Abstract
Previous studies have shown that the neurosteroid analogue, 6-Azi-pregnanolone (6-AziP), photolabels voltage-dependent anion channels and proteins of approximately 55 kDa in rat brain membranes. The present study used two dimensional electrophoresis and nano-electrospray ionization ion trap mass spectrometry (nano-ESI-MS) to identify the 55 kDa proteins (pI 4.8) as isoforms of β-tubulin. This identification was confirmed by immuno-blot and immunoprecipitation of photolabeled protein with anti-β-tubulin antibody and by the demonstration that 6-AziP photolabels purified bovine brain tubulin in a concentration-dependent pattern. To identify the photolabeling sites, purified bovine brain tubulin was photolabeled with 6-AziP, digested with trypsin, and analyzed by matrix-assisted laser desorption/ionization mass spectrometry (MALDI). A 6-AziP adduct of TAVCDIPPR (m/z=1287.77), a β-tubulin specific peptide, was detected by MALDI. High resolution LC-MS/MS analysis identified that 6-AziP was covalently bound to cysteine 354 (Cys-354), previously identified as a colchicine binding site. 6-AziP photolabeling was inhibited by 2-methoxyestradiol, an endogenous derivative of estradiol thought to bind to the colchicine site. Structural modeling predicted that neurosteroids could dock in this colchicine site at the interface between α- and β-tubulin with the photolabeling group of 6-AziP positioned proximate to Cys-354.
Keywords: tubulin, mass spectrometry, photolabeling, neurosteroids
Introduction
Neurosteroids are defined as steroids that are synthesized within the central nervous system that produce rapid actions on neuronal substrates. The endogenous neurosteroids include allopregnanolone, pregnanolone, dehydroepiandrosterone, and their sulfates [1]. Neurosteroids are known to produce an array of neurobiological effects including anxiolytic, analgesic, anticonvulsant, sedative, and hypnotic effects [2, 3]. In addition to acting via nuclear receptors [4], neurosteroids also act via ligand-gated [5, 6] and voltage-gated ion channels to enhance or depress neuronal activity and regulate developmental or regenerative processes. Neurosteroids have also been found to interact with cytoskeletal proteins, particularly microtubules and microtubule-associated proteins [7-10]
The neurosteroid analogue photolabeling reagent 6-Azi-Pregnanolone (6-AziP) was synthesized as a probe to identify neurosteroid binding proteins and to characterize the neurosteroid binding domains on these proteins. We have previously demonstrated that 6-AziP predominantly photolabels two proteins in brain, one of ≈30 kDa and one of ≈55 kDa [11]. The 30 kDa protein was identified by ESI-MS/MS as isoforms of voltage-dependent anion channel (VDAC) [11, 12]. In this study, we show that the 55 kDa protein photolabeled by 6-AziP is β-tubulin. Matrix-assisted laser desorption ionization (MALDI) with time-of-flight (TOF) mass spectrometry identified a modified peptide (TAVCDIPPR) with the 6-AziP adduct. Using high resolution LC-ESI-MS/MS and directed MS2 spectral acquisition (DMSA), we determined that 6-AziP was covalently attached to cysteine 354 (Cys-354), previously identified as a colchicine binding site. The specificity of photolabeling by 6-AziP was shown by inhibition with 2-methoxyestradiol (2-ME), an endogenous derivative of estradiol known to bind to the colchicine site.
Experimental Procedures
Chemical Synthesis
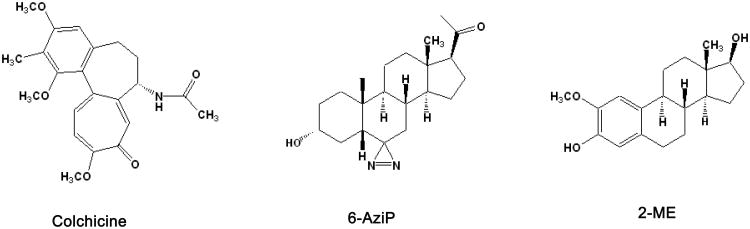
6-AziP was prepared by multi-step synthesis from commercially available progesterone as described in our previous study (1). Selective reduction of the 3-keto group in the 6-azi-3, 20-diketone precursor yields 6-AziP. [3H]-6-AziP was prepared from the same precursor with sodium borotritiide. Figure 1 shows the structures of 6-AziP, colchicine, and 2-ME.
Figure 1.
Structures of 6-AziP, 2-methoxyestradiol, and colchicine.
Membrane preparation
Rat brain membranes were prepared using previously described methods [11, 12]. Briefly, rat brain cortex was homogenized in ice cold 0.32 M sucrose (10 mL/gm) and centrifuged for 10 min at 1,500g. The supernatant was centrifuged for 30 min at 10,000g to obtain the P2 pellet, which was washed 3 times with 50 mM K-Phosphate/200 mM NaCl, pH 7.4. The pellet was re-suspended in 50 mM K-Phosphate/200 mM NaCl, pH 7.4 and recollected by centrifugation for 20 min at 10,000g. The pellet was stored at -80 °C.
Photolabeling
Ten μg of purified bovine brain tubulin was placed in a quartz cuvette (in 50 mM potassium phosphate buffer, pH 7.4 and 150 mM NaCl in a final volume of 1 mL) and incubated with 1 μM tritiated or non-tritiated 6-AziP for 1 hour at 37 °C in the dark. The samples were then irradiated for 5 minutes using a photoreactor emitting light >320 nm (1). Following irradiation, tubulin was precipitated using trichloracetic acid (TCA). The precipitated tubulin was then solubilized in 2 dimensional gel loading buffer (40 mM Tris, 7 M urea, 2 M thiourea, 4% CHAPS, 2% SB-310, 2 mM tributyl phosphine, 1 mM EDTA, 1 mM PMSF and 1 μg/mL each of pepstatin A, chymostatin, leupeptin and antipain) for 2-dimensional gel electrophoresis (2-DE), in tissue solubilization solution for quantification of incorporation of radioactivity, or in trypsin solution for proteolytic digestion. For experiments examining inhibition of photolabeling by 2-ME, 10 μg purified bovine brain tubulin was photolabeled with 0.3 μM [3H]-6-AziP in a volume of 0.1 mL for 3 minutes with or without 30 μM 2-ME; the sample was then diluted with 25 μL of 5× SDS-sample buffer (312.5 mM Tris-HCl, 5% SDS, 0.5 M dithiothreitol, 50% glycerol, and 0.1% bromphenol blue) and analyzed by SDS-PAGE.
