Abstract
A1/Bfl-1 is a NF-κB dependent, anti-apoptotic Bcl-2 family member that contains four Bcl-2 homology domains (BH) and an amphipathic C-terminal domain, and is expressed in endothelial cells (EC). Based on NF-κB reporter assays in bovine aortic EC, we have previously demonstrated that A1, like Bcl-2 and Bcl-xL, inhibits NF-κB activation. These results, however, do not fully translate when evaluating the cell’s own NF-κB machinery in human EC overexpressing A1 by means of recombinant adenovirus (rAd.) mediated gene transfer. Indeed, overexpression of full-length A1 in human umbilical vein EC (HUVEC), and human dermal microvascular EC (HDMEC) failed to inhibit NF-κB activation. However, overexpression of a mutant lacking the C-terminal domain of A1 (A1ΔC) demonstrated a potent NF-κB inhibitory effect in these cells. Disparate effects of A1 and A1ΔC on NF-κB inhibition in human EC correlated with mitochondrial (A1) versus non-mitochondrial (A1ΔC) localization. In contrast, both full-length A1 and A1ΔC protected EC from staurosporine (STS)-induced cell death, indicating that mitochondrial localization was not necessary for A1’s cytoprotective function in human EC. In conclusion, our data uncover a regulatory role for the C-terminal domain of A1 in human EC: anchoring A1 to the mitochondrion, which conserves but is not necessary for its cytoprotective function, or by its absence freeing A1 from the mitochondrion and uncovering an additional anti-inflammatory effect.
Keywords: A1/Bfl-1, Mitochondria, Nuclear Factor Kappa B, Apoptosis, C-terminal domain
1. Introduction
A1, also known as Bfl-1, is a Bcl-2 family member originally identified in hematopoietic cells, where it is abundantly expressed in response to treatment with granulocyte-macrophage colony stimulating factor [1]. Initially described as a hematopoietic-specific gene, A1 is also expressed in tumors including gastrointestinal and hematopoietic [2, 3]. Additionally, it is upregulated in response to inflammatory stimuli in numerous cell types such as endothelial cells (EC) [4, 5], renal proximal tubular epithelial cells (RPTEC) [6], and pancreatic beta cells [7].
Based on NF-κB reporter assays in bovine aortic EC (BAEC), we have previously demonstrated that A1, like Bcl-2 and Bcl-xL, inhibits NF-κB activation [5, 8], and hence could have anti-inflammatory functions in EC [9]. A1 is either not expressed or expressed at very low levels in EC at baseline, and becomes upregulated in response to pro-inflammatory stimuli in a NF-κB-dependent manner. Accordingly, A1ΔCould possibly qualify as part of the anti-inflammatory and cytoprotective regulatory response of EC to injury [5, 10].
Homology between A1 and Bcl-2 was extensively studied. Currently, it is recognized that A1ΔContains four Bcl-2 homology domains (BH1, BH2, BH3 and BH4) and an amphipathic tail anchoring peptide in its C-terminal domain that directs its binding to mitochondria, even if this domain is not a bona fide transmembrane domain like in other anti-apoptotic Bcl-2 family members [11–13]. This structural configuration, particularly the BH4 domain and the C-terminal mitochondria-anchoring domain, appears crucial for the anti-apoptotic function of Bcl-2 family members [8, 11, 12, 14–16]. In EC, we showed that the BH4 domain of Bcl-2 and Bcl-xL is also key for their NF-κB inhibitory function [8]. However, the role of the C-terminal domain in supporting Bcl-2, Bcl-xL, or A1’s NF-κB inhibitory function in EC is still not fully elucidated.
The goals of this study were to evaluate the contribution of the C-terminal domain of A1 to its anti-inflammatory function in EC, and to extend our findings to human EC. Our results indicate that full-length A1 has no anti-inflammatory effect in human EC, contrasting with what we had reported in BAEC, when using transfection based NF-κB reporter assays. However, the A1 deletion mutant lacking the C-terminal domain (A1ΔC) showed NF-κB inhibitory effect in human EC. We reproduced these data in rAd.A1 and rAd. A1ΔC tranduced BAEC, which indicated that discrepancies in results related to differences in experimental systems used. Both full-length A1 and A1ΔC protected human EC from cell death, indicating that mitochondrial localization was not necessary for the cytoprotective function of this protein, at least in this context. Together these data resolve an important regulatory role for the C-terminal domain of A1 in EC, including in human primary EC cultures. It also cautions extrapolating results from reporter-based assays without evaluating the modulation of gene expression in the context of the cell’s own signaling machinery and DNA/chromatin accessibility.
2. Material and Methods
2.1. Cell culture
Human umbilical vein EC (HUVEC) were purchased from Genlantis PrimaPure (San Diego, CA) and cultured according to manufacturer’s instructions. Human dermal microvascular EC (HDMEC) and BAEC were isolated in our laboratory and cultured as described [17, 18]. The Burkitt lymphoma cell line, BJAB was a kind of Dr. R. Khosravi-Far, and the human hepatoma cell line HepG2 was purchased from ATCC (Manassas, VA). Cells were grown at 37°C in a 5% humidified CO2 atmosphere. Experiments using EC were performed on confluent cells between passage 5 and 9 (BAEC), 5 and 7 (HDMEC), and 4 and 5 (HUVEC)
2.2. Expression plasmids
The N-terminal hemagglutinin A (HA)-tagged human A1 expression plasmid was previously constructed in our laboratory [5]. This construct was subcloned in the pAC.CMV-pLpASR+ (pAC) expression plasmid to generate recombinant adenovirus (rAd.), as previously described [19]. A1 deletion mutants were generated as follows: deletion of the C-terminal domain (A1ΔC) was achieved, with HA tagged A1ΔCDNA as template, by reverse transcription polymerase chain reaction (RT-PCR) using the following primers: sense: 5’-CGCGGGGTACCCCATGTATCCTTATGATGTT-3’ and anti-sense: 5’-CCCCAAGCTTGGTTACATCCAGCCAGATTTAGG-3’. Deletion of the BH4 domain (A1ΔBH4) was performed by site-directed mutagenesis based on overlap extension [20], using the following primers: RT-PCR reaction 1 - Sense A: 5'-CGCGGGGTACCCCATGTATCCTTATGATGTT-3' and Anti-sense B: 5'-TTCCACTTCTTTAGGTTGTGGTATCTGTAGGAC-3'; RT-PCR reaction 2 - Sense C: 5'-CCACAACCTAAAGAAGTGGAAAAGAATCTGAAG-3' and Anti-sense D: 5'-CCCCAAGCTTGGTCAACAGTATTGCTTCA-3'. Deletion of the C-terminal and BH4 domains (A1ΔΔ was achieved by RT-PCR using A1ΔBH4 cDNA as template and the same primers previously employed to generate A1ΔC RT-PCR products were verified by sequencing, and cloned in the CMV promoter driven pAC expression plasmid. Expression of HA-tagged transgenes was verified in transfected BAEC, as shown in the results section. All primers used for cloning were purchased from Gibco, Life Technologies (Grand Island, NY).
