Abstract
Epstein-Barr virus (EBV) has evolved exquisite controls over its host cells, human B lymphocytes, not only directing these cells during latency to proliferate and thereby expand the pool of infected cells, but also to survive and thereby persist for the lifetime of the infected individual. Although these activities ensure the virus is successful, they also make the virus oncogenic, particularly when infected people are immunosuppressed. Here we show, strikingly, that one set of EBV’s miRNAs both sustain BL (Burkitt’s lymphoma) cells in the absence of other viral oncogenes and promote the transformation of primary B lymphocytes. Burkitt’s Lymphoma cells were engineered to lose EBV and found to die by apoptosis and could be rescued by constitutively expressing viral miRNAs in them. Two of these EBV miRNAs were found to target Caspase 3 to inhibit apoptosis at physiological concentrations.
Keywords: EBV, BART miRNAs, apoptosis, RISC-IP, Burkitt’s Lymphoma
Introduction
Epstein-Barr virus is an extraordinary pathogen. Rather than infecting replicating cells, it infects quiescent cells and drives them to proliferate. Rather than integrating its genome into its host cell’s DNA, it maintains itself as a licensed plasmid replicon in infected, proliferating cells contributing only one protein, EBNA1, to this replicon. This latter property has a cost for the virus. Because not all copies of the viral DNA are duplicated each cell cycle 1 the virus will be lost from proliferating cells. EBV can be retained in populations of these cells only if it provides them sufficient selective advantages to outgrow those cells that lose it. Burkitt’s lymphoma was the first human cancer found to be caused by a virus, EBV. We now know that EBV is found in subsets of all major categories of lymphoma, but curiously, its best-known oncogenes – those that drive cells to survive and proliferate - are not consistently expressed in tumor cells, presumably because these viral proteins are immunogenic 2. Indeed, canonical Burkitt’s lymphomas express none of these viral oncogenes. We have previously developed a means to reveal the selective advantage the virus affords cells by evicting EBV conditionally from them 3. Specifically, lymphoma cells have been engineered to express inducibly a dominant negative derivative of EBNA1 4 (dnEBNA1), which forces the loss of EBV plasmids over time by interfering with EBNA1’s ability to foster the replication and partitioning of EBV genomes in proliferating cells. Here we have uncovered how the virus sustains Burkitt’s lymphoma cells in the absence of its well-known oncogenes. It uses some of its miRNAs, which are unlikely to be detected by the host’s immune response, to inhibit apoptosis, a developmental fate common to uninfected B cells. We have developed derivatives of EBV that do or do not encode the viral miRNAs expressed in Burkitt’s Lymphomas. These viruses differ in their transformation of primary B-cells, showing that the viral miRNAs which help maintain Burkitt’s Lymphomas also aid transformation of naïve B-cells.
Results
EBV’s BART miRNAs block apoptosis in canonical Burkitt’s lymphomas
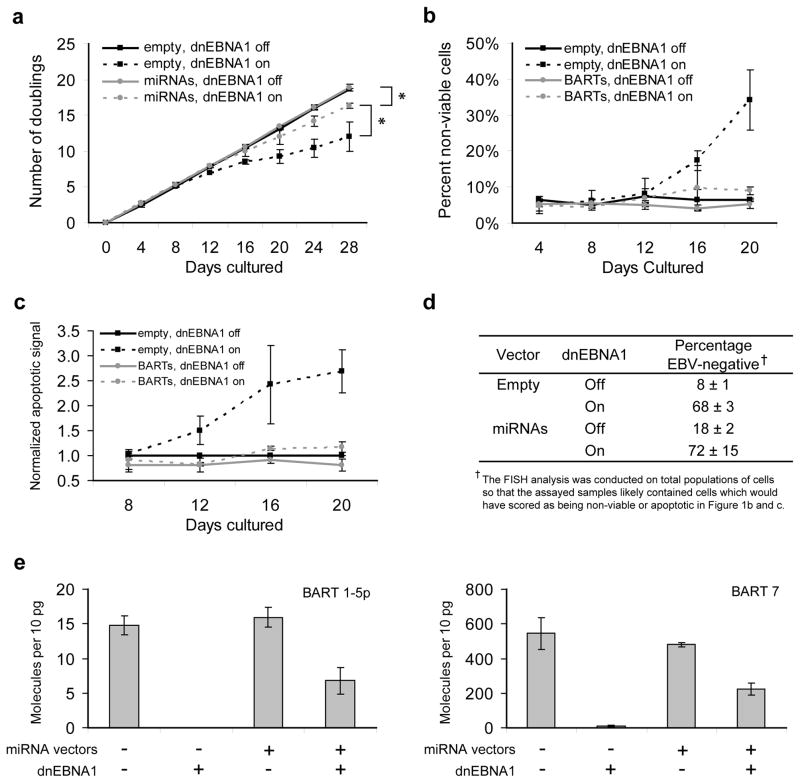
Canonical Burkitt’s lymphomas express the smallest set of EBV genes of studied tumors and, in particular, do not express viral oncogenes such as LMP1 or EBNA2 12. They do express the BART miRNAs, the EBERs and EBNA1 3. We asked if the BART miRNAs sustain these lymphoma cells. To do so, cells were engineered from two recently explanted canonical Burkitt’s lymphomas, Sav-BL and Dante-BL, to express dnEBNA1 inducibly 3 and viral miRNAs ectopically. These cell lines have not been extensively passaged in culture and thus are likely representative of BL. The BART miRNAs (specifically, portions of EBV DNA encoding BART 1, 3-20, and 22), were introduced into dnEBNA1-inducible clones of the BL cell lines Sav-BL (clone S1-1) and Dante-BL (clone D7-1) by two sequential rounds of retroviral transduction and FACS (fluorescence activated cell sorting). At this level of ectopic expression, cells losing EBV but expressing the BART miRNAs proliferated robustly when compared to control cells losing EBV; by 28 days post-induction there were 20-fold more BART-complemented than non-complemented cells (Figure 1a). The presence of the ectopic BART miRNAs reduced global cell death (Figure 1b), revealing that the BART miRNAs were promoting population growth at least in part by inhibiting cell death. The BART miRNAs prevented cell death by blocking the induction of apoptosis (Figure 1c). Importantly, the ectopic expression of the BART miRNAs did not interfere with the ability of dnEBNA1 to evict the virus (Figure 1d). In fact, the presence of ectopic BART miRNAs allows cells that spontaneously lose the virus (3–5% of the cells per generation, 1) to accumulate in the population expanding the percentage of EBV-negative cells from 8 to 18% (Figure 1D). The levels of the miRNAs in the complemented cells (surviving on ectopic miRNA expression) were found to be approximately one-half that of wildtype levels (Figure 1e), for both a scarce miRNA (BART 1-5p) and