Photolabeling of rat brain membranes was performed as previously described [11, 12]. Briefly, rat brain membranes (400 μg) were incubated in the presence of tritiated or non-tritiated 6-AziP for 90 min at 4 °C in the dark. The samples were then irradiated for 5 minutes using the photoreactor. Following irradiation, the rat brain membranes were collected by centrifugation and solubilized in immunoprecipitation (IP) lysis buffer (1% Triton X-100, 150 mM NaCl, 50 mM Tris, 1 mM EDTA, pH 7.4), SDS sample buffer or 2-DE loading buffer.
Immunoprecipitation
After photolabeling, rat brain membranes were solubilized in 1 mL IP lysis buffer for 60 minutes at 4 °C. Insoluble material and debris were collected by centrifugation at 16,000g for 15 min and discarded. The lysate (supernatant) was incubated with either anti-β-tubulin or anti-mouse IgG overnight at 4 °C, following which immune-complexes were precipitated using protein G agarose. After washing with lysis buffer 3 times, the immunoprecipitated proteins were eluted by boiling in 5× SDS-sample buffer for 10 min followed by SDS-PAGE.
Gel Slicing
After electrophoresis, the 1D gels were cut in vertical columns and sliced in 1 mm horizontal slices using a DE 113 manual gel slicer (Hoeffer Scientific Instruments, San Francisco, CA). Slices were digested with 4 mL of tissue solubilization solution consisting of 3a20TM and TS-2 (ratio 9:1) for 24 hours, and the radioactivity in each slice was determined by scintillation spectrometry [13].
Immuno-blotting
Proteins from SDS-PAGE gels were transferred onto polyvinylidene fluoride membrane using a semi-dry transfer instrument. The membranes were blocked with 5% dried milk and incubated with α-tubulin antibody (Santa Cruz, 1:200) or β-tubulin antibody (Santa Cruz, 1:200) for 1 h followed by peroxidase-conjugated secondary antibody (Santa Cruz, 1:10,000). Immunoreactive bands were visualized using the ECL-plus immuno-blotting detection system (Amersham Bioscience).
Autoradiography
SDS-PAGE gels were fixed for 30 minutes in isopropanol: water: acetic acid (25:65:10) at room temperature and then dried under vacuum. The dried gels were placed in cassettes and exposed to [3H]-sensitive ultra-film (Kodak Biomax light film) at -70 °C for periods ranging from five days to two weeks [11, 12].
2-Dimensional Electrophoresis
Rat brain membranes (0.5-2 mg) or 10 μg bovine brain tubulin photolabeled with either non-tritiated 6-AziP or [3H]6-AziP were solubilized in 2-DE lysis buffer for 60 minutes on ice and incubated with pI 3-10 isoelectric focusing strips (Amersham). The strips were then rehydrated with 5 M urea, 2 M thiourea, 2% CHAPS, 2% SB-310, 40 mM Tris, 0.2% ampholyte, 50 mM DTT. Following sequential isoelectric focusing (200 V for 30 min (gradient), 8000 V for 2.5 hr (gradient) and 8000 V for 50,000-80,000 Vhr.), the strips were incubated in equilibration buffer (50 mM Tris-Cl, pH 8.8, 6 M urea, 30% glycerol, 2% SDS, 1% DTT) and then subjected to 10% SDS-PAGE. The non-radioactive 2-D gel was stained with Coomassie blue and the gel with tritiated sample was silver-stained or analyzed by autoradiography.
Sample Preparation for Mass Spectrometry
Spots on the Coomassie blue-stained 2-DE gel were manually excised, dried in 100% acetonitrile (ACN), and digested with sequencing grade trypsin (Sigma, sequencing grade, 1 μg) in 20 μL 1 mM triethylammonium bicarbonate (TEABC) (pH 8.5) at 37 °C for 1 hour. The tryptic peptides were extracted from the gel using 60% ACN and 1% formic acid (FA) and analyzed by MALDI/TOF-TOF. For ESI/MS, the peptides were dried and resuspended in 1% ACN and 1% FA [11].
In-solution digestion
Bovine brain tubulin samples were digested with trypsin employing two methods. For identification of the photolabeled peptide, tubulin was photolabeled with 10 μM 6-AziP in 1 mM TEABC. The sample was adjusted to pH 8.5 using NH4Cl and 5 μg trypsin was added. After incubation at 37 °C for 1 hour, the digestion was terminated by adding soybean trypsin inhibitor. The samples were dried and re-suspended in 1% ACN and 1% FA for analysis by MALDI and ESI/MS. Alternatively, in-solution digestion was performed with Lys-C followed by either trypsin or chymotrypsin using a previously described protocol [14]. Briefly tubulin samples were precipitated using an SDS-cleanup kit (Amersham) [15], resolubilized in 20 μl of 8 M urea, 5% RapiGest™ (Waters, Milford MA), 100 mM Tris, pH 8.5 and incubated for 30 min at 37 °C with agitation. The proteins were then precipitated again with the SDS-cleanup kit and resuspended in 20 μl of 8 M Urea, 0.1% RapiGest™, 100 mM Tris pH 8.5. Proteins were then reduced by adding tris (2-carboxyethyl) phosphine (TCEP) to 5 mM and incubating at room temperature for 30 min and subsequently alkylated with 10 mM iodoacetamide in the dark at room temperature for 30 min. The TCEP and iodoacetamide were quenched with 5 mM dithiothreitol (DTT) at room temperature for 10 min. The samples were then digested with 2 μg Lys-C at 37°C overnight. The urea was then diluted to 2 M for trypsin digestion and 1 M for chymotrypsin digestion using 100 mM Tris (pH 8.5). Trypsin digestion was performed by adding 2 μg trypsin and incubating at 37° for 16 hours. For chymotrypsin digestion samples were incubated for 12 hours at room temperature with 5 μg chymotrypsin and 10 mM CaCl2. The digested peptides were recovered by solid phase extraction using porous graphitic carbon Nutips (Glygen, Columbia, MD) and eluted with 60% ACN and 1% FA. Peptides were then dried and resuspended in 1% ACN and 1% FA for mass spectrometric analysis using MALDI or high-resolution LC-MS/MS.