2.3. Recombinant adenoviruses
Recombinant adenoviral vectors encoding hemagglutinin A (HA)-tagged human A1 (rAd.A1) and A1ΔC (rAd.A1ΔC) were generated in our laboratory, as described [19]. The control adenoviral vector encoding nuclear β-galactosidase (rAd.βgal) was a kind gift of Dr. R. Gerard (University of Texas, Southwestern Medical Center). Recombinant adenoviruses were produced in the human embryonic 293 kidney cell line (HEK 293, American Type Culture Collection, Rockland, MD), extracted and purified using the Adenopure kit (Puresyn, Inc., Malvern, PA) and tittered by limiting dilution on 293 cells, as described [19]. Pre-confluent (80–90%) human EC were transduced with a multiplicity of infection (MOI) of 200 plaque-forming units per cell (pfu/cell), while BAEC were transduced at a MOI of 1,000 pfu/cell that we have shown to achieve transgene expression in more than 95% of EC, without causing toxicity, and within 48 h of transduction [19].
2.4. Immunofluorescence staining
Human EC cultured on glass coverslips were transduced with rAd.βgal, rAd.A1 or rAd.A1ΔC. Forty-eight hours after transduction, cells were labeled with 100 nM Mitotracker Red CMXRos, following the manufacturer’s instruction (Invitrogen, Molecular Probes, Carlsbad, CA), then incubated with rat monoclonal anti-HA-fluorescein antibody (Roche Diagnostics, Indianapolis, IN) O/N at 4°C. Cells were then washed and mounted on glass slides. Images were obtained using an Olympus BX41 microscope connected to a DP72 digital camera, using 100X oil immersion objective.
2.5. Protein extraction and Western blot analysis
Total protein cell lysates were recovered from EC before, and at different times after treatment with human recombinant tumor necrosis factor (TNF) (R&D Systems, Minneapolis, MN). Protein cell lysates were analyzed by Western blot (WB), as described [21], using the following primary antibodies: rabbit anti-IκB-α IgG, mouse anti-β-actin IgG1 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), rat anti-hemagglutinin A (HA) monoclonal antibody (Roche Molecular Biochemicals, Indianapolis, IN). Donkey anti-rabbit- or goat anti-mouse-HRP-conjugated antibodies (Thermo Scientific, Rockford, IL) were used as secondary antibodies. Protein bands were detected with enhanced chemiluminescence kit (ECL) (PerkinElmer Life Science, Waltham, MA), followed by exposure to autoradiography films.
2.6. Analysis of intercellular adhesion molecule-1 (ICAM-1) expression by fluorescence-activated cell sorting (FACS)
Forty-eight hours after rAd. transduction, EC were treated with TNF (200 U/ml) overnight, then recovered and incubated for 30 min in FACS buffer (0.5% BSA in PBS), followed by 1 h incubation with FITC-conjugated mouse anti-human ICAM-1 IgG (eBioscence, Inc., San Diego, CA) or a control isotype IgG1 (BD Pharmingen, San Jose, CA) at room temperature in the dark. Cells were then washed, fixed in 2% paraformaldehyde, and evaluated for surface expression of ICAM-1 by flow cytometry (FACSCanto; BD, Franklin Lakes, NJ). Results were analyzed using FlowJo software (Tree Star Inc., Ashland, OR) and presented as percentage of median fluorescence intensity, considering βgal-transduced cells as control (100%).
2.7. Analysis of cell death
To induce cell death, we treated HUVEC and HDMEC, 48 h after rAd. transduction, with 100 nM staurosporine (Sigma-Aldrich, St Louis, MO) for 6 h. Cells were then collected, washed, fixed in cold 70% ethanol, and stained with propidium iodide buffer (200 µL PBS, 0.1% Triton X, 0.1 mmol/L EDTA, 50 µg/mL RNAse and 50 µg/mL propidium iodide) for 1 h in the dark. DNA content analysis was then conducted using a FACScan bench top model (Becton Dickinson, San Jose, CA) with Cellquest acquisition and analysis software (Becton Dickinson, San Jose, CA). Cells with a normal DNA content (at least diploid [2N]) were scored as viable, whereas cells with a hyplodiploid DNA content (lower than 2N) were scored as nonviable.
2.8. NF-κB luciferase reporter assay
BAEC were co-transfected with the different expression plasmids encoding for A1, its mutants or the empty control plasmid, pAC (0.7 µg/well), together with a NF-κB luciferase reporter plasmid (0.6 µg/well) comprising 4 NF-κB binding sites derived from the pig E-selectin promoter, as described [17]. Rous Sarcoma Virus (RSV)-β-galactosidase reporter plasmid (RSV-βgal, 0.3 µg/well) was co-transfected with the two other plasmids to correct for transfection efficiency. Transfection of BAEC was achieved using cationic liposome-mediated gene transfer (Lipofectamine, Gibco, Life Technologies, Grand Island, NY), as described [17]. BAEC were stimulated 48 h after transfection with TNF (200 U/mL), harvested 7 h later and assayed for β-galactosidase and luciferase activities, as described [17]. Luciferase activity was normalized for βgal by using the formula: luciferase activity/βgal activity × 1,000. Normalized luciferase activity is given in relative light units (RLU).
2.9. Statistical analysis
Statistical analysis was performed on Prism 5 for Mac (GraphPad software, Inc., La Jolla, CA, USA). Data were analyzed by one-way analysis of variance (ANOVA) followed by post-hoc Tukey test. Differences between groups were rated significant at a probability error (P) of less than 0.05. Results are presented as means ± standard error of mean (SEM).
