Abstract
Aims
Aldo-ketoreductases have been implicated in the metabolism of doxorubicin. We sought to assess the influence of AKR1C3 genetic variants on doxorubicin metabolism.
Methods
We sequenced AKR1C3 exon 5 and genotyped seven functional single nucleotide polymorphisms in CBR3, ABCB1 and SLC22A16 involved in doxorubicin pharmacology in 151 Asian breast cancer patients treated with doxorubicin-containing chemotherapy, and correlated these genotypes with doxorubicin pharmacokinetics and pharmacodynamics.
Results
Two previously reported AKR1C3 intronic variants, IVS4–212 C>G and IVS4+218 G>A, were detected. The AKR1C3 IVS4–212 GG genotype was associated with significantly lower cycle 1 day 15 leucocyte (mean leucocytes 2.49 ± 1.57 × 109 vs. 3.85 ± 3.42 × 109 l−1, P = 0.007) and neutrophil counts (mean neutrophils 0.70 ± 1.01 × 109 vs. 1.56 ± 2.80 × 109 l−1, P = 0.008) and significant improvement of progression-free survival [PFS, mean PFS 49.0 (95% confidence interval 42.2–55.8) vs. 31.0 (95% confidence interval 20.7–41.2) months, P = 0.017] and overall survival [OS; mean OS 64.4 (95% confidence interval 58.3–70.5) vs. 46.3 (95% confidence interval 35.1–57.5) months, P = 0.006] compared with those carrying at least one C allele. There was no significant association between AKR1C3 IVS4–212 C>G and doxorubicin pharmacokinetics. Of the other seven single nucleotide polymorphisms genotyped, CBR3 G11A correlated with doxorubicinol area under the concentration–time curve and OS, ABCB1 G2677T/A correlated with doxorubicin clearance and platelet toxicity, while ABCB1 IVS26+59 T>G correlated with OS. The AKR1C3 IVS4–212 C<G genotype remained significantly correlated with both PFS and OS on multivariate analysis with clinical prognosticators.
Conclusions
The AKR1C3 IVS4–212 GG genotype was associated with greater haematological toxicity and longer progression-free survival and overall survival after doxorubicin-based therapy, suggesting potential interaction of this variant with doxorubicin metabolism.
Keywords: aldo-ketoreductase (AKR)1C3, ATP-binding cassette (ABCB)1, carbonyl reductase (CBR)3, doxorubicin, pharmacogenetics, solute carrier family (SLC)22A16
WHAT IS ALREADY KNOWN ABOUT THIS SUBJECT
Significant interindividual variation in doxorubicin efficacy and toxicity exists.
Aldo-ketoreductase AKR1C3 genetic variants have been identified in vitro to affect doxorubicin metabolism.
Correlation between these genetic variants and doxorubicin metabolism in vivo will advance our understanding of pharmacogenetic influences on the complex doxorubicin disposition pathway.
WHAT THIS STUDY ADDS
A common AKR1C3 intronic variant, IVS4–212 C>G, correlated with doxorubicin pharmacodynamics in breast cancer patients. The variant remains significantly correlated with patient survival on multivariate analysis with known clinical prognosticators.
This variant appears to be more common in Asians than Caucasians and may explain the previously observed greater doxorubicin-induced myelosuppression in Chinese compared with Caucasian patients.
These findings may contribute to the use of pharmacogenetic information to tailor drug therapy in cancer patients.
Introduction
Doxorubicin is an effective anticancer agent for a range of solid tumours, including breast cancer 1. Interindividual variations in doxorubicin response and toxicity are well recognized 2–4. The doxorubicin disposition pathway is complex and involves various influx and efflux transporters across the cellular membrane and metabolizing enzymes that are postulated to be responsible for the wide variability in pharmacokinetics and pharmacodynamics of doxorubicin 5.
Genetic factors have been suggested to explain the variability in therapeutic efficacy and toxicity of drugs in individual patients 6. The influence of single nucleotide polymorphisms (SNPs) from potential functional candidate gene variants across the doxorubicin disposition pathway has been investigated 5–9. These include influx [solute carrier family (SLCs)] and efflux drug transporters [ATP-binding cassette (ABCBs)] and drug metabolizing enzymes [carbonyl reductase (CBR) and aldo-ketoreductase (AKR)] SNPs. However, the results observed from these studies have still not been clearly elucidated, in particular for AKRs.
Aldo-ketoreductases have been implicated in the metabolism of anthracycline antibiotics, including doxorubicin 10. Aldo-ketoreductase 1C3 plays a significant role in the deactivation of doxorubicin to doxorubicinol, a less active metabolite 11. An in vitro study has demonstrated that two nonsynonymous SNPs in exon 5 of the AKR1C3 gene (R170C and P180S) encode for enzymes with significantly reduced metabolism of doxorubicin compared with their wild-type counterparts and may contribute to the interindividual variability in anthracycline metabolism 12.
We sought to evaluate the frequency distribution of genetic variants in and around the coding region of exon 5 in the AKR1C3 gene and to determine the correlation between identified AKR1C3 genetic variants with doxorubicin pharmacokinetics, haematological toxicities and antitumour efficacy in an Asian breast cancer population. The influence of SNPs from other functional candidate gene variants across the doxorubicin disposition pathway, namely SLC22A16 (influx transporter), ABCB1 (efflux transporter) and CBR3 (drug metabolizing enzyme), were also examined concurrently, to assess the potential interactions of these more widely studied genetic variants with AKR1C3 SNPs.