a plentiful miRNA (BART 7). The minor subpopulation of cells that fail to evict the virus fully also fail to contribute to the BART expression of the total population measurably over the times studied (the level of BART 7, for example, drops 100-fold; Figure 1E), indicating either that the residual viral genomes are inefficient templates for expression, or that the FISH analysis underestimates the number of EBV-negative cells. Similar results were found for Dante-BL clone D7-1 (Supplementary Figure 1). The forced loss of EBV in D7-1 cells is inefficient (the majority of the cells fail to lose the virus, Supplementary Figure 1b), however, even under these conditions the BART-complemented cells accumulated 6-fold more cells by day 28 post-induction of dnEBNA1 than those with empty vector controls. Taken together, these results demonstrate that the BART miRNAs maintain canonical BL tumor cells by blocking apoptosis. The complementing BART vectors do not appreciably express BART5 10, which has previously been found to regulate the transcript of the anti-apoptotic gene BBC3 (PUMA) 13 in epithelial cells. EBV’s miRNAs are thus inhibiting apoptosis in these BLs by a previously unknown mechanism. It is not now known whether a wildtype level of expression of the BART miRNAs would fully complement the loss of EBV or whether the EBERs and EBNA1 also directly contribute to sustaining these tumor cells.
Figure 1. Ectopic expression of BART miRNAs partially complements the loss of EBV in BL cells.
(a) The population growth of S1-1 cells was measured over time as EBV was evicted following induction of dominant negative EBNA1 (dnEBNA1) at day 0 with doxycycline. The average of three independent experiments, each with one technical replicate ± SD is shown. * p=0.05, one-sided Wilcoxon signed rank test, comparing total number of cell doublings. (b) Cells with or without exogenously introduced BART miRNAs plus or minus induction of dnEBNA1 to evict EBV were assayed for global cell death by staining cells with trypan blue and examining cell morphology. For each data point at least 600 cells were analyzed. The average of three independent experiments ± SD (at least 200 cells per experiment) is shown. (c) Cells as treated in B were assayed for the induction of apoptosis by detecting the activation of caspases (Caspase-Glo 3/7, Promega). Signals were normalized to homeostatic levels (cells transduced with an empty vector in which dnEBNA1 is not induced) which were arbitrarily set to one. The average of three independent experiments ± SD is shown. (d) Cells were cultured for 20 days with or without induction of dnEBNA1 and then scored for the presence of viral DNA by FISH analysis. For each experiment, at least 200 cells were counted per condition. The average percentage of EBV-negative cells ± SD from three independent experiments is shown. (e) Real-time PCR measurements of the levels of two BART miRNAs, BART 1-5p (left graph) and BART 7 (right graph), were made 20 days after induction of dnEBNA1 in S1-1 cells. The average number of miRNA molecules per 10 picograms of total RNA ± SD is shown from three independent experiments.
We took advantage of the fact that by 20 days following induction of dnEBNA1, BART miRNA expression is virtually undetected (Figure 1d and 1e) to interrogate this population of cells for molecular changes that might explain how the BART miRNAs sustain the tumor cells. Comparing these cells against ones in which the BART miRNAs are ectopically expressed provides a unique opportunity to assess the role of the miRNAs in BL cells that are dependent on these viral genes.
It was possible that the BART miRNAs exerted their effects on survival by down-regulating the pro-apoptotic protein BIM. BIM regulates some facets of B cell development and can be targeted by cellular miRNAs 14–15. The expression of BIM has been shown to be inhibited by EBV 16–17 and is upregulated in the BL cell line Oku-BL as EBV is evicted from it 3. To test this possibility, BIM levels were measured as EBV was depleted from S1-1 cells in the absence or presence of ectopic expression of BART miRNAs. No correlation between the presence of the BART miRNAs and BIM levels could be observed (Supplementary Figure 2). Canonical Burkitt’s lymphoma cells such as S1-1 express neither EBNA3A nor EBNA3C, two viral proteins found to down-regulate BIM concertedly, 16 consistent with their having evolved to regulate BIM epigenetically 18. Thus, the miRNAs likely exert their anti-apoptotic effect by means other than repressing BIM translation.
EBV’s BART miRNAs target CASP3 to inhibit apoptosis
We sought to identify mRNAs regulated by the BART miRNAs with computational algorithms 19–20 which were uninformative both because herpesviral miRNAs are poorly conserved 21–23 and the large number of BART miRNAs leads to too many predicted targets to be useful. Instead, taking an empirical approach to identify cellular mRNAs indirectly or directly regulated by the BART miRNAs, the transcriptome of S1-1 cells was examined with microarrays and the RNA induced silencing (RISC) complexes were interrogated by deep sequencing.