3. Results
3.1. Generation of A1 deletion mutants
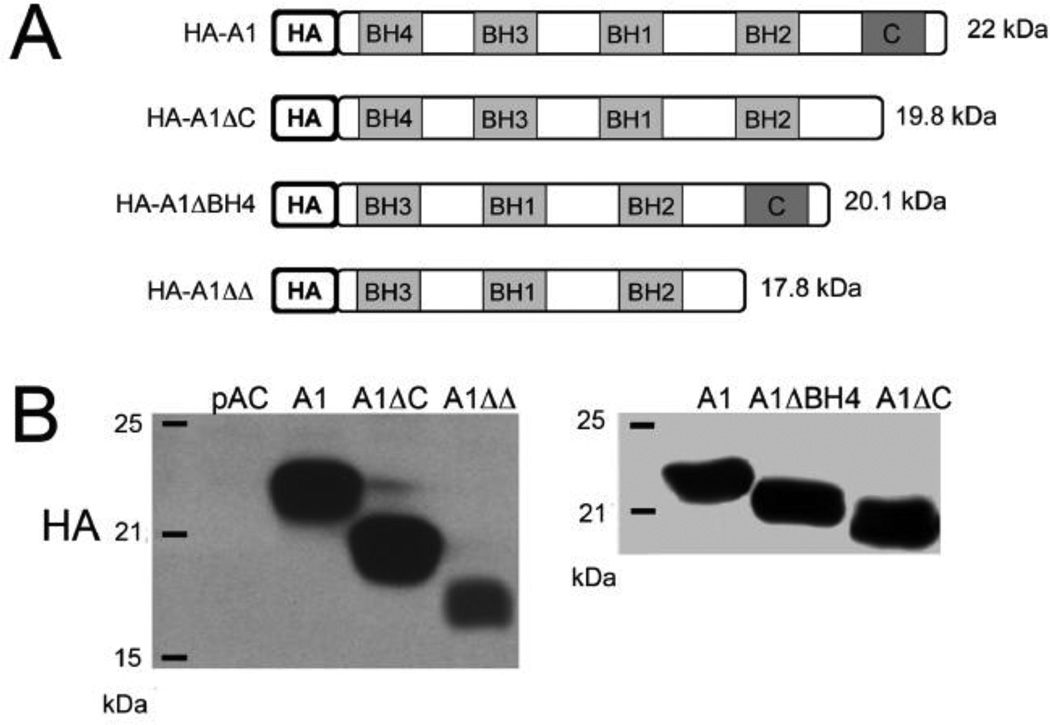
In order to perform structure/function analysis of A1 in EC, we generated A1 mutants with deletions of the C-terminal domain (pAC.A1ΔC), the BH4 domain (pAC.A1ΔBH4) or both (pAC.A1ΔΔ) (Fig. 1A). All A1 mutants were HA-tagged and cloned in the pAC expression plasmid, allowing for subsequent generation of recombinant adenoviruses (rAd.) [19]. All mutants were confirmed by DNA sequencing (Core Laboratory of Boston University). We subsequently transfected BAEC with 0.7 µg of full length A1 or the above-described A1-deletion mutant plasmids, and evaluated expression of the different transgenes by Western blot (WB) analysis using anti-HA antibody. Our data showed that all mutants were expressed and migrated at their expected molecular size (Fig. 1B). Maximal transgene expression occurred 48h after transfection (data not shown).
Fig. 1.
(A) Schematic representation of the sequences of HA-tagged full-length human A1 (22 kDa), A1ΔC (19.8 kDa) – A1 mutant with a deletion of the C-terminus domain, A1 BH4 (20.1 kDa) – A1 mutant with a deletion of the BH4 domain, A1 (17.8 kDa) – A1 mutant with a deletion of the C-terminus and the BH4 domains that were cloned in pAC plasmids. (B) Expression of A1 and A1 mutants in bovine aortic endothelial cells (BAEC) following their transfection with the respective plasmids, as demonstrated by Western blot analysis using anti-HA antibody. Molecular weight markers are labeled on the left side of the figures.
3.2. Subcellular localization of A1 and A1ΔC in human EC using recombinant adenovirus-mediated gene transfer
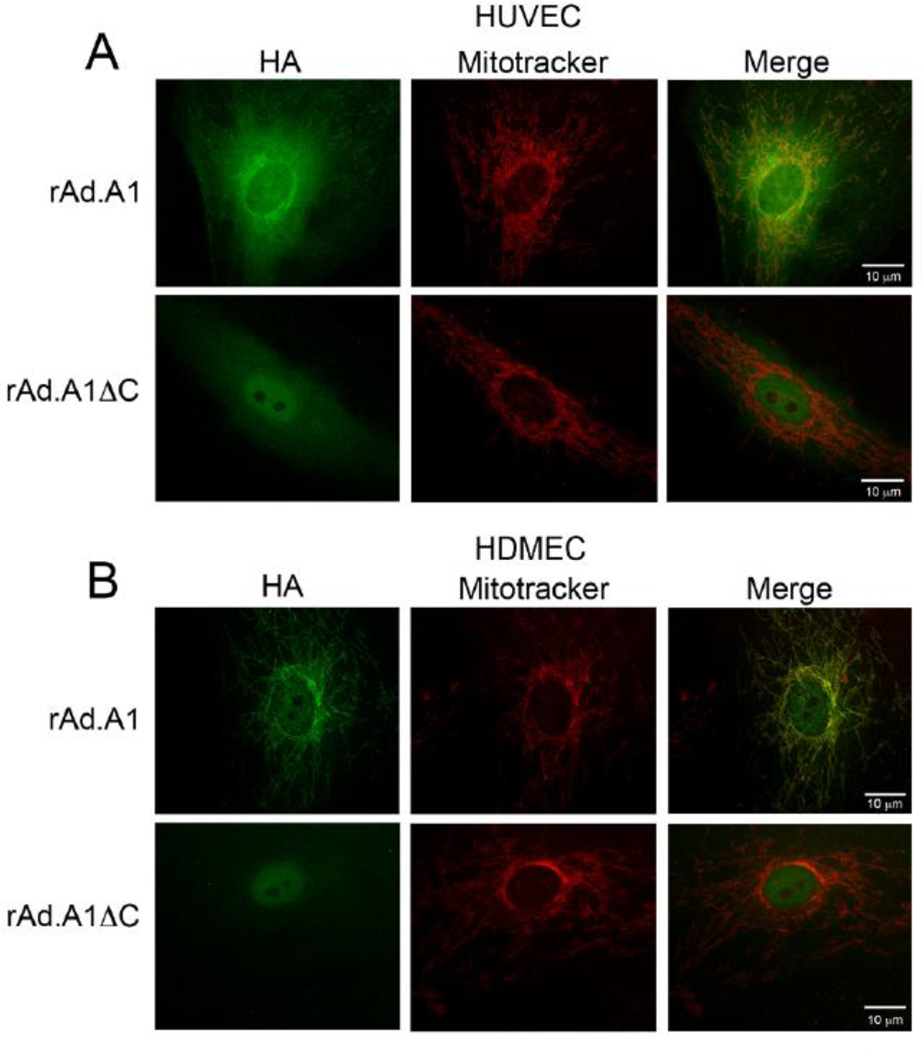
To evaluate the role of A1 and its mutant harboring a deletion in the C-terminal domain in human EC, we generated rAd. expressing A1 and A1ΔC. rAd-mediated gene transfer usually achieves significant expression of the transgene in over 95% of transduced EC, and therefore overcomes the limitation of inefficient transfection systems in these cells. Following verification of rAd.A1 and rAd.A1ΔC by sequencing, we evaluated the expression of these transgenes in human EC cultures by immunohistochemistry (IHC), using anti-HA antibody. Our results confirmed that the different transgenes were expressed in >95% of transduced EC (data not shown). Since subcellular localization of proteins is difficult by IHC, we performed co-immunofluorescence staining and showed that full-length A1 mostly co-localizes with mitotracker red in both HUVEC and HDMEC, indicating that it associated with mitochondria (Figs. 2A & 2B). This supports previous data reporting mitochondrial localization of A1 in different cell types [22, 23]. In contrast, A1ΔC did not co-localize with mitotracker red, instead it showed diffuse cytosolic distribution and some nuclear staining.
