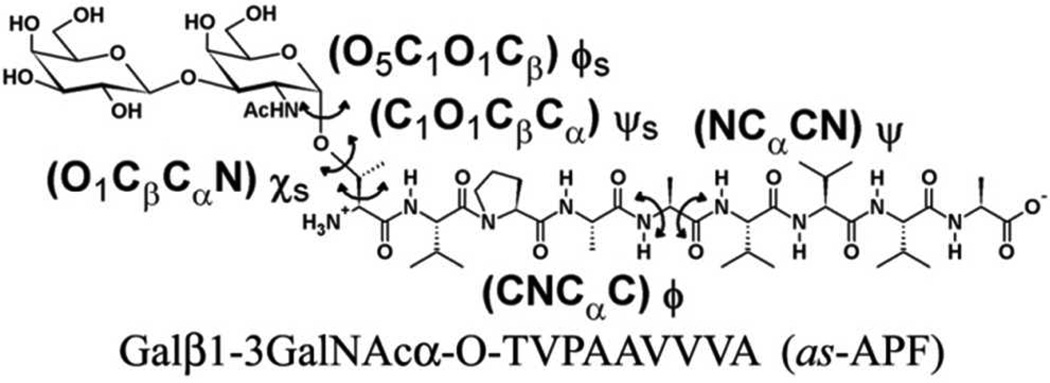
Abstract
The antiproliferative factor (APF) involved in interstitial cystitis is a glycosylated nonapeptide (TVPAAVVVA) containing a sialylated core α-O-disaccharide linked to the N-terminal threonine. The chemical structure of APF was deduced using spectroscopic techniques and confirmed using total synthesis. The synthetic APF provided a platform to study amino acid modifications and their effect on APF activity, based on which a structure-activity relationship (SAR) for APF activity was previously proposed. However, this SAR model could not explain the change in activity associated with minor alterations in the peptide sequence. Presented is computational analysis of 14 APF derivatives to identify structural trends from which a more detailed SAR is obtained. The APF activity is found to be dictated by the close interplay between carbohydrate-peptide and peptide-peptide interactions. The former involves hydrogen bond and hydrophobic interactions and the latter is dominated by hydrophobic interactions. The highly flexible hydrophobic peptide adopts collapsed conformations separated by low energy barriers. APF activity correlates with hydrophobic clustering associated with amino acids 4A, 6V and 8V. Peptide conformations are highly sensitive to single point mutations, which explain the experimental trends. The presented SAR will act as a guide for lead optimization of more potent APF analogues of potential therapeutic utility.
INTRODUCTION
Interstitial cystitis (IC) is a disease of the urinary bladder which is characterized by the thinning of the bladder epithelium. 1, 2 This painful bladder disorder affects approximately 1 million Americans, with evidence suggesting that it occurs eight times more frequently in women than in men.1, 2 While the cause of this disease still remains unknown, urine from IC patients has been shown to contain an antiproliferative factor (APF) that decreases 3H-thymidine incorporation by human bladder epithelial cells.3 Using a total synthesis approach, APF was identified as a nonapeptide (TVPAAVVVA) containing a 2,3-sialylated core 1 α-O-linked disaccharide (Galβ 1-3GalNAc: The Thomsen-Friedenreich antigen) linked to the N-terminal threonine residue (Neu5Acα2–3Galβ 1-3GalNAcα-O-TVPAAVVVA).4 Synthetic APF or its de-sialylated analogue (asialo APF, or as-APF) were shown to inhibit proliferation of normal bladder epithelial cells at nanomolar concentrations4, to increase paracellular permeability and decrease the expression of tight junction proteins.5 These effects were rescued by the D-proline and D-pipecolic acid derivatives of APF, the only two analogues that were shown to inhibit APF action on the normal bladder epithelium.6 APF was further shown to inhibit proliferation of T24 bladder carcinoma cells4, with subsequently proven similar effects on a panel of solid tumor cell lines 7, making APF a fascinating lead candidate for both anti-IC and anticancer drug design. Some insight into the mechanism of APF activity and the associated targets was recently found by the discovery that cytoskeletal-associated protein 4 (CKAP4) was a functional cellular receptor for APF in bladder epithelial cells. 8
While as-APF maintains full potency, further truncation of the resultant disaccharide results in a complete loss of activity.9 In the absence of structural information synthetic analogs of as-APF have been tested for their biological antiproliferative activity to establish structure activity relationships (SAR) in as-APF.4,9,10 Based on these studies, which targeted extensive modifications in the peptide region of APF, it was found that the smallest synthetic analog of APF that maintained full potency was the synthetic glyco-octapeptide Galβ 1-3GalNAcα-O-TVPAAAAA, where the trivaline tail was replaced by three alanine residues.9,10 The availability of biological activity data for the various synthetic analogs of as-APF with differing amino acid sequences sets the groundwork for an extensive study relating the conformational properties of APF and its analogs to the bioactivity of the glycopeptide.
In this study, we performed Hamiltonian replica exchange (HREX) molecular dynamics (MD) simulations on as-APF and selected analogs to gain insights into the structure activity relationships (SAR) in APF. The as-APF derivatives were chosen based on three criterion; (i) the availability of experimental 3J coupling and NOE data for a direct comparison between the experiments and simulations, (ii) differing biological activities (inactive to active) and (iii) differing peptide sequences to develop the SAR model. Greater understanding of the SAR for active APF derivatives will aid in the rational design of APF inhibitors that may be of utility for the treatment of IC.
METHODS
Model compounds
A total of 12 as-APF derivatives and two unglycosylated amino acid sequences were chosen for the HREX MD simulations (Table 1).4, 9, 10 The chosen derivatives including as-APF (compound 1; Scheme 1) represent a selection of as-APF derivatives with differing biological activities. The details of the synthesis of the glycopeptides have been described in detail in earlier studies.4, 9, 10 MD simulations were performed using an equilibration and production strategy as tested earlier for O-linked glycoprotein systems.11 In brief, the simulations were performed with the CHARMM program.12 The CHARMM22 protein force field13 with CMAP (dihedral correction map), 14 the CHARMM carbohydrate force field,15–19 and the modified TIP3P water model20 were used to represent the glycopeptides in solution. Initial configurations corresponding to anti (180°) and gauche+ (60°) conformations were chosen for the H–N–C2–H2 (Acetylamino side chain in carbohydrate) and O1-Cβ-Cα-N (O-linkage) dihedrals respectively, in accordance with NMR studies (Table S1 and Table S2, supporting information (SI) file). The rest of the geometries of the model compounds were generated from the topology information present in the force field. These initial geometries were subjected to a 1000-step steepest descent (SD) minimization followed by an adopted basis Newton–Raphson (ABNR) minimization to a force gradient tolerance of 10−6 kcal/mol/Å.21, 22 The minimized geometries of the glycopeptides were then immersed in a pre-equilibrated cubic water box of size 45 Å × 45 Å × 45 Å. The size of the water box was selected based on the condition that it extended at least 10 Å beyond the non-hydrogen atoms of the glycopep-tides. Water molecules with the oxygen overlapping with the non-hydrogen solute atoms within a distance of 2.8 Å were deleted. For all of the subsequent minimizations and MD simulations, periodic boundary conditions were employed using the CRYSTAL module implemented in the CHARMM program.
Table 1.
APF analogs used in the present study.
| Com. No. | Sequence† | % Activity | NMR data |
|---|---|---|---|
| 1 | Galβ1-3GalNAcα-O-TVPAAVVVA | 100% | yes |
| 2 | Galβ1-3GalNAcα-O-SVPAAVVVA | 0.01% | yes |
| 3 | Galβ1-3GalNAcα-O-SLPAAVVVA | 1% | yes |
| 4 | Galβ1-3GalNAcα-O-TLPAAVVVA | 0% | yes |
| 5 | Galβ1-3GalNAcα-O-TVPAVVVVA | 0% | no |
| 6 | Galβ1-3GalNAcα-O-TVPAAAVVA | --- | --- |
| 7 | Galβ1-3GalNAcα-O-TVPAAVAVA | --- | --- |
| 8 | Galβ1-3GalNAcα-O-TVPAAVVAA | --- | --- |
| 9 | Galβ1-3GalNAcα-O-TVPAAAAAA | 1% | no |
| 10 | Galβ1-3GalNAcα-O-TVPAAVVV | 100% | yes |
| 11 | Galβ1-3GalNAcα-O-TVPAAAAA | 100% | yes |
| 12 | TVPAAVVVA | 0% | no |
| 13 | TVPAAVVV | 0% | yes |
| 14 | Galβ1-3GalNAcα-O-TV(dP)AAVVVA | 0% | no |
Scheme 1.
Structure of as-APF.
