Abstract
Gonadotroph adenomas comprise 15–40 % of all pituitary tumors, are usually non-functioning and are often large and invasive at presentation. Surgery is the first-choice treatment, but complete resection is not always achieved, leading to high recurrence rates. As gonadotroph adenomas poorly respond to conventional pharmacological therapies, novel treatment strategies are needed. Their identification has been hampered by our incomplete understanding of the molecular pathogenesis of these tumors. Recently, we demonstrated that MENX-affected rats develop gonadotroph adenomas closely resembling their human counterparts. To discover new genes/pathways involved in gonadotroph cells tumorigenesis, we performed transcriptome profiling of rat tumors versus normal pituitary. Adenomas showed overrepresentation of genes involved in cell cycle, development, cell differentiation/proliferation, and lipid metabolism. Bioinformatic analysis identified downstream targets of the transcription factor SF-1 as being up-regulated in rat (and human) adenomas. Meta-analyses demonstrated remarkable similarities between gonadotroph adenomas in rats and humans, and highlighted common dysregulated genes, several of which were not previously implicated in pituitary tumorigenesis. Two such genes, CYP11A1 and NUSAP1, were analyzed in 39 human gonadotroph adenomas by qRT-PCR and found to be up-regulated in 77 and 95 % of cases, respectively. Immunohistochemistry detected high P450scc (encoded by CYP11A1) and NuSAP expression in 18 human gonadotroph tumors. In vitro studies demonstrated for the first time that Cyp11a1 is a target of SF-1 in gonadotroph cells and promotes proliferation/survival of rat pituitary adenoma primary cells and cell lines. Our studies reveal clues about the molecular mechanisms driving rat and human gonadotroph adenomas development, and may help identify previously unexplored biomarkers for clinical use.
Electronic supplementary material
The online version of this article (doi:10.1007/s00401-013-1132-7) contains supplementary material, which is available to authorized users.
Keywords: Pituitary gonadotroph adenoma, Transcriptome analysis, MENX model, CYP11A1, NUSAP1
Introduction
Clinically non-functioning adenomas of the pituitary gland account for about 30 % of all adenomas. About 80 % of them belong to the gonadotroph lineage. They are usually diagnosed when signs and symptoms of mass effects occur, and about 40 % of them extend to the cavernous sinus and, less commonly, invade the sellar floor, making their resection and post-operative radiotherapy challenging [14, 34]. In addition, they are usually resistant to pharmacological treatment with somatostatin analogs and dopamine agonists. Chemotherapy has been reserved as salvage therapy in aggressive cases although the results have often been disappointing [34]. Novel therapeutic approaches are therefore needed for these tumors.
A prerequisite to identify novel therapeutic targets for human tumors is a good understanding of the pathogenetic mechanisms. The molecular mechanisms underlying the development of PAs in general, and of gonadotroph adenomas in particular, have only been incompletely unraveled and this is in part due to the limited availability of suitable animal models and by the fact that those available are not always representative of human adenomas [27].
We have recently identified a multiple endocrine neoplasia syndrome occurring in a Sprague–Dawley-derived rat strain (named MENX) characterized by the occurrence of spontaneous PAs with complete penetrance. MENX is caused by a biallelic loss-of-function mutation in Cdkn1b, encoding the cell cycle inhibitor p27, which results in a highly unstable protein [29, 33]. MENX-associated PAs are histologically and ultrastructurally remarkably similar to human gonadotroph adenomas. They also show high mitotic activity and elevated Ki67 labeling index [26]. Primary cells derived from these tumors are a suitable model for pharmacological studies of PAs [24].
To elucidate the mechanisms associated with the development of human gonadotroph adenomas we exploited the MENX model and performed whole genome transcriptome analysis of the rat tumors and of normal pituitary tissues of wild-type animals. With this approach, we have identified genes differentially expressed in rat PAs that had been previously found dysregulated in human adenomas by array analysis, but not yet further validated. We here show that two such genes (i.e. CYP11A1 and NUSAP1) are highly expressed at both mRNA and protein level in human gonadotropinomas and may represent novel biomarkers of these tumors. In vitro functional assays demonstrated for the first time that the high expression of Cyp11a1 promotes proliferation/survival of PA cells (gonadotroph- and somatotroph-derived), thereby supporting a role for this gene in pituitary tumorigenesis. We also determined that Cyp11a1 expression is regulated by the steroidogenic transcription factor SF-1 in gonadotroph cells. Noteworthy, in addition to NUSAP1, other genes involved in mitosis were found overexpressed in both rat and human PAs by meta-analysis. If further validated, these genes may represent novel potential targets for the therapy of gonadotropinomas.
Materials and methods
Human pituitary tissue samples
Pituitary adenoma samples were obtained from patients at Imperial College and the University of Tübingen at the time of transsphenoidal surgery. All patients consented for research and appropriate ethical approval for the study was obtained. Fragments of the specimens that were not used for diagnostic assessment were snap-frozen at −80 °C. Gonadotroph tumors were defined as demonstrating positive immunostaining for LHβ, FSHβ or αGSU in >5–10 % of cells. Extension to the cavernous sinus and invasiveness of the sellar floor were identified at preoperative magnetic resonance imaging (MRI). The modified Hardy criteria were applied [18]. Post-mortem samples of normal adenohypophysis used for immunohistochemistry (IHC) were obtained from Imperial College. Autopsies were performed within 2–18 h from death and were formalin-fixed and routinely processed to paraffin embedding. Consent for research use of this tissue was obtained from families. RNA of normal pituitary tissues was purchased from BioChain Inc (Hayward, CA, USA).
Rat pituitary tissue samples
MENX-affected rats develop multifocal pituitary adenomas in the anterior lobe of the gland. Based on hormone expression by immunohistochemistry we could establish that these tumors mainly derive from gonadotroph cells [26]. The lesions become histologically detectable at 4 months of age and progress to become large, mature tumors that efface the gland at 7–8 months. The number of cells expressing LHβ or FSHβ decreases with tumor progression and is negligible in the largest adenomas, while tumors remain positive for the common gonadotropin alpha subunit (αGSU). Rat adenomas show mitotic activity and high Ki67 labeling (average 8 % at 7–8 months), thus resembling aggressive pituitary adenomas. Approximately 10–17 % of the pituitary lesions contain cells expressing growth hormone (GH) or prolactin (PRL) [26]. The pituitary glands used for RNA extraction (upon microdissection) derived from rats aged 7–8 months having large tumors approximately 1–2 mm in size.
RNA isolation and microarray preparation
Rat RNA was extracted from macrodissected pituitary tissues and processed for array analysis as reported [30]. Amplified cDNA (2 μg) was hybridized on Affymetrix Rat Gene 1.0 ST arrays (Santa Clara, CA, USA) containing about 26 k gene-level probe sets. Staining and scanning was done according to the Affymetrix expression protocol. RNA was extracted from frozen human pituitary tumor samples following standard protocols [30].
Biostatistical and bioinformatic analysis
Expression console (Affymetrix) was used for quality control and to obtain annotated normalized RMA gene-level data (standard settings including median polish and sketch-quantile normalization). Statistical analyses and heat maps were generated in CARMAweb [36]. Genewise testing for differential expression was done employing t test and Benjamini-Hochberg multiple testing correction (FDR < 5 %). Genes with fold-changes >2× were further analyzed using GePS software (Genomatix Software GmbH, Munich, Germany). Signaling pathways containing, or regulated by, differentially expressed genes were identified using the Ingenuity Pathway Analysis (IPA) library of canonical pathways. Array data were submitted to Gene Expression Omnibus (GSE23207). The comparison of rat and human datasets was done in Venny (http://bioinfogp.cnb.csic.es/tools/venny/index.html) based on the match of human genes to rat probe set IDs via rat Entrez IDs provided by Affymetrix. Human Entrez IDs of regulated genes were matched to rat Entrez IDs mainly based on the NCBI homologene database, supplemented with information provided by Ingenuity pathway software. Human datasets were taken from publications: Morris et al. [32], 1,723 probe sets with fold-changes >1.5×; Moreno et al. [31], Supplementary Table 2,297 probe sets with fold-changes >2×) or calculated from.cel files; Michaelis et al. [28], Acc. Number GSE26966, analysis as described above, 3,558 probe sets with ratios >2×. All human datasets were done with Affymetrix arrays and human Entrez IDs of regulated probe sets were taken from annotations provided by Affymetrix. For pairwise comparisons of rat and human datasets, probe set based gene lists from the Venn diagram were further processed: for redundant entries only the entry with the highest ratio in rat was kept, and datasets were filtered for the same direction of regulation in rat and human datasets.
