Abstract
Context
Chromosomal abnormalities (namely 13q, 17p, and 11q deletions) have prognostic implications and are recurrent in chronic lymphocytic leukemia (CLL), suggesting that they are involved in a common pathogenetic pathway; however, the molecular mechanism through which chromosomal abnormalities affect the pathogenesis and outcome of CLL is unknown.
Objective
To determine whether the microRNA miR-15a/miR-16-1 cluster (located at 13q), tumor protein p53 (TP53, located at 17p), and miR-34b/miR-34c cluster (located at 11q) are linked in a molecular pathway that explains the pathogenetic and prognostic implications (indolent vs aggressive form) of recurrent 13q, 17p, and 11q deletions in CLL.
Design, Setting, and Patients
CLL Research Consortium institutions provided blood samples from untreated patients (n=206) diagnosed with B-cell CLL between January 2000 and April 2008. All samples were evaluated for the occurrence of cytogenetic abnormalities as well as the expression levels of the miR-15a/miR-16-1 cluster, miR-34b/miR-34c cluster, TP53, and zeta-chain (TCR)–associated protein kinase 70kDa (ZAP70), a surrogate prognostic marker of CLL. The functional relationship between these genes was studied using in vitro gain- and loss-of-function experiments in celllines and primary samples and was validated in a separate cohort of primary CLL samples.
Main Outcome Measures
Cytogenetic abnormalities; expression levels of the miR-15a/miR-16-1 cluster, miR-34 family, TP53 gene, downstream effectors cyclindependent kinase inhibitor 1A (p21, Cip1) (CDKN1A) and B-cell CLL/lymphoma 2 binding component 3 (BBC3), and ZAP70 gene; genetic interactions detected by chromatin immunoprecipitation.
Results
In CLLs with13qdeletions the miR-15a/miR-16-1 cluster directly targetedTP53 (mean luciferase activity for miR-15a vs scrambled control, 0.68 relative light units (RLU) [95%confidence interval {CI}, 0.63–0.73]; P=.02;meanfor miR-16 vs scrambled control, 0.62RLU[95%CI, 0.59–0.65]; P=.02) and its downstream effectors. In leukemic cell lines and primary CLL cells, TP53 stimulated the transcription of miR-15/miR-16-1 as well as miR-34b/miR-34c clusters, and the miR-34b/miR-34c cluster directly targeted theZAP70 kinase(meanluciferase activity for miR-34a vs scrambled control, 0.33RLU [95%CI, 0.30–0.36]; P=.02;meanformiR-34bvsscrambledcontrol,0.31RLU [95%CI, 0.30–0.32];P=.01; and mean for miR-34c vs scrambled control, 0.35 RLU [95% CI, 0.33–0.37]; P=.02).
Conclusions
A microRNA/TP53 feedback circuitry is associated with CLL pathogenesis and outcome. This mechanism provides a novel pathogenetic model for the association of 13q deletions with the indolent form of CLL that involves microRNAs, TP53, and ZAP70
Chronic lymphocytic leukemia (CLL) is the most common leukemia among adults in the Western world, with an annual incidence in the United States of approximately 10,000 new cases1. The clinical staging systems devised by Rai et al. 2 and Binet et al. 3 are useful for assessing the extent of CLL in an individual patient but they fail to differentiate between the indolent and aggressive forms of CLL. Most typically these forms of CLL are characterized by low and high levels of the 70-kDa-zeta-associated protein (ZAP70)4–5, respectively. By using fluorescent in situ hybridization (FISH) of 325 patients with CLL, Döhner et al.5 found that chromosomal abnormalities occurred in 82% of cases, and that these abnormalities included the 13q deletion (55%), 11q deletion (18%), and 17p deletion (7%). CLL patients with the 17p and 11q deletions experience the aggressive form of the disease, whereas patients with the 13q deletion or with normal cytogenetics experience the indolent form of the disease 4, 6. The occurrence of common and recurring chromosomal abnormalities 4, suggests that these deletions affect thus far undefined pathways important for the pathogenesis of CLL.
MicroRNAs (miRNAs) are small, non-coding RNAs with regulatory functions 7–10, whose expression is frequently dysregulated in tumors 11–14. In most CLLs, the expression of the miR-15a/16-1 cluster, which maps within a 30-kb region of loss 15 at 13q14.1, is abolished or reduced 16–17. We have previously shown that the expression of this miRNA cluster is inversely correlated with the expression of BCL2, an anti-apoptotic gene that is overexpressed in the majority of CLLs 18. The loss of the long arm of chromosome 11 involves the region where miR-34b/34c cluster is located 19. The observation that miR-34b and miR-34c are transactivated by Tp53 (which maps at 17p) 20, suggests the possible existence of a genetic link and significant molecular interactions between 17p and 11q chromosomal deletions in CLL. At present, it is not known how the 13q, 11q, and 17p deletions contribute to CLL pathogenesis and affect the prognosis of CLL patients.
In this report, we describe a feedback circuitry involving Tp53 and miRNAs (namely, miR-15a/16-1 and miR-34b/34c), which represents the common pathogenetic pathway linking the 13q, 11q, and 17p deletions in CLL. The model that we propose also helps to explain the different expression levels of ZAP70, a surrogate prognostic marker for CLL 5, between indolent and aggressive forms and the prognostic implications of the most frequent chromosomal deletions in human CLL.
METHODS
Patient samples and cell lines
Blood samples were obtained at the CLL Research Consortium Institutions from 206 patients with B-cell CLL (Table). Written informed consent was obtained from all patients before sample analyses, and the study was in accordance with the approved institutional review board protocols (LAB07-0734 and 2005C0014).
Table.
