Abstract
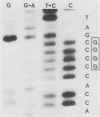
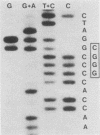
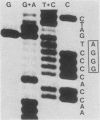
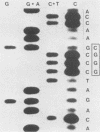
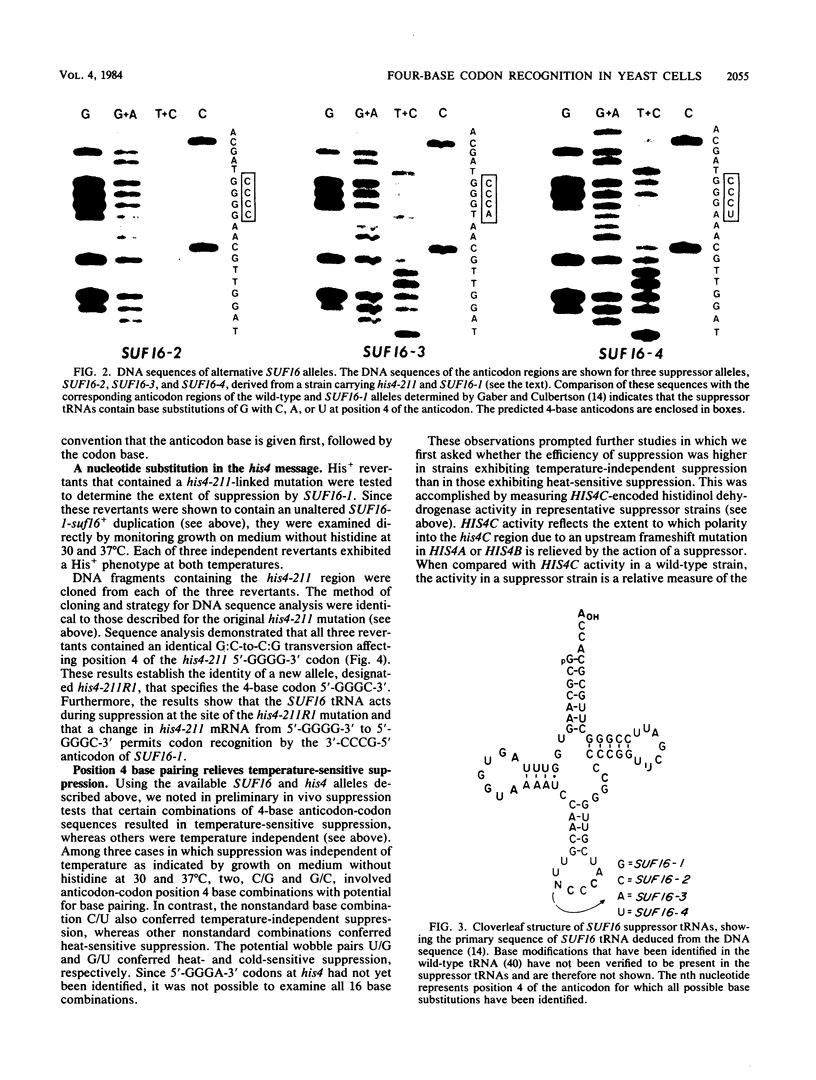
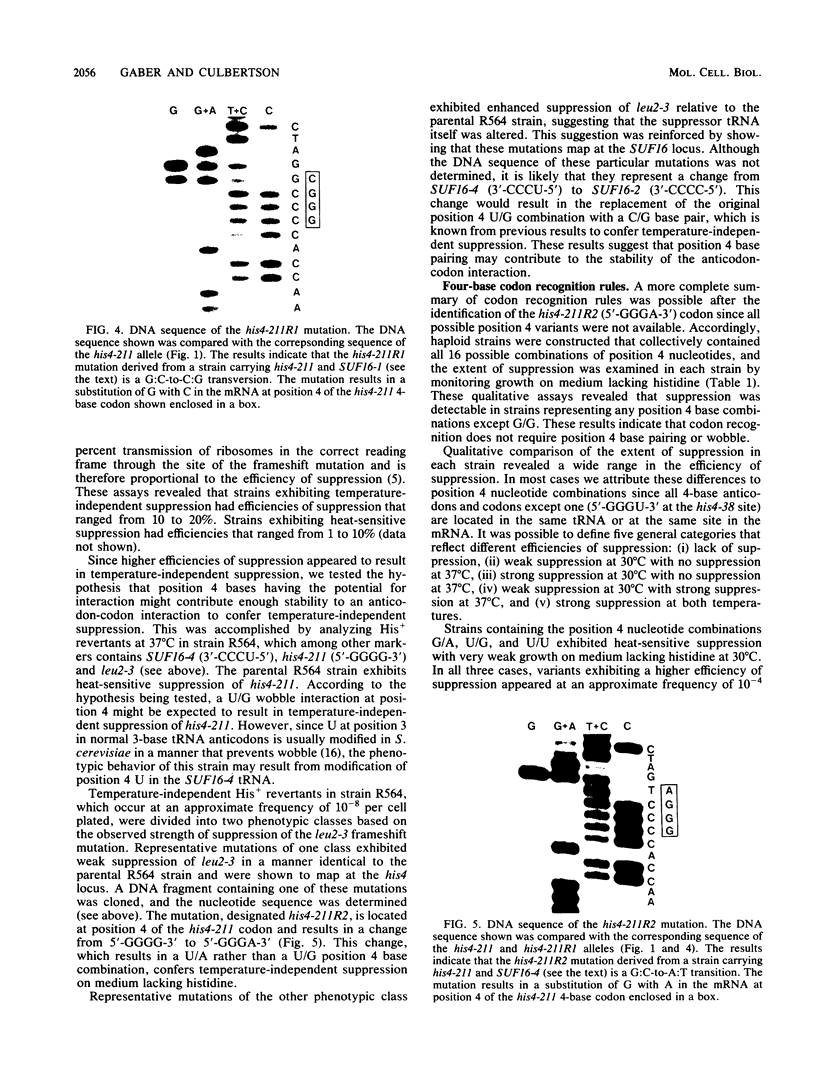
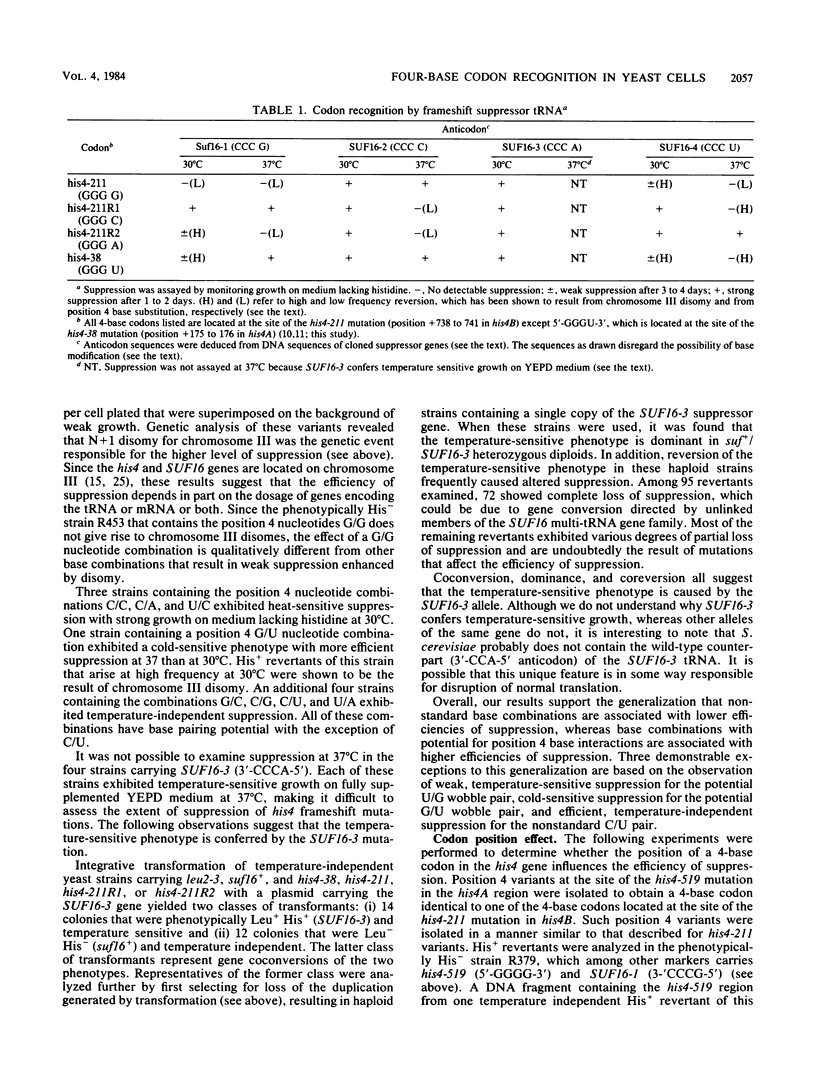
A genetic approach has been used to establish the molecular basis of 4-base codon recognition by frameshift suppressor tRNA containing an extra nucleotide in the anticodon. We have isolated all possible base substitution mutations at the position 4 (N) in the 3'-CCCN-5' anticodon of a Saccharomyces cerevisiae frameshift suppressor glycine tRNA encoded by the SUF16 gene. Base substitutions at +1 frameshift sites in the his4 gene have also been obtained such that all possible 4-base 5'-GGGN-3' codons have been identified. By testing for suppression in different strains that collectively represent all 16 possible combinations of position 4 nucleotides, we show that frameshift suppression does not require position 4 base pairing. Nonetheless, position 4 interactions influence the efficiency of suppression. Our results suggest a model in which 4-base translocation of mRNA on the ribosome is directed primarily by the number of nucleotides in the anticodon loop, whereas the resulting efficiency of suppression is dependent on the nature of position 4 nucleotides.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkins J. F., Gesteland R. F., Reid B. R., Anderson C. W. Normal tRNAs promote ribosomal frameshifting. Cell. 1979 Dec;18(4):1119–1131. doi: 10.1016/0092-8674(79)90225-3. [DOI] [PubMed] [Google Scholar]
- Bossi L., Roth J. R. Four-base codons ACCA, ACCU and ACCC are recognized by frameshift suppressor sufJ. Cell. 1981 Aug;25(2):489–496. doi: 10.1016/0092-8674(81)90067-2. [DOI] [PubMed] [Google Scholar]
- Bossi L., Ruth J. R. The influence of codon context on genetic code translation. Nature. 1980 Jul 10;286(5769):123–127. doi: 10.1038/286123a0. [DOI] [PubMed] [Google Scholar]