Fig. 2.
Subcellular localization of A1 and A1ΔC overexpressed transgenes in human EC. (A) HUVEC and (B) HDMEC transduced with rAd.A1 and rAd.A1ΔC were double-labeled with HA-FITC antibody and the mitochondrial probe Mitotracker Red and analyzed by immunofluorescence microscopy with overlay of the different fluorescence colors. Results show that A1, but not A1ΔC, co-localizes with Mitotracker Red, indicating that the C-terminal domain of A1 is required to anchor it to mitochondria. In contrast, A1ΔC fluorescence shows a cytosolic and nuclear pattern. Representative micrographs of 3 (HUVEC) and 2 (HDMEC) different experiments are shown. Original magnification 1000X.
3.3. A1ΔC, but not full length A1, exerts an anti-inflammatory effect in human EC
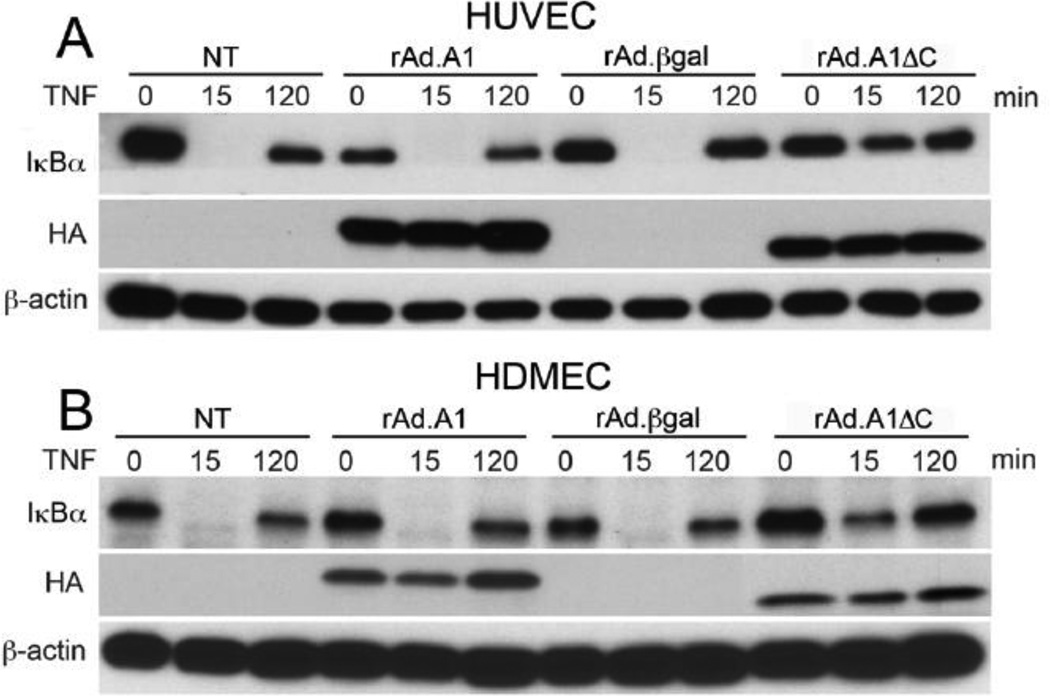
To evaluate the effect of A1 and A1ΔC on NF-κB activation in human EC cultures, we stimulated rAd.A1 and rAd.A1ΔC-transduced HUVEC and HDMEC with 200 U/mL of TNF for 15 min and 2 h, and used WB to check for IκBα degradation, a surrogate for NF-κB activation. Non-transduced (NT) and rAd.βgal-transduced EC served as controls. Overexpression of full length A1 in both HUVEC and HDMEC failed to inhibit TNFmediated IκBα degradation. However, overexpression of the A1ΔC mutant substantially reduced degradation of IκBα in these cells 15 min after addition of TNF (Fig. 3A & 3B). To ascertain that overexpression of A1ΔC was not simply delaying IκBαdegradation, we performed an extensive time course analysis of IκBα protein levels, ranging 15 to 90 min. following TNF stimulation, in HUVEC. Our data confirmed that overexpression of A1ΔC in EC inhibits, not simply delays TNF-mediated IκBα degradation, and even allows faster recovery of baseline IκBα protein levels (Supplementary Fig. 1).
Fig. 3.
Overexpression of A1ΔC but not full length A1 prevents TNF-induced IκBα degradation in HUVEC and HDMEC. Cell lysates from non-transduced (NT), rAd.A1, rAd.A1ΔC or rAd.βgal-transduced cells (A) HUVEC, and (B) HDMEC treated with TNF (200 U/ml) for 15 and 120 min were immunoblotted with anti-IκBα antibody. Results demonstrate that overexpression of A1 does not affect TNF-induced degradation and loss of IκBα in all EC tested. However, overexpression of A1ΔC inhibits TNF-induced IκBαdegradation in both HUVEC and HDMEC. Immunoblotting with anti-HA and anti-βactin antibodies was used to confirm transgene expression and correct for loading, respectively. Data shown are representative of 3 independent experiments for each endothelial cell type.