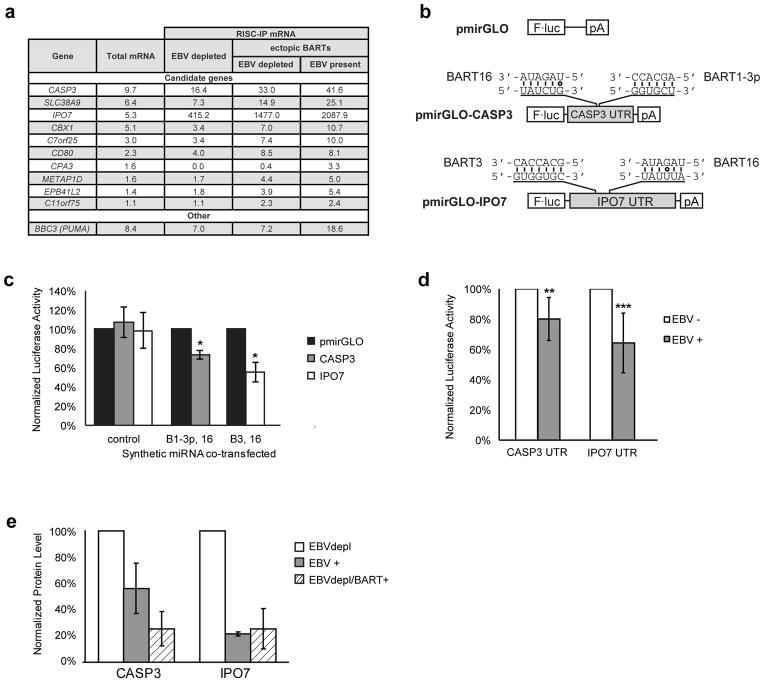
To analyze the transcriptome, levels of cellular transcripts in cells in which EBV was depleted (uncomplemented cells) were set as a baseline and compared with the levels in cells in which EBV was depleted in the presence of ectopic BART miRNAs (complemented cells). Three biological replicates of microarrays identified seven candidates (Supplementary Fig. 3) whose levels correlated with the presence of EBV and the presence of the BARTs. Three of these candidates –chemokine receptor 7 (CCR7), cluster of differentiation 2 (CD2), and chorionic gonadotrophin alpha (CGA) – could be confirmed by real-time PCR (Supplementary Fig. 3). CCR7 was inhibited, while CD2 and CGA were stimulated in the presence of the BART miRNAs. CCR7 has been found to be induced in infection of an EBV-negative Burkitt’s cell line with the B95.8 strain of EBV which lacks most of the BART miRNAs 24 making its inhibition by them intriguing. It was not clear, though, how any of these three potential targets provides an obvious role in EBV’s maintenance of Burkitt’s lymphomas. Rather, these data indicated that EBV’s miRNAs were likely to promote survival directly through the regulation of translation, and not indirectly through the regulation of transcription.
The mechanism by which miRNAs inhibit translation is generally by their incorporation into RISC. RISCs loaded with miRNAs bind target mRNAs and prevent their translation. To analyze the transcripts associated with RISCs in S1-1 cells, these complexes were immunoprecipitated and the RNAs present were measured by deep sequencing. Transcripts of interest were identified as those that 1) were enriched in RISC in EBV positive cells, 2) were lost from RISC as EBV was evicted from cells, and 3) were returned to RISC in EBV-depleted cells that ectopically expressed BART miRNAs with two-fold or greater changes for each of these criterion (Figure 2a). The transcript for BBC3 (PUMA) was not enriched in the RISC in EBV-depleted cells that ectopically expressed BART miRNAs but was enriched in the presence of endogenous EBV.
Figure 2. Identification of BART miRNA targets.
(a) RNA-seq was performed on total mRNA and RISC immunoprecipitations from S1-1 cells. The top 10 candidate genes (ranked by their expression level in the S1-1 cells) are listed with their detected expression values for each condition. As a control, the expression values of the BART5 target BBC3 (PUMA) are also listed. BART5 is inefficiently expressed from the BART retroviral vectors. (b) Reporter assays were conducted with constructs encoding the 3′-UTRs of CASP3 and IPO7 downstream of luciferase. The predicted target sites and corresponding miRNA seed sequences are shown. (c) 293 cells were transfected with luciferase vectors and mature synthetic miRNA, and the luciferase activity was normalized as described in the Materials and Methods. Data are the average of three independent experiments ± s.d. (*p<0.05, Wilcoxon rank sum test). (d) The vectors were introduced into EBV-positive or EBV-negative Oku-BL cells, and the normalized luciferase activity in the EBV-negative cells was set to 100%. Data are the average of six independent experiments ± s.d. (**p =0.037, ***p=0.006, Wilcoxon rank sum test). (e) Importin-7 and caspase-3 levels from cell lysates of S1-1 cells treated as in Figure 1 were measured by Western blot. Importin-7 and caspase-3 levels were normalized to alpha-tubulin levels, and then compared to the EBV depleted cells (cells transduced with empty vectors, dnEBNA1 turned on) whose normalized level was arbitrarily set to 100%. Data are the average of two independent experiments ± s.d. The blot images are given in Supplementary Figure 5.