Methods
Study population
We studied 151 Asian breast cancer patients who participated in two preoperative chemotherapy trials conducted at the National University Cancer Institute, Singapore (n = 101 from the first study cohort 7, and n = 50 from the second study cohort). Both study populations were composed of female chemo-naïve patients with histologically or cytologically confirmed breast cancer, for whom preoperative chemotherapy was indicated. The first study cohort was treated with six cycles of preoperative alternating sequential doxorubicin (A) and docetaxel (T) every 3 weeks, and patients were randomized to start either with doxorubicin or docetaxel (A→T→A→T→A→T or T→A→T→A→T→A), followed by surgery, followed by two additional cycles of postoperative alternating docetaxel and doxorubicin. Patients from the second study cohort were randomized to either four cycles of preoperative doxorubicin (n = 25) or four cycles of docetaxel every 3 weeks (n = 25), followed by surgery, followed by four cycles of the alternative drug (docetaxel or doxorubicin). Doxorubicin was administered at 75 mg m−2 as a slow bolus and docetaxel at 75 mg m−2 over 1 h in both studies. Routine use of prophylactic colony stimulating factor was not allowed.
Genotyping of exon 5 of the AKR1C3 gene
Germline DNA was extracted from peripheral mononuclear cells from study participants using standard methods. Exon 5 of the AKR1C3 gene, which encodes two known functional SNPs (R170C and P180S), was amplified by polymerase chain reaction (PCR). All PCRs were carried out in 25 μl volume with 10× PCR buffer, 25 mm magnesium chloride, 10 mm dNTP, 2 units Faststart Taq DNA polymerase, 12.5 μm each of forward and reverse primer, and 5 ng genomic DNA. The primer sequences were as follows (5′ to 3′, forward and reverse): CCCAGGTTCAATAGGAAAGAA and ACCTTCACCCATGCACTTTC. The size of the PCR product was 850 bases, which included 727 bases of intronic sequences. The PCR was performed with an initial denaturation step at 95°C for 5 min, followed by 36 cycles of denaturation at 95°C for 30 s, annealing at 57.5°C for 45 s, and extension at 72°C for 1 min, followed by a final extension step at 72°C for 5 min. The PCR products were purified and sequenced on the ABI 3100 automated sequence analyser (Applied Biosystems Inc., Foster City, CA, USA) with the forward or reverse PCR primer. Generated sequences were compared with the reference sequence for AKR1C3 available online at http://www.ncbi.nlm.nih.gov/nuccore/NC_000010.10?from=5136568&to=5149878&report=genbank.
Genotyping of functional SNPs from ABCB1, CBR3 and SLC22A16
A total of seven SNPs from these three genes were chosen, based on previous reports 7, 9, 13, 14. The ABCB1 G2677T/A genotype has been reported to influence doxorubicin clearance and is linked to two other ABCB1 functional variants, C1236T and C3435T, while the ABCB1 IVS26+59 T>G variant has been reported to be associated with anthracycline-induced cardiotoxicity 13. Both CBR3 G11A and G730A were selected based on our previous study, which showed significant correlation with doxorubicin pharmacokinetics and pharmacodynamics 7. Genotypes SLC22A16 T312C, T755C and T1226C were included because they were present at significant frequencies in our Asian breast cancer population, but there is a paucity of data on their influence on doxorubicin pharmacokinetics and pharmacodynamics 14. Genotype data for CBR3 G11A were obtained from our previous report of the first study cohort (n = 99) 7, while genotyping for the other six SNPs was performed in multiplexed reactions using the MassArray iPLEX Gold platform (Sequenom, San Diego, CA, USA) for both study cohorts (n = 151). In brief, PCR amplification was carried out using 20 ng of DNA in a 5μl reaction that contained 0.5 units Taq polymerase, 10× PCR buffer, 4 mm magnesium chloride, 500 μm dNTPs and 0.1 μm of primers, in the following conditions: 95°C for 2 min, 45 cycles of 95°C for 30 s, 56°C for 30 s, 72°C for 1 min and a final extension step of 72°C for 5 min. Unincorporated dNTPs were removed using 0.5 units of shrimp alkaline phosphatase dissolved in the shrimp alkaline phosphatase buffer provided. Single base extension was carried out in a 9μl reaction that contained iPLEX GOLD buffer, termination mix, extend primer mix and enzyme, using the following two cycling loop programme: 94°C for 30 s, 40 cycles of 94°C for 5 s, 52°C for 5 s and 80°C for 5 s. Within the 40 cycles, the annealing and extension step was repeated five times (i.e. 40 × 5 = 200 short cycles), before a final extension step of 3 min at 72°C. Reactions were desalted using 6 mg of clean resin and diluted with 16 μl water. A 10 nl sample of each reaction was spotted onto the 384-spot SpectroChipII using the Nanodispenser (Sequenom), followed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) analysis using the MassARRAY Compact system (Sequenom). The mass spectra analysis and genotype calls were generated using the TYPER 4.0.22 software (Sequenom).
Pharmacokinetics analyses
Whole blood (5 ml) was collected from the contralateral arm from chemotherapy administration for analysis of doxorubicin and doxorubicinol levels at 0, 1, 2, 4, 7 and 24 h after the first doxorubicin treatment. Analytical grade doxorubicin hydrochloride was purchased from Woo Shin Medics Co. (Seoul, South Korea) and doxorubicinol hydrochloride was purchased from Toronto Research Chemicals Inc. (Canada). Plasma concentrations of doxorubicin and its major metabolite, doxorubicinol, were determined in all samples by a reverse-phase high-performance liquid chromatography method with fluorescence detection modified from a published method 15. The limits of quantification for doxorubicin and doxorubicinol were 2 and 1 ng ml−1, respectively. The calibration curves were linear over a concentration range of 2–200 ng ml−1 for doxorubicin and 1–100 ng ml−1 for doxorubicinol. The average recoveries were greater than 84% for both analytes. The intraday and interday precision coefficients of variation ranged from 1.1 to 14% for doxorubicin and from 2.3 to 13% for doxorubicinol. Accuracy was 97–110% for doxorubicin and 92–105% for doxorubicinol.
Doxorubicin pharmacokinetic parameters were analysed with two-compartmental and noncompartmental methods using Kinetica software, version 4.4 (Thermo Fisher Scientific, Inc., Waltham, MA, USA). All pharmacokinetic parameters for doxorubicin were derived from two-compartmental analysis. The noncompartmental method was used to analyse the area under the concentration–time curve (AUC) of doxorubicinol because it was a metabolite. The AUC, maximal concentration (Cmax), clearance, elimination half-life and volume of distribution at steady state were calculated.