Cell culture, treatments and assays
GH3 and Y1 cells were purchased from the ATCC and grown in DMEM supplemented with 10 % (v/v) FBS and 1 % (v/v) penicillin–streptomycin. The murine gonadotroph cell line LβT2 was kindly provided by P. Mellon (University of California, San Diego, USA) and was grown in DMEM + GlutaMAX™-I plus 4.5 g/l d-glucose and pyruvate with 10 % (v/v) FBS and 1 % (v/v) penicillin–streptomycin. Primary pituitary tumor cells were isolated from mutant rat pituitaries and grown as previously described [24]. Media, serum and supplements were from Gibco/Invitrogen (Karlsruhe, Germany). Transfection with siRNA oligos was performed as previously reported [24]. For infections, GH3 and primary tumor cells were plated on 96-well plates and 24 h later cells were infected by lentiviral vectors expressing the green fluorescence protein (GFP) or GFP and shRNA against Cyp11a1. Virus production and lentiviral infection were performed according to previous protocols [2]. WST-1 colorimetric assay (Roche, Mannheim, Germany) for cell viability was performed 72 h after infection according to the manufacturer’s recommendations.
Apoptosis was measured by assessing the activity of caspase-3/7 using Caspase-Glo® 3/7 Assay kit (Promega, Mannheim, Germany). Primary cells or transfected GH3 cells were plated in 96-well plates and 36 h later were infected with sh-Cyp11a1 or mock GFP lentiviral vectors. Forty-eight h after treatment, caspase-3/7 enzymatic activity was assessed with a proluminescent caspase-3/7 substrate which contains the tetrapeptide sequence DEVD. Luminescence was measured using a luminometer (Berthold, Bad Wildbad, Germany).
For treatment with ethyl 2-[[2-[2-[(2,3-dihydro-1,4-benzodioxin-6-yl)amino]-2-oxoethyl]-1,2-dihydro-1-oxo-5-isoquinolinyl] (IsoQ) (Tocris Bioscience, Bristol, UK), 2.5 × 104 primary rat pituitary cells (2.5 × 104 cells/well) or LβT2 cells (1 × 104 cells/well) were seeded into 96-well plates and allowed to settle for 36 h. IsoQ was dissolved in DMSO and serial dilutions (10 μM–10 nM) were prepared in culture medium. Treated cells were incubated with IsoQ for 24 and 48 h, while control cells with 0.01 % (v/v) DMSO. Cell viability was assessed using the WST-1 assay as above.
Quantitative TaqMan RT-PCR
Quantititative RT-PCR was performed using TaqMan inventoried primers and probes for the genes reported in the article and for the rat beta 2-microglobulin gene, mouse beta 2-microglobulin gene or human TBP gene as internal controls (Applied Biosystem, CA, USA). RNA was extracted and TaqMan assays were set up as previously reported [30].
RNA interference
The specific Cyp11a1 shRNA sequence was cloned into the pSUPER lentiviral vector. The sequence of the complementary oligonucleotides is: sh-Cyp11a1 Fw GATCCGGATGTTGGAGGAGATCGTTTCAAGAGAACGATCTCCTCCA ACATCCTTTTTG; Rev: AATTCAAAAAGGATGTTGGAGGAGATCGTTCTCTT GAAACGATCTCCTCCAACATCCG.
Immunohistochemistry and Immunofluorescence
Immunohistochemistry (IHC) staining was performed on an automated immunostainer (Ventana Medical Systems, Tucson, AZ, USA) according to the manufacturer’s protocols with minor modifications, as previously described [30]. Primary antibodies were raised against Ass1 (Sigma, Louis, MO, USA); Ki-67 (Clone B56, Dako, Hamburg, Germany); rat P450scc (Abcam, Abcam, Cambridge, UK); human P450scc (Santacruz, Heidelberg, Germany); NuSAP (Proteintech, Chicago, IL, USA). Antibodies were diluted in Dako REALTM antibody diluent (Dako). The supersensitive detection system (BioGenex, Munich, Germany) was used and the immunoreaction was developed in the diamino-benzidine (DAB) supplied with the kit (Vector lab, Burlingame, CA). Positive controls were included in each batch. To score the P450scc staining, both the intensity of the staining and the number of positive tumor cells were taken into consideration. For number of positive cells, we used a score: 0 = 0 % (or <1 %) positive cells; 1 = up to 25 % positive cells; 2 = up to 50 %; 3 = up to 75 %; 4 = up to 100 %. For staining intensity, a 4-scaled scoring system was used: 0 = negative, 1 = weak, 2 = moderate, 3 = strong. These values were then multiplied to obtain ‘immunoreactivity scores’ (IRS). Ki67 and NuSAP nuclear immunoreactivity was determined semiquantitatively and was indicated as the percent of positive cells against all neoplastic cells in the section examined (Labeling Index, LI). Images were recorded using a Hitachi camera HW/C20 installed in a Zeiss Axioplan microscope with Intellicam software (Carl Zeiss MicroImaging GmbH, Gottingen, Germany).
For immunofluorescence (IF), we used the primary antibodies used for IHC and secondary anti-mouse Alexa Fluor® 555 Conjugate (Cell Signaling) or anti-rabbit FITC-conjugated (Invitrogen, Darmstadt, Germany) antibodies as reported [26]. Sections were then analyzed with a Zeiss Axiovert 200 epifluorescence microscope including Apotome unit (Carl Zeiss MicroImaging GmbH).
Statistical analysis
Results of the cell viability assays are shown as the mean of values obtained in independent experiments ± SEM (standard error of the mean). A paired two-tailed Student’s t test was used to detect significance between two series of data and P value (P) < 0.05 was considered statistically significant.
Results
Genetic signature of MENX-associated pituitary adenomas
In order to unravel the genetic changes associated with pituitary adenoma (PA) formation, we performed whole genome transcriptome analysis of 16 individual pituitary lesions from 11 adult MENX mutant rats (8 months of age) and compared them with 5 normal pituitaries. Using a >twofold-change cut-off for gene expression changes, 487 probe sets appeared up-regulated in adenomas versus normal wild-type gland, and 400 were down-regulated (Supplementary Fig. 1 and Dataset 1). The relative high number of differentially expressed transcripts is not unexpected since normal pituitary is composed of multiple cell types of which only 10–20 % are gonadotroph cells. Gene ontology (GO) term enrichment analysis showed that genes related to cell cycle (P = 9.85E−09), development (P = 1.2E−06), cell differentiation (P = 1.36E−06), cell proliferation (P = 2.53E−07), and lipid metabolism (P = 9.43E−04) are overrepresented in our dataset (ratios >twofold, Table 1). Genes encoding proteins involved in cell cycle regulation and especially in mitosis such as Aurka, Bub1, Bub1b, Ccna1, Ccnb1, Ccnb2, Ccne1, Cdc2, Cdc20, Cdkn3, Kif4, Kif11, Nusap1, Prc1, as well as genes involved in pituitary development or adenohypophyseal cell differentiation, such as Dax-1/Nr0b1, Egr1, Fgfr2, Neurod1, Notch2, Nr5a1, Pou1f1, Tbx19 [6, 10, 23] were differentially expressed in rat PAs (Table 1). Array data mining found that genes such as Ccnb1, Igfbp3, Nusap1, Pttg1, Racgap1, Top2a, Tpx2, which are associated with the aggressive behaviour of various human tumors, including PAs, and proto-oncogenes such as Ect2, Kit, Kras, Lyn, Ret are up-regulated in rat adenomas [12, 15, 42, 43]. This result is in agreement with the elevated proliferative activity of the tumors [26]. Interestingly, also genes belonging to the category “lipid metabolic process” were significantly overrepresented in the rat adenomas, and several of them are known targets of the transcription factor steroidogenic factor 1 (SF-1) (see below).
Table 1.