System equilibration were initiated with a 50-step SD minimization followed by a 50-step ABNR minimization cycle in which mass-weighted harmonic restraints of 1.0 kcal/mol/Å were applied on the non-hydrogen atoms of the glycopeptides,. This was followed by a 100 ps simulation in the NVT ensemble with the same harmonic restraints to equilibrate the solvent molecules around the glycopeptides. A 200 ps NPT simulation at 1 atm and 298 K followed the NVT simulation, wherein all the previous restraints were removed. In the NPT simulation the center of mass of the glycopeptide was restrained near the origin by using the MMFP module23 in CHARMM using a harmonic restraint of 1.0 kcal/mol/Å applied to the center of mass of the glycopeptides. The electrostatic interactions were treated via the particle mesh Ewald method with a real-space cutoff of 12 Å, a kappa value of 0.34 Å−1, and a sixth-order spline.24 Nonbond interaction lists were updated heuristically out to 16 Å with a force switch smoothing function from 10 to 12 Å used for the Lennard-Jones interactions. 25 The Leapfrog integrator employing an integration time step of 2 fs with the SHAKE algorithm was used to constrain all covalent bonds involving hydrogen atoms.26 The temperature was maintained at 298 K by a Nose–Hoover heat bath with a thermal piston parameter of 2000 kcal mol−1ps2. 27 Constant pressure of 1 atm was controlled using the Langevin piston with a mass calculated using the equation Pmass = integer(system mass/50.0). 28
HREX MD production simulations were performed using the REPDSTR module of a modified version of CHARMM c36a2. 29 The HREX simulations were started from the equilibrated coordinates obtained after the 200 ps unbiased NPT simulation at 1 atm and 298K. The same MMFP restraints used in the NPT runs were utilized in the HREX runs to constrain the glycopeptide at the center of the simulation box. An exchange between neighboring replicas was attempted every 1000 MD steps, and the coordinates were saved every 1 ps. Each replica was simulated for 11.4 ns, thereby amounting to a cumulative simulation time of 91.2 ns (11.4 ns × 8), and the trajectories from the final 10 ns of the first replica (unbiased, ground state replica) were used for subsequent analysis.
A combination of the two-dimensional (2D) dihedral grid-based energy correction map (CMAP) extension of the CHARMM force field and a Saxon-Wood potential were used as the biasing potential across the different replicas.14 Two CMAP biasing potentials were used, corresponding to the φs/ψs and φ/ψ dihedral pairs (Scheme 1) to sample the conformational space of the glycopeptides around the O-glycosidic linkage and the peptide backbone. Additionally, the Saxon-Wood potential, equation 1, was used to enhance the conformational sampling about the χs dihedral (Scheme 1) in the Thr/Ser side chain.
| (1) |
where h = (n × −0.75) kcal/mol, n going from 0 to 7 for replicas 1 to 8, p1 = 0.1, p2 = 0.3 and θref = 90°. The biasing potential CMAPs were obtained using an established protocol for glycopeptides O-linkages which has been reported in detail earlier and successfully applied in studying O-linked glycopeptides.11
NMR 3J coupling constants about the dihedral torsions H-N-C2-H2 (for GalNAc), N-Cα-Cβ-O1 and C-N-Cα-C were calculated from the simulations using the following Karplus equations. 30, 31, 32
| (2) |
| (3) |
| (4) |
RESULTS
Developing a SAR model for conformationally flexible molecules such as APF requires appropriate conformational sampling. NMR experiments provide indirect insight into the conformation of a molecule via information on dihedral angles and close contacts obtained from NMR 3J coupling and NOE data, respectively. In this study we use this information to test the performance of the HREX conformational sampling method. As detailed below, the HREX protocol successfully reproduces the 3J coupling and NOE data for the seven compounds with available NMR data, thus providing confidence in the conformational sampling protocol. The HREX protocol was then extended to the remainder of the model compounds (Table 1), resulting in the data set that allowed for development of a refined SAR model for APF.
3J Coupling Constants
3J coupling constants calculated from HREX simulations by applying the appropriate Karplus equations (eq 2, 3, 4) are summarized in Table S1, S2 and S3 (SI file) for the H-N-C2-H2 (for GalNAc), N-Cα-Cβ-O1 and C-N-Cα-C dihedral torsions respectively. The 3J coupling constants obtained by averaging over the simulations are in agreement with the experimentally observed values for the various dihedrals, indicating that the HREX simulations sample the experimentally observed conformational space. The H-N-C2-H2 dihedral largely samples the anti conformation, which corresponds to the large 3J value of ~10.08 Hz observed experimentally. For all the Thr O-linked carbohydrate systems the N-Cα-Cβ-O1 dihedral predominantly samples the +60°(gauche+) conformation, as has been observed experimentally for model Thr O-linkages.33–35 This conformational rigidity is lost on sub- stituting T with S (2), which also samples the −60°(gauche-) conformation for close to 20 % of the simulation time (Table S2). This loss in rigidity can be related to the removal of the −CH3 side chain on substituting T to S which results in the release of steric strain. Interestingly on substituting the subsequent amino acid SV (2) by SL (3), which amounts to introducing the −CH3 side chain at the 2nd position, the conformational rigidity is regained, with the N-Cα-Cβ-O1 dihedral preferentially sampling the +60°(gauche +) conformation (Table S2). The loss and gain in the rigidity of the N-Cα-Cβ-O1 dihedral with the number of −CH3 side chains matches well with the observed biological activity (Table 1). Substituting T (1) with S (2) resulted in a 4 orders of magnitude loss of activity, going from 100% to 0.01%. While re-substituting a −CH3 group at the 2nd position (SV to SL, 3) to create an “isosteric” substitution, resulted in a 100-fold gain “back” in activity (1%). However adding additional methyls to the system (TV to TL, 4), resulted in a complete loss of activity. These results are indicative of carbohydrate-peptide interactions controlling the observed biological activity. Finally, the 3J coupling constants corresponding to the C-N-Cα-C peptide backbone torsions are summarized in Table S3 of the SI file. The 3J coupling constants are in close agreement with the experimental values; with the average RMS difference with respect to the experimental values (1.3 Hz) being less than the RMS deviation observed in the simulations (2.3 Hz). The only exception to this pattern was the last amino acid wherein the RMS difference of 2.2 Hz was greater than the RMS deviation of 1.5 Hz. The strong agreement with the experimentally observed 3J coupling in the carbohydrate, carbohydrate-peptide linkage and peptide regions highlights the appropriate conformational sampling in the HREX simulations.
ROE (Rotating Frame Nuclear Overhauser Effect) gives insight into the non-covalent interactions in the systems based on the proximity of the interacting magnetic nuclei, in our case the protons. Experimental ROE’s can be compared to close contact probabilities obtained from HREX simulations. ROE interactions in the glycopeptides can be classified into (a) Carbohydrate-Peptide and (b) Peptide-Peptide interactions. All the observed experimental ROE’s are tabulated in Table 2.
Table 2.
NMR ROE signals in the glycopeptide systems.
| Comp. No. | Carbohydrate-Peptide | Peptide-Peptidea | |
|---|---|---|---|
| 1 | GalNAc (−H5)-1T (Hγ) | 4A (NH) – 5A (NH) m | |
| GalNAc (−CH3)-3P (Hδ) | 5A (NH) – 6V (NH) w | ||
| 6V (NH) – 7V (NH) m | |||
| 8V (NH) – 9A (NH) w | |||
| 2 | GalNAc (−CH3)-3P (Hα) | 4A (NH) – 5A (NH) m | |
| GalNAc (−CH3)-3P (Hδ) | 5A (NH) – 6V (NH) m | ||
| 6V (NH) – 7V (NH) m | |||
| 8V (NH) – 9A (NH) m | |||
| 3 | GalNAc (−CH3)-3P (Hα) | 4A (NH) – 5A (NH) w | |
| GalNAc (−CH3)-3P (Hδ) | 5A (NH) – 6V (NH) m | ||
| 6V (NH) – 7V (NH) m | |||
| 8V (NH) – 9A (NH) m | |||
| 4 | GalNAc (−H5)-1T (Hγ) | 4A (NH) – 5A (NH) m | |
| GalNAc (−CH3)-3P (Hδ) | 5A (NH) – 6V (NH) m | ||
| 6V (NH) – 7V (NH) m | |||
| 8V (NH) – 9A (NH) m | |||
| 10 | GalNAc (−H5)-1T (Hγ) | 4A (NH) – 5A (NH) w | |
| GalNAc (−CH3)-3P (Hδ) | 5A (NH) – 6V (NH) w | ||
| 6V (NH) – 7V (NH) w | |||
| 11 | GalNAc (−H5)-1T (Hγ) | 4A (NH) – 5A (NH) m | |
| GalNAc (−CH3)-3P (Hδ) | |||
| GalNAc (−CH3)-2V (Hα) | |||
| 13 | 4A (NH) – 5A (NH) w | ||
| 5A (NH) – 6V (NH) w | |||
| 6V (NH) – 7V (NH) w | |||
The intensity of the peptide-peptide ROE signal is presented in bold.
