The cytochrome P450 (CYP) 3A4*22 allelic status influences the pharmacokinetics of tacrolimus independently of an individual's CYP3A5 genotype 1, 2. In their recent publication, Passey et al. 3 described an algorithm that predicts tacrolimus apparent clearance (CL/F) by taking into account the age and CYP3A5 genotype of a patient, time after kidney transplantation, whether the transplantation was performed at a steroid-sparing centre or not and whether the patient was treated with a calcium channel blocker (CCB) (Box 1). However, as the effect of the CYP3A4*22 allele has only recently been reported, these authors did not consider this variable as a potential predictive factor for tacrolimus CL/F.
Figure 1.
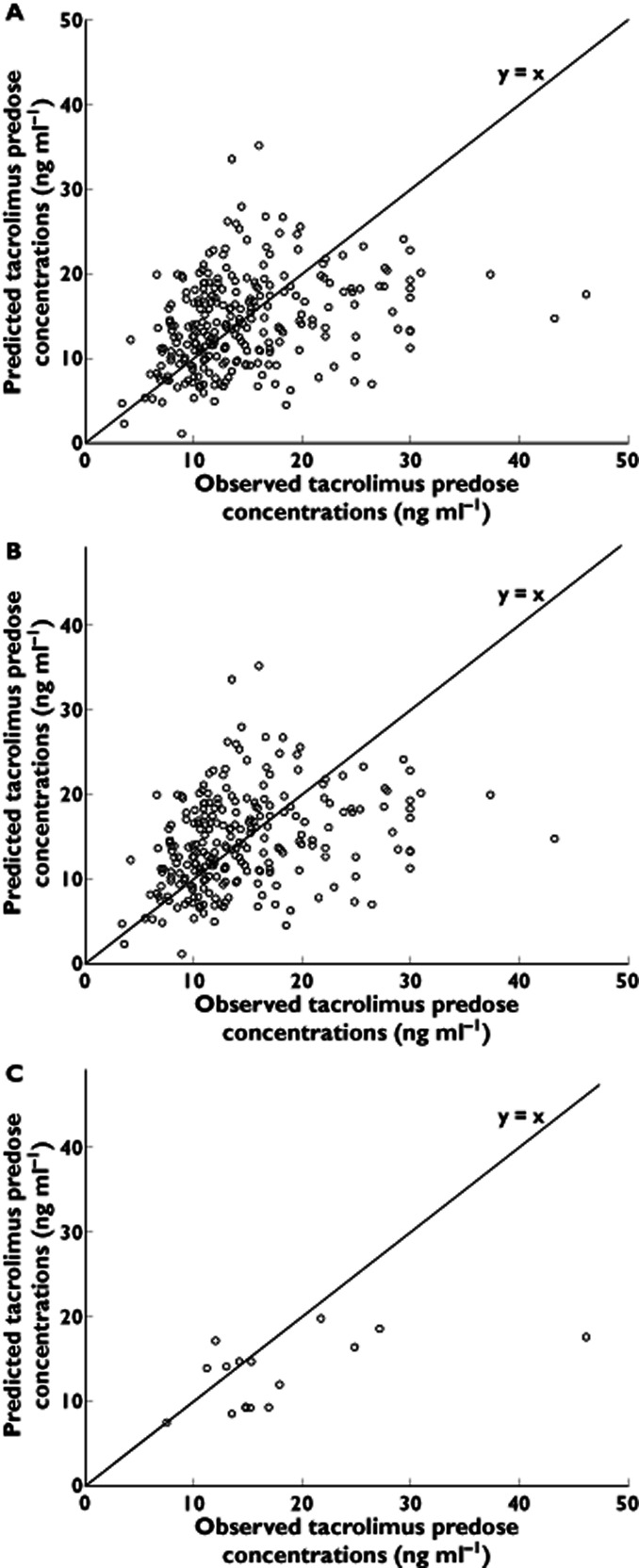
Observed tacrolimus predose concentrations vs. predicted tacrolimus predose concentrations in (A) the whole population, (B) CYP3A4*1/*1 homozygous wild-type patients and (C) in CYP3A4*22 carriers
Box 1 Tacrolimus apparent clearance (CL/F) predictive equation [3]
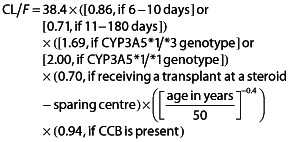 |
CL/F, apparent clearance; CCB, calcium channel blockers
The aim of the present report was to evaluate whether taking the CYP3A4*22 allelic status into account affects the predictive value of the algorithm developed by Passey et al. 3. The patients included in the present analysis were 185 de novo kidney transplant recipients treated with a tacrolimus-based regimen who participated in a trial comparing fixed-dose (FD) with concentration-controlled (CC) mycophenolate mofetil (MMF) treatment 4. All patients had been previously genotyped for CYP3A4*22 and CYP3A5*3 alleles 1. The required patient characteristics were inserted in the equation 3 to obtain the predicted tacrolimus CL/F for each individual. Predicted predose concentrations (Cpred) were then estimated according to the following equation:
where CL/F is in l h−1 and the dose administered is the total daily dose of tacrolimus in mg divided by 24 h. In the FDCC study, information on the use of CCBs was not available. The corresponding term in the equation was thus left out of the algorithm but the resulting effect is assumed to be minor as (i) the concomitant use of a CCB is associated with only a small (6%) decrease in tacrolimus CL/F (Box 1) and (ii) an estimated 30–50% of the population are CCB users, and most are on dihydropyridine CCBs, which have only a limited effect on CYP3A activity 