Enriched GO categories by GePS software from Genomatix
| GO-term | GO-term ID | P value | Observed/expected | Number of genes in GO | Selected dysregulated genes |
|---|---|---|---|---|---|
| Enriched terms within the up-regulated genes | |||||
| Cell cycle | GO: 0007049 | 9.85E−09 | 42/16 | 667 | Aurka, Bmp7, Bub1, Bublb, Ccnbl, Ccnb2, Cdc2, Cdc20, Cdknla, Cdkn2c, Cdkn3, Cenpf, Cited2, Id2, Nusapl, Pttgl |
| Developmental process | GO: 0032502 | 1.20E−06 | 98/62 | 2,597 | Angpt2, Ass1, Bmp7, Cdkn2c, Cited2, Cyp11a1, Fgfr2, Id2, Igfbp3, Igfbp7, Lyn, Neurod1, Nos1, Nr0b2, Nr0b1, Nr5a1, Ret, Sox11 |
| Cell differentiation | GO: 0030154 | 1.36E−06 | 63/34 | 1,431 | Angpt2, Apoe, Ar, Bag1, Bmp7, Ccnb1, Cdc2, Cdkn2c, Cenpf, Cited2, Cyp11a1, Fgfr2, Id2, Igfbp3, Kit, Kras, Lyn, Neurod1, Neurod4, Nr0b1, Nr5a1, Racgap1, Ret, Sox11, Top2a, Tshr, Vegfa |
| Cell proliferation | GO: 0008283 | 2.53E−07 | 45/20 | 832 | Apoe, Ar, Bub1, Ccna2, Ccnb1, Cdc2, Cdc20, Cdkn1a, Cdkn1c, Cdkn2c, Cenpf, Fgfr2, Gnrhr, Id2, Igfbp3, Kit, Kras, Lyn, Pttg1, Racgap1, Scarb1, Sox11, Tac1, Vegfa |
| Lipid metabolic process | GO: 0006629 | 9.43E−04 | 33/19 | 772 | Apoe, Apof, Cyp1b1, Cyp11a1, Cyp11b1, Cyp11b2, Fdxr, Nr0b1, Scarbl |
| Enriched terms within the down-regulated genes | |||||
| Developmental process | GO: 0032502 | 7.73E−10 | 69/34 | 2,597 | Egr1, Notch2, Nr4a3, Sema3e, Dlk1, Angptl, Nr4a2, Ghrhr, Adcyap1r1, Pou1f1, Tbx19 |
| Negative regulation of cell proliferation | GO: 0008285 | 9.29E−03 | 9/4 | 271 | Nfib, Notch2, Gjb6, Wfdd, Rbp4, Gpc3, Cdk6, Msx1, Slit2 |
Genes encoding pituitary hormones were down-regulated in rat adenomas, with the exception of Cga encoding the common alpha subunit of the gonadotropins (αGSU), which was highly expressed in all adenomas (Supplementary Fig. 1). These results are in keeping with previous immunohistochemical stainings showing that the large, older rat pituitary lesions do not express any hormone subunit except αGSU [26].
Transcription factor analysis using Ingenuity Pathway Analysis (IPA) predicted the activation of several upstream transcription factors, including Nr5a1 (z-score = +2.34; P = 5E−08) encoding SF-1 (Supplementary Table 1). The genes encoding the gonadotropin hormone subunits LHβ and FSHβ, which are transcriptionally regulated by SF-1 in gonadotroph cells, are down-regulated in the large rat tumors, as mentioned above. In contrast, several downstream targets of SF-1 were found highly expressed in the adenomas. While the mRNA level Nr5a1 was up-regulated 2.7-fold in the adenomas (Dataset 1), its downstream targets were overexpressed up to 23-fold in the tumors, including Angpt2, Cyp11a1, Cyp11b1, Cyp11b2, Giot1, Gnrhr, Nos1, Nr0b1/Dax1, Nr0b2, Scarb1. Also Fgf13 and Col18a1 were up-regulated, as reported in adrenal cancer cells [8], and so was Cited2, encoding for a co-activator of SF-1 during adrenal development [40]. Altogether, these data suggest that the activity of SF-1 is enhanced in rat PAs (Supplementary Fig. 2a). Further support to the activation of SF-1 in the rat tumors came from the validation of Cyp11b1 and Cyp11b2 overexpression in independent adenoma samples (Supplementary Fig. 2b). Several of the SF-1 targets mentioned above play a physiological role in the synthesis or transport of steroid hormones in steroidogenic tissues and have not so far been implicated in adenohypophyseal tumorigenesis.
Real-time quantitative RT-PCR (qRT-PCR) for 9 selected overexpressed genes was performed on 11 additional individual pituitary adenomas to validate the array data (Fig. 1a). These tumors were compared with five pituitaries from wild-type rats. Nr5a1 (Sf-1) and Cga (αGSU) were included as controls because the encoded proteins are expressed in these tumors [26], while the other transcripts were chosen because they have interesting functions and they span a broad range of increased fold-changes, from ~3-fold to >20-fold. The results confirmed the up-regulation of these genes in rat PAs, with Cdkn1c (p57) showing a trend toward higher levels in the tumors versus normal pituitary which did not reach the statistical significance (Fig. 1a). qRT-PCR also confirmed the down-regulation of Pomc, Lhb, Fshb in rat adenomas (Fig. 1b).
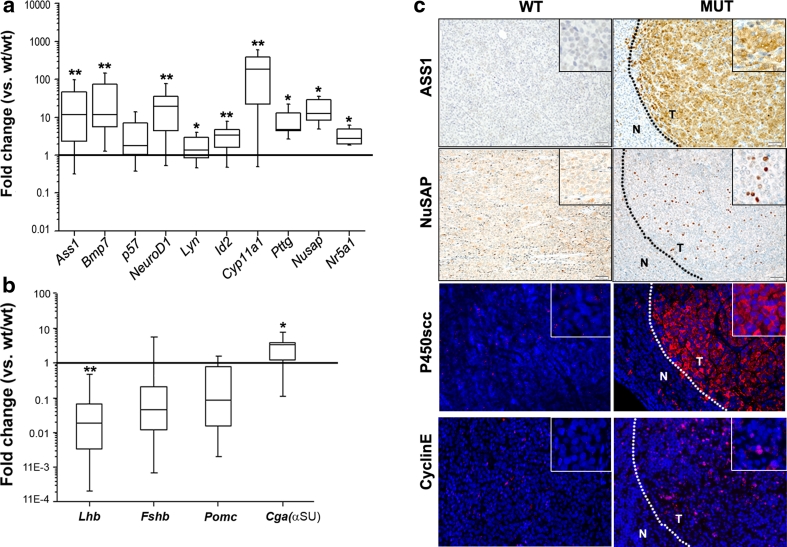
Fig. 1.
Validation of selected genes by qRT-PCR and by IHC on rat PA tissues. a, b RNA was extracted from 11 macrodissected pituitary tumors of adult mutant rats and from five normal pituitaries of wild-type (wt/wt) rats. qRT-PCR was performed using TaqMan primer and probe sets specific to a rat genes up-regulated by array analysis: Ass1, Bmp7, p57, NeuroD1, Lyn, Id2, Cpy11a1, Pttg, Nusap, Nr5a1 and b rat genes down-regulated by array analysis: Lhβ, Fshβ, Pomc, Cga (αGSU). The relative mRNA expression level of the target genes was normalized for input RNA using the housekeeping β2-microglobulin gene and a calibrator RNA always run in parallel and was calculated with the 2−ΔΔCt formula. The obtained relative value was normalized against the average level in normal pituitary, arbitrarily set to 1. The boundary of the box closest to zero indicates the 25th percentile, the line within the box marks the median, and the boundary of the box farthest from zero indicates the 75th percentile. Error bars above and below the box indicate the 99th and 1th percentiles. *P < 0.05, **P < 0.001. c Formalin-fixed, paraffin-embedded (FFPE) pituitaries from adult wild-type (WT) (left) and mutant (MUT) (right) rats were used. IHC was performed using antibodies against ASS1 and NuSAP and counterstained with hematoxylin. Immunofluorescence was performed with antibodies against P450scc or Cyclin E and nuclei were counterstained with DAPI. T tumor area, N normal adjacent area. Original magnification: ×200; insets: ×400
The differential expression of selected genes was further validated with immunohistochemistry (IHC) and immunofluorescence (IF). Stainings for argininosuccinate synthetase 1 (Ass1), NuSAP, Cyclin E, P450scc (encoded by Cyp11a1) showed higher Ass1 and P450scc levels in tumors compared with non-tumorous pituitary tissue of mutant rats or wild-type rat pituitaries (Fig. 1c). Similarly, NuSAP- and CyclinE-positive cells were found in higher number in adenomas compared with surrounding non-tumorous gland, or with normal pituitaries (Fig. 1c).
Dysregulated pathways and genes previously associated with human pituitary tumors
Some of the pathways identified as dysregulated in rat PAs by the IPA software have also been implicated in human PAs by proteomics profiling, and include cell cycle, lipid metabolism and endocrine system development [45]. In addition, the homologues of several genes up-regulated in the rat tumors had also been already implicated in human non-functioning/gonadotroph PAs. Examples are ECT2, NEUROD1, PTTG1, VEGFA and the oncogene c-fos [22, 25, 31, 32]. NOS1 and DAX1, regulated by and/or regulating the transcription factor SF-1, are highly expressed in human gonadotroph adenomas [20, 39]. Androgen receptor (AR) is expressed in 74 % of gonadotropinomas [37], and DLK1 is silenced in non-functioning adenomas, while it is highly expressed in functional adenomas [1].