MALDI/TOF-TOF and high resolution LC-MS/MS
A 4700 Proteomics Analyzer (Applied Biosystems, Foster City, CA) was used for MALDI MS1 and MS2 analyses. A 1 μL sample was mixed with 1 μL α-cyano-4-hydroxycinnamic acid. 1 μL of this mixture was spotted on the MALDI target and allowed to dry. MS analysis was performed in the positive ion mode using reflecton TOF-MS. The mass range was m/z 900 to 3000 and 2500 laser shots were acquired with fixed laser power of 4900 (arbitrary unit). High resolution MS2 spectra were acquired with an LTQ-Orbitrap XL hybrid mass spectrometer (Thermo Fisher Scientific, San Jose, CA) coupled with a NanoLC-Ultra system (Eksigent Technologies, Dublin, CA). Mobile phases were 0.1% formic acid in water (A) and 0.1% formic acid in 60% acetonitrile (B). 5 μL of sample was loaded over 15 min at 1 μL/min in 100% A onto a 20 cm × 75 μm C14 column (Jupiter Proteo, 4 μm, 90 Å, Phenomenex, Torrance, CA) and eluted at 250 μL/min using the following gradient: isocratic at 2% B (0-5 min), linear gradient from 2% B to 50% B (5-65 min), linear gradient from 50% B to 80% B (65-70 min), isocratic at 80% B (70-72 min), linear gradient from 80% B to 2% B (72-77 min), isocratic at 2% B (5 min). Total run time, including column equilibration, sample loading, and analysis was 98 min. MS analysis was performed in positive ion mode from m/z 350-2000 at 60,000 resolution. The following parameters were used: capillary temperature 125 °C, source voltage 3.2 kV, source current 100 μA, tube lens 70 V, FTMS max ion time 500 msec, and FTMS MSn max ion time 1000 msec. MS2 analysis was triggered at 15,000 resolution with an accurate inclusion mass screening of m/z 644.37, corresponding to the mass of 6-AziP-labeled TAVCDIPPR. DMSA was triggered with an isolation width of 4 Da when the signal for the ion of interest exceeded 1000 counts. The initial identification of β-tubulin from ESI/MS sequence analysis of gel plugs was performed at Harvard Microchemistry Facility by microcapillary reverse-phase HPLC nanoelectrospray tandem mass spectrometry (μLC/MS/MS) on a Finnigan LCQ DECA quadruple ion trap mass spectrometer (San Jose, CA).
Data Processing and Analysis
Database searches of in-solution digests
The LC-MS data files for both low resolution and high resolution MS2 were processed using MASCOT Distiller (Matrix Science, version 2.3.0.0) with previously described settings for both low resolution [16] and high resolution [17] MS2. MS2 centroided files were used for database searching with MASCOT (Matrix Science, version 2.1.6) against the NCBI non-redundant protein database. The search was conducted using “no enzyme”, allowing 9 missed cleavages, specifying oxidation of Met residues as a variable modification and setting a fragment ion mass tolerance of 0.8 Da and a parent ion tolerance of 50 ppm. For photolabeled samples, 6-AziP was included as a variable modification on Cys, Glu, Asp, Met, and Gln. The Protein and Peptide Prophet algorithms in Scaffold (version 3_00_03, Proteome Software Inc., Portland, OR) were used to qualify the database results. A probability of ≥80% was used to assign peptide sequences to MS2 spectra using the Peptide Prophet algorithm [18]. A probability of ≥95% was used to assign proteins with the Protein Prophet algorithm [19]. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Structure Modeling
A homology model of rat α1 and β2A subunits of tubulin was prepared by aligning the relevant sequences to a 3.5 Å bovine α and β tubulin structure (1JFF) obtained from the Protein Data Bank [20]. The alignment was produced using the program MUSCLE [21]; the aligned sequences were then used with the program Modeller [22] to produce three dimensional models with both subunits modeled simultaneously. The best model was then read into Sybyl 7.2, where hydrogens were added, the heavy atoms were fixed and all hydrogens optimized to a 0.01 gradient.
Colchicine has previously been shown to bind to Cys-354 on β tubulin. In this experiment, docking studies were performed to dock both colchicine and 6-AziP to α-β tubulin dimers using the program Autodock. The protein model was read into Autodock Tools where all non-polar hydrogens were merged into the heavy atoms and Kollman charges added (total charge = -49.624). The docking grid was centered near Cys-354 using a grid spacing of 0.375 Å. Models of both colchicine (5 rotatable bonds) and 6-AziP (2 rotatable bonds) were built as mol2 files, then prepared for use in Autodock using Autodock Tools. Each molecule was docked into the site using the Lamarkian Genetic Algorithm and 100 docking runs with 25,000,000 evaluations per run.
Molecular structure comparison of 6-AziP and colchicine
The program Omega (OpenEye Scientific Software, Santa Fe, NM) was used to generate conformational libraries of both colchicine and 6-AziP which were then exported as a multi-molecule file for use in similarity alignment using the ROCS (OpenEye Scientific Software, Santa Fe, NM) program. 6-AziP was found to give only one conformation within the energy constraints utilized, while colchicine was found to give ten conformations. The colchicine conformations were then superposed against 6-AziP using a combination of the best shape match and chemical features.