The set of transcripts potentially targeted by the ectopic BART miRNAs include IPO7 and CASP3. Transcripts for both of these genes have recently been either detected or been found to be enriched in RISCs in the presence of the BARTs, 25–27 but the functional consequences of this detection or enrichment remain unknown. CASP3 is a particularly tantalizing possible target because of its well-documented role in the induction of apoptosis, a phenotype prevented by the presence of the BART miRNAs in S1-1 cells. To test if CASP3 and IPO7 are targets of the BART miRNAs, luciferase reporter assays were performed. The 3′ UTRs of CASP3 and IPO7 were cloned downstream of luciferase (Figure 2b) and introduced into 293 cells along with synthetic mature BART miRNAs. A reproducible inhibition of luciferase was observed in the presence of miRNAs predicted to bind the UTRs (Figure 2c). To determine whether the constructs were targeted by miRNAs expressed at physiological levels, the vectors were introduced into the EBV-positive Burkitt’s lymphoma cell line Oku-BL and its engineered EBV-negative counterpart (Kuzembayeva and Sugden, unpublished). The presence of EBV correlated with a reproducible inhibition of luciferase activity for constructs containing the 3′-UTR of either CASP3 or IPO7; inhibition by the CASP3 3′-UTR was dependent on its binding sites for BART16 and BART 1-3p (Figure 2d, Supplementary Figure 4). If EBV’s BART miRNAs do regulate the expression of CASP3 and IPO7 directly in BL cells, the levels of these proteins should increase as EBV is evicted from them. Indeed, the levels of CASP3 and IPO7 proteins did increase as EBV was depleted from S1-1 cells and the ectopic expression of the BART miRNAs in these cells reduced those levels (Figure 2e, Supplementary Figure 5). In fact, the observed suppression of CASP3 and IPO7 proteins was more robust than the reporter assays predicted, revealing that the reporters likely underestimated the inhibitory capacity of the miRNAs. Taken together, these data indicate that IPO7 and CASP3 are bonafide targets for direct regulation by the BART miRNAs. Furthermore, they indicate that EBV’s miRNAs function to promote the survival of S1-1 cells at least in part by regulating the pro-apoptotic CASP3 transcript.
EBV’s BART miRNAs promote proliferation of newly infected B cells
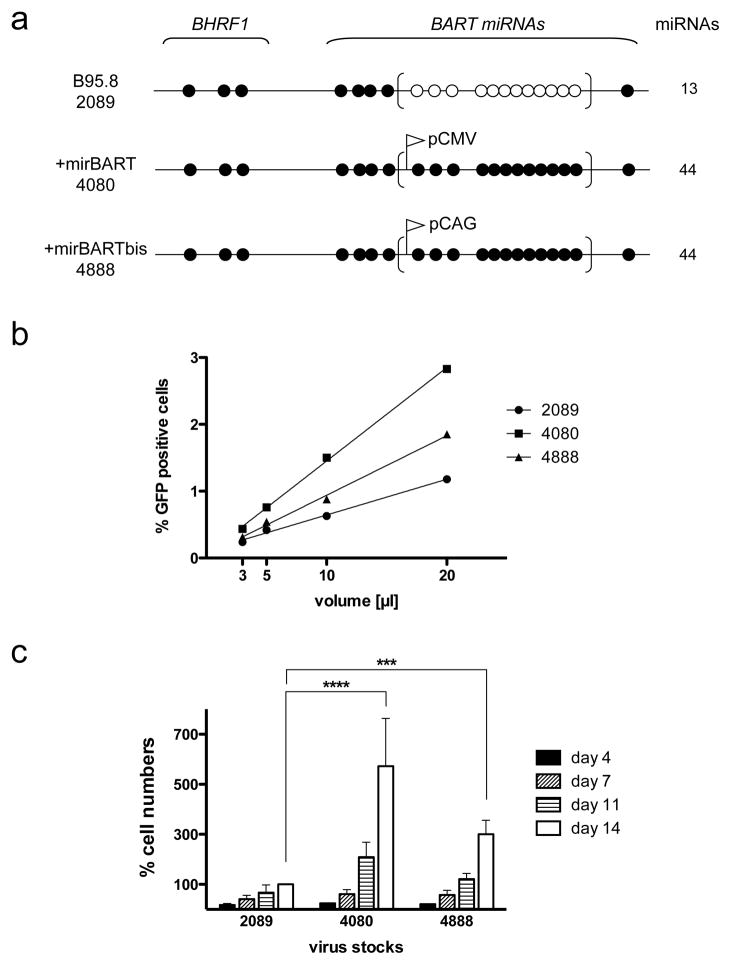
Because we had uncovered such a central role for EBV’s miRNAs in sustaining Burkitt’s lymphoma cells, we tested the hypothesis that they also contribute detectably to the transformation of primary B-lymphocytes. Variants of EBV were used to test this hypothesis. Because the laboratory strain of EBV, B95-8, and its recombinant, 2089, express only 5 of the 22 known BART pre-miRNAs, two reconstituted variants were generated which ectopically express all the known BART miRNAs (Figure 3a). The 22 pre-miRNAs of the BART locus were assembled either under the control of the CMV promoter and introduced into the BALF1 gene of EBV, which acts redundantly (26), to yield the strain termed 4080 or under the control of the CAG promoter 28 and inserted into the prokaryotic backbone of 2089 to yield the strain termed 4888. The levels of selected BART miRNAs expressed in cells infected with 4080 were compared with those in cells infected with a field strain of EBV that also encodes all the known BART miRNAs. While miRNAs encoded by 2089 (BART 1-5p and 2-5p) were expressed at comparable levels, miRNAs not encoded by 2089 (BART 8-5p and 22) were expressed over a range from wild-type (BART 22) or 15% (BART 8-5p) of levels of the field strain 9.
Figure 3. BART miRNAs promote the transformation of primary human B cells.