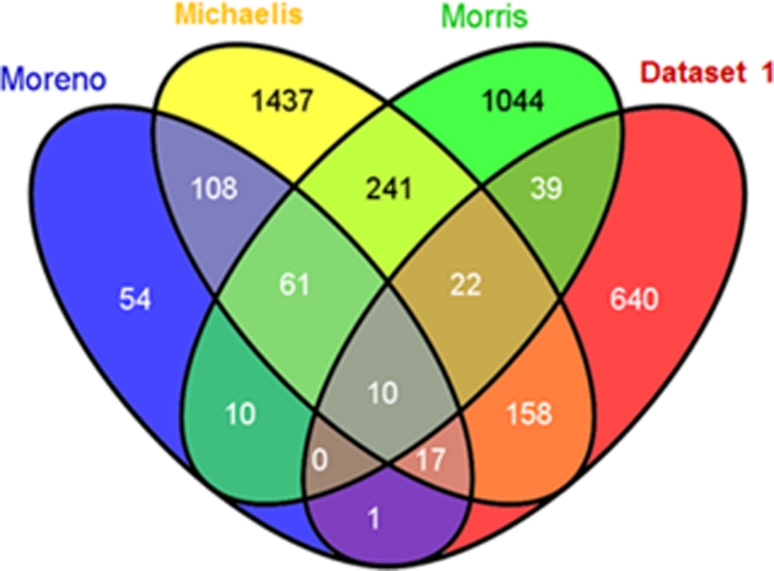
Studies comparing the transcriptome of human gonadotroph adenomas with that of normal pituitary tissues [1, 9, 11, 28, 31] or of non-invasive versus invasive gonadotroph adenomas [12, 19] have been performed. We therefore compared our Dataset 1 with the available lists of significantly differentially expressed genes in human non-functioning/gonadotroph adenomas. Examples of concordantly dysregulated genes in both species are reported in Supplementary Table 2. Remarkably, the degree of overlap of genes differentially expressed among different human sample sets is similar to the overlap between rat and human datasets, as estimated using Venn diagrams (Fig. 2). The 10 genes differentially expressed in all the datasets we compared are: Ptprk, Atp8a1, Lgals3, Nr4a2, Pon3, Fos, Angpt1, Amigo2, Rcan2, Maob (for details, please refer to “Materials and methods”). We found several common dysregulated genes in the rat tumors and in the dataset recently published by Michaelis et al. [28] analyzing 14 human gonadotroph adenomas. Among the genes dysregulated in both rat and human adenomas, we found genes associated with pituitary function or pituitary tumorigenesis and mentioned earlier in the article, such as AR, CCNB1, DLK1, ECT2, FOS, NEUROD1, NR0B1/DAX1, VEGF. Genes encoding transcription factors and hormones important in other pituitary cell lineages (i.e. POU1F1/PIT-1, TBX19, GH, PRL, POMC) are down-regulated in rat and human gonadotroph-derived adenomas, as expected. BAG1, coding for an anti-apoptotic protein, was found overexpressed in human non-functioning PAs by array analysis [32], and is also up-regulated in MENX-associated adenomas. Similarly to the rat tumors, several downstream targets of SF-1 were found to be highly expressed in human gonadotroph adenomas by array analysis (Supplementary Fig. 3). Interestingly, several genes dysregulated in both human and rat array datasets, although not yet evaluated in PAs, have been shown to play a role in other human malignancies, such as BUB1, BUB1B, GALNT12, GPC3, HMMR, KIF4, NR4A3, NUSAP1, PDE4B, PRC1 and thus warrant follow-up studies.
Fig. 2.
Overlap of differentially expressed genes. Venn diagram displaying the overlap among lists of differentially expressed genes from gene expression studies in human gonadotroph/non-functioning PAs [28, 31, 32] and rat PAs (Dataset 1). For details, see “Materials and methods”
CYP11A1 and NUSAP1 are overexpressed in human PAs
Among the genes overexpressed in the rat pituitary adenomas, predicted to be up-regulated in at least 2 of the 3 human expression array studies by data mining (Supplementary Table 2, in bold), and never before associated with pituitary adenomas, are Cyp11a1 and Nusap1. Thus, these two genes were selected for further analyses. Cyp11a1 encodes P450scc, the mitochondrial enzyme that catalyzes the conversion of cholesterol to pregnenolone in steroidogenic tissues. Nusap1 encodes a protein that plays a role in spindle microtubule organization and is necessary for cell division [35]. To verify whether these genes are truly up-regulated in human gonadotroph adenomas, we performed qRT-PCR in a series of 39 samples. NR5A1 and NEUROD1 were also included in the analysis as they are expected to be up-regulated in these tumors. We found that CYP11A1 was up-regulated in 77 % (27 out of 35) and NUSAP1 in 95 % (37 out of 39) of the human PAs (Fig. 3a). NR5A1 and NEUROD1 were overexpressed in 82 % of the cases.
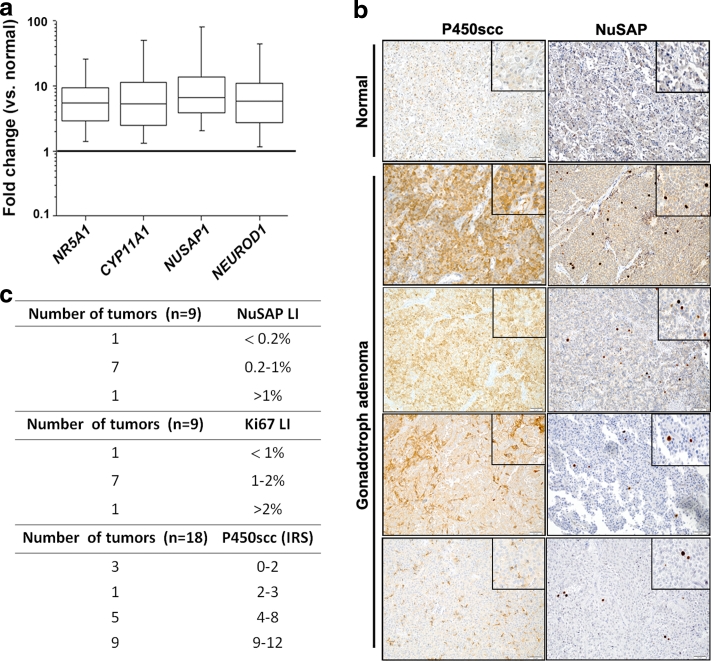
Fig. 3.
Overexpression of CYP11A1 and NUSAP1 and their encoded proteins in human gonadotroph tumors. a RNA was extracted from frozen pituitary tumors obtained after transsphenoidal surgery. qRT-PCR was performed using TaqMan primer and probe sets specific to human NR5A1, CYP11A1, NUSAP1 and NEUROD1. The relative mRNA expression level of the target genes was normalized for input RNA using human TBP gene expression (housekeeping gene) and a calibrator human brain RNA always run in parallel and was calculated with the 2−ΔΔCt formula. The obtained relative value was normalized against the average expression of normal pituitary arbitrarily set to 1. The boundary of the box closest to zero indicates the 25th percentile, the line within the box marks the median, and the boundary of the box farthest from zero indicates the 75th percentile. Error bars above and below the box indicate the 99th and 1th percentiles. b Human normal pituitary (top) and gonadotroph adenoma samples were used. IHC was performed using antibodies against P450scc (left) or against NuSAP (right) and counterstained with hematoxylin. Original magnification: ×200; insets: ×400. c Scoring of the expression of NuSAP, Ki-67 and P450scc in a series of human gonadotroph adenomas
To determine whether gene up-regulation translates into higher levels of the encoded protein, we obtained formalin-fixed, paraffin-embedded (FFPE) tissue blocks from 18 of the adenomas analyzed by qRT-PCR, and stained them for P450scc and NuSAP. Normal anterior pituitary cells had only weak cytoplasmic positivity for P450scc in sparse hypophyseal cells, while P450scc staining was more intense, although often uneven, in adenomas (Fig. 3b). Immunoreactivity score (IRS) were calculated and showed that 13 of 18 cases (72 %) had a IRS ≥6, indicating moderate to strong staining in up to 75 % of the tumor cells, and 5 cases (28 %) showed weaker staining (IRS ≤4) (Fig. 3c). Of the human gonadotropinomas used for IHC, 9 were invasive and 8 non-invasive. No significant difference in the degree of positivity for P450scc was observed between the two groups of tumors.