Results
6-AziP labels a 55 kDa protein
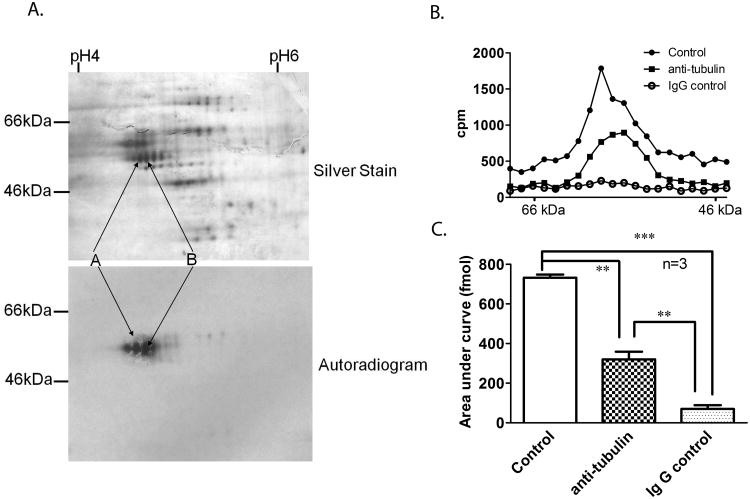
Our previous studies demonstrated that [3H]6-AziP photolabels proteins of ≈30 kDa and ≈55 kDa molecular weight [11]. The ≈30 kDa protein was identified by ESI-MS/MS as isoforms of VDAC [11, 12]. To investigate the identity of the 55 kDa protein, rat brain membrane was incubated with either 10μM non-tritiated or tritiated 6-AziP and irradiated for 5 min. The two photolabeled samples were analyzed by 2-D electrophoresis (2-DE), with the isoelectric focusing strips and the SDS-PAGE being run simultaneously, “back-to-back”, under the same conditions. As shown in Figure 2A lower panel, autoradiography confirmed that protein spots of ≈55 kDa and isoelectric point (pI) about 4.8 were labeled by [3H]6-AziP. The silver stained spots (Figure 2A, upper panel, labeled as A and B) on the non-tritiated 6-AziP labeled gel, corresponding to the radioactive spots on the autoradiogram, were excised and digested with trypsin. The digested peptides were applied to a single micro-capillary reverse-phase HPLC coupled to a nano-electrospray ionization ion trap mass spectrometer. MS/MS unambiguously showed that the proteins identified in the spots are isoforms of β-tubulin including (in the order of abundance): β3-tubulin (42.7% sequence coverage), β5-tubulin (23.8 % sequence coverage), β15-tubulin (24.9 % sequence coverage), β2c-tubulin (6% sequence coverage), and β4-tubulin (6% sequence coverage); α2 -tubulin was also detected (34.5 % sequence coverage).
Figure 2. Identification of the 55 kDa protein photolabeled by 6-AziP as tubulin.
A. Rat brain membranes (250 μg) were photolabeled with 10 μM non-tritiated or tritiated 6-AziP. These two samples were separated by 2-dimensional electrophoresis with both isoelectric focusing and SDS-PAGE being run simultaneously under the same conditions followed by either silver staining (upper panel, for non-tritiated 6-AziP) or autoradiography (lower panel, for [3H]6-AziP). A train of proteins observed in silver staining, corresponding to the radioactive spots (labeled as A and B), were excised from the gel, digested with trypsin and identified as isoforms of α- and β-tubulin using mass spectrometry. B. A representative gel slicing experiment illustrating that most of the photolabeled 55 kDa protein is immunoprecipitated by anti β-tubulin antibody. Rat brain membranes photolabeled with 10 μM [3H]6-AziP were immunoprecipitated with anti β-tubulin antibody or anti-mouse IgG, followed by SDS-PAGE and gel slicing. The control is photolabeled rat brain membrane without immunoprecipitation. Gel slices were analyzed by scintillation spectrometry. C. The areas under the curve of the 55 kDa proteins from 3 replicate gel slicing experiments. They are: 733 ± 15 fmol for control membranes; 321 ± 39 fmol for the anti-β-tubulin immunoprecipitation group, and 71± 19 fmol for the IgG immunoprecipitation group.
Immunoprecipitation of the [3H]6-AziP labeled ≈55 kDa protein by anti β-tubulin antibody
To confirm that the labeled 55 kDa protein is β-tubulin, rat brain membranes (60 μg) were photolabeled with 10 μM [3H]6-AziP and detergent-solubilized. Aliquots of the solubilized proteins were then immunoprecipitated with either an anti-β-tubulin antibody (labeled as anti-tubulin in Figure 2B) or a non-specific anti-Ig-G antibody (labeled as IgG control in Figure 2B). In the control, photolabeled membranes were solubilized in SDS sample buffer but not immunoprecipitated. Proteins in each sample were then separated by SDS-PAGE and analyzed by scintillation spectrometry of gel slices (Figure 2B). The labeled protein peak at ≈55 kDa was present in both the control rat brain membranes and in the anti-β-tubulin immunoprecipitated complex. The area under the curve from 3 replicates was: 732.5± 15.5 fmol for control membranes; 320.5.5± 38.5 fmol for the anti-β-tubulin immunoprecipitation; and 70.5± 18.5fmol for the non-specific IgG immunoprecipitation (Figure 2C, p <0.001 anti β-tubulin vs control; p< 0.01 anti β-tubulin vs. non-specific IgG). These data indicate that half of the [3H]6-AziP labeled 55 kDa protein was present in the anti-β-tubulin immunoprecipitated complex. Very little radioactivity was detected in the anti-IgG immunoprecipitated group, indicating that the immunoprecipitation was β-tubulin specific. Of note the shoulder of radioactivity at 48 – 56 kDa was not immunoprecipitated by anti-β-tubulin, indicating that more than one protein may be photolabeled by 6-AziP.
[3H]6-AziP preferentially photolabels β-tubulin
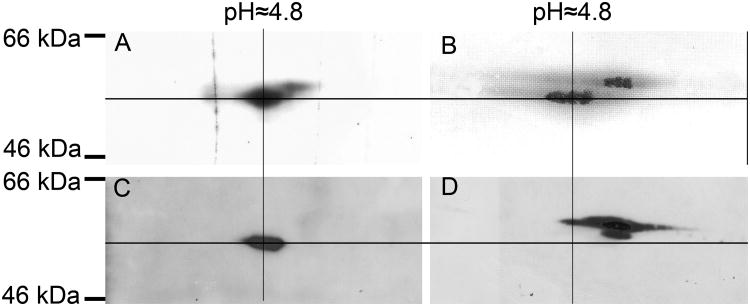
To determine whether the α-, β- or both subunits of tubulin are photolabeled by 6-AziP, bovine brain tubulin (10 μg) was photolabeled with either 3 μM tritiated or 3 μM non-tritiated 6-AziP and then simultaneously separated by 2-DE. The 2-DE gel of the tritiated sample was analyzed by autoradiography and the 2-DE gels of the non-tritiated samples were analyzed by immunoblotting with antisera directed against the α- or β-subunits of tubulin or by silver staining. All these gels were aligned according to molecular weight and pI. The most abundant radioactive spots on the autoradiogram (Figure 3A) corresponded to the lower sets of spots on the silver staining (Figure 3B). These are β-tubulin isoforms as shown by the immunoblot using specific anti-β-tubulin antisera (Figure 3C). These spots were located at ≈55 kDa and pI 4.8, similar to the 2-DE of 6-AziP photolabeled crude rat brain membranes. The upper sets of spots on the silver stained gel (Figure 3B), corresponding to the less radioactive spots in Figure 3A, are α-tubulin isotypes as shown by the specific anti-α-tubulin immunoblot (Figure 3D). These results indicate that 6-AziP preferentially photolabels the β- subunit of tubulin, with modest photolabeling of the α-subunit.