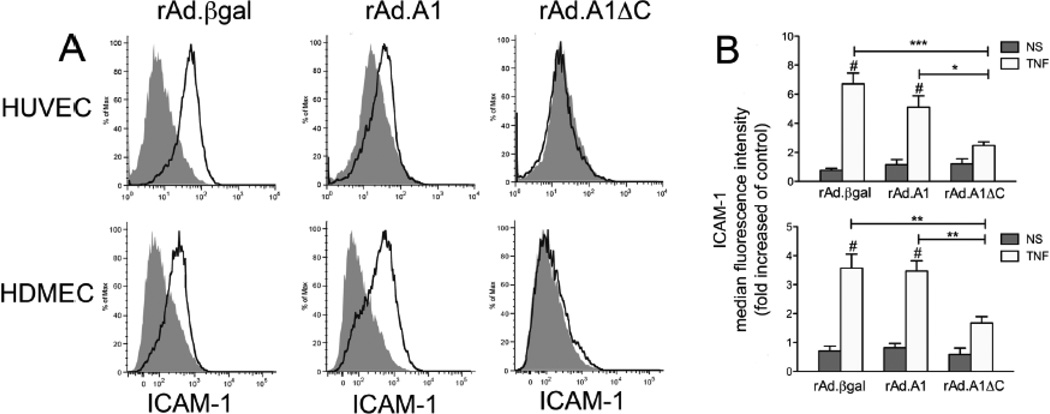
Downstream of IκBα degradation, we checked by flow cytometry for the expression of NF-κB-dependent intercellular adhesion molecule 1 (ICAM-1), 16 h following treatment with TNF. Our results indicated that overexpression of A1ΔC, but not full length A1, in HUVEC and HDMEC significantly inhibit TNF-induced upregulation of ICAM-1 (Fig. 4). Altogether, our data unveil a novel role for A1’s C-terminus in regulating the anti-inflammatory function of this protein in human EC, i.e. the C-terminal domain that anchors A1 to the mitochondria interferes with its ability to inhibit NF-κB activation in human EC. These results contradict our previous data showing that full length human A1 inhibits TNF-mediated activation of a NF-κB luciferase reporter in BAEC [5]. Given this discrepancy, we wanted to confirm our previous data using the same BAEC based co-transfection assay and evaluate in this system the effect of A1ΔC, and also A1ΔBH4, and A1ΔΔ . Accordingly, we co-transfected BAEC with 0.7 µg of pAC empty plasmid or pAC encoding full length A1 or A1 deletion mutants (A1ΔC, A1ΔBH4, or A1ΔΔ, together with a NF-κB luciferase reporter (0.7 µg) and a RSV-βgal reporter (0.3 µg), used to correct for transfection efficiency. Forty-eight hours following transfection, we checked for luciferase and β-gal activities in control non-treated cells, and 7 h after treatment with 200 U/ml of TNF. Our results confirmed that overexpression of full-length A1 inhibits TNF-induced activation of the NF-κB reporter in BAEC (Supplementary Fig. 2A). Inhibition of TNF-induced activation of the NF-κB reporter was maintained in BAEC transfected with A1ΔC, indicating that this mutant has similar propensity to inhibit NF-κB reporter activity in bovine EC and IκBα degradation in human EC transduced with rAd. A1ΔC (Supplementary Fig. 2A). In contrast, deletion of the BH4 domain from either full length A1 or the A1ΔC mutant abolished A1’s NF-κB inhibitory effect (Supplementary Fig. 2A), which implies that, akin to Bcl-2, the BH4 domain is essential for A1’s anti-inflammatory function, at least in reporter assay based evaluation of NF-κB activity in BAEC. To further explore the mechanism(s) behind the discrepancies in A1’s ability to inhibit NF-κB luciferase reporter in BAEC transfected with an A1 plasmid, but not IκBα degradation/NF-κB activation in human EC transduced with rAd.A1, we used the same rAd.based transduction protocol to overexpress A1 and A1ΔC in BAEC, then checked for TNF-induced IκBα degradation. Interestingly, our results fully reproduced our data in human primary EC. i.e. overexpression A1ΔC but not A1 significantly precluded TNF-induced IκBα degradation (Supplementary Fig. 2B). We excluded differences in expression levels of the A1 transgene as the reason for discrepancy between transduced (rAd.A1) and transfected (A1 plasmid) in terms of NF-κB activation. Indeed, either expression method resulted in comparable levels of HA-tagged A1 protein as shown by Western blot analysis (Fig. 1 and supplementary Fig. 2B).
Fig. 4.
Overexpression of A1ΔC but not full length A1 prevents TNF-induced ICAM-1 upregulation in HUVEC and HDMEC. (A) FACS analysis of ICAM-1 surface expression, rAd.A1, rAd.A1ΔC, or rAd.βgal-transduced HUVEC, and HDMEC 16 h following treatment with TNF (200 U/ml). Our results demonstrate that overexpression of A1 does not affect TNF-induced upregulation of ICAM-1 in all EC tested. However, overexpression of A1ΔC significantly inhibits TNF-induced ICAM-1 upregulation in HDMEC and HUVEC. Histograms shown are representative of 3 different experiments. Gray shaded areas represent baseline ICAM-1 expression in non-stimulated (NS) EC, while empty black-lined areas indicate the shift in ICAM-1 surface expression following TNF stimulation. (B) Bar charts depicting fold increase of median fluorescence intensity SEM ± of 3 experiments performed. *p<0.05, **p<0.01, ***p<0.001 indicate significance among different rAd. treatment groups, whereas # p 0.001 indicate significance between NS and TNF-treated cells within the same experimental group.
In gaining insight into the physiologic role of A1 in EC’s inflammatory response, we performed loss of function experiments by transducing HUVEC with A1-specific siRNA, achieving >80% knockdown of A1 mRNA (Supplementary Fig. 3A). A1 knockdown in HUVEC was particularly significant when compared to cells transfected with control AllStar negative siRNA that showed a 2 to 3 fold increase in A1 mRNA, a likely consequence of the usual toxicity of the transfection reagents triggering a survival response. None of the commercially available A1 antibodies was able to detect endogenous A1 protein levels in HUVEC. However, given the documented short half-life of the A1 protein (15 min), we believe that A1’s protein knockdown should be at least commensurate to its mRNA knockdown, even if we can not formally prove it [24]. On the sole basis of mRNA levels, A1mRNA knockdown did not impact IκBα degradation following TNF (Supplementary Fig. 3B), which could suggest that A1 is not a physiologic regulator of NF-κB activation. This contrasts with knockdown of the NF-κB inhibitory gene A20 that causes prolonged NF-κB activation with delayed recovery of IκBα protein levels in different cell types [25], including EC (data not shown). These data support our gain of function experiments in rAd.A1 transduced EC, as they indicate that full length A1 does not have a significant physiologic role in down-regulating NF-κB activation in these cells.