Clinical Data From All Patients With B-Cell Chronic Lympriocytic Leukemia in Molecular Studies
| Deletion (n = 188) |
Additional Deletion (n = 18) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristic | 13q Alone | 11 q Alone | 17p Alone | 11q +13q | 17p + 13q | 11q + 17p | 11q +17p+ 13q | Normal Cytogenetic Profiles |
Total | 11q Alone | Total |
| Patients. No.(%) | 115 (61.2) | 5 (2.6) | 5 (2.6) | 18 (9.6) | 15 (8.0) | 0 | 2 (1.1) | 28 (14.9) | |||
| Men/women | 68/47 | 4/1 | 3/2 | 12/6 | 10/5 | 0/0 | 1/1 | 19/9 | 117/71 | 14/4 | 131/75 |
| Age, mean (SD),y | 57.4 (14.6) | 58.9 (12.3) | 47.1 (10.4) | 55.8 (13.7) | 54.1 (15.1) | 0 | 55.4 (4.3) | 53.2 (9.5) | 54.6 (12.1) | 54.5 (10.4) | 54.6 (11.3) |
| Rai stage | |||||||||||
| 0 | 60 | 2 | 4 | 5 | 6 | 0 | 1 | 6 | 84 | 7 | 91 |
| 1 | 39 | 2 | 1 | 10 | 1 | 0 | 0 | 18 | 71 | 9 | 80 |
| 2 | 10 | 1 | 0 | 3 | 4 | 0 | 1 | 3 | 22 | 2 | 24 |
| 3 | 2 | 0 | 0 | 0 | 3 | 0 | 0 | 1 | 6 | 0 | 6 |
| 4 | 4 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 5 | 0 | 5 |
| Patients with ZAP70 positive cells >20%. No.(%) | 28 (24.4) | 2 (40.0) | 2 (40.0) | 12 (66.7) | 8 (53.3) | 0 | 2 (100) | 12 (43.1) | 10 (55.6) | ||
Abbreviation: ZAP70, zeta-chain (TCR)–associated protein kinase 70kDa.
Measured by flow cytometry.
We first evaluated the occurrence of chromosomal abnormalities in 188 consecutive patients with untreated CLL. Then, to test our hypothesis that the miR-34b/miR-34c cluster modulates ZAP70 (GenBank 7535) expression in CLL, we analyzed 18 additional patients with B-CLL who had the 11q deletion alone. Details about the isolation of mononuclear cells are provided in the eMethods. MEG-01, K562, H1299, and A549 cell lines were obtained from the American Type Culture Collection.
Genes
Two genes were considered downstream effectors of the TP53 pathway signaling: cyclin-dependent kinase inhibitor 1A (p21, Cip1) (CDKN1A [GenBank1026], the expression of which is tightly regulated by TP53, which induces a TP53-dependent cell cycle G1 phase arrest in response to various stress stimuli); and BCL2 binding component 3 (BBC3 [GenBank 27113], formerly named PUMA, a p53 up-regulated modulator of apoptosis, which binds to BCL2 and is a proapoptotic gene). Procaspase 3 is the precursor of caspase 3 apoptosis-related cysteine peptidase (CASP3), a key protein involved in apoptosis. The cleavage of procaspase 3 to CASP3 is one of the strongest indicators of apoptosis.
Luciferase Reporter Assays
MicroRNAs interact with their target genes by means of a “seed” region sequence between the mature microRNA and the target gene messenger RNA (mRNA). A luciferase reporter assay is performed to demonstrate that the microRNA-mRNA interaction is direct (meaning by complementarity). In this assay, the microRNA binding site on the target mRNA is cloned on a plasmid carrying the gene for luciferase, just downstream of the luciferase gene. Cells are then cotransfected with this plasmid and the microRNA of interest (or a scrambled microRNA as a control). If the microRNA targets the cloned binding site, the overall luciferase reporter activity in the cells cotransfected with the microRNA of interest is reduced. The destruction (and/or mutation) of the microRNA binding site on the target mRNA abolishes the reduction in the luciferase reporter activity, which indicates that the targeting is direct.
A luciferase reporter assay also was used to determine the effects of TP53 on the expression of the microRNAs of interest. TP53, like many other transcription factors, recognizes a specific binding sequence and binds to it. As a result, any gene located downstream of the binding site can be activated or suppressed. The predicted TP53 binding sites were cloned upstream of the luciferase gene in a luciferase-expressing reporter plasmid, and H1299 cells were transfected with this plasmid and a TP53-expressing plasmid or an empty plasmid. The effect (either activation or suppression) of TP53 on the expression of the microRNAs of interest was expressed as increased or decreased luciferase reporter activity of the TP53-treated group vs that of the empty vector–treated group (see eMethods for details).
Assessment of ZAP70 and Cytogenetic Data
Expression of ZAP70 was assessed by immunoblotting and flow cytometry analysis. Cytogenetic data were available for all 206 patients in this study. The following probes were used to perform FISH analyses: ataxia telangiectasia mutated (ATM) (11q22.3), D13S319 (13q14.3), and TP53 (17p13.1, henceforth designated 17p13). A commercial probe set (CLL Panel; Vysis Inc, Downers Grove, Illinois) was used to perform FISH analyses on peripheral blood samples that had been cultured for 24 hours without stimulation.
Statistical Analysis
Results are presented as means with 95% confidence intervals (CIs). A probability value of P < .05 by 2-sided t test was considered statistically significant. Allelic distributions for the 3 chromosomal abnormalities (ie, 13q, 17p, and 11q deletions) in all patients were tested with a χ2 goodness-of-fit test for compliance with Hardy-Weinberg equilibrium. Relationships between microRNA and TP53 or ZAP70 mRNA expression determined by qRT-PCR were calculated as Pearson correlations. Based on our previous qRT-PCR data for the expression of miR-15a and miR-16,15 we calculated that by using a cohort of 206 patients, the statistical power would be greater than 95% to detect an effect, if one actually exists, with a minimal detectable effect of 9.73 (calculated as the difference between the lowest and the largest detected means) and an α of .05 by 2-sided test (see eMethods for details).
RESULTS
13q-17p Molecular Link in Patients With B-CLL
The clinical characteristics of the 188 consecutive patients with untreated B-CLL are shown in the Table. No departures from Hardy-Weinberg equilibrium were detected for any of the 3 chromosomal abnormalities investigated. The 13q deletion was the chromosomal abnormality that occurred most often in patients with CLL and was frequently the only cytogenetic abnormality in these patients, whereas the other 2 most common chromosomal abnormalities in CLL (11q deletion and 17p deletion) were often associated with the 13q deletion, but they rarely occurred together in 1 clone.