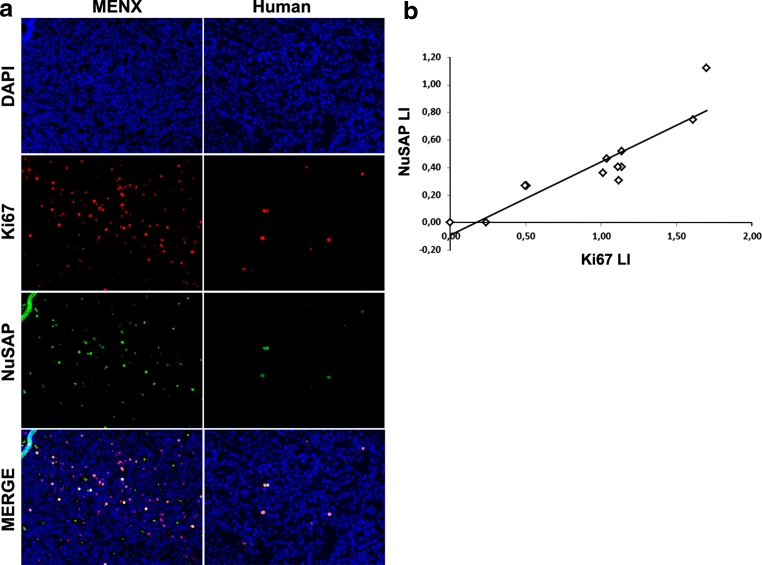
We also performed IHC to detect NuSAP expression and we could detect tumor cells having a strong, nuclear staining in every human PA sample (Fig. 3b). Being NuSAP associated with mitosis, we determined the relationship between its expression and that of the Ki67 antigen, an established marker of cell division. Quantification of the IHC results demonstrated that Ki-67-positive cells were more numerous than NuSAP-positive cells (Fig. 3c). NuSAP-positive pituitary tumor cells always co-expressed Ki-67 (Fig. 4a). Co-expression of the two markers was also seen in rat PAs, which have elevated proliferation rates (Fig. 4a) [26]. A good correlation between the Labeling Index (LI) of NuSAP and of Ki-67 was seen in the human tumor tissue samples (R 2 = 0.8131) (Fig. 4b).
Fig. 4.
Expression of Ki-67 and NuSAP in rat and human pituitary adenoma cells. a Example of immunofluorescent staining with antibodies against Ki-67 (red) and NuSAP (green) performed on FFPE pituitary tumor tissues from a MENX rat (MENX) and from a patient with gonadotroph adenoma (Human). Nuclei were counterstained with DAPI. Only the tumor area is shown. Original magnification: ×100. b Correlation between NuSAP and Ki-67 labeling index (LI) in 12 human gonadotroph adenoma samples
Cyp11a1 plays a pro-proliferative role in tumorigenesis
We have established that Cyp11a1 is highly expressed in rat and human gonadotroph tumors at mRNA and protein levels. To clarify the role of this gene in tumorigenesis, we first used Y1 cancer cells derived from a steroidogenic tissue and expressing Cyp11a1 at high level. siRNA-mediated knock-down of Cyp11a1 reduced the ability of Y1 cells to proliferate when compared with cells transfected with scrambled siRNA oligos (Supplementary Fig. 4b). Efficient knock-down of the Cyp11a1 gene was verified by qRT-PCR (Supplementary Fig. 4a). Encouraged by these findings we pursued the role of Cyp11a1 overexpression in pituitary tumorigenesis. We used somatotroph adenoma-derived GH3 cells, which express relatively high levels of Cyp11a1 (Supplementary Fig. 5), to evaluate the effect of modulating gene expression on pituitary tumor cell proliferation. We found that siRNA-mediated silencing of Cyp11a1 reduced GH3 cells proliferation, while scrambled siRNA oligos elicited no effect (Supplementary Fig. 4c, d).
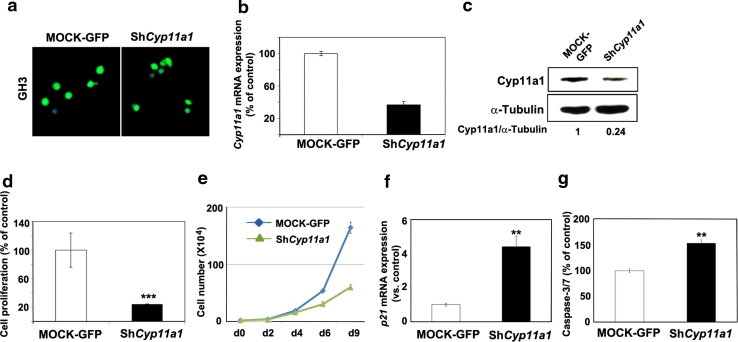
qRT-PCR showed that rat primary PA cells maintain the high Cyp11a1 expression level observed in the tumor tissues from which they derive (Supplementary Fig. 5), and thus represent a suitable model for analyzing the effect of gene knock-down. As these cells were difficult to transfect, we generated a lentiviral vector expressing both a small hairpin (sh) RNA molecule directed against rat Cyp11a1 and the green fluorescent protein (GFP) to monitor infection efficiency (ca. 80 %, data not shown). Two RNA sequences were tested but only one gave a consistent and robust gene down-regulation and was then used on further experiments (data not shown). The sh-Cyp11a1 lentiviral vector was first tested in GH3 cells (Fig. 5a), where it caused a very strong decrease in Cyp11a1 expression (Fig. 5b, c) and, concomitantly, a reduction in cell proliferation when compared with mock infected cells (GFP) (Fig. 5d). In growth curve experiments, viral-mediated Cyp11a1 knock-down decreased the growth rate of GH3 cells, while infection with the mock vector had not effect on cell growth (Fig. 5e).
Fig. 5.
Effect of Cyp11a1 on tumor cell proliferation. a Infection of GH3 cells with lentiviral vectors expressing shCyp11a1-GFP or GFP only (MOCK). b GH3 cells were infected with the lentiviral vectors as in “a” and the level of Cyp11a1 was analyzed by qRT-PCR 72 h later. c In samples parallel to “b”, proteins were extracted, and Western blotting was performed to monitor Cyp11a1 expression. The ratios of the band intensities for P450scc versus α-tubulin, normalized against the ratio in the MOCK control (ratio = 1), is indicated. d In samples parallel to “b”, cell proliferation was assessed 72 h after infection using the WST-1 assay. Data were analyzed independently with six replicates each and were expressed as the mean ± SEM. e Growth curve of GH3 cells infected with lentiviral vectors expressing shCyp11a1-GFP or GFP only (MOCK). Cells were trypsinized, stained with Trypan Blue and viable cells were counted. Values are the mean of 3 independent experiments ± SD. f In samples parallel to “d”, the expression of p21 was assessed by qRT-PCR as indicated in the legend of Fig. 1, and is reported relative to the expression level in mock-transfected cells arbitrarily set to 1. g In samples parallel to “d”, caspase 3/7 activity was measured to monitor apoptosis 72 h post-infection. Data were analyzed independently with six replicates each and were expressed as the mean ± SEM. **P < 0.01, ***P < 0.001, versus MOCK
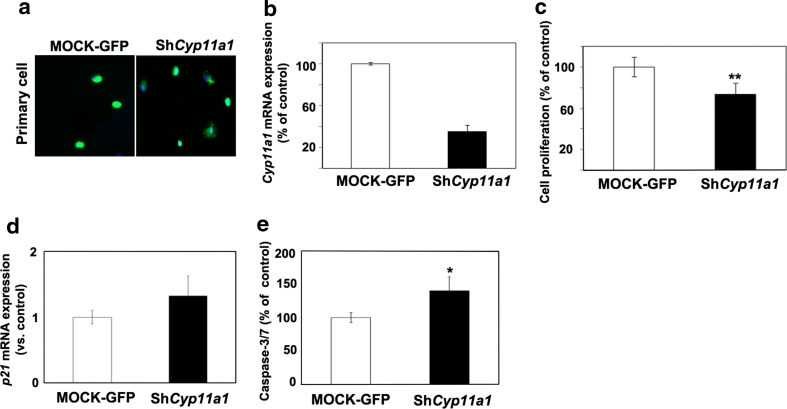
To determine what caused the reduction in the proliferation capacity of GH3 cells, we checked for the induction of cell cycle inhibitors following Cyp11a1 gene knock-down. We determined the level of expression of p16, p18 and p21 after infection by qRT-PCR and we found that while the amount of p16 and p18 transcripts does not change (data not shown), p21 expression increases significantly upon reduction of Cyp11a1 levels (Fig. 5f). We also assessed caspase-3/7 activity as a measure of apoptosis in GH3 cells infected with GFP or with sh-Cyp11a1. Caspase-3/7 activity is higher in cells infected by sh-Cyp11a1 compared with mock infected cells, suggesting that this process might also explain part of the reduced cell growth (Fig. 5g). We then went back to our original model (MENX rats) and established primary PA cultures from three independent mutant rats. Cells were infected with sh-Cyp11a1 vector, or with the mock GFP vector (Fig. 6a) and cell viability was measured. Upon infection of the primary cells with sh-Cyp11a1 vector, but not with mock vector, we observed a reduction in Cyp11a1 expression (Fig. 6b), which was accompanied by a decrease in cell viability (−30 % versus control GFP) (Fig. 6c). Similarly to what we observed in GH3 cells, also in primary rat PA cells there was an increase in both p21 expression and caspase-3/7 activity, although less pronounced than in GH3 cells (Fig. 6d, e). In conclusion, high expression of Cyp11a1 associates with increased proliferation and survival not only in adrenocortical carcinoma cells (Y1), but also in rat pituitary tumor cells from different lineages (somatotroph and gonadotroph).