Carbohydrate-Peptide interactions
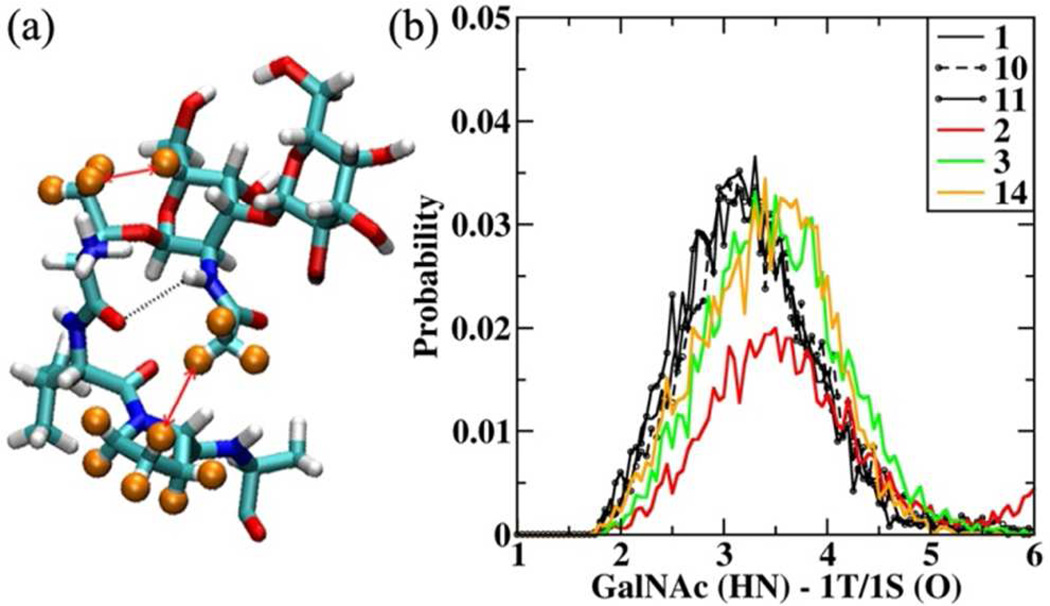
NMR studies show the presence of two medium strength carbo-hydrate-peptide ROE signals (Table 2) in the glycopeptides systems. The first is between the H5 of GalNAc and the −CH3 side chain of 1T in all the T substituted glycopeptides (1, 4 – 11, 14), and the second is between the −CH3 in the N-acetyl group of GalNAc and the Hγ' hydrogen of 3P. It is of interest to note that no ROE signal was observed between H5 of GalNAc and 1S in 2 and 3. An analysis of the simulation of 1 revealed the presence of a strong hydrogen bond (H-bond) between the acetylamino side chain of GalNAc and the amide backbone of 1T/1S, which locks the sugar conformation with respect to the peptide (Figure 1a). Such H-bonds between the acetylamino side chain of GalNAc and the amide backbone have been observed previously in mucin architecture.36, 37 This H-bond between GalNAc and 1T (NH-O) brings the methyl group in the acetylamino (−NHCOCH3) side chain to the proximity of the proline ring, resulting in the GalNAc (−CH3) - 3P (Hδ) ROE signal. In Table 3 the H-bond occupancy corresponding to the GalNAc (HN) – 1T/1S (O) H-bond is presented. The probability distribution associated with this H-bond for all the systems is presented in Figure S1 of the SI file. In Figure 1b we present the H-bond distribution for 2, 3 and 14, which show reduced H-bond occupancy (Table 3), and for compounds 1, 10 and 11, which show 100% activity for direct comparison.
Figure 1.
(a) Representative structure of 1 from HREX simulations showing the carbohydrate unit and the first three amino acids (TVP). The H-bond is shown in black dotted line. The hydrogen’s involved in the ROE signals are identified using orange spheres and the interaction is highlighted using red arrows. (b) GalNAc (HN) − 1T/1S (O) H-bond distribution for 1, 10, 11, 2, 3, 14. Distances presented in Å.
Table 3.
Carbohydrate-peptide and peptide-peptide hydrogen bond occupancies.a
| Comp. No. | Carbohydrate-Peptide H-bond Occupancy |
Peptide-Peptide H-bond Occupancy |
|||
|---|---|---|---|---|---|
| GalNAc (HN) – 1T/1S (O) |
GalNAc (O) – 4A (HN) |
4A (O)- 7V/7A (HN) |
3P (O)- 6V/6A (HN) |
5A/5V (O)- 8V/8A (HN) |
|
| 1 | 0.672 | 0.092 | 0.035 | 0.103 | 0.050 |
| 2 | 0.292 | 0.059 | 0.020 | 0.106 | --- |
| 3 | 0.491 | 0.122 | 0.092 | 0.282 | 0.102 |
| 4 | 0.653 | 0.064 | --- | 0.177 | --- |
| 5 | 0.638 | 0.098 | --- | 0.026 | --- |
| 6 | 0.706 | 0.127 | 0.164 | 0.070 | 0.039 |
| 7 | 0.664 | 0.117 | 0.011 | 0.034 | 0.034 |
| 8 | 0.527 | 0.059 | 0.015 | 0.019 | 0.081 |
| 9* | 0.629 | 0.103 | 0.625 | 0.623 | 0.492 |
| 10 | 0.621 | 0.082 | --- | --- | 0.093 |
| 11* | 0.681 | 0.091 | 0.481 | 0.531 | 0.247 |
| 12 | --- | 0.015 | 0.014 | ||
| 13 | 0.043 | 0.096 | 0.023 | ||
| 14 | 0.520 | 0.465 | --- | --- | ---- |
Occupancies calculated using a H-bond cutoff distance of 3.5 Å.
Additional H-bonds between 3P(O)-7A(HN) (0.589 and 0.436) and 4A(O)-8A(HN) (0.543 and 0.209) found in 9 and 11.
From the probability distribution it is observed that as-APF (1) presents a unimodal distribution centered on 2.9 Å. This distribution gets affected on substituting T (1) with S (2), with the latter exhibiting a bimodal distribution (Figure S1). This bimodal distribution is consistent with the sampling of the −60°(gauche-) conformation for the N-Cα-Cβ-O1 dihedral in 2 (Table S2). However, this distribution becomes unimodal on substituting −CH3 at the 2nd position (SV to SL, 3), albeit the H-bond is elongated in both 2 and 3, with the distribution centered on 3.1 Å. From the occupancy data presented in Table 3 it can be seen that the H-bond occupancy is affected for 2 (0.292), 3 (0.491) and 14 (0.520). From this data it becomes clear that the H-bond is necessary to lock the conformation of the sugar unit with respect to the peptide and may explain the observed activity pattern for the TV (1) versus SV (2) or SL (3) mutations. However the H-bond analysis fails to provide insight into the activity profile observed in the other T variants, starting from TL (4) which showed 0% activity but has the same H-bond profile as 1 (Figure S1).
These results suggest that secondary interactions involving the peptide unit affect the activity. To gain insight into the peptide-peptide interactions, the peptide region of as-APF (1) was closely scrutinized, including comparison with ROE data.
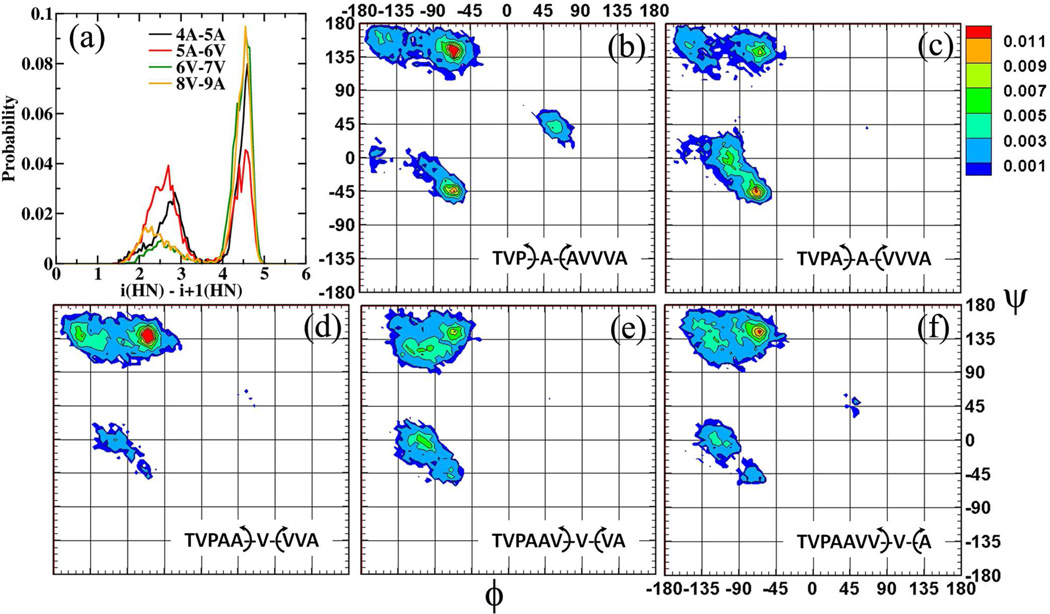
In the standard NMR study of peptides, sequential ROE signals between the amide protons (-NH) on consecutive amino acids are analyzed for indications of bending/kinking in the peptide backbone. For as-APF (1) medium strength signals were observed between 4A-5A and 6V-7V, while weak signals were observed between 5A-6V and 8V-9A. The strengths of these signals are altered to varying degrees for the other as-APF derivatives as tabulated in Table 2. This experimental evidence provides the incentive to analyze these distance distributions to establish a potential structural descriptor of activity. The distance distribution for these amide pairs for 1 is presented in Figure 2a. It is seen that the amide protons pairs 4A-5A remain in close contact for close to 40% of the simulation time (d4A-5A< 3.5 Å), while the 5A-6V pair remains in close contact for close to 57% of the simulation time (d5A-6V < 3.5 Å). The proximity of these particular proton pairs is traced back to the conformations sampled by the underlying peptide backbone. The φ/ψ dihedral distributions associated with 4A, 5A, 6V, 7V and 8V for 1 are also presented in Figure 2. The peptide backbone samples both the a-helical and extended conformations (p and PPII) around the alanines (4A and 5A) while it predominantly samples only the extended conformations around the three valines (6V, 7V, 8V). This observation is in line with the large 3JHNHαcoupling constants (between 7.0 Hz and 8.9 Hz) experimentally observed for the trivaline region.4, 9, 10 Thus, while the alanines also sample the folded α-helical conformations the trivaline tail does not. The presence of the commonly found α-helical AXXXA (X=V in as-APF) motif38 in APF had initially encouraged the hypothesis of a well formed α-helical tail region in APF in the original SAR proposal.9 However, further structural characterization using circular dichroism (CD) spectroscopy and chemical shift data suggested an extended tail like region around the trivalines (6V, 7V, 8V) and a folded region around the alanines (4A and 5A).10
Figure 2.