Statistical analysis
Identified genetic variants were correlated with antitumour efficacy, haematological toxicity and doxorubicin and doxorubicinol pharmacokinetic (PK) data. The PK data, which included doxorubicin and doxorubicinol AUC, doxorubicin Cmax, clearance and terminal half-life, were available only for 98 patients recruited into the first trial. Efficacy parameters of treatment included the objective clinical response rate after completing four to six cycles of preoperative chemotherapy, which was classified according to the World Health Organization criteria 16, progression-free survival (PFS) and overall survival (OS). Analysis of the objective clinical response rate was limited to the 125 patients who were treated with four to six cycles of preoperative chemotherapy containing doxorubicin, excluding the 25 patients in the second study cohort who were randomized to receive doxorubicin postoperatively. The haematological parameters evaluated included weekly complete blood counts, nadir blood counts and febrile neutropenia rates during the first cycle of doxorubicin.
The Mann–Whitney U-test was used to compare means of continuous variables, while the chi-squared or Fisher's exact test was used to compare categorical variables. Differences in survival distributions between different genotype variants in each assessed gene were compared using the Kaplan–Meier method and the logrank test. In addition, multivariate analysis using Cox regression was performed by including genetic variants and known clinical parameters that influenced survival at P < 0.1 in univariate analysis. These prognostic clinical parameters include metastatic status, histology grade and hormonal receptor status. All tests were two sided at the 5% level of significance.
The Lewontin's D′ was used to determine linkage disequilibrium between two genetic variants and calculated using the software PowerMarker (version 3.25, http://statgen.ncsu.edu/powermarker/).
Results
The median age of the entire study cohort was 51 years (range 26–74 years), and the study population mainly comprised of Chinese (57%) and Malay women (34%). Forty-seven patients (31%) had metastatic disease at diagnosis.
AKR1C3
Genetic variants identified in exon 5 of AKR1C3 and their allelic frequencies are listed in Table 1. Both coding region variants of interest (R170C and P180S) were not identified in this study cohort. However, two common intronic variants were detected, IVS4–212 C>G and IVS4+218 G>A, which have been previously reported [National Center for Biotechnology Information, Reference SNP (ref. SNP) cluster report: rs1937840; National Center for Biotechnology Information, ref. SNP cluster report: rs1937841]. The IVS4–212 C>G variant was common in this Asian population, with an overall variant frequency of 0.86; 76% of the study population was homozygous for the variant. The IVS4+218 G>A variant was less common, with an overall variant frequency of 0.11 in this population. Strong linkage dysequilibrium was observed between AKR1C3 IVS4–212 C and IVS4+218 A (D′ = 0.9274, P < 0.001) using Lewontin's D′ measure. There was no ethnic difference (Chinese vs. Malay vs. Indian vs. others) in the distribution for IVS4+218 G>A (P = 0.63) and IVS4–212 C>G (P = 0.23).
Table 1.
Frequency distribution of AKR1C3 genetic variants (n = 151)
| Genetic variants (SNP) | Genotype frequency | Allele frequency | |||
|---|---|---|---|---|---|
| C/C | C/T | T/T | C | T | |
| Exon 5 508 C>T (rs35575889) | 100% | 0% | 0% | 1.00 | 0 |
| Exon 5 538 C>T (rs34186955) | 100% | 0% | 0% | 1.00 | 0 |
| IVS4–212 C>G (rs1937840) | C/C | C/G | G/G | C | G |
| Chinese | 0% (0/86) | 20% (17/86) | 80% (69/86) | 0.10 | 0.90 |
| Malay | 8% (4/52) | 21% (11/52) | 71% (37/52) | 0.18 | 0.82 |
| Indian | 11% (1/9) | 33% (3/9) | 56% (5/9) | 0.28 | 0.72 |
| Others | 0% (0/4) | 0% (0/4) | 100% (4/4) | 0 | 1.00 |
| Total | 3% (5/151) | 21% (31/151) | 76% (115/151) | 0.14 | 0.86 |
| IVS4+218 G>A (rs1937841) | G/G | G/A | A/A | G | A |
| Chinese | 79% (68/86) | 21% (18/86) | 0% (0/86) | 0.90 | 0.10 |
| Malay | 77% (40/52) | 21% (11/52) | 2% (1/52) | 0.88 | 0.12 |
| Indian | 89% (8/9) | 11% (1/9) | 0% (0/9) | 0.94 | 0.06 |
| Others | 100% (4/4) | 0% (0/4) | 0% (0/4) | 1.00 | 0 |
| Total | 79% (120/151) | 20% (30/151) | 1% (1/151) | 0.89 | 0.11 |
Correlation of AKR1C3 intronic variants with doxorubicin pharmacokinetics and pharmacodynamics
IVS4–212 C>G
After completing four to six cycles of preoperative chemotherapy, patients who were homozygous for AKR1C3 IVS4–212 GG (n = 95; Table 2) had a higher objective response rate than patients who carried at least one C allele (n = 30), although the difference was not statistically significant (85 vs. 77%, P = 0.27). At a median follow-up of 36 months, patients who were IVS4–212 GG homozygotes had significantly longer PFS compared with those who carried at least one C allele {mean PFS 49.0 [95% confidence interval (CI) 42.2–55.8] vs. 31.0 [95% CI 20.7–41.2] months, P = 0.017; Figure 1A}. Overall survival was statistically longer in patients who were IVS4–212 GG homozygotes compared with those who carried at least one C allele [mean OS 64.4 (95% CI 58.3–70.5) vs. 46.3 (95% CI 35.1–57.5) months, P = 0.006; Figure 1B].
Table 2.
