem. 267, 6743–6749 [PubMed] [Google Scholar]
- 26. Tobimatsu Y., Elumalai S., Grabber J. H., Davidson C. L., Pan X., Ralph J. (2012) Hydroxycinnamate conjugates as potential monolignol replacements. In vitro lignification and cell wall studies with rosmarinic acid. ChemSusChem. 5, 676–686 [DOI] [PubMed] [Google Scholar]
- 27. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 28. Stein N. (2008) CHAINSAW. A program for mutating pdb files used as templates in molecular replacement. J. Appl. Crystallogr. 41, 641–643 [Google Scholar]
- 29. Matera I., Ferraroni M., Kolomytseva M., Golovleva L., Scozzafava A., Briganti F. (2010) Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP. Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts. J. Struct. Biol. 170, 548–564 [DOI] [PubMed] [Google Scholar]
- 30. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Painter J., Merritt E. A. (2006) Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 33. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity. All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera. A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 35. Hura G. L., Menon A. L., Hammel M., Rambo R. P., Poole F. L., 2nd, Tsutakawa S. E., Jenney F. E., Jr., Classen S., Frankel K. A., Hopkins R. C., Yang S. J., Scott J. W., Dillard B. D., Adams M. W., Tainer J. A. (2009) Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nat. Methods 6, 606–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Putnam C. D., Hammel M., Hura G. L., Tainer J. A. (2007) X-ray solution scattering (SAXS) combined with crystallography and computation. Defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 40, 191–285 [DOI] [PubMed] [Google Scholar]
- 37. Konarev P. V., Petoukhov M. V., Volkov V. V., Svergun D. I. (2006) ATSAS 2.1, a program package for small-angle scattering data analysis. J. Appl. Crystallogr. 39, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Svergun D. I., Petoukhov M. V., Koch M. H. (2001) Determination of domain structure of proteins from x-ray solution scattering. Biophys. J. 80, 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ikegami T., Okada T., Hashimoto M., Seino S., Watanabe T., Shirakawa M. (2000) Solution structure of the chitin-binding domain of Bacillus circulans WL-12 chitinase A1. J. Biol. Chem. 275, 13654–13661 [DOI] [PubMed] [Google Scholar]
- 40. Pelikan M., Hura G. L., Hammel M. (2009) Structure and flexibility within proteins as identified through small angle x-ray scattering. Gen. Physiol. Biophys. 28, 174–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ohlendorf D. H., Lipscomb J. D., Weber P. C. (1988) Structure and assembly of protocatechuate 3,4-dioxygenase. Nature 336, 403–405 [DOI] [PubMed] [Google Scholar]
- 42. Whittaker J. W., Lipscomb J. D. (1984) Transition state analogs for protocatechuate 3,4-dioxygenase. Spectroscopic and kinetic studies of the binding reactions of ketonized substrate analogs. J. Biol. Chem. 259, 4476–4486 [PubMed] [Google Scholar]
- 43. Whittaker J. W., Lipscomb J. D., Kent T. A., Münck E. (1984) Brevibacterium fuscum protocatechuate 3,4-dioxygenase. Purification, crystallization, and characterization. J. Biol. Chem. 259, 4466–4475 [PubMed] [Google Scholar]
- 44. Vetting M. W., Ohlendorf D. H. (2000) The 1.8-Å crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker. Structure 8, 429–440 [DOI] [PubMed] [Google Scholar]
- 45. Ferraroni M., Solyanikova I. P., Kolomytseva M. P., Scozzafava A., Golovleva L., Briganti F. (2004) Crystal structure of 4-chlorocatechol 1,2-dioxygenase from the chlorophenol-utilizing Gram-positive Rhodococcus opacus 1CP. J. Biol. Chem. 279, 27646–27655 [DOI] [PubMed] [Google Scholar]