(A) A simplified diagram of the pre-miRNAs of the three recombinant viruses studied is shown. The upper diagram represents 2089, which is the recombinant version of the prototypic EBV strain B95-8. Closed circles represent encoded pre-miRNAs; open circles represent deleted pre-miRNAs. EBV field strains other than the reference strain B95.8 encode up to 25 pre-miRNAs, which result in four mature BHRF1 miRNAs and 40 BART miRNAs. Two reconstituted EBV mutants that ectopically express the full set of 40 BART miRNAs were assembled from sub-genomic fragments as described 9. To construct the mutant EBV +mirBART (4080), the BART miRNA locus was inserted into the BALF1 gene of EBV, where it is expressed under the control of the CMV immediate early promoter. To construct the mutant EBV +mirBARTbis (4888), the assembled BART miRNA locus was inserted into the prokaryotic plasmid backbone of the prototypic 2089 strain, where it is expressed from the composite CAG promoter 28.
(B) Titration of virus stocks. 1×105 Raji cells were infected with different volumes of virus stocks as shown. Three days post infection, the percentage of GFP-positive Raji cells was determined on a FACS machine, calculated and plotted. The virus stocks 2089, 4080, and 4888 contained 7.1×104±1×104, 1.5×105±4.6×103, and 1×105±9.3×103 green Raji units [GRU]/ml, respectively. (C) Primary human B cells isolated from adenoids from five different donors were infected with the three virus stocks at a multiplicity of infection of 1×10−3, incubated for 18 hours, and seeded in fresh medium at a density of 1.5×105 cells per ml. At the days indicated the cells were harvested and their proliferation analyzed by FACS. To determine the absolute number of cells counted, a volume standard was added prior to FACS analysis as described 9, 30. To correct for variations between donors’ B cells, the raw cell counts were normalized to those infected with 2089 at day 14 (whose value was arbitrarily set to 100%). The average of five different donors ± SD is shown. p-values (p<0.001 ***, p<0.0001 ****) for the time points are indicated (two way ANOVA linked to Bonferroni post-tests; www.graphpad.com, Prism vers. 5.0).
The recombinant EBV strains 2089, 4080 and 4888 were carefully titered (Figure 3b) and used to infect primary B cells to identify contributions the BART miRNAs make to affecting survival and/or proliferation of infected cells in the presence of the other viral oncogenes. When freshly isolated B cells from five donors were infected under conditions to mimic infections in vivo (e.g. a low MOI), the primary B cells infected with the reconstituted strains 4080 and 4888 had increasingly higher numbers of activated cells than those infected with 2089 (Figure 3c), a consequence of an enhanced rate of proliferation or survival, or both. This increased efficiency of transformation occurred in B-cells isolated from both adenoids and peripheral blood (Supplementary Figure 6). That both viral strains expressing the BART miRNAs from different promoters in different genomic contexts enhance transformation of primary B-cells demonstrates that the BART miRNAs act to promote their survival and/or proliferation.
Discussion
EBV drives the proliferation and survival of infected B-cells by expressing multiple oncogenes. While the virus is clearly associated with lymphoma, its identified viral oncogenes often are no longer expressed in these lymphomas. In addition to oncogenes, EBV expresses many miRNAs which, given the size of its genome, are encoded at a much higher frequency per genome length than by the human genome. The role of these miRNAs in the viral life cycle has remained elusive, though. We have found that tumor cells which do not express viral oncogenes but otherwise retain the virus are sustained by the BART miRNAs. The miRNAs’ survival functions include blocking apoptosis. We favor a mechanism in which the BARTs exert their anti-apoptotic effects by repressing the translation of the pro-apoptotic transcript of CASP3 because: 1. we detect activated caspases in BL tumor cells in the absence of EBV; their activation is prevented by the introduction of the miRNAs (Figure 1c); 2. the levels of a CASP3 luciferase reporter are reduced in the presence of EBV or in the presence of BART 1-3p and 16 alone (Figure 2c and d); and 3. the protein levels of CASP3 decrease in the presence of EBV or with the introduction of the BART miRNAs (Figure 2e). CASP3 was also recently found to be targeted by miRNAs encoded in another human tumor herpesvirus KSHV 29, suggesting a conserved requirement for these viruses to inhibit expression of CASP3 during their life cycle. However, other targets of the BART miRNA (such as IPO7) may contribute to the survival of BL tumor cells by as yet unknown mechanisms. The role of these uncharacterized targets are thus a focus for further investigation, and our system using inducible dnEBNA1 provides a tractable means to do so. Surprisingly, BART miRNAs also contribute to the survival and proliferation of newly infected B-cells, at the time when potent viral oncogenes begin to be expressed, when the B-cells are infected under conditions likely similar to infection in vivo. This contribution by the BART miRNAs occurred when they were expressed from two different recombinant viruses and used to infect B-cells isolated from the adenoids and peripheral blood of eight donors. It is clear that EBV’s miRNAs are significant viral contributors to the survival and proliferation of both infected normal and tumor cells. Its miRNAs encoded in the BHRF1 transcript have been found recently to inhibit apoptosis and foster proliferation of newly infected B-cells, too 9. Strikingly, these two newly identified phenotypes place EBV’s miRNAs in the center of the virus’s regulation of its host cell and underscore the wealth of functions EBV encodes and exploits during tumorigenesis.