Patients (n = 22) with CLLs with a homozygous 13q deletion (13q−/−) had significantly lower expression levels of miR-15a (mean fold induction, 0.04 [95% CI, 0.035–0.045]; P = .03) and miR-16 (mean fold induction, 0.18 [95% CI, 0.16–0.20]; P = .01) than patients (n = 28) with CLLs with normal cytogenetic profiles (defined as no abnormality detected by FISH) (set as fold induction, 1 [95% CI, 0.94–1.05] for both microRNAs). Conversely, patients with 13q−/− CLLs had significantly higher TP53 expression levels than patients with CLLs with normal cytogenetic profiles, both at the mRNA level (mean fold induction, 4.83 [95% CI, 4.36–5.30] vs 1 [95% CI, 0.91–1.09]; P = .03) and at the protein level (mean fold induction, 2.06 [95% CI, 1.98–2.14] vs 1 [95% CI, 0.96–1.04]; P = .04) (eFigure 2). Also, patients with CLLs with a heterozygous 13q deletion (13q+/−) had significantly lower expression levels of miR-15a and miR-16 than patients with CLLs with normal cytogenetic profiles (mean fold induction, 0.48 [95% CI, 0.46–0.50]; P = .03 and 0.64 [95% CI, 0.62–0.66]; P = .04 vs 1 [95% CI, 0.97–1.03], respectively) (eFigure 3A). Patients with 13q+/− CLLs had significantly higher TP53 expression levels than those with CLLs with normal cytogenetic profiles both at the mRNA level (mean fold induction, 2.32 [95% CI, 2.23–2.41]; P = .03 vs 1 [95% CI, 0.97– 1.03]) and at the protein level (mean fold induction, 1.48 [95% CI, 1.45–1.51]; P = .04 vs 1 [95% CI, 0.97– 1.03]), although TP53 expression in patients with 13q+/− CLLs was not as high as that in patients with 13q−/− CLLs (eFigure 3A and B).
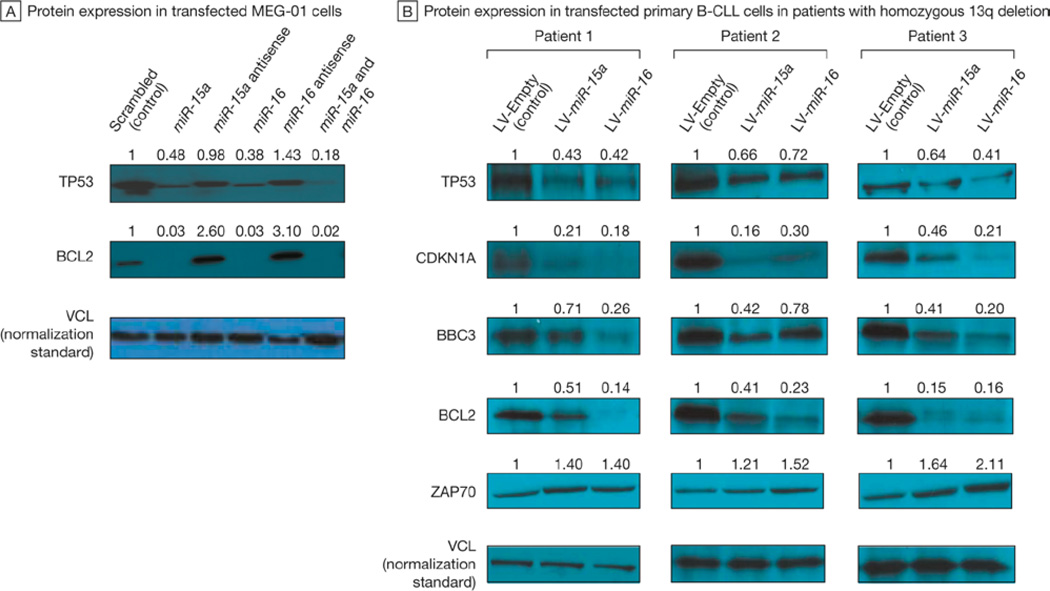
A slight inverse correlation was found between expression of miR-15a/miR-16 and TP53 mRNA (r = −0.31; P < .04) in patients with 13q−/− CLLs compared with patients with CLLs with normal cytogenetic profiles. In MEG-01 cells, TP53 protein expression was highly reduced when the miR-15a/miR-16-1 cluster was overexpressed (eFigure 4). Overexpression of each microRNA of the cluster was associated with reduced TP53 expression (52% for miR-15a and 62% for miR-16) compared with a scrambled oligonucleotide control, and the combination of both miR-15a and miR-16 almost completely repressed TP53 expression (TP53 protein expression was reduced by 82%) (Figure 1A). No TP53 silencing effect was observed with the antisense oligonucleotides. Both miR-15a and miR-16 were found to target BCL2 (Figure 1A), which is consistent with our previous findings.18 In MEG-01 cells stably expressing miR-16, TP53 expression was reduced, as were the protein levels of its downstream effectors CDKN1A, BBC3, and BCL2 (eFigure 5). Additionally, cleavage of procaspase 3 was observed in MEG-01 cells stably expressing miR-15a or miR-16-1, confirming that the miR-15a/miR-16-1 cluster has a caspase-dependent proapoptotic role in this cell line (eFigure 5). No effect on CDKN1A and BBC3 was observed in TP53-negative H1299 cells (eFigure 6), which suggests that the effects of miR-15a and miR-16 on CDKN1A and BBC3 are mediated by their effects on TP53. Conversely, the targeting effect of miR-15a and miR-16 on BCL2 persisted also in TP53-null cells (eFigure 6). These results were confirmed in primary B-CLL cells collected from 3 patients with 13q−/− CLL. When miR-15a and miR-16 were overexpressed in these primary B-CLL cells, the expression of TP53, CDKN1A, B BC3, a nd BCL2 was reduced b oth a t t he p rotein and mRNA level (Figure 1B and eFigure 7 [left panels]).
FIGURE 1. Targeting of TP53 by miR-15a and miR-16 and Effects on TP53 Downstream Effectors in Cell Lines and Primary B-Cell Chronic Lymphocytic Leukemia (B-CLL) Samples.
A, Immunoblots showing the protein expression of tumor protein p53 (TP53), B-cell CLL/lymphoma 2 (BCL2), and vinculin (VCL) in MEG-01 cells transfected with microRNA 15a (miR-15a), microRNA 16 (miR-16), their combination, or their antisense oligonucleotides. Cotransfection of miR-15a and miR-16-1 was performed at the same concentration of oligonucleotides per each; therefore, the total amount of transfected microRNAs was doubled with respect to the other lanes. VCL is the normalization standard used to normalize the amount of proteins loaded to each well. The numbers above the blots indicate the intensity of the band expressed as a ratio “gene product (TP53 or BCL2)/VCL” and normalized to “scrambled.” B, Immunoblots showing the protein expression of TP53, cyclin-dependent kinase inhibitor 1A (p21, Cip 1) (CDKN1A), BCL2 binding component 3 (BBC3), BCL2, zeta-chain (TCR)–associated protein minase 70 kDa (ZAP70), and VCL in primary B-cell CLL cells of 3 patients with CLL with a homozygous 13q deletion. Primary leukemic cells were stably infected with a lentiviral vector expressing miR-15a (LV- miR-15a), a lentiviral vector expressing miR-16-1 (LV- miR-16), or an empty lentiviral vector (LV-Empty). VCL is the normalization standard used to normalize the amount of proteins loaded to each well. The numbers above the blots indicate the intensity of the band expressed as ratio “gene product (TP53, CDKN1A, BBC3, BCL2 or ZAP70)/VCL” and normalized to “LV-Empty.