- Cieslà Z., Salvatore F., Broach J. R., Artz S. W., Ames B. N. Histidine regulation in Salmonella typhimurium. XVI. A sensitive radiochemical assay for histidinol dehydrogenase. Anal Biochem. 1975 Jan;63(1):44–55. doi: 10.1016/0003-2697(75)90187-6. [DOI] [PubMed] [Google Scholar]
- Culbertson M. R., Charnas L., Johnson M. T., Fink G. R. Frameshifts and frameshift suppressors in Saccharomyces cerevisiae. Genetics. 1977 Aug;86(4):745–764. doi: 10.1093/genetics/86.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson M. R., Henry S. A. Genetic analysis of hybrid strains trisomic for the chromosome containing a fatty acid synthetase gene complex (fas1) in yeast. Genetics. 1973 Nov;75(3):441–458. doi: 10.1093/genetics/75.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson M. R., Underbrink K. M., Fink G. R. Frameshift suppression Saccharomyces cerevisiae. II. Genetic properties of group II suppressors. Genetics. 1980 Aug;95(4):833–853. doi: 10.1093/genetics/95.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins C. M., Donahue T. F., Culbertson M. R. Nucleotide sequence of the SUF2 frameshift suppressor gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3565–3569. doi: 10.1073/pnas.79.11.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins C. M., Gaber R. F., Culbertson M. R., Mann R., Fink G. R. Frameshift suppression in Saccharomyces cerevisiae. III. Isolation and genetic properties of group III suppressors. Genetics. 1980 Aug;95(4):855–879. doi: 10.1093/genetics/95.4.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Farabaugh P. J., Fink G. R. Suppressible four-base glycine and proline codons in yeast. Science. 1981 Apr 24;212(4493):455–457. doi: 10.1126/science.7010605. [DOI] [PubMed] [Google Scholar]
- Donahue T. F., Farabaugh P. J., Fink G. R. The nucleotide sequence of the HIS4 region of yeast. Gene. 1982 Apr;18(1):47–59. doi: 10.1016/0378-1119(82)90055-5. [DOI] [PubMed] [Google Scholar]