Comparison of tumour efficacy, haematological toxicity parameters and doxorubicin pharmacokinetic parameters in patients with AKR1C3 IVS4–212 C>G genotype
| GG | GC/CC | P value | |
|---|---|---|---|
| Objective response rate (%; n = 125) | 84 | 76 | 0.25 |
| Progression-free survival (months; n = 151) [mean (95% CI)] | 49.0 (42.2–55.8) | 31.0 (20.7–41.2) | 0.017* |
| Overall survival (months; n = 151) [mean (95% CI)] | 64.4 (58.3–70.5) | 46.3 (35.1–57.5) | 0.006* |
| Haematological toxicities after cycle 1 doxorubicin (n = 142†; mean ± SD) | P value | 95% CI of difference | ||
|---|---|---|---|---|
| Day 8 neutrophils (×109 l−1) | 3.76 ± 1.79 | 3.53 ± 2.27 | 0.19 | −0.60 to 1.07 |
| Day 15 neutrophils (×109 l−1) | 0.70 ± 1.01 | 1.56 ± 2.80 | 0.008* | −1.80 to 0.10 |
| Nadir neutrophils (×109 l−1) | 0.41 ± 0.52 | 0.49 ± 0.48 | 0.23 | −0.26 to 0.11 |
| Day 8 leucocytes (×109 l−1) | 5.34 ± 1.87 | 5.04 ± 2.48 | 0.21 | −0.60 to 1.21 |
| Day 15 leucocytes (×109 l−1) | 2.49 ± 1.57 | 3.85 ± 3.42 | 0.007* | −2.54 to −0.18 |
| Nadir leucocytes (×109 l−1) | 1.93 ± 1.05 | 2.05 ± 1.12 | 0.58 | −0.54 to 0.32 |
| Day 8 haemoglobin (g dl−1) | 11.43 ± 1.59 | 11.48 ± 1.94 | 0.54 | −0.76 to 0.68 |
| Day 15 haemoglobin (g dl−1) | 10.78 ± 1.41 | 11.01 ± 1.93 | 0.33 | −0.93 to 0.46 |
| Nadir haemoglobin (g dl−1) | 10.11 ± 1.51 | 10.19 ± 1.88 | 0.73 | −0.78 to 0.61 |
| Day 8 platelets (×109 l−1) | 240 ± 76 | 252 ± 92 | 0.58 | −46.65 to 21.49 |
| Day 15 platelets (×109 l−1) | 264 ± 139 | 303 ± 141 | 0.06 | −92.96 to 14.55 |
| Nadir platelets (×109 l−1) | 189 ± 70 | 198 ± 69 | 0.41 | −36.22 to 17.45 |
| Febrile neutropenia (%) | 18.3 | 13.5 | 0.51 | — |
| Pharmacokinetic parameters (n = 98) | ||||
|---|---|---|---|---|
| Median doxorubicin dose 115 mg (range 90–140 mg) | ||||
| Mean ± SD (range) | Mean ± SD (range) | P value | 95% CI of difference | |
| Doxorubicin AUC (mg l−1 h) | 1.42 ± 0.55 (0.67–3.86) | 1.47 ± 0.63 (0.58–3.60) | 0.73 | −0.34 to 0.24 |
| Doxorubicin clearance (l h−1) | 90.02 ± 26.83 (28.12–178.84) | 88.55 ± 38.88 (36.13–214.46) | 0.49 | −15.94 to 18.87 |
| Doxorubicin Cmax (mg l−1) | 0.57 ± 1.36 (0.09–10.26) | 0.37 ± 0.32 (0.09–1.12) | 0.67 | −0.14 to 0.54 |
| Doxorubicin half-life (terminal phase; h) | 16.24 ± 5.78 (6.70–35.45) | 14.13 ± 3.73 (9.81–24.35) | 0.09 | 0.04 to 4.17 |
| Doxorubicinol AUC (mg l−1 h) | 1.99 ± 1.66 (0.83–12.08) | 1.74 ± 0.68 (0.88–3.43) | 0.88 | −0.22 to 0.72 |
Abbreviations: AUC, area under the concentration–time curve; CI, confidence interval; Cmax, maximal plasma concentration.
Significant. †n = 142 because nine patients who were randomized to start with docetaxel in the first cycle (T→A→T→A→T→A) in the first study cohort (n = 2) or to postoperative doxorubicin in the second study cohort (n = 7) did not receive doxorubicin due to study withdrawal.
Figure 1.