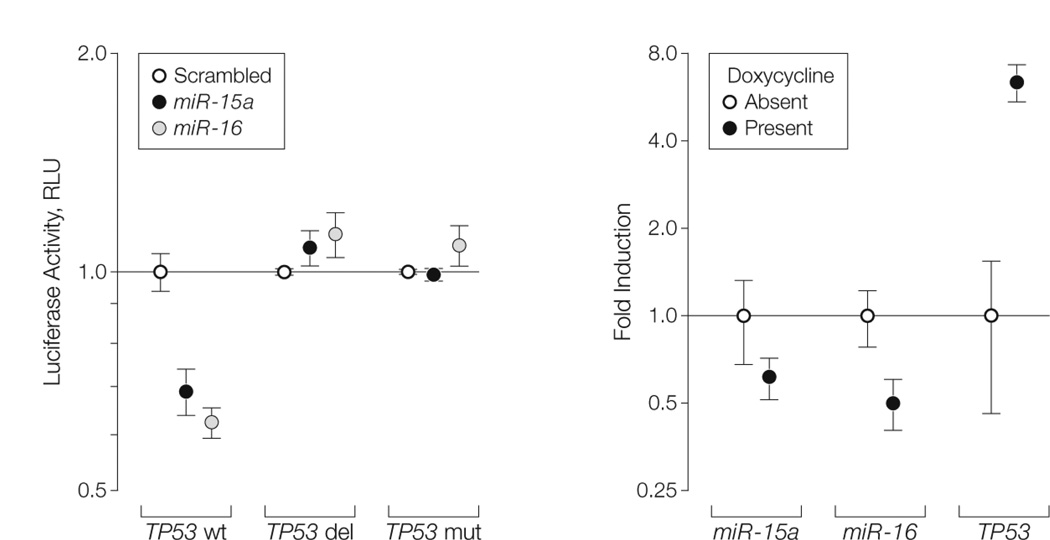
A binding site for both miR-15a and miR-16 inside the 3′-UTR (untranslated region) of TP53 was identified using the sequencer software (eFigure 8). A luciferase reporter assay showed that both miR-15a and miR-16 directly target the identified TP53 binding site and significantly reduced the luciferase reporter activity compared with a scrambled oligonucleotide-negative control (mean luciferase activity, 0.68 relative light units [RLU] [95% CI, 0.63–0.73]; P = .02 for miR-15a and 0.62 RLU [95% CI, 0.59–0.65]; P = .02 for miR-16 vs 1 [95% CI, 0.94–1.06] for scrambled). This effect was completely abolished when the binding site was either deleted or mutated (Figure 2A). Lastly, by using a miR-15a/miR-16-1 doxycycline-inducible HeLa cell line when miR-16 was down-regulated in presence of doxycycline, the expression of TP53 mRNA was significantly increased (mean fold induction, 6.39 [95% CI, 5.46–7.32] vs 1 [95% CI, 0.46– 1.54]; P = .01) (Figure 2B).
FIGURE 2. Targeting of TP53 by miR-15a and miR-16.
A, Luciferase reporter assay (as means [error bars indicate 95% confidence intervals] of experiments conducted in sextuplicate) in cells cotransfected with wild-type tumor protein p53 (TP53) 3′-UTR (TP53 wt) and microRNA 15a (miR-15a) or 16 (miR-16). Luciferase activity normalized to scrambled; RLU indicates relative light units. TP53 del indicates deletion of miR-15a/miR-16 binding site on TP53 3′-UTR; TP53 mut indicates mutation of miR-15a/miR-16 binding site on TP53 3′-UTR. P values calculated for miR-15a and miR- 16 vs scrambled; values were statistically significant (P < .05) for TP53 wt comparisons only. B, Expression of miR-15a, miR-16, and TP53 messenger RNA (mRNA) in Tet-Off miR-15a/miR-16-1 – inducible HeLa cells as detected by quantified real-time polymerase chain reaction. Results presented as means (error bars indicate 95% confidence intervals) of experiments performed in triplicate. P values calculated for the cells in the presence of doxycycline (indicates reduced expression of the miR-15a/miR-16-1 cluster) vs the cells in the absence of doxycycline (indicates increased expression of the miR-15a/miR-16-1 cluster); all values were statistically significant (P < .05).