Fig. 6.
Effect of Cyp11a1 on primary pituitary tumor cell proliferation. a Infection of rat primary pituitary tumor cells with lentiviral vectors expressing shCyp11a1-GFP or GFP only (MOCK). b Primary pituitary tumor cells from three mutant rats were infected with the lentiviral vectors as in “a” and the level of Cyp11a1 was analyzed by qRT-PCR as indicated in the legend of Fig. 1, and is reported relative to the expression level in mock-transfected cells arbitrarily set to 100. c In samples parallel to “b”, cell proliferation was assessed 72 h after infection using the WST-1 assay. Data were analyzed independently with six replicates each and were expressed as the mean ± SEM. d In samples parallel to “c”, the expression of p21 was assessed by qRT-PCR as indicated in the legend of Fig. 1, and is reported relative to the expression level in mock-transfected cells arbitrarily set to 1. e Caspase 3/7 activity was measured in samples in parallel to “c” to monitor apoptosis in GH3 cells 72 h post-infection. *P < 0.05, **P < 0.01 versus MOCK
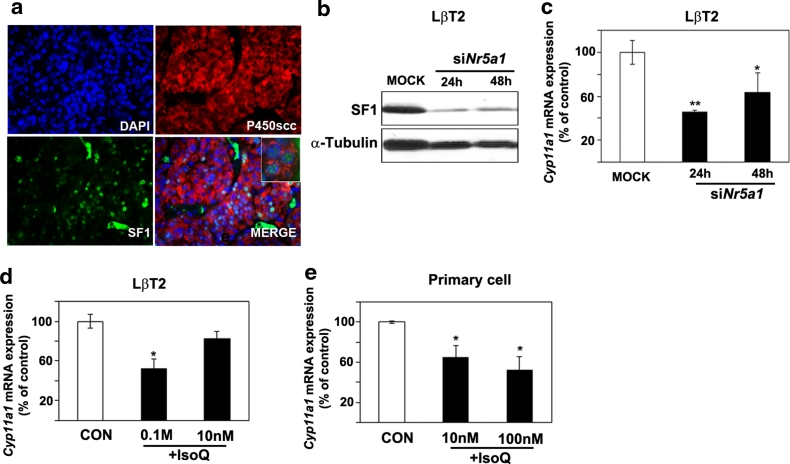
Rat PA cells express both Nr5a1 and Cyp11a1 and, noteworthy, in these cells SF-1 and P450scc co-localize, as determined by dual co-immunofluorescence (Fig. 7a). SF-1 regulates Cyp11a1 transcription in steroidogenic cells but whether it also does so in adenohypophyseal cells is not known. Therefore, we silenced Nr5a1 expression by means of siRNA oligos in gonadotroph LβT2 cells and then assessed the level of Cyp11a1 mRNA by qRT-PCR. As experimental control, we used again Y1 adrenocortical cancer cells, where Nr5a1 gene down-regulation caused a reduction of Cyp11a1 mRNA expression (Supplementary Fig. 6). We observed that knock-down of Nr5a1 in LβT2 cells, as demonstrated by western blotting (Fig. 7b), associates with a decrease in Cyp11a1 expression (Fig. 7c). To further confirm that SF-1 transcriptional activity plays a role in regulating Cyp11a1 expression in these cells, we treated LβT2 cells with a small molecule inhibitor of SF-1, isoquinolinone (IsoQ) [8]. This compound caused a significant dose-dependent reduction of Cyp11a1 expression (Fig. 7d). Similar results were also obtained exposing rat primary PA cells (with high endogenous levels of Cyp11a1) to IsoQ (Fig. 7e). Altogether, these data provide indirect evidence that SF-1 is involved in regulating Cyp11a1 expression in gonadotroph cells.
Fig. 7.
Co-localization of P450scc and SF-1 in rat PAs and reduction of Cyp11a1 expression following down-regulation or inhibition of SF-1 in gonadotroph cells. a Immunofluorescent staining with antibodies against P450scc (red) and SF-1 (green) was performed on FFPE pituitary tumor tissues from MENX mutant rats. Nuclei were counterstained with DAPI. The inset in MERGE shows the co-localization of SF-1 and P450scc in rat tumor cells. Original magnification: ×200; inset: ×400. b LβT2 cells were transfected with scrambled (MOCK) or siRNA oligos against the mouse Nr5a1 gene (siNr5a1). SF-1 and α-tubulin expression levels were assessed by Western blotting 24 and 48 h after transfection. c In samples parallel to “b”, Cyp11a1 expression level was assessed by qRT-PCR as indicated in the legend of Fig. 1, and is reported relative to the expression level in mock-transfected cells arbitrarily set to 100. d LβT2 cells were treated with different concentrations of the SF-1 inhibitor IsoQ (0.1 M and 10 nM) or left untreated (CON). After 24 h, we determined Cyp11a1 expression levels as in “c”. e Primary pituitary tumor cells from mutant rats (n = 3) were incubated with IsoQ as in “d”. After 24 h, we assessed Cyp11a1 expression levels by qRT-PCR as in “c”. Data from primary cultures were analyzed independently with six replicates each and were expressed as the mean ± SEM. *P < 0.05, ***P < 0.001 versus MOCK or CON
Discussion
We recently demonstrated that PAs developing in MENX-affected rats are pathologically similar to human gonadotroph adenomas [26]. The current study confirms this observation and shows that common genetic signatures exist between MENX-associated and human gonadotropinomas. In addition, novel putative markers of human PAs were identified, suggesting that MENX-associated neoplasms may be exploited as to gene discovery.
In silico analysis of gene expression array data of human gonadotroph adenomas identified several genes differentially expressed between tumors and normal pituitaries in both patients and MENX rats. An example of a gene down-regulated in both rats and human PAs is Dlk1, encoding a non-canonical ligand of Notch. DLK1 was found to be selectively silenced in NFPAs but not in other types of PA [1, 5]. DLK1 is located in the DLK1/MEG3 imprinted locus on human chromosome 14q32.3 and MEG3, coding for a maternally imprinted long noncoding RNA with tumor suppressive function, was also found to be specifically silenced through methylation in NFPAs, but not in other hypophyseal adenoma types [13]. In rat PAs, Gtl2 (Meg3) is down-regulated (−1.95) but did not reach statistical significance.
Previous studies have compared human non-invasive and invasive/aggressive PAs with the aim to unveil the molecular mechanisms leading to an aggressive behavior and to identify markers predictive of invasiveness [12, 19, 43]. Some genes or gene families we found up-regulated in MENX adenomas had previously been identified in human aggressive-invasive PAs, e.g., PTTG1, RACGAP1, CCNB1, CENPE, AURKB [12]. PTTG1, the homologue of yeast securin, controls the faithful separation of sister chromatids [46], while deregulated expression of centrosome proteins (CENP) and Aurora kinases has been linked to defects in the mitotic machinery [41, 44]. Differential expression of the five genes mentioned above was observed among benign, pre-malignant and malignant tumors, with a tendency to higher expression in the more malignant ones, with Pttg1 being expressed only in the latter group. Altogether, these findings further support our initial hypothesis that MENX-associated PAs resemble aggressive tumors [26], and suggest that these lesions may be exploited to identify novel targets for specific therapies of aggressive PAs, currently orphan of specific molecular-targeted treatment strategies. Moreover, our data further confirm the critical role of Pttg1 in pituitary tumorigenesis across different species and genetic backgrounds.