Figure 3. 6-AziP preferentially photolabels the β- subunit of tubulin.
Purified bovine brain tubulin (10 μg) was photolabeled with either 3 μM [3H]6-AziP or non-tritiated 6-AziP and analyzed by 2-DE. (A). Autoradiogram demonstrating that [3H]6-AziP labels proteins at ≈55 kDa and pI 4.8. (B). Silver staining showing 2 trains of spots representing α- and β-tubulin. (C). Immuno-blot with anti-β-tubulin antibody. (D). Immuno-blot with anti-α-tubulin antibody. Cross marking and alignment of all the gels based on MW and pI indicates that β-tubulin is preferentially photolabeled by 6-AziP.
MALDI-TOF identified a β-tubulin peptide with 6-AziP adduct
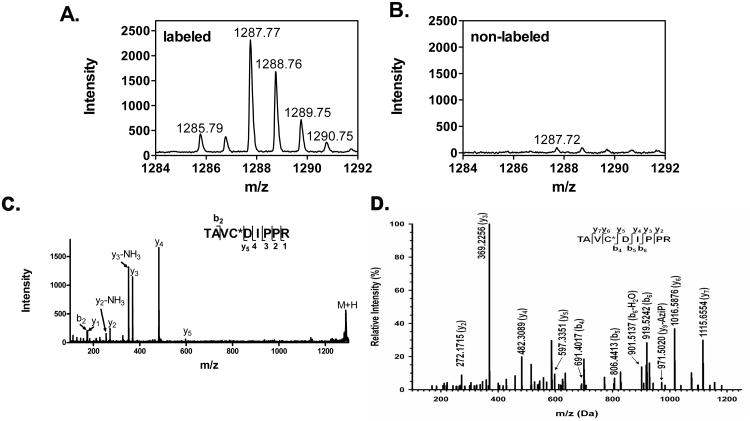
To identify the peptides(s) photolabeled by 6-AziP, bovine brain tubulin was photolabeled with 10 μM 6-AziP, digested with trypsin and analyzed by MALDI. The MALDI spectrum of photolabeled tubulin (Figure 4A) was compared to the spectrum of unlabeled tubulin (Figure 4B) to detect MS1 features different between the two groups. One peptide (m/z=1287.77) was detected only in the photolabeled sample. MALDI-TOF tandem mass spectrometry identified the peptide as TAVCDIPPR, a β-tubulin specific peptide bearing a single 6-AziP moiety (Figure 4C). The m/z of this singly charged ion is the sum of the m/z of TAVCDIPPR (m/z=917.49) and the m/z of the 6-AziP adduct (m/z=316.24), resulting from covalent binding of 6-AziP to the peptide, with the expected loss of two nitrogens during photolysis [23]. The MS2 spectrum contains b2 and y1 to y4 ions as the adduct-free fragment ions. The observed unmodified y5 ion suggests that the photolabeling site is either on Cys or Val, but no fragment ions diagnostic of the precise site of 6-AziP incorporation were observed.
Figure 4. Mass spectrometry identifies Cys-354 of β-tubulin as the site of 6-AziP incorporation.
Comparison of the MALDI mass spectra of photolabeled (A) and non-labeled tubulin (B), identified a peptide with m/z =1287.77 as unique to the photolabeled sample. (C). MALDI tandem mass spectrum identified the peptide as TAVCDIPPR bearing a single 6-AziP adduct. (D). A high-resolution MS2 spectrum (OrbiTrap) demonstrated Cys-354 as the site of 6-AziP photo-incorporation in the peptide TAVC*DIPPR.
ESI-LC-Mass Spectrometry identifies Cys-354 of β-tubulin as the site of 6-AziP incorporation
To determine the amino acid residue modified by 6-AziP, high resolution ESI-LC-MS (LTQ-Orbitrap) was used to analyze the tryptic digest of photolabeled tubulin. An MS1 feature consistent with the 6-AziP adduct of TAVCDIPPR (m/z=644.372, z=2) was identified. Because this MS1 feature had a low intensity, no MS2 spectrum was generated for this peptide using data-dependent acquisition. We therefore used directed MS2 spectral acquisition to obtain a high resolution MS2 spectrum. The high resolution MS2 spectrum shown in Figure 4D shows the diagnostic pairs of y5 and y6 ions, indicating that 6-AziP is specifically incorporated at Cys-354 of β-tubulin, a site previously identified as the colchicine binding site [24]. It should be noted that fragment ions associated with loss of 6-AziP (e.g. y9 – 6-AziP) are observed in the MS2 spectrum, indicating some modest loss of the 6-AziP adduct during ion fragmentation for MS2.
The sequence coverage provided by tryptic and chymotrypsin digestion and analysis using the LTQ-Orbitrap was 91% (Supplemental Figure 1 and Supplemental Table 1). A MASCOT search of the MS2 data from these digestions using 6-AziP as a variable modification failed to identify any additional 6-AziP-modified peptides other than TAVCDIPPR.