Materials and methods
Cell lines and culture
The dnEBNA1-inducible Burkitt’s lymphoma cell lines have previously been described 3. All lymphoma cell lines were grown in RPMI 1640 (Gibco, Grand Island, NY) supplemented with 10% fetal bovine serum (R10F). The cell line 293T was grown in Dulbecco’s modified Eagle medium (Gibco) supplemented with 10% fetal bovine serum (D10F). When propagating retroviruses in 293T cells, 10% bovine calf serum was often substituted for fetal bovine serum. All cell culture media were supplemented with 200 U/mL penicillin and 200 μg/mL streptomycin. All cells were grown at 37°C in a 5% CO2 humidified atmosphere.
Retroviral transduction
Retroviral vectors (Supplementary Table 1) were generated in 293T cells as previously described with modifications. 5 Briefly, retrovirus was generated by cotransfecting 293T in a 70% confluent 10-cm dish with 3 μg of a plasmid encoding the Gag-Pol element, 1 μg of a plasmid encoding the vesicular stomatitis virus G protein, 1 μg of a plasmid encoding a derivative of NF-κB, and 10 μg of a plasmid carrying the retroviral backbone containing either intronic portions of the BART locus encoding BART 1, 3-20, and 22 or empty vector control (as described in Supplementary Table 1) using polyethylenimine (PEI; Sigma-Aldrich, St Louis, MO). A total of 40μg PEI per 10-cm dish was used. Twenty-four hours after transfection, the culture medium was replaced with Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum and 50 mM HEPES then the culture was irradiated (3 000 rads). Lymphoma cells were then co-cultivated on top of the irradiated 293T cells in D10F for 16–24 hours.
FISH (Fluorescent in situ hybridization)
Cells were stained for FISH analysis as previously described 1. To determine the percentage of EBV-negative cells, at least 200 cells per experiment were assayed at each time point.
Cell sorting
Single cells were sorted on FACS-Vantage SE with the FACS-DIVAoption (Becton Dickinson, San Jose, CA). To obtain cells that efficiently expressed the particular transgene, cells were sorted for highest fluorescence intensity (the top 10–20%) of the marker protein (mRFP or CFP) because the intensity of the marker correlates with levels of the co-expressed miRNA (data not shown).
RNA isolation
Total RNA was isolated using TRIzol reagent (Invitrogen, Grand Island, NY), following the manufacturer’s protocols, except RNA precipitation was conducted in the presence of approximately 5 μg/mL linear acrylamide (Ambion, Grand Island, NY) to enhance efficiency of precipitation as previously described 6. In some cases the isolated RNA was then treated with Turbo DNAse (Ambion) following the manufacturer’s instructions and re-purified with TRIzol.
Reverse transcription and real-time PCR
Measurements were conducted as described elsewhere 3, except in some cases SuperScript II reverse transcriptase (Invitrogen) was used. Probes were labeled with 5′-FAMRA and 3′-TAMRA or 3′-Iowa Black FQ (IDT, Integrated DNA Technologies, Coralville, IA). Primer and probe sequences are listed in Supplementary Table 2.
Stem-loop realtime PCR
EBV BART miRNAs were detected as described 7. They were specifically reverse transcribed using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Stem-loop primers to the assayed miRNAs were designed in a similar manner to those designed for reverse transcription of human miRNAs. Two modifications were made to the protocol. For each miRNA assayed, 250 ng of total RNA was reverse transcribed as described by the manufacturer. In addition, half the amount of the enzyme (Multiscribe RT) was often used.
Growth curves and cell viability measurements
Cells were diluted to 3–5×104 cells/mL in culture medium and treated with either 10 ng/mL doxycycline or the vehicle, ethanol. Live cell concentrations were measured every two or four days with a hemocytometer. Cells stained with trypan blue (with a 1:10 dilution of 0.3% trypan blue dissolved in 1x PBS) or exhibiting an aberrant morphology were considered non-viable. After counting, if necessary cells were diluted in fresh medium back to approximately the starting concentration (3–5×104 cells/mL) and doxycycline or ethanol was added.
Western blotting
Western blotting was performed as described 8; the blots were probed with rabbit polyclonal anti-Bim/BOD (AAP-330, Stressgen (Enzo Life Sciences), Farmingdale, NY) at 1:1 000 dilution or mouse monoclonal anti-alpha-tubulin (Sigma, St. Louis, MO) at 1:5 000 or 1:10 000 dilution followed by alkaline phosphatase-labeled secondary antibodies (Jackson ImmunoResearch, West Grove, PA). The signals were quantified using ImageQuant software.
For quantification of caspase-3 and importin-7 by Western blotting, the above methods were used with some modifications. Cell lysates were transferred to PVDF membranes, blocked in TBS/BLOTTO (5% nonfat milk, 0.1% Tween-20 in 1XTBS) and probed with rabbit polyclonal anti-caspase-3 (9662, Cell Signaling Technology, Danvers, MA) at 1:1 000 dilution or mouse monoclonal anti-importin-7 (SAB1402521, Sigma) at 1:1 000 dilution or mouse monoclonal anti-alpha-tubulin (Sigma) at 1:20 000 dilution followed by Horseradish Peroxidase-labeled secondary antibodies (Promega, Madison, WI). Blots were incubated with Promega ECL Western Blotting Substrate (Promega) and exposed to GeneMate Blue Basic Autorad Film (BioExpress, Kaysville, UT) for detection. After scanning the film, signals were quantified using ImageJ. All signals used for quantification were shown to be in the linear range of detection by determining that their intensities were within the linear range of a standard curve of dilutions on the same blot.