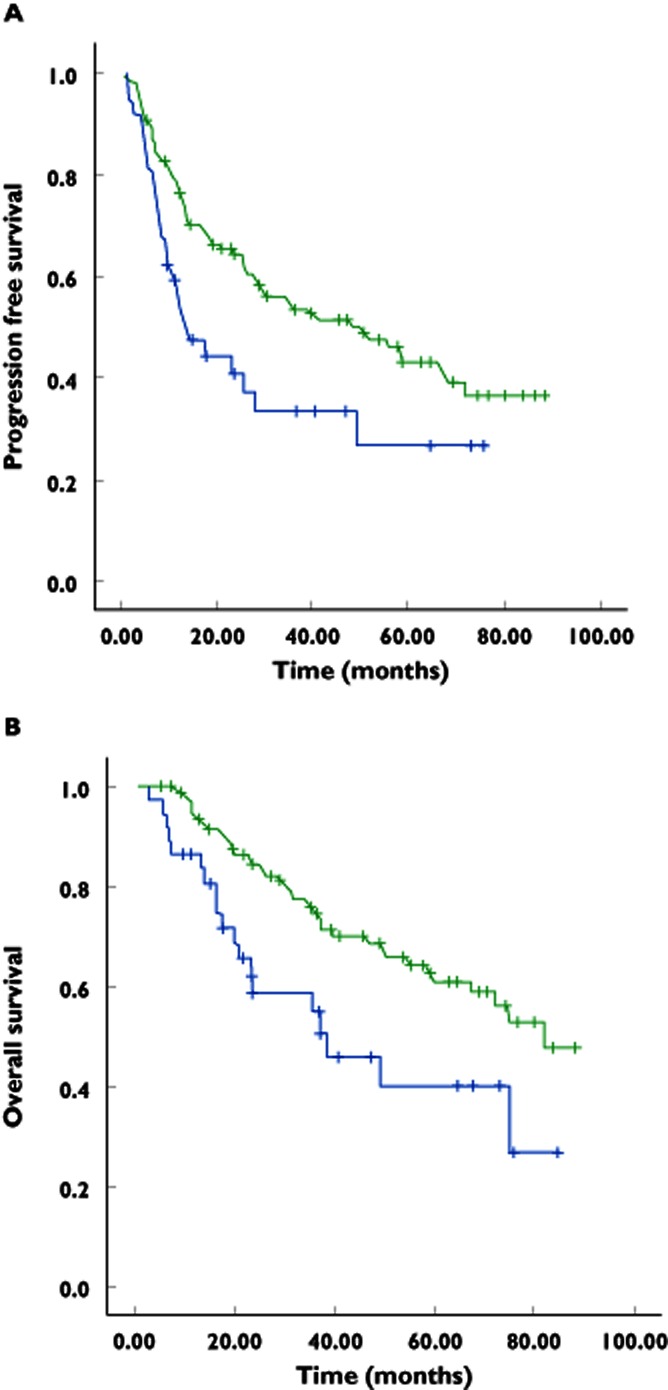
(A) Kaplan–Meier estimate of progression-free survival.  , IVS4–212 GG, mean progression-free survival 49.0 (42.2–55.8) months;
, IVS4–212 GG, mean progression-free survival 49.0 (42.2–55.8) months;  , IVS4–212 GC/CC, mean progression-free survival 31.0 (20.7–41.2) months; P = 0.017. (B) Kaplan–Meier estimate of overall survival.
, IVS4–212 GC/CC, mean progression-free survival 31.0 (20.7–41.2) months; P = 0.017. (B) Kaplan–Meier estimate of overall survival.  , IVS4–212 GG, mean overall survival 64.4 (58.3–70.5) months;
, IVS4–212 GG, mean overall survival 64.4 (58.3–70.5) months;  , IVS4–212 GC/CC, mean overall survival 46.3 (35.1–57.5) months; P = 0.006
, IVS4–212 GC/CC, mean overall survival 46.3 (35.1–57.5) months; P = 0.006
After the first cycle of doxorubicin, patients who were IVS4–212 GG homozygotes developed significantly lower absolute leucocyte and neutrophil counts on day 15 compared with those who carried at least one C allele (mean leucocyte count 2.49 ± 1.57 × 109 vs. 3.85 ± 3.42 × 109 l−1, P = 0.007; mean neutrophil count 0.70 ± 1.01 × 109 vs. 1.56 ± 2.80 × 109 l−1, P = 0.008). There was no significant difference in other haematological parameters between the two groups, including febrile neutropenia rates. No significant difference in doxorubicin pharmacokinetics were observed between the two genotype groups, including doxorubicin and doxorubicinol AUC, doxorubicin Cmax and doxorubicin terminal half-life.
IVS4+218 G>A
Despite the significant linkage dysequilibrium between AKR1C3 IVS4–212 CC or CG and IVS4+218 GA or AA, the only significant association for this intronic variant with doxorubicin pharmacokinetics or pharmacodynamics was lower absolute neutrophil counts on day 15 after the first cycle of doxorubicin for IVS4+218 GG compared with those who carried at least one A allele (mean neutrophil count 0.76 ± 1.12 × 109 vs. 1.53 ± 2.93 × 109 l−1, P = 0.038). Otherwise, there was no statistically significant correlation observed between this intronic variant with other doxorubicin pharmacokinetics or pharmacodynamic parameters, including the objective clinical response rate, PFS or OS.
Correlation between CBR3, ABCB1 and SLC22A16 genetic variants with doxorubicin pharmacokinetics and pharmacodynamics
The distribution of the genotypes is summarized in Table 3. Patients who carried at least one A allele for CBR3 G11A had significantly lower doxorubicinol AUC (mean AUC 1.78 ± 1.53 vs. 2.18 ± 1.37 mg l−1 h, P = 0.004) and longer OS [mean OS 64.4 (95% CI 56.9–71.9) vs. 51.2 (95% CI 40.4–62.0) months, P = 0.05]. Patients carrying at least one A allele for CBR3 G730A had significantly higher doxorubicinol AUC (mean AUC 2.15 ± 1.79 vs. 1.56 ± 0.60 mg l−1 h, P = 0.031) compared with GG homozygotes, but there was no correlation with pharmacodynamics parameters, including survival. For ABCB1 G2677T/A, patients who carried at least one T allele had significantly higher doxorubicin clearance (93.29 ± 29.22 vs. 84.46 ± 30.86 l h−1, P = 0.027) and significantly higher day 8 and nadir platelet counts compared with those who did not have T alleles (mean day 8 platelet count 255 ± 79 × 109 vs. 227 ± 79 × 109 l−1, P = 0.031; mean nadir platelet count 201 ± 66 × 109 vs. 176 ± 74 × 109 l−1, P = 0.031) after the first cycle of doxorubicin. Patients who carried at least one G allele for ABCB1 IVS26+59 T>G had significantly higher day 8 neutrophil and leucocyte counts (neutrophil count 4.03 ± 2.10 × 109 vs. 3.14 ± 1.41 × 109 l−1, P = 0.019; leucocyte count 5.56 ± 2.22 × 109 vs. 4.74 ± 1.55 × 109 l−1, P = 0.032) and significantly longer OS than those who did not [mean OS 64.6 (95% CI 58.0–71.2) vs. 50.6 (95% CI 41.5–59.6) months, P = 0.026].
Table 3.