(a) Distance distribution between the amide proton pairs for 1. φ/ψ dihedral distributions for (b) 4A, (c) 5A, (d) 6V, (e) 7V and (f) 8V for 1. Distances are presented in Å and the dihedral angles are presented in degrees°.
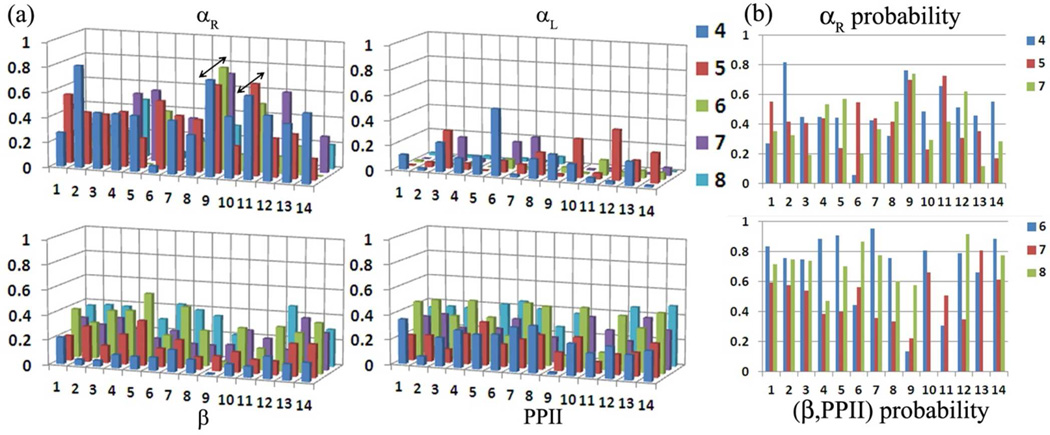
Presented in Figure 3a are the population distributions of the φ/ψ dihedrals in the four commonly observed backbone geometry regions for the amino acids in the tail region 4 to 8 or 9 depending on the number of amino acids for all the systems.39 The same for amino acids at position 2 and 3 (VP) is presented in Figure S2 of the SI file. The amino acids prefer the extended conformation around 2 and 3 (Figure S2). The only exception to this trend is for 3P in 14, wherein the D-proline also samples 50 % a-helical conformation. Other than this the head regions of the glycopeptides (TVP) generally remain only in extended conformations. Variations in the population distribution are observed from 4A onwards (Figure 3a). At the outset it is seen that only 9 (Galβ1–3GalNAcα-O-TVPAAAAAA) and 11 (Galβ1–3GalNAcα-O-TVPAAAAA) exhibit a complete α-helical fold with all the amino acids in positions 4 to 8 sampling the a-helical region. This is also established by the large H-bond occupancies between 4A-7A, 3P-6A and 5A-8A in 9 and 11 (Table 3), which are not seen in the other systems. In all the other cases amino acids at 4, 5 and 7 sampled the α-helical region substantially, while the rest of the amino acids predominantly sample the extended conformations (β-sheet or polyproline II (PPII)). In Figure 3b we present the probability distributions in the αR region for amino acids 4, 5 and 7; and the summed probability in the extended regions (β and PPII) for amino acids 6, 7 and 8 for all the systems.
Figure 3.
(a) Population distribution of the φ/ψ dihedrals in the four commonly observed backbone geometry regions as defined in the Ramachandran surface for amino acids 4 to 8/9. Arrows are used to highlight the formation of an α-helical fold in 9 and 11. (b) Probability distribution in the αR region for amino acids 4, 5, 7 and the summed probability in the extended regions for amino acids 6, 7, 8. Backbone geometry regions are defined as αR (−160° < φ< −20° and −120° < ψ< 50°), αL (20° < φ< 160° and −50° <ψ< 120°), β (−180° < φ< −90° and 110° < ψ< 180°) and PPII (−90° < φ< −20° and 110° < ψ< 180°)
It is observed that in all the systems a valine at position 6 (6V) disrupts the formation of the a-helical fold by predominantly favoring the extended geometries (probability > 0.6). Substituting 6V by 6A results in a reduction in the number of extended conformations (6: 0.4, 9: 0.1 and 11: 0.3). However the remainder of the tail regions (6 to 8/9) do not favor a truly extended β-sheet or PPII geometry, with the peptide backbone sampling both the folded and extended conformations at position 7, irrespective of amino acid (A or V) type. Thus, it would be incorrect to classify the tail region as being in a truly extended geometry conformation. These results are indicative of stronger long range interactions in the peptide region. For example, indications of this long-range effect are seen on the population of the folded geometry at 4A in 2 and 6. In 2 the αR probability increases to 0.8 while the substitution is at position 1 (SVPAAVVVA), while in 6 the αR probability decreases to 0.05 while the substitution is at position 6 (TVPAAAVVA). Analyzing the simulation revealed the formation of various hydrophobic-collapsed structures brought about by the interaction of the side chains. Hence, to identify the structural descriptors of as-APF activity it is important to probe the long-range interactions between the side chains. This is first attempted and described in detail for as-APF (1), while the interactions in all the other systems are compared to as-APF.
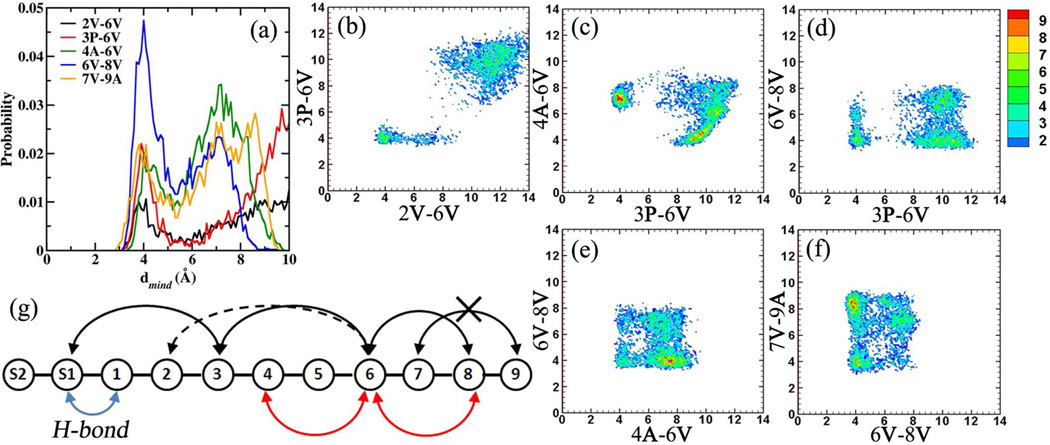
Close contact distances involving the side chains in as-APF (1) were evaluated between all the amino acid side chain pairs separated by one amino acid. The contact probability was evaluated using the minimum distance (mind) between the non-hydrogen atoms in the two respective selections. Five significant close contact (dmind < 6.0 Å) probabilities were observed between the amino acid pairs 2V-6V, 3P-6V, 4A-6V, 6V-8V and 7V-9A, with the distance distributions presented in Figure 4a. It is seen that the 6V-8V pair remians in close contact for a significant portion of the simulation followed by 3P-6V, 4A-6V and 7V-9A, with the 2V-6V pair being in the least contact. Following the identification of these close-contact pairs, two-dimensional (2D) distributions were evaluated between all the contact pairs to identify the occurrence of folded geometries. The analysis of the 2D distributions revealed that the 2V-6V and 3P-6V contact pairs occur simultaneously (Figure 4b), while the 2V-6V or 3P-6V and 4A-6V pairs are not formed simultaneously (Figure S3a/Fig 4c). Interestingly, both 3P-6V and 4A-6V contact pairs can form simultaneously with the 6V-8V contact pair (Figure 4d and 4e), while they do not form in tandem with the 7V-9A contact pair (Fig S3b and Fig S3c). However, the tail region from 6V to 9A favors the simultaneous formation of the 6V-8V and 7V-9A contact pairs (Fig 4f). The occurrence of a few of these close-contact pairs revealed the formation of two well defined folded geometries in as-APF; the first in which the hyrobhobic clustering of 2V-6V, 3P-6V and 6V-8V side chains lead to the formation of folded structures and, alternatively the second in which the hydrophobic clustering of 4A-6V and 6V-8V leads to the formation of folded structures. These situtations are schematically presented in Figure 4g. It is important to note that in all these collapsed geometries the −CH3 side chain of α-GalNAc interacts with the 3P proline ring via hydrophobic interactions, and with the 1T/1S backbone via H-bonding. Working back from the data it was found that the various folded geometries of the peptide backbone could be classified into three distinct regions using the 3P-6V/4A-6V 2D distribution.
Figure 4.
(a) Close contact probabilities observed between the amino acids in as-APF (1). Selected 2D distributions of the significant close contact probabilities for (b) 2V-6V/3P-6V, (c) 3P-6V/4A-6V, (d) 3P-6V/6V-8V, (e) 4A-6V/6V-8V and (f) 6V-8V/7V-9A. (g) Schematic representation of the various interactiosn in as-APF (1). Distances are presented in Å.