5. As in the validation study of Passey et al. 6, the evaluation of the predictive performance of the equation was restricted to initial predose concentrations (days 3 and 10). The predictive values of the algorithm were estimated as described in this validation study 6. The predictive performance for the estimation of predose concentrations was thus evaluated with individual prediction errors (PEi). PEi was calculated as the difference between Cpred and the corresponding observed predose concentration (Cobs):
The bias was defined as the median prediction error (MPE = median PEi) and the precision as the median absolute prediction error (MAPE = median PEi). The significance of the inaccuracy of the prediction was assessed with one-sample Wilcoxon single rank test under the null hypothesis that the MPE or MAPE do not deviate from 0. To evaluate the difference in the performance parameters obtained in both CYP3A4*22 carriers and CYP3A4*1/*1 groups, we used a Mann-Whitney test under the null hypothesis that there is no difference in bias or precision between both CYP3A4 genotype groups.
For the entire population, Cobs was plotted against Cpred (Figure 1A). We observed a bias (MPE) of +0.09 ng ml−1 and a precision (MAPE) of 4.30 ng ml−1. In their validation study, Passey et al. reported a bias of +0.2 ng ml−1 6, which is in line with our data. However, they reported a precision of 1.8 ng ml−1 which reflects a lower relative scattering of their predicted values compared with ours. The MAPE in our study deviates significantly from 0 (P < 0.001). This lack of precision in our dataset could be due to the lower number of predose concentrations available in our study (n = 265) in comparison with the study of Passey et al. (n = 412) and/or the non-consideration of the use of CCBs. Nevertheless, a good accuracy of the equation was still observed and the slight Cpred overestimation (i.e. +0.09 ng ml−1) is suggested to be minor as it was not significant (P = 0.87). When the analysis was performed by taking CYP3A4*22 carriership into account (Figure 1B and C), in CYP3A4*1/*1 homozygous patients the bias was +0.36 ng ml−1 but not significantly different from 0 (P = 0.70). By contrast, in CYP3A4*22 carriers, a negative bias of −5.24 ng ml−1 was observed, which did significantly deviate from 0 (P = 0.013). The difference in bias observed in both groups was significant (0.36 vs. −5.24 ng ml−1, P = 0.011).