- Fink G. R., Styles C. A. Gene conversion of deletions in the his4 region of yeast. Genetics. 1974 Jun;77(2):231–244. doi: 10.1093/genetics/77.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox T. D., Weiss-Brummer B. Leaky +1 and -1 frameshift mutations at the same site in a yeast mitochondrial gene. Nature. 1980 Nov 6;288(5786):60–63. doi: 10.1038/288060a0. [DOI] [PubMed] [Google Scholar]
- Gaber R. F., Culbertson M. R. Frameshift suppression in Saccharomyces cerevisiae. IV. New suppressors among spontaneous co-revertants of the Group II his4-206 and leu 2-3 frameshift mutations. Genetics. 1982 Jul-Aug;101(3-4):345–367. doi: 10.1093/genetics/101.3-4.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaber R. F., Culbertson M. R. The yeast frameshift suppressor gene SUF16-1 encodes an altered glycine tRNA containing the four-base anticodon 3'-CCCG-5'. Gene. 1982 Sep;19(2):163–172. doi: 10.1016/0378-1119(82)90002-6. [DOI] [PubMed] [Google Scholar]
- Hicks J., Fink G. R. Identification of chromosomal location of yeast DNA from hybrid plasmid p Yeleu 10. Nature. 1977 Sep 15;269(5625):265–267. doi: 10.1038/269265a0. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastelein R. A., Remaut E., Fiers W., van Duin J. Lysis gene expression of RNA phage MS2 depends on a frameshift during translation of the overlapping coat protein gene. Nature. 1982 Jan 7;295(5844):35–41. doi: 10.1038/295035a0. [DOI] [PubMed] [Google Scholar]
- Keesey J. K., Jr, Bigelis R., Fink G. R. The product of the his4 gene cluster in Saccharomyces cerevisiae. A trifunctional polypeptide. J Biol Chem. 1979 Aug 10;254(15):7427–7433. [PubMed] [Google Scholar]
- Losson R., Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Olson M. V., Loughney K., Hall B. D. Identification of the yeast DNA sequences that correspond to specific tyrosine-inserting nonsense suppressor loci. J Mol Biol. 1979 Aug 15;132(3):387–410. doi: 10.1016/0022-2836(79)90267-5. [DOI] [PubMed] [Google Scholar]
- Petes T. D., Broach J. R., Wensink P. C., Hereford L. M., Fink G. R., Botstein D. Isolation and analysis of recombinant DNA molecules containing yeast DNA. Gene. 1978 Sep;4(1):37–49. doi: 10.1016/0378-1119(78)90013-6. [DOI] [PubMed] [Google Scholar]