Microarray analysis
Total RNA (DNAse treated) was reverse-transcribed, labeled with Cy3 or Cy5, and hybridized to arrays (AMADID 014850 Whole Human Genome, Agilent, Santa Clara, CA) in biological triplicates (a service provided by the laboratory of Dr. Chris Bradfield). The reference samples (co-hybridized to arrays with experimental samples) were cDNAs from Sav-BL S1-1 cells transduced with empty vectors and dnEBNA1 uninduced. Data were analyzed using EDGE3 (Dr. Chris Bradfield). Analysis parameters included a minimum induction of 1.5, a minimum repression of -1.5, processed signal values of at least 100 (p-value cutoff of 0.01), and employed a statistical t-test (p-value cutoff of 0.05 and an rFDR correction). Genes were often returned by EDGE3 in which most of the processed signal values were less than 100 (even though the cutoff was set at 100) or the average fold change was less than 1.5 (even though the cutoff was set at ±1.5). These incorrectly identified genes were discarded. Sequences that were probed on the array but not defined as a gene or a hypothetical gene by NCBI were also discarded. The array dataset is available under the GEO accession number GSE22586.
Construction and preparation of recombinant EBV
All recombinant viruses as well as the methods to prepare and quantify infectious viral stocks have been described elsewhere 9.
Isolation, infection, and analysis of human primary B lymphocytes
Human primary B cells from adenoids were separated from T cells by rosetting with sheep erythrocytes and purified by Ficoll-Hypaque density gradient centrifugation. B cells isolated from human peripheral blood mononuclear cells (PBMC) by Ficoll-Hypaque gradient centrifugation were purified using the B-cell isolation kit II (Miltenyi Biotec, Gladbach, Germany) and MACS separators (Miltenyi Biotec). For virus infection, primary B cells were incubated with each virus stock for 18 hrs. Virus stocks were titered on Raji cells (8) with their standard deviations varying from 3 to 14%. After replacement with fresh medium, the infected cells were seeded at an initial density of 4.5×105 cells per ml.
RISC immunoprecipitation and RNA isolation
RISC was immunoprecipitated and RNA isolated from1×108 cells as described 10.
Deep sequencing
Total mRNA and RISC-immunoprecipitated mRNA samples were reverse transcribed and prepared for sequencing by the UWBC DNA Sequencing Facility following protocols provided by Illumina (San Diego, CA). Sequencing was carried out on an Illumina GAIIx. Sequence analysis was conducted using CLC Genomics Workbench Version 4.9. Sequences from each sample were mapped to the hg18 genome build, annotated as previously described 11. From the mapped reads, the unique exon reads for each gene were identified. These reads were then used to create expression values for each gene with the following formula:
Luciferase reporter assays
The 3′ UTRs of CASP3 and IPO7 were cloned downstream of FLuc (firefly luciferase) in the expression plasmid pmirGLO (Promega). PmirGLO contains a separate expression cassette encoding RLuc (renilla luciferase) as an internal control. The sequences of the UTRs were verified and are available upon request. The pmirGLO reporters were electroporated into EBV-positive and EBV-negative Oku-BL cells. Two days later, 2.5×106 cells were harvested, lysed for 15 minutes at RT in 200uL of Passive Lysis Buffer (Promega), then FLuc and RLuc activity in clarified lysate from 2.5×105 cell equivalents were measured using a Dual-Luciferase Assay Kit (Promega) on a Monolight 3010 according to the manufacturer’s protocol. To control for transfection efficiencies and cell viability, FLuc activity was normalized to RLuc activity. To control for differences among cell lines in their base luciferase activities, signals from pmirGLO constructs containing UTRs were normalized to signals from pmirGLO alone. The construct with mutated BART miRNA binding sites was constructed using QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene, Santa Clara, CA).
Synthetic BART Luciferase reporter assays
The constructs from the luciferase reporter assays were assayed with synthetic BART miRNAs. 293 cells were co-transfected with the constructs previously described and synthetic mature miRNAs (IDT). Nearly confluent 6-well dishes were transfected with Lipofectamine 2000 with a modified version of a protocol from Invitrogen. Briefly, 10 ng of pmirGLO or pmirGLO containing either CASP3 or IPO7 UTR downstream of FLuc were cotransfected with a total of 3 ug of synthetic miRNA or control. In wells where more than one miRNA was transfected, an equal mass of each miRNA was transfected, in a total of 3 ug per well. Twenty four hours after transfection, cells were harvested and analyzed for FLuc and RLuc activity as described above.
Statistical Analysis
The program Mstat 5.10 was used for all statistical analysis unless otherwise stated (N. Drinkwater, McArdle Laboratory for Cancer Research, School of Medicine and Public Health, University of Wisconsin) and is available for downloading at http://www.mcardle.wisc.edu/mstat.