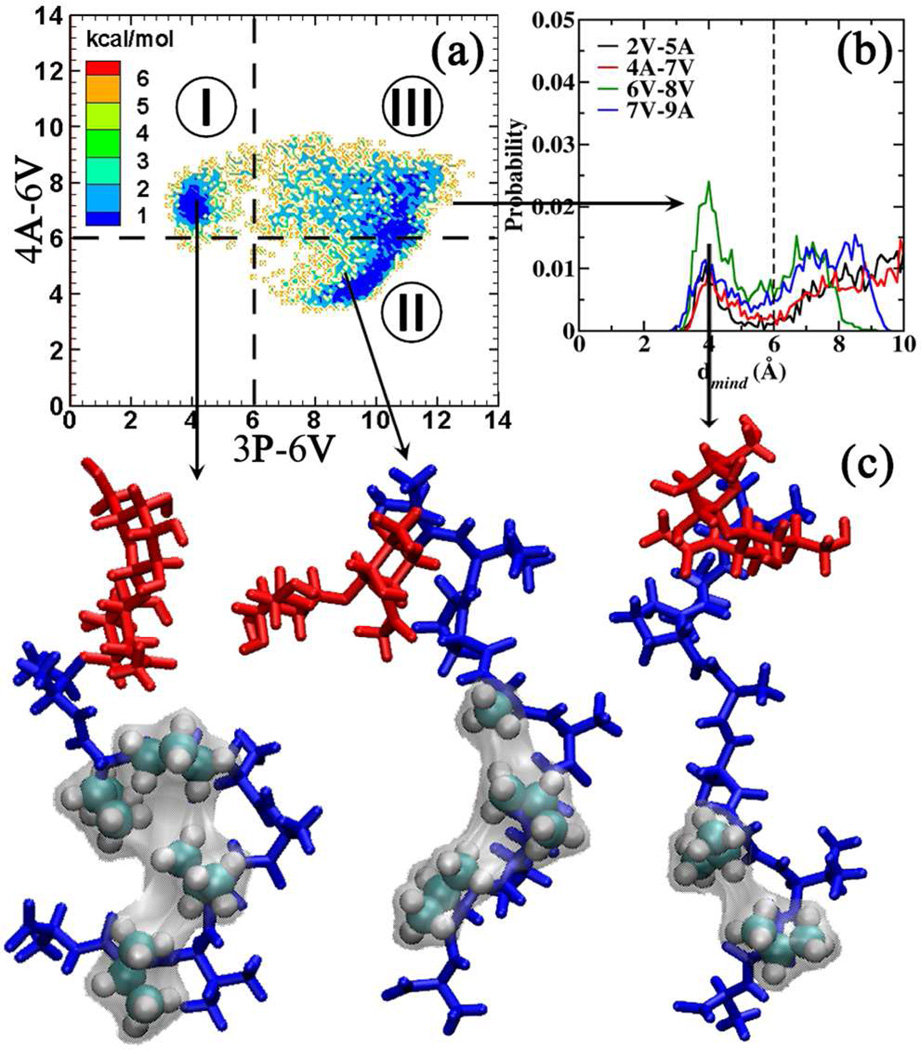
Presented in Figure 5a is the Boltzmann inverted 3P-6V/4A-6V 2D distribution. Inspection reveals a free energy surface that contains two minima separated by a low free energy barrier (< 2 kcal/mol). The free energy surface can be classified into three well-defined regions:
Region I: d3P-6V < 6.0 Å and d4A-6V > 6.0 Å. This region favors the hydrophobic clustering of 2V, 3P, 6V and 8V.
Region II: d3P-6V > 6.0 Å and d4A-6V < 6.0 Å. This region favors the hydrophobic clustering of 4A, 6V and 8V.
Region III: d3P-6V > 6.0 Å and d4A-6V > 6.0 Å. This region favors the extended geometries for the tail region PAAVVVA. Analysis of the conformations belonging to this region revealed significant close contacts between 6V-8V (Figure 5b). This region can be further classified into the 6V-8V close contact (d6V-8V < 6.0 Å) and 6V-8V extended regions (d6V-8V > 6.0 Å).
Figure 5.
(a) Boltzmann inverted 3P-6V/4A-6V 2D distribution for 1. Regions identified using distance cutoffs are distinguished using dotted lines. (b) Close contact probabilities observed between conformations belonging to region III. (c) Representative structures obtained by clustering the conformations belonging to each region using the backbone φ/ψ dihedrals. Color Coding: Carbohydrate region is presented in red and the peptide region in blue. Amino acids involved in the hydrophobic clustering are presented in VDW representation. The surface representation is used to highlight the hydrophobic clustering.
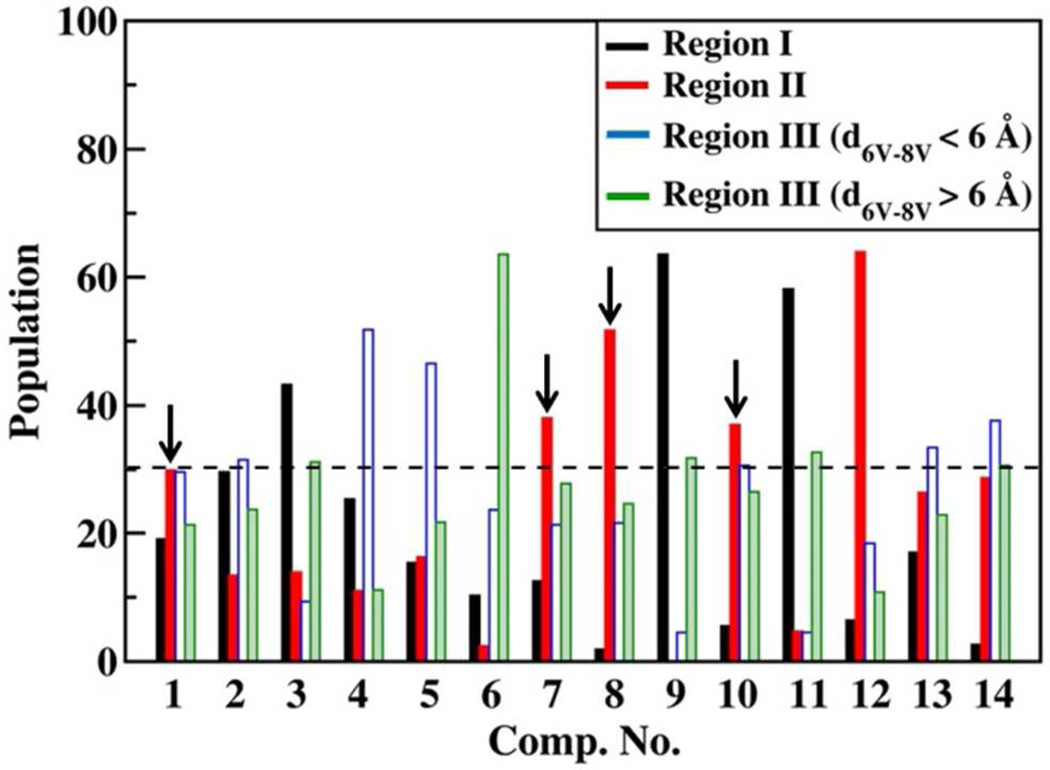
The population distribution of the conformations in these three regions for all the systems is presented in Table 4. From the population distributions it is seen that the population of the folded conformations in region II (plotted for clarity in Figure 6), which corresponds to the hydrophobic clustering of amino acids at positions 4, 6 and 8, shows correlation with the observed activity profile (Table 1). Both 1 and 10, which exhibit 100% activity, favor the hydrobhobic clustering of amino acids at 4, 6 and 8 (population ~30%). The derivatives which show 0% activity (4 and 5) show an appreciable reduction in population in region II (population ~15%). However, this descriptor is limited as it fails to describe the 0% activity observed in amino acid only derivatives (12, 13) and the D-proline derivative (14), which show appreciable population in region II. Thus, the observed activity in as-APF is a combination of various structural features. This becomes clearer on attempting a case-by-case analysis of the various as-APF derivatives. Accordingly, the as-APF derivatives are divided into various subclasses based on the position of the subsitution.
Table 4.
Population distribution of the conformations in the regions identified by the 3P-6V/4A-6V 2D distribution.
| Comp. No. | Region I | Region II | Region III | Outliers | |
|---|---|---|---|---|---|
| d6V-8V< 6.0 Å | d6V-8V> 6.0 Å | ||||
| 1 | 19.1% | 29.9% | 29.6% | 21.3% | 0.2% |
| 2 | 29.6% | 13.4% | 31.5% | 23.8% | 1.7% |
| 3 | 43.2% | 13.9% | 9.3% | 31.2% | 2.3% |
| 4 | 25.4% | 11.0% | 51.8% | 11.1% | 0.7% |
| 5 | 15.4% | 16.3% | 46.5% | 21.7% | 0.0% |
| 6 | 10.3% | 2.4% | 23.7% | 63.6% | 0.0% |
| 7 | 12.5% | 38.0% | 21.3% | 27.8% | 0.4% |
| 8 | 1.9% | 51.7% | 21.6% | 24.7% | 0.1% |
| 9 | 63.6% | 0.1% | 4.5% | 31.8% | 0.0% |
| 10 | 5.5% | 37.0% | 30.6% | 26.5% | 0.3% |
| 11 | 58.1% | 4.7% | 4.5% | 32.7% | 0.0% |
| 12 | 6.4% | 64.0% | 18.4% | 10.8% | 0.4% |
| 13 | 17.0% | 26.4% | 33.4% | 22.9% | 0.2% |
| 14 | 2.6% | 28.7% | 37.6% | 30.6% | 0.4% |
Figure 6.