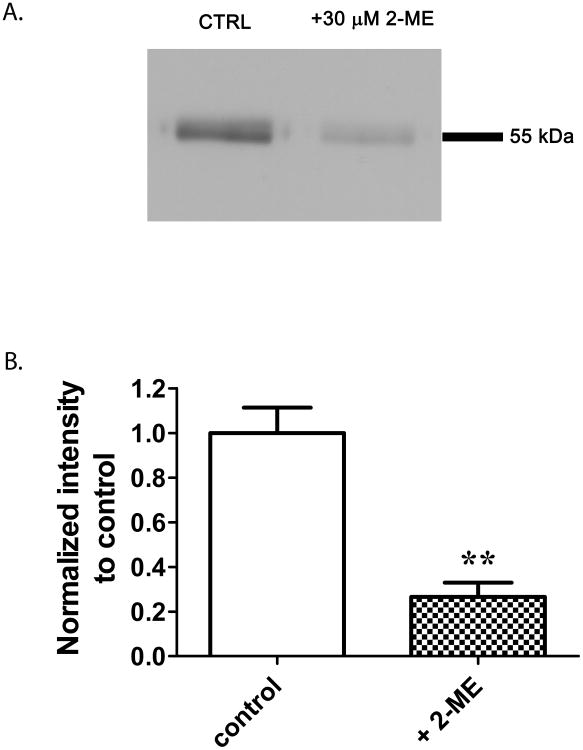
6-AziP photolabeling of tubulin is prevented by 2-ME
Cys-354, the site of 6-AziP photoincorporation, has previously been identified as a binding site for colchicine, a natural product inhibitor of tubulin polymerization. 2-ME is a chemotherapeutic agent thought to act at the colchicine binding site. To confirm that the major site of 6-AziP photoincorporation is Cys-354 (i.e. this is not merely one of many non-specific sites of photoincorporation), we examined the ability of 2-ME (30 μM) to prevent tubulin photolabeling by 0.3 μM [3H]6-AziP. (n.b. colchicine was not used to prevent photolabeling because it absorbs light at 354 nm and would thus prevent photolabeling by an independent mechanism). The autoradiogram in Figure 5A shows that 2-ME largely prevents photolabeling of tubulin. The densitometric analysis of 5 replicates (Figure 5B) indicates that 30 μM 2-ME prevents 6-AziP photolabeling by about 73%. 6-AziP photolabeling of tubulin was minimally and inconsistently prevented by pregnanolone (30 μM) or allopregnanolone (30 μM), suggesting that neither of these endogenous neurosteroids is a high affinity ligand for the colchicine site (data not shown).
Figure 5. 2-ME prevents 6-AziP photolabeling of bovine brain tubulin.
10 μg of purified bovine brain tubulin was photolabeled with 0.3 μM [3H]6-AziP in the presence or absence of 2-ME (30 μM). (A). A representative autoradiogram demonstrating that 6-AziP photolabeling is inhibited by 2-ME. (B) Densitometric analysis from 5 replicate experiments indicates that 2-ME prevents 6-AziP photolabeling of bovine brain tubulin by about 75% (student's t-test, **p<0.01). .
Docking 6-AziP to Cys-354 using structural modeling
To further analyze the interaction of 6-AziP with the colchicine binding site, modeling studies were performed using a structural model of an α-β tubulin dimer. Colchicine and 6-AziP were located randomly outside of the α-β tubulin dimer and allowed to move freely in order to dock to their binding site. Colchicine and 6-AziP both docked near Cys 354 of β tubulin in each docking run. The azi-group of 6-AziP was positioned proximate to Cys-354, indicating that Cys-354 was the photolabeling site. At the end of each docking run, a large number of possible molecular orientations (poses) are produced. The resultant poses were clustered into groupings of similar poses using an rmsd tolerance of 2.0 Å for the atomic coordinates. The 6-AziP results were clustered into a total of ten clusters and the lowest energy conformation from cluster one was predicted to have a mean binding energy of -5.24 kcal/mol. In comparison, colchicine was clustered into a total of fifteen clusters, and the lowest energy conformation from cluster one was predicted to have a mean binding energy of -3.51 kcal/mol. These results indicate that 6-AziP would favorably compete with colchicine for this site. The initial docking results for Cys 354 were then re-analyzed using the program Dockres [25], allowing refinement of ligand docking predictions utilizing the method of Ruvinsky [26]. Upon reanalysis of the docking results, 6-AziP was predicted to have an improved binding energy of -6.6 kcal/mol and colchicine -5.6 kcal/mol.
Discussion
The initial finding motivating this study was the demonstration that the neurosteroid analogue photolabeling reagent 6-AziP photolabels a ∼55-kDa protein in rat brain with a pI of 4.8 [11, 12]. In the current work we have identified this photolabeled protein as the β-subunit of tubulin. Subsequent analysis using mass spectrometry identified the major site of 6-AziP incorporation as Cys-354, previously identified as a colchicine binding site [27]. The photolabeling was inhibited by 2-ME, confirming that Cys-354 is the predominant site of photo-induced labeling. Molecular modeling studies identified the existence of a high affinity neurosteroid binding pocket near Cys-354.
The potential importance of neurosteroid binding to colchicine binding sites on β-tubulin derives from the fact that colchicine is a highly efficacious modulator of tubulin polymerization that has major therapeutic and investigational applications [28]. Colchicine is a plant alkaloid derived from the autumn crocus (colchicum) that has long been used as a laxative, as an emetic and in the treatment of gout. Its acts by preventing polymerization of tubulin dimers and is thus useful experimentally as an inhibitor of microtubule assembly and as an anti-mitotic [28]. Colchicine binds at Cys-354 near the interface of the α- and β-subunits of tubulin [24]. Our photolabeling data with 6-AziP and subsequent modeling data indicate that 6-AziP also binds to the same colchicine site. Our modeling results show that the diazirine moiety of 6-AziP is positioned proximate to Cys-354, which can facilitate photolabeling of this amino acid. The diazirine is not predicted to be near any reactive amino acids at the α-tubulin interface, explaining the minimal labeling of the α-subunit.
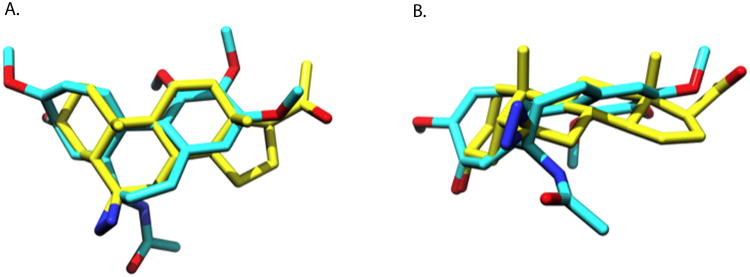
The observation that neurosteroids interact with the colchicine binding sites seems unlikely at first due to the large structural differences between the compounds. However, colchicine and 6-AziP have quite similar molecular volumes, with 6-AziP occupying 339.82 Å3 while colchicine occupies 350.82 Å3. The program ROCS provides a method for obtaining molecular alignments optimizing both steric and chemical features such as charge and hydrogen bond donors/acceptors. Figure 6 shows the optimal alignment of colchicine and 6-AziP providing a large overlap of the core ring systems and of several oxygen atoms capable of serving as hydrogen bond acceptors. These similarities in structure may explain the observation that colchicine and its analogues (e.g. thiocolchicines) also act on GABAA receptors and glycine receptors [29-32], known targets of neurosteroid binding and action [1]. To date, the only other endogenous compound thought to bind to or act via the colchicine binding site is 2-ME [7]. 2-ME is a competitive inhibitor of [3H]colchicine binding to β-tubulin and like colchicine, inhibits polymerization of microtubules. It also inhibits the photolabeling of 6-AziP on tubulin as shown in Figure 5.