Frequency distribution of ABCB1, CBR3 and SLC22A16 genetic variants (n = 151)
| Genetic variants (SNPs) | Genotype frequency | Allele frequency | |||||||
|---|---|---|---|---|---|---|---|---|---|
| ABCB1 | |||||||||
| G2677T/A* | GG | GT | GA | TT | AA | TA | G | T | A |
| 37 (25%) | 56 (37%) | 18 (12%) | 23 (15%) | 4 (3%) | 12 (8%) | 0.49 | 0.38 | 0.13 | |
| IVS26+59 T>G | TT | TG | GG | T | G | ||||
| 56 (37%) | 64 (42%) | 31 (21%) | 0.37 | 0.63 | |||||
| CBR3 | |||||||||
| G11A† | GG | GA | AA | G | A | ||||
| 36 (37%) | 30 (30%) | 33 (33%) | 0.52 | 0.48 | |||||
| G730A* | GG | GA | AA | G | A | ||||
| 31 (21%) | 56 (37%) | 63 (42%) | 0.39 | 0.61 | |||||
| SLC22A16 | |||||||||
| T312C* | TT | TC | CC | T | C | ||||
| 63 (42%) | 65 (43%) | 22 (15%) | 0.64 | 0.36 | |||||
| T1226C* | TT | TC | CC | T | C | ||||
| 145 (97%) | 5 (3%) | 0 (0%) | 0.98 | 0.02 | |||||
| T755C* | TT | TC | CC | T | C | ||||
| 123 (82%) | 27 (18%) | 0 (0%) | 0.91 | 0.09 | |||||
Genotype data are available for only 150 patients because of failed assays in one patient.
Genotype data are available for only 99 patients of the first study cohort.
Regarding the SLC22A16 T312C variant, patients carrying at least one C allele had lower day 15 platelet counts (mean platelet count 250 ± 107 × 109 vs. 307 ± 171 × 109 l−1, P = 0.036) than patients with the TT genotype, while patients with the SLC22A16 T1226C TT genotype had lower nadir platelet counts (mean platelet count 189 ± 70 × 109 vs. 244 ± 36 × 109 l−1, P = 0.044) than patients with at least one C allele. However, there was no correlation between these two variants with other doxorubicin pharmacokinetics or pharmacodynamics parameters. There was also no significant correlation between the SLC22A16 T755C variant with doxorubicin pharmacokinetics and pharmacodynamics.
Multivariate analysis
Three genetic variants, namely AKR1C3 IVS4–212 C>G (P = 0.006), ABCB1 IVS26+59 T>G (P = 0.026) and CBR3 G11A (P = 0.05), and two clinical parameters, namely metastatic status (metastatic vs. nonmetastatic, P < 0.001) and histological grade (grade 1 and 2 vs. 3, P = 0.005), influenced OS on univariate analysis and were included in multivariate Cox regression analysis. All five factors remained significantly predictive of OS (AKR1C3 IVS4–212 C>G, P = 0.017; ABCB1 IVS26+59 T>G, P = 0.036; CBR3 G11A, P = 0.031; metastatic status, P < 0.001; histological grade, P = 0.000). Both AKR1C3 IVS4–212 GG (P = 0.017) and metastatic status (P < 0.001) significantly influenced PFS on univariate analysis, and both remained significant on multivariate analysis (P = 0.044 and P < 0.001, respectively).
Discussion
Aldo-ketoreductases play an important role in the metabolism of a broad array of carbonyl-containing compounds, including anthracycline antibiotics, and functional AKR1C3 genetic variants have been previously described to affect doxorubicin metabolism 11. The two previously described functional SNPs in the AKR1C3 gene were not present in this Asian population. However, two common AKR1C3 intronic variants were identified, and we report, for the first time, correlations between the AKR1C3 IVS4–212 GG genotype and doxorubicin pharmacodynamics. Patients with the GG genotype developed significantly lower day 15 leucocyte and neutrophil counts after one cycle of doxorubicin, and had significantly longer progression-free and overall survival following doxorubicin-containing chemotherapy treatment. In concordance, these patients had slightly higher objective response rates, although the difference was not statistically significant. Given these observations, we postulate that the AKR1C3 IVS4–212 GG intronic variant may influence AKR1C3 activity and thus doxorubicin metabolism. Indeed, a previous in vitro study had demonstrated a high proportion of noncoding intronic SNPs to contribute towards chemotherapeutic drug sensitivity 17. Influence of intronic SNPs in polymorphic expression of cytochrome P450 3A5 has also been reported to cause variability in drug metabolisms 18, 19. Postulated molecular mechanisms include alteration of the upstream regulatory regions, resulting in alternative splicing and protein truncation of gene products.
The insignificant pharmacokinetics correlation that was demonstrated in our study in contrast to the significant pharmacodynamics findings may be related to sample size, because pharmacokinetic data were available in only a subset of patients; furthermore, the relationship between drug pharmacokinetics and pharmacodynamics is often complex and nonlinear, and pharmacokinetics correlations may be difficult to demonstrate clinically with analysis of only the parent compound and selected metabolites 20–22. For doxorubicin, factors contributing to the differences in pharmacokinetics end-points and the clinical pharmacodynamics outcomes may be attributed to tumour heterogeneity, pre-existing physiological conditions, age, co-morbid disease states and co-medications 23. This phenomenon of inconsistency observed in our study between the biological effects and pharmacological parameters is likely to be multifactorial and needs to be explored further.