NUSAP1 is among the genes overexpressed in both rat and human PAs but has not so far been investigated in these tumors. We here report that 90 % of human gonadotroph adenomas express the NUSAP1 gene at high level. Concordantly, the number of cells immunoreactive for NuSAP was higher in tumors compared with normal pituitaries. NuSAP is a microtubule-associated protein that bundles and stabilizes microtubules thereby linking chromosomes to the mitotic spindle [35]. Recently, overexpression of NUSAP1 has been found in various human cancers by array analysis, including glioblastomas, hepatocellular carcinomas, pancreatic adenocarcinoma (reviewed in [21]). The high expression of the NUSAP1 gene has been associated with poor prognosis in melanoma patients [3], with a malignancy-risk genetic signature in breast cancer [4] and with recurrence in prostate cancer [15]. The study on prostate cancer has validated gene up-regulation seen by array analysis with immunostaining for the NuSAP protein: tumors with Gleason grading scores ≥7 had more NuSAP-positive cells than tumors with lower scores [15]. We estimated the percentage of NuSAP-positive cells in human gonadotroph tumors (average 0.5 %, range 0.2–1 %) and found a correlation with that of Ki-67-positive cells (average 1.2 %, range 0.5–2 %). Further studies are required to determine whether NuSAP labeling index correlates with the outcome of patients with PA and whether it provides a more accurate estimate of aggressive behavior than Ki67. Noteworthy, in addition to NUSAP1, our meta-analysis of human PA array data has identified the up-regulation of genes encoding other proteins playing a critical role in mitosis, mitotic spindle checkpoint and dynamics, or cytokinesis such as BUB1, CCNB1, CDC2, KIF4, KIF11, PRC1, in gonadotroph adenomas (Supplementary Table 2). These genes, if experimentally verified to be involved in PAs, may represent novel therapeutic targets for these tumors. While classical antimitotic compounds have limited clinical applications because of severe side effects, a new generation of drugs has been developed that targets kinesins (KIF proteins) and kinases with unique function in mitosis. These antitumor agents could be tested for the treatment of gonadotroph adenomas.
We observed that SF-1 and P450scc are co-expressed in PA cells of MENX-affected animals, and both siRNA-mediated silencing of the Nr5a1 gene and treatment with the SF-1 inhibitor IsoQ resulted in the down-regulation of Cyp11a1 in rat primary tumor cells and in LβT2 gonadotroph cells. These results indicate that SF-1 regulates Cyp11a1 expression in gonadotroph cells. The transcriptional regulation of Cyp11a1 in steroidogenic tissues is complex and involves the concerted action of several tissue-specific trans-regulators such as SF-1, DAX1, TReP-132, LBP, and GATA together with transcription factors having a more widespread expression, including AP-1, Sp1, and AP-2 [16]. Further studies are needed to identify the additional factors involved in the transcriptional regulation of Cyp11a1 in PA cells.
CYP11A1 was up-regulated in 75 % of human gonadotropinomas and P450scc protein expression was high in these tumors, while barely detectable in normal pituitaries. P450scc has been detected in the developing mouse at embryonic day 9.5 in Rathke’s pouch and at day 18.5 in the pituitary primordium [17]. However, its potential role in pituitary development or in the adult pituitary has not been addressed. Here we show that Cyp11a1 plays a pro-proliferative role in Y1 adrenocortical cancer cells, but also in GH3 somatotroph adenoma cells and in rat primary pituitary gonadotroph cells. To the best of our knowledge, only two studies have so far addressed the putative role of Cyp11a1 in regulating cell proliferation or viability. The first one showed that overexpression of CYP11A1 in tumorigenic and non-tumorigenic mammalian cell lines can either inhibit or enhance cell viability in a cell type-specific manner [7]. In cells sensitive to CYP11A1, gene overexpression led to decreased cell proliferation through increase of p21 expression and induction of apoptosis [7]. The authors did not investigate the molecular processes associated with CYP11A1-mediated increase in cell proliferation. Based on our results, when the reduction of CYP11A1 level decreases cell proliferation (as seen in PA cells), both apoptosis and p21 expression are also involved, indicating that these are common pathways downstream of CYP11A1 signaling. In a more recent study, a truncated isoform of P450scc was found to suppress osteoblast proliferation in physiological conditions [38]. Unlike the full length P450scc isoform, which is located in the mitochondria, this small isoform localizes to cytoplasm and nucleus, and might therefore be involved in still unidentified molecular pathways. Our studies show that Cyp11a1 plays a pro-proliferative role in adrenocortical cancer and PA cells, thereby providing additional evidence supporting a role for this gene in tumorigenesis.
In conclusion, we here provide experimental evidence that the MENX animal model can be exploited as experimental tool to shed light into the pathomechanisms involved in human pituitary tumorigenesis. Several of the dysregulated genes here reported have never been associated with PAs, and may represent novel biomarkers for future clinical applications.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank E. Samson, D. Mörzl and E. Pulz for excellent technical assistance, and Dr. P.L. Mellon for providing the LβT2 cell line. This study was supported by grant SFB824 from the Deutsche Forschungsgemeinschaft (DFG) and grant #107973 from the Deutsche Krebshilfe to N.S.P., and also in part by a research grant from Novartis Pharma GmbH, Nürnberg, Germany.
Footnotes
M. Lee and I. Marinoni contributed equally to the paper.
References
- 1.Altenberger T, Bilban M, Auer M, et al. Identification of DLK1 variants in pituitary- and neuroendocrine tumors. Biochem Biophys Res Commun. 2006;340:995–1005. doi: 10.1016/j.bbrc.2005.12.094. [DOI] [PubMed] [Google Scholar]
- 2.Anastasov N, Klier M, Koch I, et al. Efficient shRNA delivery into B and T lymphoma cells using lentiviral vector-mediated transfer. J Hematopathol. 2009;2:9–19. doi: 10.1007/s12308-008-0020-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogunovic D, O’Neill DW, Belitskaya-Levy I, et al. Immune profile and mitotic index of metastatic melanoma lesions enhance clinical staging in predicting patient survival. Proc Natl Acad Sci USA. 2009;106:20429–20434. doi: 10.1073/pnas.0905139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen DT, Nasir A, Culhane A, et al. Proliferative genes dominate malignancy-risk gene signature in histologically-normal breast tissue. Breast Cancer Res Treat. 2010;119:335–346. doi: 10.1007/s10549-009-0344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheunsuchon P, Zhou Y, Zhang X, et al. Silencing of the imprinted DLK1-MEG3 locus in human clinically nonfunctioning pituitary adenomas. Am J Pathol. 2011;179:2120–2130. doi: 10.1016/j.ajpath.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis SW, Castinetti F, Carvalho LR, et al. Molecular mechanisms of pituitary organogenesis: in search of novel regulatory genes. Mol Cell Endocrinol. 2010;323:4–19. doi: 10.1016/j.mce.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derouet-Humbert E, Roemer K, Bureik M. Adrenodoxin (Adx) and CYP11A1 (P450scc) induce apoptosis by the generation of reactive oxygen species in mitochondria. Biol Chem. 2005;386:453–461. doi: 10.1515/BC.2005.054. [DOI] [PubMed] [Google Scholar]
- 8.Doghman M, Karpova T, Rodrigues GA, et al. Increased steroidogenic factor-1 dosage triggers adrenocortical cell proliferation and cancer. Mol Endocrinol. 2007;21:2968–2987. doi: 10.1210/me.2007-0120. [DOI] [PubMed] [Google Scholar]
- 9.Elston MS, Gill AJ, Conaglen JV, et al. Wnt pathway inhibitors are strongly down-regulated in pituitary tumors. Endocrinology. 2008;149:1235–1242. doi: 10.1210/en.2007-0542. [DOI] [PubMed] [Google Scholar]
- 10.Ericson J, Norlin S, Jessell TM, Edlund T. Integrated FGF and BMP signaling controls the progression of progenitor cell differentiation and the emergence of pattern in the embryonic anterior pituitary. Development. 1998;125:1005–1015. doi: 10.1242/dev.125.6.1005. [DOI] [PubMed] [Google Scholar]