Population distribution corresponding to the various regions of the 3P-6V/4A-6V distribution.
DISCUSSION
A case-by-case summary is presented of the various as-APF derivatives which lead to an improved understanding of as-APF and a refined SAR model.
Serine derivatives (2, 3)
The serine derivatives form the first class of as-APF derivatives, wherein the modifications directly affect the carbohydrate-peptide interactions. The observed activity profile of these derivatives can be explained by the effect of the modification on the strength of the intramolecular H-bond (α-GalNAc (HN) – 1T/1S (O)) and hydrobhobic interactions (α-GalNAc (−CH3) – 3P (Proline ring)) between the carbohydrate and the peptide regions (Figure 1). It is also observed that the weakening of the carbohydrate-peptide interactions affects the peptide-peptide interactions, as an appreciable reduction is seen in the conformations belonging to region II (Table 4) of the 3P-6V/4A-6V 2D distribution (population ~ 13%). Thus, it is seen that the interactions in the head region propagate to the tail region of as-APF.
Compound 4
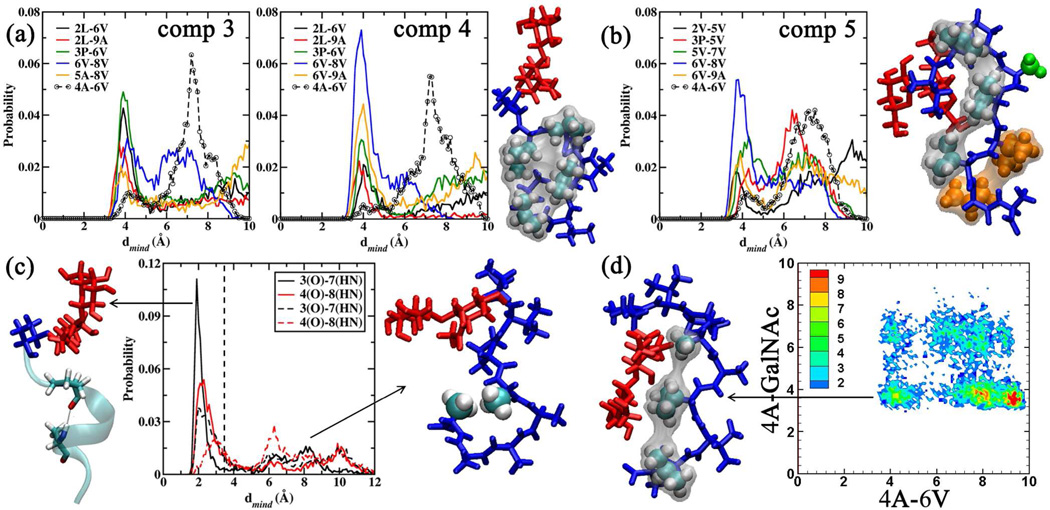
The modification of 2V to 2L in 4 leads to a complete loss of activity. This effect can be traced to the depeletion of the population in Region II of the 3P-6V/4A-6V 2D distribution (Table 4 and Figure 6) to 11.0%. Analyzing the close contacts in 4 revealed that increasing the length of the hydrophobic side chain at position 2 led to an increase in the number of collapsed structures in region I (population: 25.4 %) via stronger hydrophobic interactions between 2L, 3P, 6V and 8V, an effect that is also seen in 3 (region I population: 43.2 %), the other 2L variant. The strong hydrophobic clustering induced due to 2L in 4 is depicted in the representative structure for region I presented in Figure 7a. The difference in the APF activity between 3 and 4 is thus due to the reduction in the 4A-6V contacts in 4 when compared to 3 (Figure 7a dotted lines), which results in the lowering of the population in region II.
Figure 7.
(a) Close contact probabilities observed between the amino acids in 3 and 4. Representative structure for 4 obtained by clustering the conformations belonging to region I. Amino acids 2L, 3P, 6V and 8V are shown in VWD representation to highlight the hydrophobic clustering. (b) Close contact probabilities in 5. Representative structure for 5 obtained upon clustering conformations in region III. Amino acids 2V, 5V and 7V are presented in VWD representation with the corresponding surface to highlight the hydrophobic clustering. The other amino acids 4A (green VDW representation), 6V and 8V (orange VDW representation) highlight other features. (c) H-bond distribution for 3P(O)-7A(HN) (black lines) and 4A(O)-8A(HN) (red lines) for 9 (solid lines) and 11 (dashed lines). Representative structures for 9 and 11 depicting the favored α-helical folded and extended geometries. The peptide tail region is presented in new cartoon representation to highlight the α-helical fold for 9. Amino acids 4A and 8A are presented in VDW representation to highlight the hydrophobic clustering for 11. (d) 4A-6V/4A-GalNAc 2D distribution for 14.The representative structure corresponding to the collapsed region is presented. Amino acids 4A, 6V and 8V are presented in VDW representation. General Color Coding: Carbohydrate region is presented in red and the peptide region in blue. All distances are presented in Å.
Compound 5
The substitution of 5A to 5V also led to a complete loss in activity. This substitution leads to the stiffening of the underlying peptide backbone resulting in the loss of folded αR conformations (Figure 3b), which are necessary to bring about the hydrophobic interactions between 4A and 6V (population in region II: 16.3%). The stiffening was induced by the increase in the number of hydrophobic contacts with 5V, due to an increase in the hydrophobic nature of the amino acid. Figure 7b presents the result of the close contact analysis for 5 along with the representative structure for region III (d6V-8V < 6.0 Å) (population 46.5%) to illustrate the effect of the substitution.
Trivaline modifications (compounds 6, 7, 8)
With 6V being involved in the formation of the hydrophobic collapsed structures (Region II), it is of interest to study the site-by-site modifications of the trivaline region. From the population distribution presented in Figure 6 it is observed that the substitution of 6V to 6A (comp 6) adversely affects the formation of the 4–6–8 collapsed structures with the population in region II going down to 2.4%. In fact the substitution resulted in the formation of extended geometries, with region III (d6V-8V > 6.0 Å) being populated for close to 63.6 %. It is seen that the modification of 7V to 7A or 8V to 8A in fact improves the 4A-6V interactions (population in region II increasing to 38.0% and 51.7% for 7 and 8 respectively). Thus, reducing the hydrophobic content at positions 7 and 8 improves the hydrophobic collapse involving amino acids at position 4 and 6. Here, it must also be mentioned that an attempt to increase the hydrophobicity at position 9 adversely affected the biological activity. Two attempts at this, TVPAAVVVL and TVPAAVVAV, resulted in activities of 0.01% and inactive variants.9 This could be due to this substitution leading to competing hydrophobic collapse involving position 6 which would disrupt the clustering of 4A with 6V.
All alanine modifications (9 and 11)
The substitution of all the valines in the tail region leads to a separate class of as-APF derivatives. From the analysis thus far it is observed that both 7V and 8V modifications to 7A and 8A are favored by as-APF provided the hydrophobicity of the tail amino acid remained unchanged, i.e., 9A. However, changing 6V to 6A has an adverse effect on the population in region II (population being lowered to 2.4% for 6). All the above observations were made for the cases wherein a valine at position 6 (6V) acted as an α-helix breaker. However, the replacement of 6V to 6A with the subsequent amino acids also favoring the α-helix geometry opens up a new conformational profile wherein the tail region of as-APF (TVPAAAAAA or TVPAAAAA) may favor an α-helical fold (Figure 3a and Table 3). Interestingly, the shortening of the tail region by one alanine (9 to 11) led to an increase in activity of two orders of magnitude back to “full” potency (1% to 100%). Analyzing the H-bond profiles for these two systems indicates that the change in one amino acid resulted in the weakening of the α-helical geometry as is evident by the lower H-bond occupancies between i and i+4 H-bond pairs, 3P-7A (0.589 to 0.436) and 4A-8A (0.543 to 0.209) on comparing 9 with 11 (Table 3). This is also illustrated by the H-bond distribution for these two H-bonds presented in Figure 7c. The shortening of the tail by one amino acid has a drastic effect on the strong 3P-7A H-bond. In 9 this H-bond was observed in close to 58.9% of the conformations (population calculated using a distance cutoff of 3.5 Å), while in 11 this dropped to 43.6%. It is noticed that the weakening of the H-bond adds flexibility to the peptide, which resulted in hydrophobic interactions being favored over H-bonding. Thus, even in the case of a strong α-helix “inducer,” the reduction in the helical content resulted in a potent derivative. Interestingly, the attempt to replace alanine with β-alanine in 11, a modification which induces helical content,40 resulted in a three order of magnitude loss in activity (0.1%).10 Also the replacement of the trivaline tail by triglycine in 11 resulted in a complete loss of activity which is consistent with the observation described above, viz., a triglycine tail would not favor any hydrophobic clustering and would instead favor a α-helical fold, thereby lowering the APF activity of this derivative.10 These results consistently highlight the fact that enhancement of the helical content in fact adversely affects the activity.