Other SNPs in the genes for the CBR3 drug metabolizing enzyme as well as SLC22A16 influx and ABCB1 efflux transporters may potentially affect doxorubicin pharmacokinetics and pharmacodynamics 5. Thus, we took further steps to examine their effects in the context of AKR1C3 genotype variants. As previously reported, both CBR3 G11A and G730A variants influenced doxorubicinol AUC, with the former variant also correlating with overall survival. The well-studied ABCB1 G2677T/A correlated with both doxorubicin clearance and platelet counts but not survival in our study, while an ABCB1 intronic variant previously reported to be associated with cardiotoxicity was found to influence overall survival and white cell toxicity, although not in a concordant way, because survival was longer and white cell toxicity lower for patients with the G allele. Two of three SLC22A16 variants correlated with only platelet toxicities, with no other pharmacokinetics and pharmacodynamics effects observed. Interestingly, ABCB1 IVS26+59 T>G and CBR3 G11A variants correlated only with OS but not with PFS, contrary to what was observed with AKR1C3 IVS4–212 GG genotype, which prolonged both PFS and OS on both univariate and multivariate analysis that included traditional prognostic clinical parameters, such as metastatic status and histological grade of tumour. The discrepancy between the effects on OS and PFS for ABCB1 IVS26+59 T>G and CBR3 G11A variants suggested that the observed effects on OS were unlikely to be contributed solely by their influence on the doxorubicin disposition pathway but may be related to potential effects on subsequent lines of treatment or even tumour biology.
Another interesting finding from our study is the higher frequency of AKR1C3 IVS4–212 C>G GG homozygotes in our study population compared with those who carried at least one C allele (76 vs. 24%), which is in contrast with Caucasian populations, which have higher frequency of individuals carrying at least one C allele, ranging from 55.5 to 85% [National Center for Biotechnology Information, Reference SNP (ref. SNP) cluster report: rs1937840]. These interpopulation differences in genotype frequencies of SNPs may exert effects on drug outcomes in different populations. For doxorubicin pharmacodynamics, there are data suggesting interethnic differences, with Asians being more susceptible to doxorubicin-induced myelosuppression compared with Caucasians 24, 25. These findings are consistent with the significantly lower day 15 absolute leucocyte and neutrophil counts in patients who were IVS4–212 GG homozygotes, which is commoner in our Asian study population.
The strength of our study was its prospective nature, with systematic and prospective collection of efficacy and toxicity data. In addition, pharmacokinetics data were available for a significant proportion of the study cohort, allowing pharmacogenetics correlation for both doxorubicin pharmacokinetics and pharmacodynamics. However, as all the patients in the study cohort received docetaxel in addition to doxorubicin in the preoperative or postoperative setting, and many went on to receive other forms of anticancer therapy, such as surgery, radiotherapy and endocrine therapy, while doxorubicin contributed to the survival data, they could not be attributed solely to it.
Conclusions
We found a positive correlation between an AKR1C3 intronic variant with doxorubicin-induced haematological toxicity and survival, suggesting potential interaction with doxorubicin metabolism. These findings underpin the importance of pharmacogenetic factors on doxorubicin metabolism in explaining the interindividual variation of its efficacy and toxicity across various ethnic populations worldwide. Such findings could potentially translate into a better personalized treatment for different individuals in the future.
Acknowledgments
We are grateful to all patients who participated. This study was supported by grants from the National Medical Research Council, Singapore (NMRC/CSI/0009/2006, NMRC/CSI/0015/2009 and NMRC/CG/NCIS/2010) and the Cancer Science Institute Singapore (R-713-001-011–271). ClinicalTrials.gov ID: NCT00212082, NCT00669773.
Competing Interests
All authors have completed the Unified Competing Interest form and declared that there was no support from any organization for the submitted work, no financial relationships with any organization that might have an interest in the submitted work in the previous 3 years and no other relationships or activities that could appear to have influenced the submitted work.
References
- 1.Corte's-Funes H, Coronado C. Role of anthracyclines in the era of targeted therapy. Cardiovasc Toxicol. 2007;7:56–60. doi: 10.1007/s12012-007-0015-3. [DOI] [PubMed] [Google Scholar]
- 2.Barry E, Alvarez JA, Scully RE, Miller TL, Lipshultz SE. Anthracycline induced cardiotoxicity: course, pathophysiology, prevention and management. Expert Opin Pharmacother. 2007;8:1039–1058. doi: 10.1517/14656566.8.8.1039. [DOI] [PubMed] [Google Scholar]
- 3.Deng S, Wojnowskil L. Genotyping the risk of anthracycline-induced cardiotoxicity. Cardiovasc Toxicol. 2007;7:129–134. doi: 10.1007/s12012-007-0024-2. [DOI] [PubMed] [Google Scholar]
- 4.Menna P, Recalcati S, Cairo G, Minotti G. An introduction to the metabolic determinants of anthracycline cardiotoxicity. Cardiovasc Toxicol. 2007;7:80–85. doi: 10.1007/s12012-007-0011-7. [DOI] [PubMed] [Google Scholar]
- 5.Lal S, Mahajan A, Chen WN, Chowbay B. Pharmacogenetics of target genes across doxorubicin disposition pathway: a review. Curr Drug Metab. 2010;11:115–128. doi: 10.2174/138920010791110890. [DOI] [PubMed] [Google Scholar]