- 11.Evans CO, Young AN, Brown MR, et al. Novel patterns of gene expression in pituitary adenomas identified by complementary deoxyribonucleic acid microarrays and quantitative reverse transcription-polymerase chain reaction. J Clin Endocrinol Metab. 2001;86:3097–3107. doi: 10.1210/jc.86.7.3097. [DOI] [PubMed] [Google Scholar]
- 12.Galland F, Lacroix L, Saulnier P, et al. Differential gene expression profiles of invasive and non-invasive non-functioning pituitary adenomas based on microarray analysis. Endocr Relat Cancer. 2010;17:361–371. doi: 10.1677/ERC-10-0018. [DOI] [PubMed] [Google Scholar]
- 13.Gejman R, Batista DL, Zhong Y, et al. Selective loss of MEG3 expression and intergenic differentially methylated region hypermethylation in the MEG3/DLK1 locus in human clinically nonfunctioning pituitary adenomas. J Clin Endocrinol Metab. 2008;93:4119–4125. doi: 10.1210/jc.2007-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenman Y, Stern N. Non-functioning pituitary adenomas. Best Pract Res Clin Endocrinol Metab. 2009;23:625–638. doi: 10.1016/j.beem.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Gulzar ZG, McKenney JK, Brooks JD. Increased expression of NuSAP in recurrent prostate cancer is mediated by E2F1. Oncogene. 2013;32:70–77. doi: 10.1038/onc.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo IC, Shih MC, Lan HC, Hsu NC, Hu MC, Chung BC. Transcriptional regulation of human CYP11A1 in gonads and adrenals. J Biomed Sci. 2007;14:509–515. doi: 10.1007/s11373-007-9177-z. [DOI] [PubMed] [Google Scholar]
- 17.Hammer F, Compagnone NA, Vigne JL, Bair SR, Mellon SH. Transcriptional regulation of P450scc gene expression in the embryonic rodent nervous system. Endocrinology. 2004;145:901–912. doi: 10.1210/en.2003-0125. [DOI] [PubMed] [Google Scholar]
- 18.Hardy J. The transsphenoidal surgical approach to the pituitary. Hosp Pract. 1979;14:81–89. doi: 10.1080/21548331.1979.11707562. [DOI] [PubMed] [Google Scholar]
- 19.Hussaini IM, Trotter C, Zhao Y, et al. Matrix metalloproteinase-9 is differentially expressed in nonfunctioning invasive and noninvasive pituitary adenomas and increases invasion in human pituitary adenoma cell line. Am J Pathol. 2007;170:356–365. doi: 10.2353/ajpath.2007.060736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikuyama S, Mu YM, Ohe K, et al. Expression of an orphan nuclear receptor DAX-1 in human pituitary adenomas. Clin Endocrinol. 1998;48:647–654. doi: 10.1046/j.1365-2265.1998.00477.x. [DOI] [PubMed] [Google Scholar]
- 21.Iyer J, Moghe S, Furukawa M, Tsai MY. What’s Nu(SAP) in mitosis and cancer? Cell Signal. 2011;23:991–998. doi: 10.1016/j.cellsig.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 22.Jiang Z, Gui S, Zhang Y. Analysis of differential gene expression in plurihormonal pituitary adenomas using bead-based fiber-optic arrays. J Neurooncol. 2012;108:341–348. doi: 10.1007/s11060-011-0792-1. [DOI] [PubMed] [Google Scholar]
- 23.Kelberman D, Rizzoti K, Lovell-Badge R, Robinson IC, Dattani MT. Genetic regulation of pituitary gland development in human and mouse. Endocr Rev. 2009;30:790–829. doi: 10.1210/er.2009-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee M, Theodoropoulou M, Graw J, Roncaroli F, Zatelli MC, Pellegata NS. Levels of p27 sensitize to dual PI3 K/mTOR inhibition. Mol Cancer Ther. 2011;10:1450–1459. doi: 10.1158/1535-7163.MCT-11-0188. [DOI] [PubMed] [Google Scholar]
- 25.Machiavelli GA, Rivolta CM, Artese R, Basso A, Burdman JA. Expression of c-myc and c-fos and binding sites for estradiol and progesterone in human pituitary tumors. Neurol Res. 1998;20:709–712. doi: 10.1080/01616412.1998.11740588. [DOI] [PubMed] [Google Scholar]
- 26.Marinoni I, Lee M, Mountford S, et al. Characterization of MENX-associated pituitary tumours. Neuropathol Appl Neurobiol. 2013;39:256–269. doi: 10.1111/j.1365-2990.2012.01278.x. [DOI] [PubMed] [Google Scholar]
- 27.Melmed S. Pathogenesis of pituitary tumors. Nat Rev Endocrinol. 2011;7:257–266. doi: 10.1038/nrendo.2011.40. [DOI] [PubMed] [Google Scholar]
- 28.Michaelis KA, Knox AJ, Xu M, et al. Identification of growth arrest and DNA-damage-inducible gene beta (GADD45beta) as a novel tumor suppressor in pituitary gonadotrope tumors. Endocrinology. 2011;152:3603–3613. doi: 10.1210/en.2011-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molatore S, Kiermaier E, Jung CB, et al. Characterization of a naturally-occurring p27 mutation predisposing to multiple endocrine tumors. Mol Cancer. 2010;9:116. doi: 10.1186/1476-4598-9-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Molatore S, Liyanarachchi S, Irmler M, et al. Pheochromocytoma in rats with multiple endocrine neoplasia (MENX) shares gene expression patterns with human pheochromocytoma. Proc Natl Acad Sci USA. 2010;107:18493–18498. doi: 10.1073/pnas.1003956107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreno CS, Evans CO, Zhan X, Okor M, Desiderio DM, Oyesiku NM. Novel molecular signaling and classification of human clinically nonfunctional pituitary adenomas identified by gene expression profiling and proteomic analyses. Cancer Res. 2005;65:10214–10222. doi: 10.1158/0008-5472.CAN-05-0884. [DOI] [PubMed] [Google Scholar]
- 32.Morris DG, Musat M, Czirjak S, et al. Differential gene expression in pituitary adenomas by oligonucleotide array analysis. Eur J Endocrinol/Eur Fed Endocr Soc. 2005;153:143–151. doi: 10.1530/eje.1.01937. [DOI] [PubMed] [Google Scholar]
- 33.Pellegata NS, Quintanilla-Martinez L, Siggelkow H, et al. Germ-line mutations in p27Kip1 cause a multiple endocrine neoplasia syndrome in rats and humans. Proc Natl Acad Sci USA. 2006;103:15558–15563. doi: 10.1073/pnas.0603877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pereira AM, Biermasz NR. Treatment of nonfunctioning pituitary adenomas: what were the contributions of the last 10 years? A critical view. Ann Endocrinol. 2012;73:111–116. doi: 10.1016/j.ando.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 35.Raemaekers T, Ribbeck K, Beaudouin J, et al. NuSAP, a novel microtubule-associated protein involved in mitotic spindle organization. J Cell Biol. 2003;162:1017–1029. doi: 10.1083/jcb.200302129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rainer J, Sanchez-Cabo F, Stocker G, Sturn A, Trajanoski Z. CARMAweb: comprehensive R- and bioconductor-based web service for microarray data analysis. Nucleic Acids Res. 2006;34:W498–W503. doi: 10.1093/nar/gkl038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheithauer BW, Kovacs K, Zorludemir S, Lloyd RV, Erdogan S, Slezak J. Immunoexpression of androgen receptor in the nontumorous pituitary and in adenomas. Endocr Pathol. 2008;19:27–33. doi: 10.1007/s12022-007-9012-0. [DOI] [PubMed] [Google Scholar]
- 38.Teplyuk NM, Zhang Y, Lou Y, et al. The osteogenic transcription factor runx2 controls genes involved in sterol/steroid metabolism, including CYP11A1 in osteoblasts. Mol Endocrinol. 2009;23:849–861. doi: 10.1210/me.2008-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ueta Y, Levy A, Powell MP, et al. Neuronal nitric oxide synthase gene expression in human pituitary tumours: a possible association with somatotroph adenomas and growth hormone-releasing hormone gene expression. Clin Endocrinol. 1998;49:29–38. doi: 10.1046/j.1365-2265.1998.00399.x. [DOI] [PubMed] [Google Scholar]
- 40.Val P, Martinez-Barbera JP, Swain A. Adrenal development is initiated by Cited2 and Wt1 through modulation of Sf-1 dosage. Development. 2007;134:2349–2358. doi: 10.1242/dev.004390. [DOI] [PubMed] [Google Scholar]
- 41.Vigneron S, Prieto S, Bernis C, Labbe JC, Castro A, Lorca T. Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol Biol Cell. 2004;15:4584–4596. doi: 10.1091/mbc.E04-01-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang SM, Ooi LL, Hui KM. Upregulation of Rac GTPase-activating protein 1 is significantly associated with the early recurrence of human hepatocellular carcinoma. Clin Cancer Res: Off J Am Assoc Cancer Res. 2011;17:6040–6051. doi: 10.1158/1078-0432.CCR-11-0557. [DOI] [PubMed] [Google Scholar]
- 43.Wierinckx A, Auger C, Devauchelle P, et al. A diagnostic marker set for invasion, proliferation, and aggressiveness of prolactin pituitary tumors. Endocr Relat Cancer. 2007;14:887–900. doi: 10.1677/ERC-07-0062. [DOI] [PubMed] [Google Scholar]
- 44.Yao X, Abrieu A, Zheng Y, Sullivan KF, Cleveland DW. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat Cell Biol. 2000;2:484–491. doi: 10.1038/35019518. [DOI] [PubMed] [Google Scholar]
- 45.Zhan X, Desiderio DM. Signaling pathway networks mined from human pituitary adenoma proteomics data. BMC Med Genomics. 2010;3:13. doi: 10.1186/1755-8794-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.