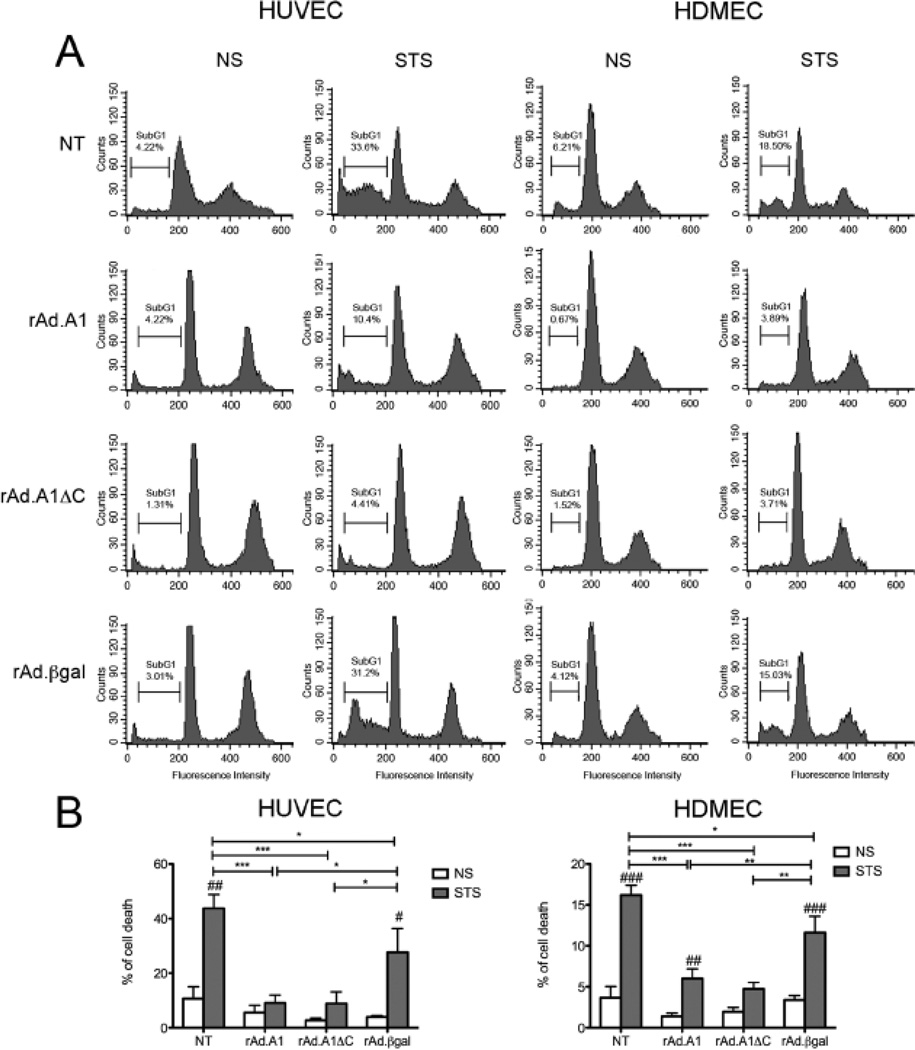
3.4. Full length A1 and A1ΔC equally protect human EC from Staurosporine-induced cell death
Classically, A1 is described as an anti-apoptotic Bcl-2 family member. However, several reports challenge this statement, demonstrating that A1ΔCould be anti-apoptotic, or proapoptotic, depending on cell type and apoptotic stimulus [26–28]. In a microvascular endothelial cell line, A1 predominantly localized at the mitochondrial membrane, where it functioned to inhibit Cytochrome c release and Caspase 9 activation, maintaining mitochondrial transmembrane potential and promoting temporary survival of these cells in response to TNF [22]. To examine the contribution of A1 mitochondrial anchoring to its cytoprotective properties in human EC derived from primary cell cultures, we checked for the effect of A1 and A1ΔC overexpression in HUVEC and HDMEC on STS-induced cell death. Our results showed that both A1 and A1ΔC significantly protect HUVEC and HDMEC from STS-induced cell death, compared to control NT and rAd.βgal transduced EC (Fig. 5; n=3). These results indicate that localization of A1 to mitochondria is not required for its cytoprotective function in primary human EC cultures. These results are completely concordant with our data in BAEC transfected with A1 and A1 mutants expression plasmids (Supplementary Fig. 2C).
Fig. 5.
Full-length A1 and A1ΔC mutant protect HUVEC and HDMEC from cell death induced by staurosporine (STS). Non-transduced (NT), rAd.A1, rAd.A1ΔC, and rAd.βgaltransduced (A) HUVEC and HDMEC were treated with STS (100 nM) for 6 h, then labeled with propidium iodide. Apoptosis was evaluated by FACS analysis of DNA content. Data shown are representative of 3 independent experiments. The percentage of cells undergoing cell death, as evidenced by DNA<2N (subG1 phase) is reported. (B) Bar chart representation of mean percentage of cell death ± SEM of the 3 experiments performed. *p<0.05, **p<0.01, ***p<0.001 indicate significance among different treatment groups, i.e. non tranduced (NT) EC and EC transduced with different rAd. #p<0.05, ##p<0.01, ###p<0.001 indicate significance between non stimulated (NS) and STS-treated HUVEC and HDMEC within the same experimental group.
4. Discussion
A1/Bfl-1 is classically described as a TNF-inducible, NF-κB-dependent, anti-apoptotic Bcl-2 family member in EC [4, 5, 22]. We had previously shown that A1 also exerts anti-inflammatory effects in BAEC by inhibiting NF-κB activation [5]. In this study, we performed a structure-function analysis of A1 in order to delineate the role of its C-terminal domain upon its NF-κB inhibitory and cytoprotective effects in EC, and extended this study to human EC.