As the CYP3A4*22 allele has been previously associated with a reduced tacrolimus clearance 1, 2, it is not surprising that an algorithm that does not take into account the CYP3A4*22 genotype will overrate the individual's ability to metabolize the drug. Indeed, predose concentrations are thus underestimated and, as a result, a negative bias is observed. This information is important as using this algorithm for CYP3A4*22 carriers may lead to prescription of a too high tacrolimus dosage, which could predispose these patients to drug overexposure and increase their risk of adverse events. The difference in precision was not significant between CYP3A4*22 genotype groups (4.2 ng ml−1 vs. 5.3 ng ml−1 for CYP3A4*1/*1 and CYP3A4*22 carriers, respectively, P = 0.97). In conclusion, this report confirms that the previously published algorithm predicting tacrolimus clearance is able to determine early predose concentrations in the general population but might be inaccurate for patients carrying the CYP3A4*22 decrease-of-function allele. Consequently, we believe that the equation should be refined and include the CYP3A4*22 allelic status of the patient.
Acknowledgments
Laure Elens is a post-doctoral researcher with the Fonds National de la Recherche Scientifique (FRS-FNRS), Belgium.
Competing Interests
All authors have completed the Unified Competing Interest form and that they are available on request from the corresponding author. All authors declare no support from any organization for the submitted work, no financial relationships with any organizations that might have an interest in the submitted work in the previous 3 years and no other relationships or activities that could appear to have influenced the submitted work
Editor's note
The authors (Passey et al.) of the cited BJCP article were offered the opportunity to comment on the letter but declined to do so.
References
- 1.Elens L, Bouamar R, Hesselink DA, Haufroid V, van der Heiden IP, van Gelder T, van Schaik RH. A new functional CYP3A4 intron 6 polymorphism significantly affects tacrolimus pharmacokinetics in kidney transplant recipients. Clin Chem. 2011;57:1574–1583. doi: 10.1373/clinchem.2011.165613. [DOI] [PubMed] [Google Scholar]
- 2.Elens L, van Schaik RH, Panin N, de Meyer M, Wallemacq P, Lison D, Mourad M, Haufroid V. Effect of a new functional CYP3A4 polymorphism on calcineurin inhibitors' dose requirements and trough blood levels in stable renal transplant patients. Pharmacogenomics. 2011;12:1383–1396. doi: 10.2217/pgs.11.90. [DOI] [PubMed] [Google Scholar]
- 3.Passey C, Birnbaum AK, Brundage RC, Oetting WS, Israni AK, Jacobson PA. Dosing equation for tacrolimus using genetic variants and clinical factors. Br J Clin Pharmacol. 2011;72:948–957. doi: 10.1111/j.1365-2125.2011.04039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Gelder T, Silva HT, de Fijter JW, Budde K, Kuypers D, Tyden G, Lohmus A, Sommerer C, Hartmann A, Le Meur Y, Oellerich M, Holt DW, Tonshoff B, Keown P, Campbell S, Mamelok RD. Comparing mycophenolate mofetil regimens for de novo renal transplant recipients: the fixed-dose concentration-controlled trial. Transplantation. 2008;86:1043–1051. doi: 10.1097/TP.0b013e318186f98a. [DOI] [PubMed] [Google Scholar]
- 5.Ma B, Prueksaritanont T, Lin JH. Drug interactions with calcium channel blockers: possible involvement of metabolite-intermediate complexation with CYP3A. Drug Metab Dispos. 2000;28:125–130. [PubMed] [Google Scholar]
- 6.Passey C, Birnbaum AK, Brundage RC, Schladt DP, Oetting WS, Leduc RE, Israni AK, Guan W, Matas AJ, Jacobson PA. Validation of tacrolimus equation to predict troughs using genetic and clinical factors. Pharmacogenomics. 2012;13:1141–1147. doi: 10.2217/pgs.12.98. [DOI] [PMC free article] [PubMed] [Google Scholar]