Peptide only derivatives (12, 13)
The experimental results clearly indicate that the carbohydrate moiety, at least Galβ1-3GalNAc is required for the activity. The analysis of the peptide only fragments is insightful as it reveals the influence of the amino acid sequence on the conformations adopted by the peptide region. From the population distribution presented in Table 4 and Figure 6 it is seen that the peptide only fragments significantly populate region II of the 3P-6V/4A-6V distribution, with the population being 64.0% and 26.4% for 12 and 13 respectively. Thus, the hydrophobic clustering of amino acids at position 4, 6 and 8 is inherent to the peptide. This observation is consistent with the CD results which found the structure of the TVPAAVVV peptide-only fragment (comp 13) to be more organized than the glycosylated fragment (comp 10).10 In all the CD results for the APF derivatives, the presence of the carbohydrate induces structural disorder into the systems. These results also highlight the fact that there is interplay of the carbohydrate-peptide and peptide-peptide interactions in AFP systems which governs the activity.
D-proline (14)
The substitution of proline with D-proline led to a complete loss of activity. However, it was found that of all the inactive derivatives of as-APF, the D-proline derivative was one of only two that blocked the antiproliferative activity of as-APF, thereby acting as an antagonist.6 Structural analysis of 14 reveals that it retains the peptide conformation observed in 1 and 10 (Table 4 and Figure 6), favoring the hydrophobic interactions between 4A, 6V and 8V. However, the change in the conformation at the proline ring exposes the peptide to the carbohydrate with 4A interacting with GalNAc. This is illustrated by the distance distribution between GalNAc and 4A for all the systems, which is presented in Figure S4 of the SI file. It is observed that 4A interacts with GalNAc significantly in 14 and for an appreciable time in 2 (where T is substituted by S). For all other systems GalNAc interacts only with 3P. In Figure 7d, the 4A-6V/4A-GalNAc 2D distribution is presented which highlights the fact that 14 can accommodate both close contacts in tandem with each other. This feature highlights the influence of the proline ring on the activity of APF wherein the proline ring acts as a barrier to curb the interactions between the peptide head region (TV) and the rest of the peptide (AAVVVA).
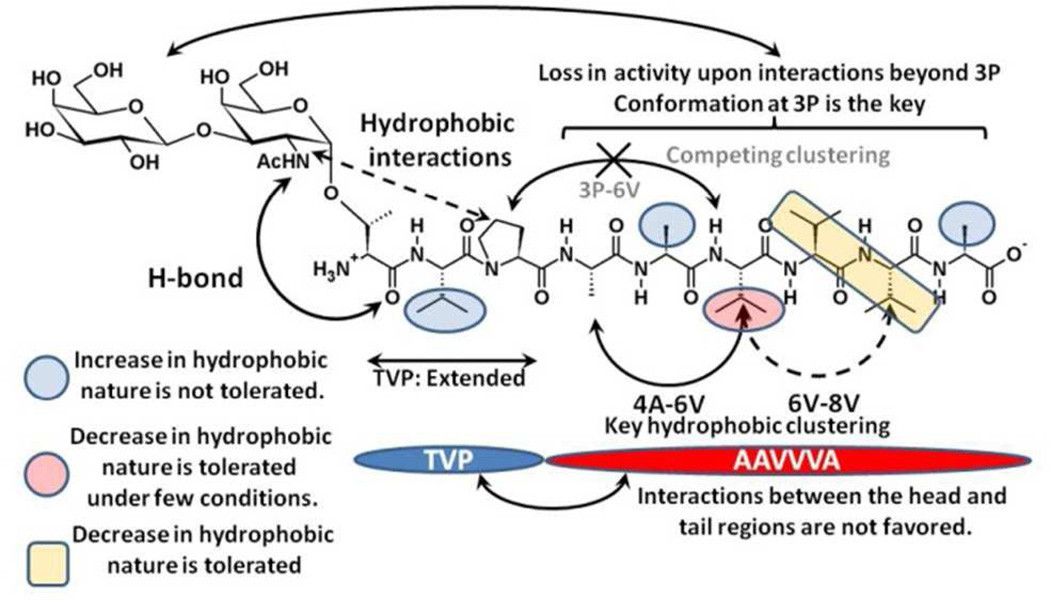
CONCLUSIONS
Presented is an overview of the structural features of as-APF and analogs that impact biological activity. Scheme 2 and the following text summarize the key findings.
The carbohydarte interacts with the peptide backbone via strong H-bond (α-GalNAc (HN) – 1T/1S (O)) and hydrobhobic interactions (α-GalNAc (−CH3) – 3P (Proline ring). The presence of the carbohydrate is required for the activity as is demonstrated by 0% activity for the peptide only compounds 12 and 13. Interactions of the carbohydrate with the rest of the peptide can cause a significant loss in activity, as seen for 14 and 2.
The head region of the peptide portion (TVP) adopts an extended conformation and generally does not interact with the tail region (AAVVVA).
The hydrophobic clustering of amino acids at position 4, 6 and 8 leads to the formation of a pseudo-extended geometry that is predicted to be the active as-APF conformation as its presence correlates with the experimental trends. While the clustering of 4 and 6 is seen to be imporant for the activity, the clusterting of 6 and 8 is found to be auxillary to the activity profile.
The hydrophobic clustering between positions 3 and 6 competes with the 4–6 clustering. Thus, subsitutions which favor these interactions adversly affect the activity as observed in 2, 3 and 4.
Increasing the hydrophobic nature at amino acids 2, 5 and 9 adversly affects the activity as it disrupts the hydrophobic clustering of 4–6–8. An increase in the hydrophobic nature at position 2 leads to increased 3–6 clustering and interactions between the head (TVP) and the tail regions of the peptide (AAVVVA). Changes at amino acids 5 and 9 lead to the disruption of the 4–6–8 clustering and favor the formation of alternate peptide conformations in the tail region (AAVVVA).
The activity of as-APF is highly sensitive to the hydrophobic content of the amino acid at position 6. A valine at this position acts as a α-helix breaker. Substituting valine with alanine is favored only if the subsequent amino acids are also substituted to alanines.
Substitution of valine to alanine at positions 7 and 8 is favored as it enhances the 4–6–8 clustering.
The all alanine tail derivatives (9, 11) form a separate class of as-APF analogs which cannot be described by the 3P-6A/4A-6A 2D distribution profile (Table 4 and Figure 6). On comparing the activities of these two derivatives a two orders of magnitude increase in the activity was observed upon a loss in the α-helix character, indicating that conformations which favor hydrophobic clustering over H-bonding are more potent.
Scheme 2.
Schematic illustration of the structural requirements for biological activity.
In summary, our analysis shows that the hydrophobic clustering of amino acids at position 4–6–8 leads to potent APF glycopeptides derivatives provided the conformation at 3P is maintained. This hydrophobic clustering is found to be very sensitive to single point mutations. However the analysis reveals regions that can tolerate both an increase as well as decrease in the hydrophobic nature of a substitution, indicating that appropriate modifications may lead to more potent as-APF derivatives. With regards to the antagonist activity, it is found that the conformational transition brought about by substituting proline with D-proline (antagonist analog of as-APF) is also partially observed in T to S. Comprehensive NMR and CD analysis has been performed on many of these APF analogues,4, 9, 10 However, the disordered nature of APF glycopeptides precludes determination of the conformational properties of the peptides with these techniques alone. The computational sampling methods utilized here produce a more complete picture of the glycopeptide conformational properties to be obtained allowing for determination of a SAR model that should greatly assist in the development of glycopeptide-mimetics as viable therapeutic lead compounds for IC and cancer.
Supplementary Material
ACKNOWLEDGEMENTS
Financial support from the NIH (GM070855) is acknowledged. The modified REPDSTR module to perform periodic boundary simulations with CHARMM c36a2 was kindly provided by Prof. Benoit Roux and Dr. Wei Jiang (University of Chicago).