17p-13q Molecular Interaction in Patients With B-CLL
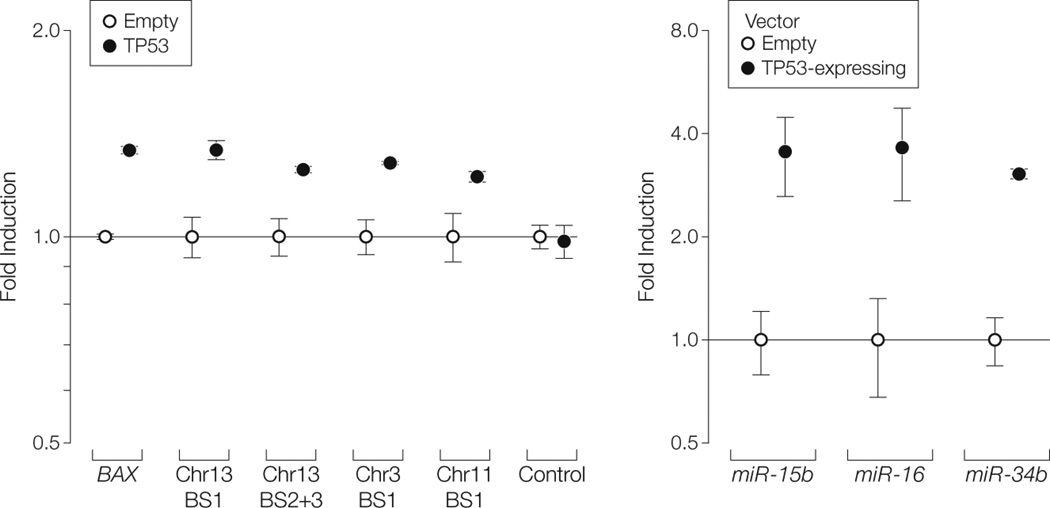
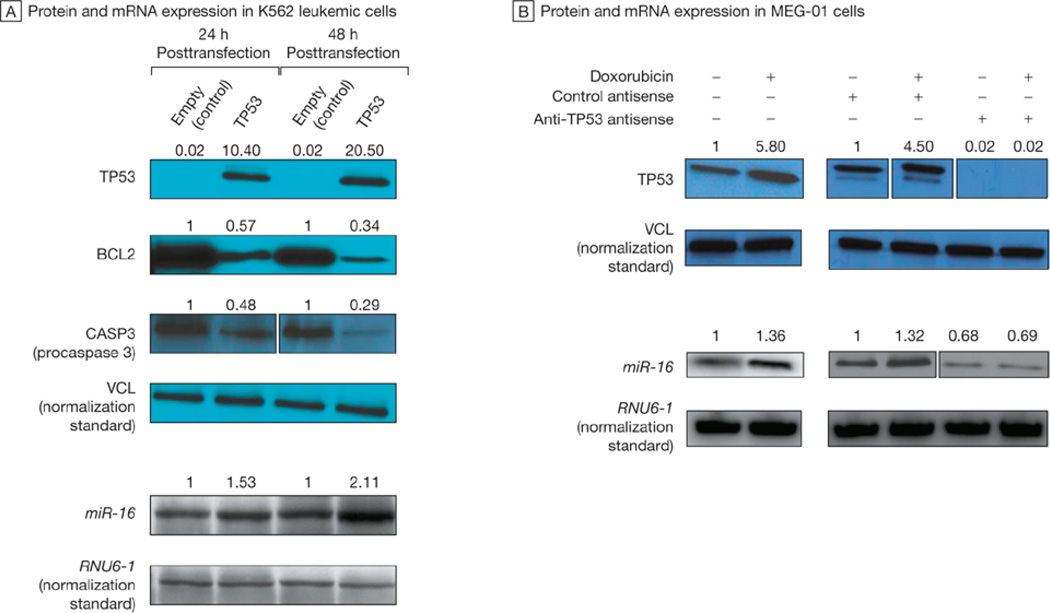
Several TP53 binding sites were found upstream of the 2 homologous miR-15/miR-16 loci located on chromosome 13 (miR-15a/miR-16-1) and on chromosome 3 (microRNA 15b [miR-15b ] [GenBank 406949]/ microRNA 16-2 [miR-16-2] [GenBank406951]), which was also analyzed because it encodes for a similar cluster of genes (eFigure 9). Chromatin immunoprecipitation analysis revealed that TP53 directly binds to its predicted binding sites on both chromosome 13 and chromosome 3, both in cell lines and in primary CLLs with normal cytogenetic profiles (eFigure 10 and eFigure 11). A luciferase reporter assay showed that TP53 significantly increased the luciferase reporter activity of all the binding site–containing vectors (Figure 3A). In MEG-01 cells, TP53 transactivation of the miR-15/miR-16 cluster was also confirmed by qRT-PCR (Figure 3B); this transactivating effect also occurred in TP53-mutated K562 cells, in which it was associated with a reduction of procaspase 3 protein levels (Figure 4A). Similarly, doxorubicin-mediated TP53 activation in MEG-01 cells increased the expression of miR-16, an effect that was abolished when TP53 was silenced by an anti-TP53 oligonucleotide (Figure 4B). Although the effect of TP53 on the miR-15/miR-16 cluster was relatively mild (highest fold induction of 2.11 after 48 hours), it still had functional consequences and was associated with 66% reduction of BCL2 protein levels after 48 hours.
FIGURE 3. Transactivation of miR-15a/miR-16-1 and miR-34b/miR-34c Clusters by TP53.
A, Promoter luciferase assay in tumor protein p53 (TP53)-null H1299 cells reported as means (error bars indicate 95% confidence intervals) of experiments performed in sextuplicate. The indicated TP53 binding site (BS) numbers correspond to those shown in red in eFigures 9 and 12. Control indicates the results obtained using the promoter vector with no binding site cloned in it. BAX indicates BCL2-associated X protein; Chr, chromosome. P values were calculated for TP53 vs empty for each group; all values were statistically significant (P < .05). B, Quantified real-time polymerase chain reaction (error bars indicate 95% confidence intervals) for microRNA 15b (miR-15b), microRNA 16 (miR-16), and microRNA 34b (miR-34b) performed on MEG-01 cells 24 hours after transfection with empty or TP53-expressing vectors. P values were calculated for TP53 vs empty for each group; all values were statistically significant (P < .05).
FIGURE 4. Transactivation of MicroRNA miR-16 Affecting Expression of miR-16 Targets.
A, Northern blots showing messenger RNA (mRNA) expression of microRNA 16 (miR-16) and immunoblots showing the protein expression of tumor protein p53 (TP53), B-cell CLL/lymphoma 2 (BCL2), caspase 3, apoptosis-related cysteine peptidase (CASP3), and vinculin (VCL) in K562 leukemic cells 24 or 48 hours after transfection with an empty or TP53-expressing vector (TP53). VCL is the normalization standard used to normalize the amount of proteins loaded to each well. The numbers above the blots indicate the intensity of the band expressed as ratio “gene product (TP53, BCL2 or CASP3)/VCL” and normalized to “empty.” B, Immunoblots showing the protein expression of TP53 and VCL, and the Northern blots showing mRNA expression of miR-16 and RNA, U6 small nuclear 1 (RNU6-1 ), after doxorubicin-induced TP53 activation in MEG-01 cells. VCL and RNU6-1 are the normalization standards used to normalize the amount of proteins and RNA loaded to each well, respectively. The numbers above the blots indicate the intensity of the band expressed as ratio “TP53/VCL” or “ miR-16/RNU6-1 ” and normalized to untreated cells (left panels) and to anti-CTRL treated cells (right panels)..