Figure 6. Structural similarity of 6-AziP and colchicine.
A top (A) and side view (B) of 6-AziP and colchicine. The two superimposed molecules represent the maximal similarity alignment of 6-AziP and colchicine as determined by the program ROCS. In both molecules hydrogens are removed for clarity, the carbons of 6-AziP are depicted in yellow and colchicine in cyan, with oxygens in red and nitrogens in blue. Colchicine and 6-AziP have similar molecular volumes with 6-AziP occupying 339.82 Å3 while colchicine occupies 350.82 Å3.
To examine the functional effects resulting from the direct interaction between neurosteroids and the colchicine binding site, we explored the effect of endogenous neurosteroids on in vitro tubulin polymerization. Both allopregnanolone and pregnanolone (1-30 μM) inhibited tubulin polymerization (data not shown). However, studies using neurosteroid to prevent 6-AziP photolabeling suggested that these endogenous neurosteroids bind with low affinity (>30 μM) to the Cys-354 site (data not shown), making it uncertain whether the functional effects of the neurosteroids on tubulin polymerization are mediated by binding to the Cys-354 site. Future studies will address whether neurosteroid inhibition of tubulin polymerization is mediated by actions at the colchicine site or a different site that is not photolabeled by 6-AziP.
One potential limitation of our study is based on the photochemistry of diazirines. Irradiation of a diazirine can, via elimination of molecular nitrogen, generate a carbene, which then participates in either an internal rearrangement reaction (intramolecular) or a bimolecular insertion reaction. Carbenes can insert into any C-H bond and are thus not selective for specific amino acids. However, if an intramolecular reaction is energetically favored, the carbene that is generated will be unavailable for bimolecular insertion. Irradiation can also convert a diazirine to a diazo intermediate that can lose nitrogen to form a carbonium ion. The carbonium ion would selectively label only those amino acids having nucleophilic side chains [23]. Recent work using azi-etomidate [33] and azi-octanol [34] as photolabeling reagents indicates that the labeling mechanism for these aliphatic diazirines is through the diazo intermediate. In the case of 6-AziP, both carbene and carbonium ion intermediates are likely formed during the photolysis reaction. The carbene generated by photolysis appears to preferentially participate in intramolecular reactions. When we examined the ability of 6-AziP to form a unique adduct with cyclohexane (a solvent in which the only bond available for attack is a C-H), we found that the photolysis products were indistinguishable from those formed when [3H]6-AziP was photolyzed in ethanol (data not shown). This indicated that the products arose from intramolecular rearrangement rather than from bimolecular reactions with solvent. Based on these findings, we concluded that the major photolysis products of 6-AziP result from intramolecular rearrangement and that photolysis of 6-AziP generates a diazo intermediate (as a minor product) that is responsible for the photolabeling of tubulin. Since the diazo intermediate can only react with nucleophilic amino acids, it is possible that we have failed to identify additional neurosteroid sites on tubulin because of unfavorable photochemistry.
A second caveat of this study is based on the incomplete amino acid sequence coverage that was deduced from the LC-MS/MS data. We identified 91% of the sequence of β-tubulin. A single labeled peptide (TAVCDIPPR; m/z=1287.77) was identified, and extensive database searching failed to identify additional labeled sites. The 9% of the β-tubulin sequence that was not identified results from the location of the trypsin and chymotryspin cleavage sites because the resulting peptides (m/z) were either too small or too large to be detected. In addition, this missing sequence also contains some post-translational modification sites, e. g. C-terminal glycylation and polyglutamylation sites [35], which can only be observed after enrichment or enzyme cleavage. Thus, it is possible that we failed to identify additional neurosteroid binding sites on tubulin because they were in the 9% of the sequence that was non-detectable.
In summary, this study demonstrates that the neurosteroid analogue photolabeling reagent, 6-AziP, forms a photo-adduct with Cys-354, a colchicine binding site on the β-subunit of tubulin. Photolabeling of tubulin is selective for this site as evidenced by the ability of 2-ME to prevent 6-AziP incorporation. While the inability of allopregnanolone or pregnanolone to prevent 6-AziP photolabeling suggests that neither of these endogenous neurosteroids are biologically important ligands for the colchicine site, other related endogenous compounds should be considered as candidate endogenous ligands. Most importantly, these data demonstrate the utility of 6-AziP, used in conjunction with directed high-resolution mass spectrometry, as a tool for identifying neurosteroid binding proteins and providing precise definition of the sites of neurosteroid incorporation. The methodology for preparation and analysis of low abundance peptides with a hydrophobic neurosteroid adduct should be generally applicable to identify photolabeling sites of other protein and ligands.