Using co-immunofluorescence staining, we showed that full length A1 mostly localizes to mitochondria, which confirms that the amphipathic C-terminus of A1 drives its localization to this organelle, tempering some of the controversy about A1’s cellular localization [11]. This result is consistent with data reporting that A1 and other anti-apoptotic Bcl-2 family members preferentially localize to mitochondria, where they directly (Bcl-2, Bcl-xL), or indirectly (A1/Bfl-1 through binding to Bid and tBid) [29] inhibit the activation of pro-apoptotic Bcl-2 family members (Bax) [30, 31].
In contrast, A1ΔC was excluded from mitochondria, showing a nuclear and cytosolic pattern, similar to that described in MCF-7 cells overexpressing a Bfl-1 deletion mutant lacking the C-terminal domain [29]. While we expected cytosolic expression for a protein devoid of its C-terminus, we questioned the nuclear staining of A1ΔC. By Western blot analysis, we demonstrated that only a negligible fraction of A1ΔC is detected within the pool of soluble nuclear proteins, suggesting that A1ΔC was mainly cytoplasmic. Even though, a fraction of the A1ΔC protein may still be attached to the inner nuclear membrane, which would explain the peri-nuclear distribution that we detect by IF (Supplementary Fig. 4A). In fact, deletion of the A1ΔC-terminus should not result in a nuclear localization sequence, contrasting with the short splice variant of human A1 (Bfl-1S), that in addition to lacking the C-terminus domain, incorporates a 56-bp exon-II into its sequence that creates a nuclear localization sequence [32]. Using Bfl-1 S specific primers, we demonstrated the presence of mRNA encoding for this short variant of A1 in HUVEC, although its levels were less abundant at baseline than that of full length A1, but alike A1 increased after addition of TNF (Supplementary Fig. 4B).
Next, we evaluated the effects of A1 and A1ΔC on NF-κB activation in human EC, using TNF-mediated IκBα degradation and ICAM-1 upregulation as surrogates. Surprisingly, full length A1 did not maintain its NF-κB inhibitory function in human EC, similar to our data in human RPTEC [6]. Furthermore, A1 silencing, at least as determined by mRNA knockdown, did not amplify TNF-induced pro-inflammatory responses of EC, suggesting that full length A1 does not influence inflammatory responses in EC. This is consistent with the absence of a pro-inflammatory phenotype in mice with a knockdown of the mouse A1a isoform, as these mice only suffer from enhanced neutrophil apoptosis [33]. In striking contrast, A1ΔC showed an anti-inflammatory function in HUVEC and HDMEC, similar to what we had reported in HUVEC transduced with a rAd.Bcl-2 adenovirus lacking its trans-membrane domain [8, 34]. Inhibition of NF-κB signaling only happens when A1 looses its amphipathic C-terminal domain and/or gets freed from the mitochondria. Additional experiments are underway to test this hypothesis. Intriguingly, these results partially disagree with our NF-κB reporter assays in BAEC, where full length A1 and A1ΔC were as effective in inhibiting TNF-induced NF-κB reporter activity. Discrepancy between results obtained in BAEC and human EC were not due to differences in the expression level of the transgene but are likely related to differences in experimental systems used (reporter assays vs. direct probing of components of the NF-κB signaling pathway in rAd. tranduced EC), since rAd.A1 and rAd.A1ΔC transduced BAEC gave similar results to what we reported in primary EC (Supplementary Figure 2B ).
On the basis of NF-κB reporter assays, we also have evidence that the BH4 domain of A1/Bfl-1 is responsible for its NF-κB inhibitory effect. This result agrees with what we had already described for the prototypic Bcl-2 and Bcl-xL family members [8]. Future work in the laboratory aims to confirm this finding in human EC, and in systems evaluating the EC’s own NF-κB machinery.
In addition, we showed that full-length A1 protected EC from cell death triggered by the pan-protein kinase inhibitor, staurosporine. This added to previously published data showing that A1 protects EC from CHX/TNF-induced apoptosis [22, 35] emphasizes a broader anti-apoptotic effect of this protein in EC. This result is important because the cytoprotective effect of A1 is far from ubiquitous. Indeed, while A1 maintains its anti-apoptotic function in skeletal and cardiac myocytes and in several cancer cells [36–38], it promotes TNF-induced apoptosis of spinal cord motor neurons [28], B cells [26] and even 293T cells [27]. Surprisingly “anti-inflammatory” A1ΔC equally protected EC from cell death, proving that mitochondrial localization is not required for the pro-survival function of A1 in EC. This result is analogous to data showing that C-terminus deletion mutants of human A1/Bfl-1 (A1ΔC and Δα9-Bfl-1) were still able to protect FL5.12 pro-B cells, and NIH-3T3 cells from STS and serum-starvation-induced apoptosis, respectively [11, 26]. However, this result contrasts with results obtained in the murine B cell lymphoma cell line, WEHI 231, where deletion or mutation of the C-terminus of A1 eliminated its ability to protect these cells from etoposide-induced cell death [24]. All these evidence stress cell type specific functions of full-length A1/Bfl-1 and its C-terminus-deletion mutant.
We, and others have shown that the anti-apoptotic function of A1 and prototypic Bcl-2 family members in EC and other cell types relies on intact BH1, BH2, and especially BH4 domains [8, 12, 14, 15]. In the case of A1/Bfl-1, an intact BH3 domain that associates with pro-apoptotic Bcl-2 family members, Bid and Bim, is also required [23, 24]. We established in BAEC transfected to express full length A1 or more importantly an A1ΔC mutant where the BH4 domain was deleted (A1ΔBH4, and A1ΔCΔBH4), that the BH4 domain of A1 and A1ΔC was required for both their anti-inflammatory and anti-apoptotic functions. We presume that specific binding partners and/or functions associated with this domain support A1/A1ΔC cytoprotective function without necessarily requiring its anchoring to the mitochondrial membrane. We recognize that this assumption challenges previously established concepts, at least in EC. Hence, future studies are underway to confirm these results in human EC, check their validity on the cell own NF-κB machinery, and potentially discover their molecular basis.
In conclusion, our data exposes an important regulatory role for the amphipathic C-terminal domain of A1 in human EC. This domain is essential for A1 anchoring to the mitochondrion, but is not a pre-requisite for its cytoprotective function in EC. The C-terminal domain of A1, or alternatively mere anchoring of this protein at mitochondria interferes with its anti-inflammatory function in EC. This function only emerges when the C-terminus of A1/Bfl-1 is deleted, releasing it from the mitochondrion. Future work aims to determine whether this domain is ever cleaved under physiologic or pathologic conditions, in response to select EC activators, to transform A1 from a sole anti-apoptotic to a multi-functional anti-apoptotic and anti-inflammatory molecule. The fact that the C-terminal domain of A1 is also involved in its ubiquitination and protein turnover supports such an endeavor [24, 26] that could redefine A1’s role in modulating EC homeostasis and phenotype. Ultimately, A1-ΔC-based EC therapies have the potential to treat vascular inflammation while providing a survival advantage.