17p-11q- ZAP70 Molecular Link in Patients With B-CLL
Chromatin immunoprecipitation analysis revealed that TP53 binds directly to a pre–miR-34b/miR-34c TP53 binding site on chromosome 11 (eFigure 10 and eFigure 12). A luciferase reporter assay showed a statistically significant transactivating effect of TP53 on the miR-34b/miR- 34c binding site (Figure 3A), which was confirmed in TP53-transfected MEG-01 cells, which had increased miR-34b expression levels (Figure 3B). Taken together, these results indicate that TP53 is a positive transcriptional regulator of the miR-34b/miR-34c cluster in leukemic cells, which is consistent with findings observed in epithelial cells.20
Patients with 11q+/− CLLs (n = 23) had significantly lower levels of miR-34b (mean fold induction, 0.37 [95% CI, 0.35–0.39]; P < .01) and miR-34c (mean fold induction, 0.39 [95% CI, 0.37–0.41]; P < .01) than patients with CLLs with normal cytogenetic profiles (n = 28) (set as fold induction, 1 [95% CI, 0.96–1.04] and 1 [95% CI, 0.97–1.03], respectively). Conversely, patients with 11q+/− CLLs had significantly higher levels of ZAP70 than patients with CLLs with normal cytogenetic profiles, both at the mRNA level (mean fold induction, 2.02 [95% CI, 1.98–2.06] vs 1 [95% CI, 0.97–1.03]; P = .01) and at the protein level (mean fold induction, 2.47 [95% CI, 2.43– 2.51] vs 1 [95% CI, 0.94–1.06]; P = .005). Patients with 11q+/− CLLs and high levels of ZAP70 experienced poorer overall survival than patients with CLLs with normal cytogenetic profiles and lower levels of ZAP70 (mean, 72.4 [95% CI, 35.4–79.2] months vs 114.7 [95% CI, 98.9–130.5] months; P = .02). A binding site for the miR-34 family was detected in the ZAP70 opening reading frame (eFigure 13A). Reduced ZAP70 expression (both at the protein and mRNA level) was observed in primary B-CLL cells from a patient with an 11q+/− deletion, in which miR-34a, miR- 34b, and miR-34c were overexpressed (eFigure 13B and eFigure 14).
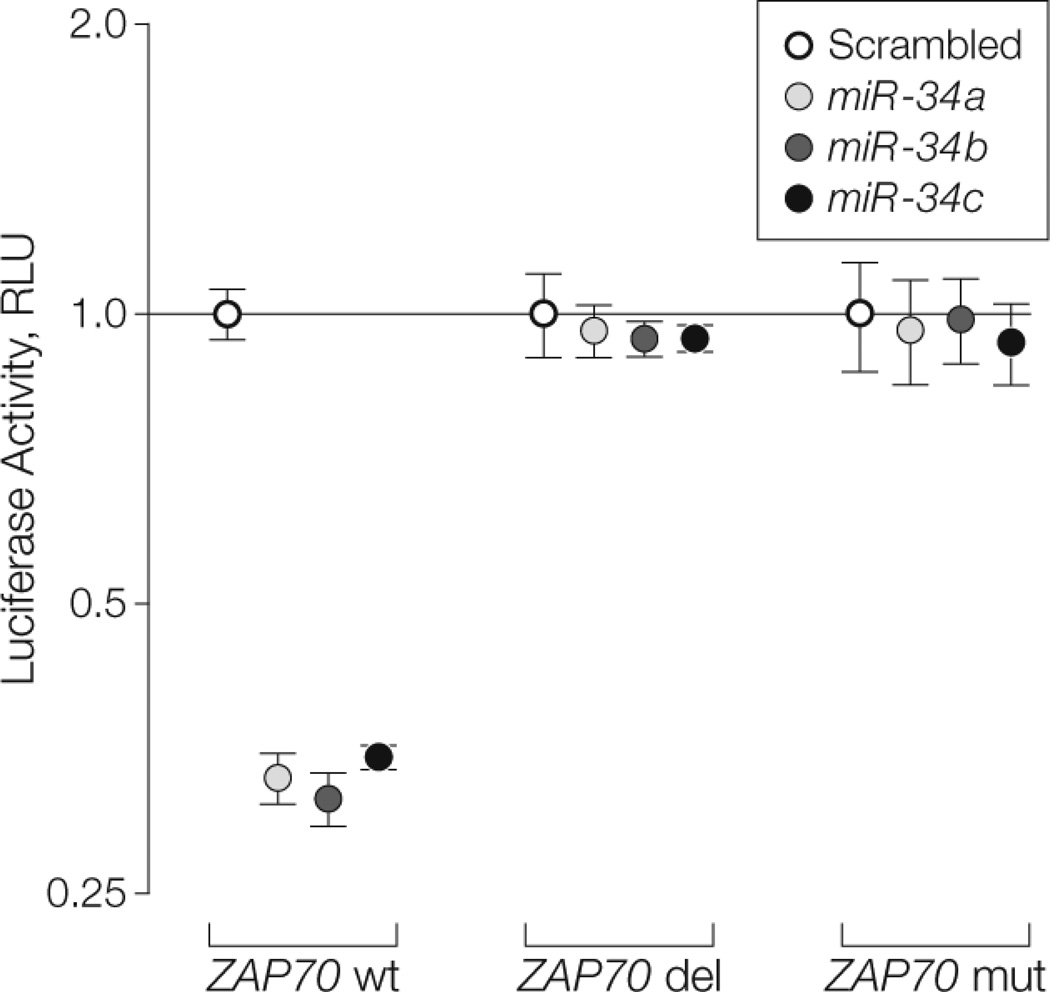
No effect on cell growth and cell proliferation was observed up to 72 hours in MEG-01 (ZAP70-negative) cells or in K562 (ZAP70-positive) cells overexpressing miR-34b or miR-34c (eFigure 15). A luciferase reporter assay showed that all 3 microRNAs directly target the predicted region on ZAP70 (mean luciferase activity, 0.33 RLU [95% CI, 0.30–0.36]; P = .02 for miR-34a, 0.31 RLU [95% CI, 0.30–0.32]; P = .01 for miR-34b, and 0.35 RLU [95% CI, 0.33–0.37]; P = .02 for miR- 34c vs 1 [95% CI, 0.94–1.06] for scrambled). This effect was abolished when the predicted site on ZAP70 was either deleted or mutated (Figure 5). In K562 cells as well as primary B-CLL lymphocytes, increased levels of ZAP70 mRNA were observed after the miR-34 family was silenced (eFigure 13C and D).
FIGURE 5. Targeting of ZAP70 by miR-34a, miR-34b, and miR-34c.
Luciferase reporter assay (as means [error bars indicate 95% confidence intervals] of experiments conducted in sextuplicate) in MEG-01 cells cotransfected with wild-type zeta-chain (TCR)–associated protein kinase 70kDa (ZAP70) binding site for microRNA 34 (miR-34) family (wt) and miR-34a, miR-34b, or miR-34c. Luciferase activity normalized to scrambled; RLU indicates relative light units. ZAP70 del indicates deletion of miR-34 binding site on ZAP70 coding region; ZAP70 mut indicates mutation of miR-34 binding site on ZAP70 coding region. P values calculated for miR-34a, miR- 34b, and miR-34c vs scrambled; values were significant (P < .05) for ZAP70 wt (wild-type) only.