- Riddle D. L., Carbon J. Frameshift suppression: a nucleotide addition in the anticodon of a glycine transfer RNA. Nat New Biol. 1973 Apr 25;242(121):230–234. doi: 10.1038/newbio242230a0. [DOI] [PubMed] [Google Scholar]
- Riddle D. L., Roth J. R. Frameshift suppressors. 3. Effects of suppressor mutations on transfer RNA. J Mol Biol. 1972 May 28;66(3):495–506. doi: 10.1016/0022-2836(72)90429-9. [DOI] [PubMed] [Google Scholar]
- Riddle D. L., Roth J. R. Suppressors of frameshift mutations in Salmonella typhimurium. J Mol Biol. 1970 Nov 28;54(1):131–144. doi: 10.1016/0022-2836(70)90451-1. [DOI] [PubMed] [Google Scholar]
- Roeder G. S., Fink G. R. DNA rearrangements associated with a transposable element in yeast. Cell. 1980 Aug;21(1):239–249. doi: 10.1016/0092-8674(80)90131-2. [DOI] [PubMed] [Google Scholar]
- Roth J. R. Frameshift suppression. Cell. 1981 Jun;24(3):601–602. doi: 10.1016/0092-8674(81)90086-6. [DOI] [PubMed] [Google Scholar]
- Scherer S., Davis R. W. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer B., Brearley I., Littlewood R., Fink G. R. A stable aneuploid of Saccharomyces cerevisiae. Genetics. 1971 Apr;67(4):483–495. doi: 10.1093/genetics/67.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. R., Calvo J. M. Nucleotide sequence of the E coli gene coding for dihydrofolate reductase. Nucleic Acids Res. 1980 May 24;8(10):2255–2274. doi: 10.1093/nar/8.10.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcomb D. T., Thomas M., Kelly J., Selker E., Davis R. W. Eukaryotic DNA segments capable of autonomous replication in yeast. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4559–4563. doi: 10.1073/pnas.77.8.4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. Molecular mechanics of translation: a reciprocating ratchet mechanism. Nature. 1970 May 30;226(5248):817–820. doi: 10.1038/226817a0. [DOI] [PubMed] [Google Scholar]
- Yoshida M. The nucleotide sequency of tRNA Gly from yeast. Biochem Biophys Res Commun. 1973 Feb 5;50(3):779–784. doi: 10.1016/0006-291x(73)91312-0. [DOI] [PubMed] [Google Scholar]