Supplementary Material
Abbreviations
- CNS
Central nervous system
- VDAC
voltage dependent anion channels
- pI
isoelectric point
- MS2
tandem mass spectrometry
- MALDI
matrix-assisted laser desorption/ionization mass spectrometry
- DHEA
dehydroepiandrosterone
- GABAA
gamma aminobutyric acid type A
- PS
pregnenolone sulfate
- PregaS
(3α, 5β)-3-hydroxy-pregnan-20-one sulfate
- MAP
microtubule-associated protein
- 6-AziP
6-Azi-pregnanolone
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- 2-DE
2-Dimensional Electrophoresis
- ESI
electrospray ionization
- ACN
acetonitrile
- FA
formic acid
- TEABC
triethylammonium bicarbonate
- DMSA
directed MS2 spectral acquisition
References
- 1.Smith SS. Neurosteroid effects in the central nervous system: the role of the GABAA receptor. CRC Press LLC; 2004. [Google Scholar]
- 2.Gasior M, Carter RB, Witkin JM. Trends Pharmacol Sci. 1999;20:107–112. doi: 10.1016/s0165-6147(99)01318-8. [DOI] [PubMed] [Google Scholar]
- 3.Rupprecht R. Psychoneuroendocrinology. 2003;28:139–168. doi: 10.1016/s0306-4530(02)00064-1. [DOI] [PubMed] [Google Scholar]
- 4.Ekins S, Reschly EJ, Hagey LR, Krasowski MD. BMC Evol Biol. 2008;8:103. doi: 10.1186/1471-2148-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hosie AM, Wilkins ME, da Silva HM, Smart TG. Nature. 2006;444:486–489. doi: 10.1038/nature05324. [DOI] [PubMed] [Google Scholar]
- 6.Majewska MD, Harrison NL, Schwartz RD, Barker JL, Paul SM. Science. 1986;232:1004–1007. doi: 10.1126/science.2422758. [DOI] [PubMed] [Google Scholar]
- 7.D'Amato RJ, Lin CM, Flynn E, Folkman J, Hamel E. Proc Natl Acad Sci U S A. 1994;91:3964–3968. doi: 10.1073/pnas.91.9.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fontaine-Lenoir V, Chambraud B, Fellous A, David S, Duchossoy Y, Baulieu EE, Robel P. Proc Natl Acad Sci U S A. 2006;103:4711–4716. doi: 10.1073/pnas.0600113103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laurine E, Lafitte D, Gregoire C, Seree E, Loret E, Douillard S, Michel B, Briand C, Verdier JM. J Biol Chem. 2003;278:29979–29986. doi: 10.1074/jbc.M303242200. [DOI] [PubMed] [Google Scholar]
- 10.Murakami K, Fellous A, Baulieu EE, Robel P. Proc Natl Acad Sci U S A. 2000;97:3579–3584. doi: 10.1073/pnas.97.7.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darbandi-Tonkabon R, Hastings WR, Zeng CM, Akk G, Manion BD, Bracamontes JR, Steinbach JH, Mennerick SJ, Covey DF, Evers AS. J Biol Chem. 2003;278:13196–13206. doi: 10.1074/jbc.M213168200. [DOI] [PubMed] [Google Scholar]
- 12.Darbandi-Tonkabon R, Manion BD, Hastings WR, Craigen WJ, Akk G, Bracamontes JR, He Y, Sheiko TV, Steinbach JH, Mennerick SJ, Covey DF, Evers AS. J Pharmacol Exp Ther. 2004;308:502–511. doi: 10.1124/jpet.103.058123. [DOI] [PubMed] [Google Scholar]
- 13.Bureau MH, Olsen RW. J Neurochem. 1993;61:1479–1491. doi: 10.1111/j.1471-4159.1993.tb13643.x. [DOI] [PubMed] [Google Scholar]
- 14.Chen ZW, Fuchs K, Sieghart W, Townsend RR, Evers AS. Molecular & cellular proteomics : MCP. 2011 doi: 10.1074/mcp.M111.011445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Washburn MP. Curr Protoc Protein Sci. 2008;Chapter 23 doi: 10.1002/0471140864.ps2306s53. Unit 23 26 21-23 26 11. [DOI] [PubMed] [Google Scholar]
- 16.Nittis T, Guittat L, LeDuc RD, Dao B, Duxin JP, Rohrs H, Townsend RR, Stewart SA. Molecular & cellular proteomics : MCP. 2010;9:1144–1156. doi: 10.1074/mcp.M900490-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer MR, Lichti CF, Townsend RR, Rao AG. Biochemistry. 2011;50:2170–2186. doi: 10.1021/bi101935x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Analytical chemistry. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 19.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. Analytical chemistry. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 20.Lowe J, Li H, Downing KH, Nogales E. J Mol Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- 21.Edgar RC. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sali A, Blundell TL. Journal of Molecular Biology. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 23.Brunner J. Annu Rev Biochem. 1993;62:483–514. doi: 10.1146/annurev.bi.62.070193.002411. [DOI] [PubMed] [Google Scholar]
- 24.Ravelli RB, Gigant B, Curmi PA, Jourdain I, Lachkar S, Sobel A, Knossow M. Nature. 2004;428:198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- 25.Mezei M. Mt Sinai School of Medicine. New York: 2007. [Google Scholar]
- 26.Ruvinsky AM. J Comput Chem. 2007;28:1364–1372. doi: 10.1002/jcc.20580. [DOI] [PubMed] [Google Scholar]
- 27.Bai R, Pei XF, Boye O, Getahun Z, Grover S, Bekisz J, Nguyen NY, Brossi A, Hamel E. J Biol Chem. 1996;271:12639–12645. doi: 10.1074/jbc.271.21.12639. [DOI] [PubMed] [Google Scholar]
- 28.Bhattacharyya B, Panda D, Gupta S, Banerjee M. Med Res Rev. 2008;28:155–183. doi: 10.1002/med.20097. [DOI] [PubMed] [Google Scholar]
- 29.Weiner JL, Buhler AV, Whatley VJ, Harris RA, Dunwiddie TV. J Pharmacol Exp Ther. 1998;284:95–102. [PubMed] [Google Scholar]
- 30.Whatley VJ, Brozowski SJ, Hadingham KL, Whiting PJ, Harris RA. J Neurochem. 1996;66:1318–1321. doi: 10.1046/j.1471-4159.1996.66031318.x. [DOI] [PubMed] [Google Scholar]
- 31.van Zundert B, Alvarez FJ, Tapia JC, Yeh HH, Diaz E, Aguayo LG. J Neurophysiol. 2004;91:1036–1049. doi: 10.1152/jn.00364.2003. [DOI] [PubMed] [Google Scholar]
- 32.Balduini W, De Angelis V, Mazzoni E, Depoortere H, Cattabeni F, Cimino M. Neuropharmacology. 2001;40:1044–1049. doi: 10.1016/s0028-3908(01)00023-5. [DOI] [PubMed] [Google Scholar]
- 33.Li GD, Chiara DC, Sawyer GW, Husain SS, Olsen RW, Cohen JB. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2006;26:11599–11605. doi: 10.1523/JNEUROSCI.3467-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forman SA, Zhou QL, Stewart DS. Biochemistry. 2007;46:11911–11918. doi: 10.1021/bi701287a. [DOI] [PubMed] [Google Scholar]
- 35.Redeker V. Methods in cell biology. 2010;95:77–103. doi: 10.1016/S0091-679X(10)95006-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.