COMMENT
In this study, we identified a microRNA/protein functional circuitry that likely underlies the pathogenesis and natural history of a major subset of human CLL. This pathway is perturbed at different levels by distinct CLL chromosomal abnormalities, which could explain the occurrence of the same disease but with distinct clinical features. This novel pathogenetic model for CLL is summarized in Figure 6.
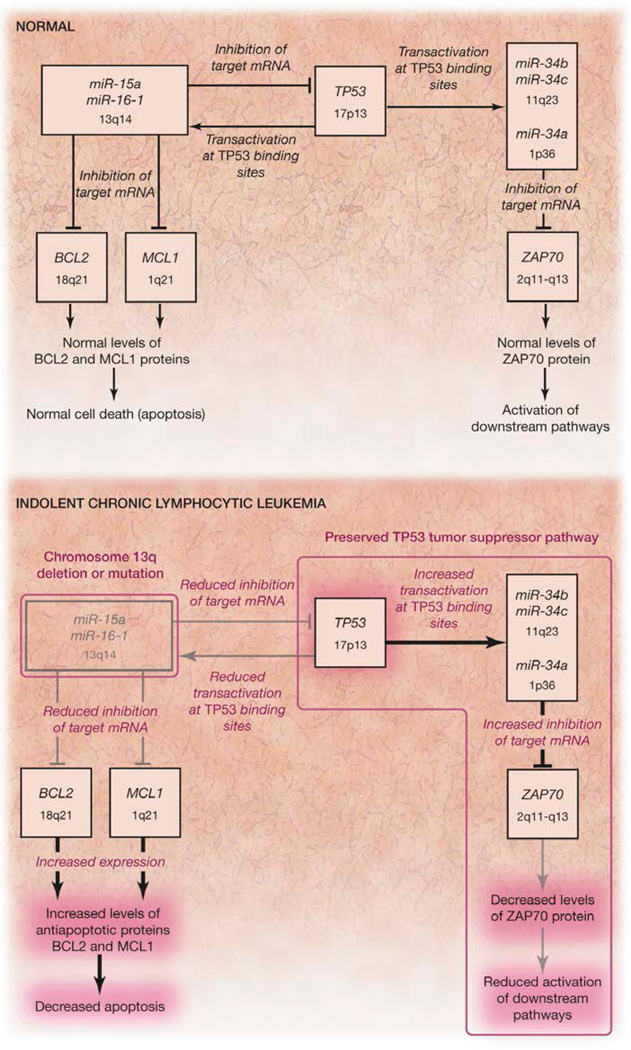
FIGURE 6. MicroRNA/TP53 Pathogenetic Model for Human CLL.
A novel pathogenetic model for chronic lymphocytic leukemia (CLL) showing a pathway of microRNAs and protein coding genes that are involved in the development of CLL. The microRNA 15a (miR-15a)/microRNA 16-1 (miR-16-1) cluster, the microRNA 34b (miR-34b)/microRNA 34c (miR-34c) cluster, and the genes tumor protein p53 (TP53), B-cell CLL/lymphoma 2 (BCL2), myeloid cell leukemia sequence 1 (BCL2-related) (MCL1), and zeta-chain (TCR)–associated protein kinase 70kDa (ZAP70) are the main partners in this model. mRNA indicates messenger RNA.
In this model, TP53 (located on chromosome 17p) represents the molecular link between the miR-15a/miR-16-1 (located on chromosome 13q) and miR-34b/miR-34c (located on chromosome 11q) clusters. In fact, the tumor suppressor protein TP53 is directly regulated by miR-15a/miR-16- 1. The loss of miR-15a/miR-16-1 expression, represented by CLLs with 13q deletions, not only shifts the balance toward higher levels of the antiapoptotic proteins BCL2 and myeloid cell leukemia sequence 1 (BCL2-related) (MCL1), as we have previously demonstrated,12,16 but also toward higher levels of the tumor suppressor protein TP53. Consequently, in patients with CLLs with 13q deletions, while the number of apoptotic cells may decrease because of the increased levels of antiapoptotic proteins, the TP53 tumor suppressor pathway remains intact, thus keeping the increase in tumor burden relatively low (eComments).
This novel finding explains how 13q deletions are associated with the indolent form of CLL, as first identified by Döhner et al.4 Moreover, increased TP53 levels, as found in patients with CLLs with 13q deletions, are associated with transactivation of miR-34b /miR-34c and reduced levels of ZAP70, a tyrosine kinase relevant in the initial step of T-cell receptor–mediated signal transduction.17 Low expression levels of ZAP70 have been found to be positively correlated with survival in patients with CLL,18 further explaining the indolent course of CLL carrying 13q deletions.
Here we showed that, in primary B cells from patients with B-CLL, use of viral infection to restore expression of the miR-15a/miR-16-1 cluster is associated with reduced expression levels of TP53, miR-34a, miR-34b, and miR-34c and increased protein levels of ZAP70. These findings also demonstrate that in primary B-CLLs, restoring the expression of a microRNA cluster (namely, the miR-15a/miR-16-1 cluster) indirectly affects the expression of another family of microRNAs (miR-34 family) by modulating the levels of TP53. Some of our results indicate that these effects also occur in non-CLL leukemic cells (such as the acute myelogenous leukemic cell line K562) and in nonhematologic cell lines (such as H1299, A549, and HeLa cells), which suggests that the proposed microRNA-TP53 loop is relevant to tumor types other than CLL. More studies are necessary to validate this statement.
In conclusion, we found that a microRNA/TP53 feedback circuitry is associated with the pathogenesis and prognosis of CLL. Our findings reveal a new pathogenetic model for human CLL that involves microRNAs (miR-15a/miR-16-1 and miR-34b/miR-34c) and protein-coding genes (such as TP53 and ZAP70) with well-known prognostic significance in CLL.
Supplementary Material
Acknowledgments
Funding/Support: Dr. Fabbri is supported by a 2009 Kimmel Scholar Award; Dr. Calin is supported as a Fellow at The University of Texas M. D. Anderson Research Trust, as a Fellow of The University of Texas System Regents Research Scholar and by the CLL Global Research Foundation. Work in Dr Calin’s laboratory is supported in part by NIH, by DOD, and by a Leukemia SPORE Developmental Research Award.; Dr. Kay, and Dr. Croce are supported by the NIH grant R01 CA111953.