- 46. Ferraroni M., Seifert J., Travkin V. M., Thiel M., Kaschabek S., Scozzafava A., Golovleva L., Schlömann M., Briganti F. (2005) Crystal structure of the hydroxyquinol 1,2-dioxygenase from Nocardioides simplex 3E, a key enzyme involved in polychlorinated aromatics biodegradation. J. Biol. Chem. 280, 21144–21154 [DOI] [PubMed] [Google Scholar]
- 47. Ferraroni M., Kolomytseva M. P., Solyanikova I. P., Scozzafava A., Golovleva L. A., Briganti F. (2006) Crystal structure of 3-chlorocatechol 1,2-dioxygenase key enzyme of a new modified ortho-pathway from the Gram-positive Rhodococcus opacus 1CP grown on 2-chlorophenol. J. Mol. Biol. 360, 788–799 [DOI] [PubMed] [Google Scholar]
- 48. Micalella C., Martignon S., Bruno S., Pioselli B., Caglio R., Valetti F., Pessione E., Giunta C., Rizzi M. (2011) X-ray crystallography, mass spectrometry and single crystal microspectrophotometry. A multidisciplinary characterization of catechol 1,2-dioxygenase. Biochim. Biophys. Acta 1814, 817–823 [DOI] [PubMed] [Google Scholar]
- 49. Holm L., Rosenström P. (2010) Dali server. Conservation mapping in 3D. Nucleic Acids Res. 38, W545-W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Orville A. M., Elango N., Lipscomb J. D., Ohlendorf D. H. (1997) Structures of competitive inhibitor complexes of protocatechuate 3,4-dioxygenase. Multiple exogenous ligand binding orientations within the active site. Biochemistry 36, 10039–10051 [DOI] [PubMed] [Google Scholar]
- 51. Akagi K., Watanabe J., Hara M., Kezuka Y., Chikaishi E., Yamaguchi T., Akutsu H., Nonaka T., Watanabe T., Ikegami T. (2006) Identification of the substrate interaction region of the chitin-binding domain of Streptomyces griseus chitinase C. J. Biochem. 139, 483–493 [DOI] [PubMed] [Google Scholar]
- 52. Vaaje-Kolstad G., Westereng B., Horn S. J., Liu Z., Zhai H., Sørlie M., Eijsink V. G. (2010) An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330, 219–222 [DOI] [PubMed] [Google Scholar]
- 53. Forsberg Z., Vaaje-Kolstad G., Westereng B., Bunæs A. C., Stenstrøm Y., MacKenzie A., Sørlie M., Horn S. J., Eijsink V. G. (2011) Cleavage of cellulose by a CBM33 protein. Protein Sci. 20, 1479–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Beeson W. T., Phillips C. M., Cate J. H., Marletta M. A. (2012) Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J. Am. Chem. Soc. 134, 890–892 [DOI] [PubMed] [Google Scholar]
- 55. Brown C. K., Vetting M. W., Earhart C. A., Ohlendorf D. H. (2004) Biophysical analyses of designed and selected mutants of protocatechuate 3,4-dioxygenase1. Annu. Rev. Microbiol. 58, 555–585 [DOI] [PubMed] [Google Scholar]
- 56. Orville A. M., Lipscomb J. D., Ohlendorf D. H. (1997) Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase. Endogenous Fe3+ ligand displacement in response to substrate binding. Biochemistry 36, 10052–10066 [DOI] [PubMed] [Google Scholar]
- 57. Ralph J. (2010) Hydroxycinnamates in lignification. Phytochem. Rev. 9, 65–83 [Google Scholar]
- 58. Vanholme R., Morreel K., Darrah C., Oyarce P., Grabber J. H., Ralph J., Boerjan W. (2012) Metabolic engineering of novel lignin in biomass crops. New Phytol. 196, 978–1000 [DOI] [PubMed] [Google Scholar]
- 59. Grabber J. H., Hatfield R. D., Lu F., Ralph J. (2008) Coniferyl ferulate incorporation into lignin enhances the alkaline delignification and enzymatic degradation of cell walls. Biomacromolecules 9, 2510–2516 [DOI] [PubMed] [Google Scholar]
- 60. Brun E., Moriaud F., Gans P., Blackledge M. J., Barras F., Marion D. (1997) Solution structure of the cellulose-binding domain of the endoglucanase Z secreted by Erwinia chrysanthemi. Biochemistry 36, 16074–16086 [DOI] [PubMed] [Google Scholar]