- 6.Evans WE, McLeod HL. Pharmacogenomics-drug disposition, drug targets, and side effects. N Engl J Med. 2003;348:538–549. doi: 10.1056/NEJMra020526. [DOI] [PubMed] [Google Scholar]
- 7.Fan L, Goh BC, Wong CI, Sukri N, Lim SE, Tan SH, Guo JY, Lim R, Yap HL, Khoo YM, Iau P, Lee HS, Lee SC. Genotype of human carbonyl reductase CBR3 correlates with doxorubicin disposition and toxicity. Pharmacogenet Genomics. 2008;18:623–631. doi: 10.1097/FPC.0b013e328301a869. [DOI] [PubMed] [Google Scholar]
- 8.Bray J, Sludden J, Griffin MJ, Cole M, Verrill M, Jamieson D, Boddy AV. Influence of pharmacogenetics on response and toxicity in breast cancer patients treated with doxorubicin and cyclophosphamide. Br J Cancer. 2010;102:1003–1009. doi: 10.1038/sj.bjc.6605587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lal S, Wong ZW, Sandanaraj E, Xiang X, Ang PCS, Lee EJD, Chowbay B. Influence of ABCB1 and ABCG2 polymorphisms on doxorubicin disposition in Asian breast cancer patients. Cancer Sci. 2008;99:816–823. doi: 10.1111/j.1349-7006.2008.00744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin Y, Penning TM. Aldo-ketoreductases and bioactivation/detoxification. Annu Rev Pharmacol Toxicol. 2007;47:263–292. doi: 10.1146/annurev.pharmtox.47.120505.105337. [DOI] [PubMed] [Google Scholar]
- 11.Veitch ZW, Guo B, Hembruff SL, Bewick AJ, Heibein AD, Eng J, Cull S, Maclean DA, Parissenti AM. Induction of 1C aldo-ketoreductases and other drug dose-dependent genes upon acquisition of anthracycline resistance. Pharmacogenet Genomics. 2009;19:477–488. doi: 10.1097/FPC.0b013e32832c484b. [DOI] [PubMed] [Google Scholar]
- 12.Bains OS, Grigliatti TA, Reid RE, Riggs KW. Naturally occurring variants of human aldo-ketoreductases with reduced in vitro metabolism of daunorubicin and doxorubicin. J Pharmacol Exp Ther. 2010;335:533–545. doi: 10.1124/jpet.110.173179. [DOI] [PubMed] [Google Scholar]
- 13.Visscher H, Ross CJ, Rassekh SR, Barhdadi A, Dubé MP, Al-Saloos H, Sandor GS, Caron HN, van Dalen EC, Kremer LC, van der Pal HJ, Brown AM, Rogers PC, Phillips MS, Rieder MJ, Carleton BC, Hayden MR. Pharmacogenomic prediction of anthracycline-induced cardiotoxicity in children. J Clin Oncol. 2012;30:1422–1428. doi: 10.1200/JCO.2010.34.3467. [DOI] [PubMed] [Google Scholar]
- 14.Lal S, Wong ZW, Jada SR, Xiang X, Chen Shu X, Ang PC, Figg WD, Lee EJ, Chowbay B. Novel SLC22A16 polymorphisms and influence on doxorubicin pharmacokinetics in Asian breast cancer patients. Pharmacogenomics. 2007;8:567–575. doi: 10.2217/14622416.8.6.567. [DOI] [PubMed] [Google Scholar]
- 15.Andersen A, Warren DJ, Slørdal L. A sensitive and simple high-performance liquid chromatographic method for the determination of doxorubicin and its metabolites in plasma. Ther Drug Monit. 1993;15:455–461. doi: 10.1097/00007691-199310000-00016. [DOI] [PubMed] [Google Scholar]
- 16.James K, Eisenhauer E, Christian M, Terenziani M, Vena D, Muldal A, Therasse P. Measuring response in solid tumors: unidimensional versus bidimensional measurement. J Natl Cancer Inst. 1999;91:523–528. doi: 10.1093/jnci/91.6.523. [DOI] [PubMed] [Google Scholar]
- 17.Gamazon ER, Huang RS, Cox NJ, Dolan ME. Chemotherapeutic drug susceptibility associated SNPs are enriched in expression quantitative trait loci. PNAS. 2010;107:9287–9292. doi: 10.1073/pnas.1001827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, Maurel P, Relling M, Brimer C, Yasuda K, Venkataramanan R, Strom S, Thummel K, Boguski MS, Schuetz E. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27:383–391. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 19.Giacomini KM, Brett CM, Altman RB, Benowitz NL, Dolan ME, Flockhart DA, Johnson JA, Hayes DF, Klein T, Krauss RM, Kroetz DL, McLeod HL, Nguyen AT, Ratain MJ, Relling MV, Reus V, Roden DM, Schaefer CA, Shuldiner AR, Skaar T, Tantisira K, Tyndale RF, Wang L, Weinshilboum RM, Weiss ST, Zineh I. The Pharmacogenetics Research Network: from SNP Discovery to Clinical Drug Response. Clin Pharmacol Ther. 2007;8:328–345. doi: 10.1038/sj.clpt.6100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lennard I, Lilleyman JS. Variable mercaptopurine metabolism and treatment outcome in childhood lymphoblastic leukemia. J Clin Oncol. 1987;7:1816–1823. doi: 10.1200/JCO.1989.7.12.1816. [DOI] [PubMed] [Google Scholar]
- 21.Rodman JH, Abromowitch M, Sinkule JA, Hayes FA, Rivera GK, Evan WE. Clinical pharmacodynamics of continuous infusion teniposide: systemic exposure as a determinant of response in a phase I trial. J Clin Oncol. 1987;5:1007–1014. doi: 10.1200/JCO.1987.5.7.1007. [DOI] [PubMed] [Google Scholar]
- 22.Jodrell DI, Egorin MJ, Canetta RM, Langenberg P, Goldbloom EP, Burroughs JN, Goodlow JL, Tan S, Wiltshaw E. Relationships between carboplatin exposure and tumor response and toxicity in patients with ovarian cancer. J Clin Oncol. 1992;10:520–528. doi: 10.1200/JCO.1992.10.4.520. [DOI] [PubMed] [Google Scholar]
- 23.Ackland SP, Ratain MJ, Vogelzang NJ, Choi KE, Ruane M, Sinkule JA. Pharmacokinetics and pharmacodynamics of long-term continuous-infusion doxorubicin. Clin Pharmacol Ther. 1989;45:340–347. doi: 10.1038/clpt.1989.39. [DOI] [PubMed] [Google Scholar]
- 24.Ma B, Yeo W, Hui P, Ho WM, Johnson PJ. Acute toxicity of adjuvant doxorubicin and cyclophosphamide for early breast cancer- a retrospective review of Chinese patients and comparison with an historic Western series. Radiother Oncol. 2002;62:185–189. doi: 10.1016/s0167-8140(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 25.Beith JM, Goh BC, Yeo W, Sullivan A, Lim S, Zhong S, Rivory LP. Inter-ethnic differences in the myelotoxicity of adriamycin/ cyclophosphamide (AC) or adjuvant breast cancer. Proc Am Soc Clin Oncol. 2002;21:252. abstract. [Google Scholar]