Role of the Sponsors: The funding agencies had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data, and preparation, review, or approval of the manuscript.
Footnotes
Author Contributions: Drs Calin and Croce had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Fabbri, Bottoni, Calin, Croce.
Acquisition of data: Fabbri, Bottoni, Spizzo, Shimizu, Nicoloso, Barbarotto, Cimmino, Adair, Wojcik, Calore, Valeri, Sampath, Fanini, Vannini, Musuraca, Davuluri, Alder, Negrini, Amadori, Calin, Croce.
Analysis and interpretation of data: Fabbri, Bottoni, Rossi, Rassenti, Dell’Aquila, Kipps, Negrini, Calin, Croce.
Drafting of the manuscript: Fabbri, Bottoni, Nakamura, Kay, Rai, Keating, Kipps, Calin, Croce.
Critical revision of the manuscript for important intellectual content: Fabbri, Bottoni, Nicoloso, Nakamura, Kay, Rai, Keating, Kipps, Negrini, Calin, Croce.
Statistical analysis: Fabbri, Bottoni, Rossi.
Obtained funding: Fabbri, Calin, Croce.
Administrative, technical, or material support: Shimizu, Adair, Calore, Alder, Barbarotto, Cimmino, Valeri.
Study supervision: Calin, Croce.
Financial Disclosures: None reported.
Additional Contributions: We thank Dr. Kay Huebner, Ph.D. from the Ohio State University, for critical discussion and reading of the manuscript, and Dr. Lionel Santibanez, Ph.D. from the Scientific Publication Department at the M.D. Anderson Cancer Center for English editing. Neither of the Contributors received compensation for their role in the study.
REFERENCES
- 1.Call TG, Phyliky RL, Noel P, et al. Incidence of chronic lymphocytic leukemia in Olmsted County, Minnesota: 1935 through 1989, with emphasis on changes in initial stage at diagnosis. Mayo Clin Proc. 1994 Apr;69(4):323–328. doi: 10.1016/s0025-6196(12)62215-0. [DOI] [PubMed] [Google Scholar]
- 2.Rai KR, Sawitsky A, Cronkite EP, Chanana AD, Levy RN, Pasternack BS. Clinical staging of chronic lymphocytic leukemia. Blood. 1975 Aug;46(2):219–234. doi: 10.1182/blood-2016-08-737650. [DOI] [PubMed] [Google Scholar]
- 3.Binet JL, Lepoprier M, Dighiero G, et al. A clinical staging system for chronic lymphocytic leukemia: prognostic significance. Cancer. 1977 Aug;40(2):855–864. doi: 10.1002/1097-0142(197708)40:2<855::aid-cncr2820400239>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 4.Dohner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. The New England Journal of Medicine. 2000 Dec 28;343(26):1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 5.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. The New England journal of medicine. 2005 Feb 24;352(8):804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 6.Dewald GW, Brockman SR, Paternoster SF, et al. Chromosome anomalies detected by interphase fluorescence in situ hybridization: correlation with significant biological features of B-cell chronic lymphocytic leukaemia. Br J Haematol. 2003 Apr;121(2):287–295. doi: 10.1046/j.1365-2141.2003.04265.x. [DOI] [PubMed] [Google Scholar]
- 7.Ambros V, Lee RC. Identification of microRNAs and other tiny noncoding RNAs by cDNA cloning. Methods Mol Biol. 2004;265:131–158. doi: 10.1385/1-59259-775-0:131. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004 Jul;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 10.Plasterk RH. Micro RNAs in animal development. Cell. 2006 Mar 10;124(5):877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 11.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature Reviews Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 12.Esquela-KerscherA, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 13.Fabbri M, Croce CM, Calin GA. MicroRNAs. Cancer J. 2008 Jan-Feb;14(1):1–6. doi: 10.1097/PPO.0b013e318164145e. [DOI] [PubMed] [Google Scholar]
- 14.Fabbri M, Garzon R, Andreeff M, Kantarjian HM, Garcia-Manero G, Calin GA. MicroRNAs and noncoding RNAs in hematologicalmalignancies: molecular, clinical and therapeutic implications. Leukemia. 2008 Mar 6; doi: 10.1038/leu.2008.30. [DOI] [PubMed] [Google Scholar]
- 15.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sc U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calin GA, Ferracin M, Cimmino A, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005 Oct 27;353(17):1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 17.Pfeifer D, Pantic M, Skatulla I, et al. Genome-wide analysis of DNA copy number changes and LOH in CLL using high-density SNP arrays. Blood. 2007 Feb 1;109(3):1202–1210. doi: 10.1182/blood-2006-07-034256. [DOI] [PubMed] [Google Scholar]
- 18.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proceedings of the National Academy of Sciences of the United States of America. 2005 Sep 27;102(39):13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auer RL, Riaz S, Cotter FE. The 13q and 11q B-cell chronic lymphocytic leukaemia-associated regions derive from a common ancestral region in the zebrafish. Br J Haematol. 2007 Jun;137(5):443–453. doi: 10.1111/j.1365-2141.2007.06600.x. [DOI] [PubMed] [Google Scholar]
- 20.Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c Are Targets of p53 and Cooperate in Control of Cell Proliferation and Adhesion-Independent Growth. Cancer Res. 2007 Sep 15;67(18):8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 21.Lall S, Grun D, Krek A, et al. A genome-wide map of conserved microRNA targets in C. elegans. Curr Biol. 2006 Mar 7;16(5):460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 22.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 23.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sindelarova L, Michalova K, Zemanova Z, et al. Incidence of chromosomal anomalies detected with FISH and their clinical correlations in B-chronic lymphocytic leukemia. Cancer Genet Cytogenet. 2005 Jul 1;160(1):27–34. doi: 10.1016/j.cancergencyto.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Calin GA, Cimmino A, Fabbri M, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci U S A. 2008 Mar 24; doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neumeister EN, Zhu Y, Richard S, Terhorst C, Chan AC, Shaw AS. Binding of ZAP-70 to phosphorylated T-cell receptor zeta and eta enhances its autophosphorylation and generates specific binding sites for SH2 domain-containing proteins. Molecular and Cellular Biology. 1995 Jun;15(6):3171–3178. doi: 10.1128/mcb.15.6.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rassenti LZ, Huynh L, Toy TL, et al. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. The New England Journal of Medicine. 2004 Aug 26;351(9):893–901. doi: 10.1056/NEJMoa040857. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009 Jul 23;460(7254):529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.