Abbreviations
- APF
Antiproliferative factor
- SAR
Structure-activity relationship
- IC
Interstitial cystitis
- as-APF
Asialo synthetic derivative of APF
- CKAP4
cytoskeletal-associated protein 4
- HREX
Hamiltonian replica exchange
- MD
Molecular dynamics
- ROE
Rotating Frame Nuclear Overhauser Effect
- Gal
Galactose
- GalNAc
N-cetylgalactosamine
- Neu5Ac
N-Acetylneuraminic acid;
Footnotes
Supporting Information Available 3J coupling constants calculated from HREX simulations for the H-N-C2-H2 (for GalNAc), N-Cα-Cβ-O1 and C-N-Cα-C. GalNAc (HN) - 1T/1S (O) H-bond distribution. Population distribution of the φ/ψ dihedrals for 2V/L and 3P. 2D distributions of the close contact probabilities for 2V-6V/4A-6V, 3P-6V/7V-9A, and 4A-6V/7V-9A. Close contact probabilities between GalNAc and 4A. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Clemens JQ, Meenan RT, Rosetti MCOK, Gao SY, Calhoun EA. Prevalence and Incidence of Interstitial Cystitis In a Managed Care Population. J. Urol. 2005;173:98–102. doi: 10.1097/01.ju.0000146114.53828.82. [DOI] [PubMed] [Google Scholar]
- 2.Parsons JK, Parsons CL. The Historical Origins of Interstitial Cystitis. J. Urol. 2004;171:20–22. doi: 10.1097/01.ju.0000099890.35040.8d. [DOI] [PubMed] [Google Scholar]
- 3.Keay S, Zhang CO, Trifillis AL, Hise MK, Hebel JR, Jacobs SC, Warren JW. Decreased sup 3 H-Thymidine Incorporation By Human Bladder Epithelial Cells Following Exposure To Urine From Interstitial Cystitis Patients. J. Urol. 1996;156:2073–2078. [PubMed] [Google Scholar]
- 4.Keay SK, Szekely Z, Conrads TP, Veenstra TD, Barchi JJ, Zhang CO, Koch KR, Michejda CJ. An antiproliferative factor from interstitial cystitis patients is a frizzled 8 protein-related sialoglycopeptide. Proc. Natl. Acad. Sci. USA. 2004;101:11803–11808. doi: 10.1073/pnas.0404509101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang CO, Wang JY, Koch KR, Keay S. Regulation of tight junction proteins and bladder epithelial paracellular permeability by an antiproliferative factor from patients with interstitial cystitis. J. Urol. 2005;174:2382–2387. doi: 10.1097/01.ju.0000180417.11976.99. [DOI] [PubMed] [Google Scholar]
- 6.Keay S, Kaczmarek P, Zhang CO, Koch K, Szekely Z, Barchi JJ, Michejda C. Normalization of Proliferation and Tight Junction Formation in Bladder Epithelial Cells from Patients with Interstitial Cystitis/Painful Bladder Syndrome by d-Proline and d-Pipecolic Acid Derivatives of Antiproliferative Factor. Chem. Biol. Drug. Des. 2011;77:421–430. doi: 10.1111/j.1747-0285.2011.01108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koch K, Zhang CO, Kaczmarek P, Barchi J, Jr, Guo L, Shahjee H, Keay S. The effect of a novel frizzled 8-related antiproliferative factor on in vitro carcinoma and melanoma cell proliferation and invasion. Invest. New Drugs. 2012;30:1849–1864. doi: 10.1007/s10637-011-9746-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conrads TP, Tocci GM, Hood BL, Zhang CO, Guo L, Koch KR, Michejda CJ, Veenstra TD, Keay SK. CKAP4/p63 Is a Receptor for the Frizzled-8 Protein-related Antiproliferative Factor from Interstitial Cystitis Patients. J. Biol. Chem. 2006;281:37836–37843. doi: 10.1074/jbc.M604581200. [DOI] [PubMed] [Google Scholar]
- 9.Kaczmarek P, Keay SK, Tocci GM, Koch KR, Zhang CO, Barchi JJ, Grkovic D, Guo L, Michejda CJ. Structure-activity relationship studies for the peptide portion of the bladder epithelial cell antiproliferative factor from interstitial cystitis patients. J. Med. Chem. 2008;51:5974–5983. doi: 10.1021/jm8002763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaczmarek P, Tocci GM, Keay SK, Adams KM, Zhang CO, Koch KR, Grkovic D, Guo L, Michejda CJ, Barchi JJ. Structure-Activity Studies on Antiproliferative Factor (APF) Glycooctapeptide Derivatives. ACS Med. Chem. Lett. 2010;1:390–394. doi: 10.1021/ml100087a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mallajosyula SS, MacKerell AD., Jr Influence of Solvent and Intramolecular Hydrogen Bonding on the Conformational Properties of O-Linked Glycopeptides. J. Phys. Chem. B. 2011;115:11215–11229. doi: 10.1021/jp203695t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brooks BR, Brooks CL, 3rd, MacKerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 14.MacKerell AD, Jr, Feig M, Brooks CL. Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 15.Guvench O, Greene SN, Kamath G, Brady JW, Venable RM, Pastor RW, MacKerell AD., Jr Additive empirical force field for hexopyranose monosaccharides. J. Comput. Chem. 2008;29:2543–2564. doi: 10.1002/jcc.21004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guvench O, Hatcher ER, Venable RM, Pastor RW, MacKerell AD., Jr CHARMM Additive All-Atom Force Field for Glycosidic Linkages between Hexopyranoses. J. Chem. Theory Comput. 2009;5:2353–2370. doi: 10.1021/ct900242e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatcher E, Guvench O, MacKerell AD., Jr CHARMM Additive All-Atom Force Field for Acyclic Polyalcohols, Acyclic Carbohydrates and Inositol. J. Chem. Theory Comput. 2009;5:1315–1327. doi: 10.1021/ct9000608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatcher E, Guvench O, MacKerell AD., Jr CHARMM additive all-atom force field for aldopentofuranoses, methyl-aldopentofuranosides, and fructofuranose. J. Phys. Chem. B. 2009;113:12466–76. doi: 10.1021/jp905496e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raman EP, Guvench O, MacKerell AD., Jr CHARMM additive all-atom force field for glycosidic linkages in carbohydrates involving furanoses. J. Phys. Chem. B. 2010;114:12981–94. doi: 10.1021/jp105758h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 21.Becker OMMJAD, Roux B, Watanabe M, editors. Computational Biochemistry and Biophysics. New York: Marcel-Dekker, Inc; 2001. [Google Scholar]
- 22.Brooks CL, III, Karplus M, Pettitt BM. Proteins: A Theoretical Perspective of Dynamics, Structure, and Thermodynamics. New York: John Wiley &, Sons; 1988. [Google Scholar]
- 23.Beglov D, Roux B. An Integral Equation To Describe the Solvation of Polar Molecules in Liquid Water. J. Phys. Chem. B. 1997;101:7821–7826. [Google Scholar]
- 24.Darden T, York D, Pedersen L. Particle mesh Ewald: An N-log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 25.Steinbach PJ, Brooks BR. New spherical-cutoff methods for long-range forces in macromolecular simulation. J. Comput. Chem. 1994;15:667–683. [Google Scholar]
- 26.Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 27.Nose S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984;81:511–519. [Google Scholar]
- 28.Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 29.Woodcock HL, Hodošček M, Gilbert ATB, Gill PMW, Schaefer HF, Brooks BR. Interfacing Q-Chem and CHARMM to perform QM/MM reaction path calculations. J. Comput. Chem. 2007;28:1485–1502. doi: 10.1002/jcc.20587. [DOI] [PubMed] [Google Scholar]
- 30.Mobli M, Almond A. N-Acetylated amino sugars: the dependence of NMR 3J(HH)-couplings on conformation, dynamics and solvent. Org. Biomol. Chem. 2007;5:2243–2251. doi: 10.1039/b705761j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Demarco A, Llinás M, Wüthrich K. Analysis of the 1H-NMR spectra of ferrichrome peptides. I. The non-amide protons. Biopolymers. 1978;17:617–636. [Google Scholar]
- 32.Vuister GW, Bax A. Quantitative J correlation: a new approach for measuring homonuclear three-bond J(HNH.alpha.) coupling constants in 15N–enriched proteins. J. Am. Chem. Soc. 1993;115:7772–7777. [Google Scholar]
- 33.Corzana F, Busto JH, Engelsen SB, Jimenez-Barbero J, Asensio JL, Peregrina JM, Avenoza A. Effect of beta-O-glucosylation on L-Ser and L-Thr diamides: A bias toward alpha-helical conformations. Chem. Eur. J. 2006;12:7864–7871. doi: 10.1002/chem.200600128. [DOI] [PubMed] [Google Scholar]
- 34.Corzana F, Busto JH, Jimenez-Oses G, Asensio JL, Jimenez-Barbero J, Peregrina JM, Avenoza A. New insights into alpha-GalNAc-Ser motif: Influence of hydrogen bonding versus solvent interactions on the preferred conformation. J. Am. Chem. Soc. 2006;128:14640–14648. doi: 10.1021/ja064539u. [DOI] [PubMed] [Google Scholar]
- 35.Corzana F, Busto JH, Jimenez-Oses G, de Luis MG, Asensio JL, Jimenez-Barbero J, Peregrina JM, Avenoza A. Serine versus threonine glycosylation: The methyl group causes a drastic alteration on the carbohydrate orientation and on the surrounding water shell. J. Am. Chem. Soc. 2007;129:9458–9467. doi: 10.1021/ja072181b. [DOI] [PubMed] [Google Scholar]
- 36.Coltart DM, Royyuru AK, Williams LJ, Glunz PW, Sames D, Kuduk SD, Schwarz JB, Chen X-T, Danishefsky SJ, Live DH. Principles of Mucin Architecture: Structural Studies on Synthetic Glycopeptides Bearing Clustered Mono-, Di-, Tri-, and Hexasaccharide Glycodomains. J. Am. Chem. Soc. 2002;124:9833–9844. doi: 10.1021/ja020208f. [DOI] [PubMed] [Google Scholar]
- 37.Hashimoto R, Fujitani N, Takegawa Y, Kurogochi M, Matsushita T, Naruchi K, Ohyabu N, Hinou H, Gao XD, Manri N, Satake H, Kaneko A, Sakamoto T, Nishimura S-I. An Efficient Approach for the Characterization of Mucin-Type Glycopeptides: The Effect of O-Glycosylation on the Conformation of Synthetic Mucin Peptides. Chem. Eur. J. 2011;17:2393–2404. doi: 10.1002/chem.201002754. [DOI] [PubMed] [Google Scholar]
- 38.Kleiger G, Grothe R, Mallick P, Eisenberg D. GXXXG and AXXXA: Common aplha-Helical Interaction Motifs in Proteins, Particularly in Extremophiles. Biochemistry. 2002;41:5990–5997. doi: 10.1021/bi0200763. [DOI] [PubMed] [Google Scholar]
- 39.Best RB, Buchete N-V, Hummer G. Are Current Molecular Dynamics Force Fields too Helical? Biophys. J. 2008;95:L07–L09. doi: 10.1529/biophysj.108.132696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seebach D, Beck AK, Bierbaum DJ. The World of β,- and γ,-Peptides Comprised of Homologated Proteinogenic Amino Acids and Other Components. Chem. Biodiversity. 2004;1:1111–1239. doi: 10.1002/cbdv.200490087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.