Supplementary Material
Acknowledgments
We thank Alan Rickinson for kindly providing the early passage BL cell lines and Dagmar Pich for her invaluable support. This work was funded by grants from the National Cancer Institute, National Institutes of Health (grant P01 CA022443, grants R01 CA133027 and R01 CA070723) and Deutsche Forschungsgemeinschaft (grants SFB455 and SFBTR5). David Vereide was supported by a predoctoral fellowship from the National Cancer Center. Takanobu Tagawa is supported by a predoctoral grant of DAAD, German Academic Exchange Service. Bill Sugden is an American Cancer Society Research Professor.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- 1.Nanbo A, Sugden A, Sugden B. The coupling of synthesis and partitioning of EBV’s plasmid replicon is revealed in live cells. EMBO J. 2007;26:4252–62. doi: 10.1038/sj.emboj.7601853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vereide D, Sugden B. Insights into the evolution of lymphomas induced by Epstein-Barr virus. Advances in cancer research. 2010;108:1–19. doi: 10.1016/B978-0-12-380888-2.00001-7. [DOI] [PubMed] [Google Scholar]
- 3.Vereide DT, Sugden B. Lymphomas differ in their dependence on Epstein-Barr virus. Blood. 2011;117:1977–85. doi: 10.1182/blood-2010-05-285791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirchmaier AL, Sugden B. Dominant-negative inhibitors of EBNA-1 of Epstein-Barr virus. J Virol. 1997;71:1766–75. doi: 10.1128/jvi.71.3.1766-1775.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee DY, Sugden B. The LMP1 oncogene of EBV activates PERK and the unfolded protein response to drive its own synthesis. Blood. 2008;111:2280–9. doi: 10.1182/blood-2007-07-100032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaillard C, Strauss F. Ethanol precipitation of DNA with linear polyacrylamide as carrier. Nucleic Acids Res. 1990;18:378. doi: 10.1093/nar/18.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pratt ZL, Kuzembayeva M, Sengupta S, Sugden B. The microRNAs of Epstein-Barr Virus are expressed at dramatically differing levels among cell lines. Virology. 2009;386:387–97. doi: 10.1016/j.virol.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pratt ZL, Zhang J, Sugden B. The latent membrane protein 1 (LMP1) oncogene of Epstein-Barr virus can simultaneously induce and inhibit apoptosis in B cells. J Virol. 2012;86:4380–93. doi: 10.1128/JVI.06966-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seto E, Moosmann A, Gromminger S, Walz N, Grundhoff A, Hammerschmidt W. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS pathogens. 2010;6:e1001063. doi: 10.1371/journal.ppat.1001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuzembayeva M, Chiu YF, Sugden B. Comparing proteomics and RISC immunoprecipitations to identify targets of Epstein-Barr viral miRNAs. PLoS One. 2012;7:e47409. doi: 10.1371/journal.pone.0047409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic acids research. 2009;37:D32–6. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelly G, Bell A, Rickinson A. Epstein-Barr virus-associated Burkitt lymphomagenesis selects for downregulation of the nuclear antigen EBNA2. Nat Med. 2002;8:1098–104. doi: 10.1038/nm758. [DOI] [PubMed] [Google Scholar]
- 13.Choy EY, Siu KL, Kok KH, Lung RW, Tsang CM, To KF, et al. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med. 2008;205:2551–60. doi: 10.1084/jem.20072581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–86. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliver PM, Vass T, Kappler J, Marrack P. Loss of the proapoptotic protein, Bim, breaks B cell anergy. J Exp Med. 2006;203:731–41. doi: 10.1084/jem.20051407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderton E, Yee J, Smith P, Crook T, White RE, Allday MJ. Two Epstein-Barr virus (EBV) oncoproteins cooperate to repress expression of the proapoptotic tumour-suppressor Bim: clues to the pathogenesis of Burkitt’s lymphoma. Oncogene. 2008;27:421–33. doi: 10.1038/sj.onc.1210668. [DOI] [PubMed] [Google Scholar]
- 17.Clybouw C, McHichi B, Mouhamad S, Auffredou MT, Bourgeade MF, Sharma S, et al. EBV infection of human B lymphocytes leads to down-regulation of Bim expression: relationship to resistance to apoptosis. Journal of immunology. 2005;175:2968–73. doi: 10.4049/jimmunol.175.5.2968. [DOI] [PubMed] [Google Scholar]
- 18.Paschos K, Smith P, Anderton E, Middeldorp JM, White RE, Allday MJ. Epstein-barr virus latency in B cells leads to epigenetic repression and CpG methylation of the tumour suppressor gene Bim. PLoS Pathog. 2009;5:e1000492. doi: 10.1371/journal.ppat.1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38 (Suppl):S8–13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 21.Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA. 2006;12:733–50. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schafer A, Cai X, Bilello JP, Desrosiers RC, Cullen BR. Cloning and analysis of microRNAs encoded by the primate gamma-herpesvirus rhesus monkey rhadinovirus. Virology. 2007;364:21–7. doi: 10.1016/j.virol.2007.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walz N, Christalla T, Tessmer U, Grundhoff A. A global analysis of evolutionary conservation among known and predicted gammaherpesvirus microRNAs. J Virol. 2010;84:716–28. doi: 10.1128/JVI.01302-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Birkenbach M, Josefsen K, Yalamanchili R, Lenoir G, Kieff E. Epstein-Barr virus-induced genes: first lymphocyte-specific G protein-coupled peptide receptors. J Virol. 1993;67:2209–20. doi: 10.1128/jvi.67.4.2209-2220.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riley KJ, Rabinowitz GS, Yario TA, Luna JM, Darnell RB, Steitz JA. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. EMBO J. 2012 doi: 10.1038/emboj.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skalsky RL, Corcoran DL, Gottwein E, Frank CL, Kang D, Hafner M, et al. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 2012;8:e1002484. doi: 10.1371/journal.ppat.1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dolken L, Malterer G, Erhard F, Kothe S, Friedel CC, Suffert G, et al. Systematic analysis of viral and cellular microRNA targets in cells latently infected with human gamma-herpesviruses by RISC immunoprecipitation assay. Cell host & microbe. 2010;7:324–34. doi: 10.1016/j.chom.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–9. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 29.Suffert G, Malterer G, Hausser J, Viiliainen J, Fender A, Contrant M, et al. Kaposi’s sarcoma herpesvirus microRNAs target caspase 3 and regulate apoptosis. PLoS pathogens. 2011;7:e1002405. doi: 10.1371/journal.ppat.1002405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altmann M, Hammerschmidt W. Epstein-Barr virus provides a new paradigm: a requirement for the immediate inhibition of apoptosis. PLoS Biol. 2005;3:e404. doi: 10.1371/journal.pbio.0030404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.