Supplementary Material
Highlights.
Deletion of the amphipathic C-terminal domain of A1/Bfl1 (A1ΔC) is performed.
A1ΔC frees A1 from mitochondrial anchorage.
Both full-length A1 and A1ΔC are cytoprotective in human endothelial cells.
rAd. mediated overexpression of A1ΔC but not A1 inhibits NF-κB in human EC.
Acknowledgments
This work was supported by NIH/NHLBI and NIH/NIDDK; Contract grant numbers: R01 HL080130, R01 DK063275 to CF; Contract grant sponsor: Juvenile Diabetes Research Foundation; Contract grant number: 1-2007-567 to CF. RPG and ER are the recipient of a fellowship award from the National Council for Scientific and Technological Development (CNPq), Brazil. CRP and CRL are the recipient of T32 NRSA grant T32 HL00734, and JMT is the recipient of a Summer Von Liebig Research Fellowship for Medical Students.
We would also like to acknowledge Mr. Alon Neidich and Dr. Ashley Rogers for their critical reading of the Manuscript.
ABBREVIATIONS
- A1ΔBH4
Deletion of the BH4 domain
- A1ΔC
mutant lacking the C-terminal domain of A1
- A1ΔΔ
Deletion of the C-terminal and BH4 domains
- ANOVA
analysis of variance
- BAEC
bovine aortic EC
- Bfl-1S
short form of A1/Bfl-1
- BH
Bcl-2 homology domains
- CHX
cycloheximide
- EBM-2
endothelial basal medium-2
- EC
endothelial cells
- ECL
enhanced chemiluminescence kit
- FACS
fluorescence-activated cell sorting
- HA
hemagglutinin A
- HDMEC
dermal microvascular EC
- HEK 293
Human embryonic 293 kidney cell line
- HUVEC
human umbilical vein EC
- ICAM-1
intercellular adhesion molecule-1
- IHC
immunohistochemistry
- MOI
multiplicity of infection
- NS
non-stimulated
- NT
non-transduced
- P
probability error
- pAC.A1ΔBH4
plasmid encoding A1 mutant with deletion of the BH4 domain
- pAC.A1ΔC
plasmid encoding A1 mutant with deletion of the C-terminal domain
- pAC
pAC.CMV-pLpASR+ plasmid
- pCA.A1ΔΔ
plasmid encoding A1 mutant with deletion of the C-terminal and BH4 domains
- rAd.A1
Recombinant adenovirus human A1
- rAd.A1ΔC
Recombinant adenovirus A1ΔC
- rAd.βgal
recombinant adenovirus β-galactosidase
- RLU
relative light units
- RPTEC
renal proximal tubular epithelial cells
- RSV
Rous Sarcoma Virus
- RSV-βgal
Rous Sarcoma Virus-β-galactosidase
- RT-PCR
reverse transcription polymerase chain reaction
- SEM
standard error of mean
- STS
staurosporine
- TNF
tumor necrosis factor
- VEGF
Vascular Endothelial Growth Factor
- WB
Western blot
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Lin EY, Orlofsky A, Berger MS, Prystowsky MB. Characterization of A1, a novel hemopoietic-specific early-response gene with sequence similarity to bcl-2. J Immunol. 1993;151:1979–1988. [PubMed] [Google Scholar]
- 2.Choi SS, Park IC, Yun JW, Sung YC, Hong SI, Shin HS. A novel Bcl-2 related gene, Bfl-1, is overexpressed in stomach cancer and preferentially expressed in bone marrow. Oncogene. 1995;11:1693–1698. [PubMed] [Google Scholar]
- 3.Jourdan M, Reme T, Goldschmidt H, Fiol G, Pantesco V, De Vos J, Rossi JF, Hose D, Klein B. Gene expression of anti- and pro-apoptotic proteins in malignant and normal plasma cells. Br J Haematol. 2009;145:45–58. doi: 10.1111/j.1365-2141.2008.07562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karsan A, Yee E, Kaushansky K, Harlan JM. Cloning of human Bcl-2 homologue: inflammatory cytokines induce human A1 in cultured endothelial cells. Blood. 1996;87:3089–3096. [PubMed] [Google Scholar]
- 5.Stroka DM, Badrichani AZ, Bach FH, Ferran C. Overexpression of A1, an NF-kappaB- inducible anti-apoptotic bcl gene, inhibits endothelial cell activation. Blood. 1999;93:3803–3810. [PubMed] [Google Scholar]
- 6.Kunter U, Daniel S, Arvelo MB, Choi J, Shukri T, Patel VI, Longo CR, Scali ST, Shrikhande G, Rocha E, Czismadia E, Mottley C, Grey ST, Floege J, Ferran C. Combined expression of A1 and A20 achieves optimal protection of renal proximal tubular epithelial cells. Kidney Int. 2005;68:1520–1532. doi: 10.1111/j.1523-1755.2005.00564.x. [DOI] [PubMed] [Google Scholar]
- 7.Sarkar SA, Kutlu B, Velmurugan K, Kizaka-Kondoh S, Lee CE, Wong R, Valentine A, Davidson HW, Hutton JC, Pugazhenthi S. Cytokine-mediated induction of anti-apoptotic genes that are linked to nuclear factor kappa-B (NF-kappaB) signalling in human islets and in a mouse beta cell line. Diabetologia. 2009;52:1092–1101. doi: 10.1007/s00125-009-1331-x. [DOI] [PubMed] [Google Scholar]
- 8.Badrichani AZ, Stroka DM, Bilbao G, Curiel DT, Bach FH, Ferran C. Bcl-2 and Bcl-XL serve an anti-inflammatory function in endothelial cells through inhibition of NF-kappaB. J Clin Invest. 1999;103:543–553. doi: 10.1172/JCI2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pober JS, Sessa WC. Evolving functions of endothelial cells in inflammation. Nature Reviews Immunology. 2007;7:803–815. doi: 10.1038/nri2171. [DOI] [PubMed] [Google Scholar]
- 10.Bach FH, Hancock WW, Ferran C. Protective genes expressed in endothelial cells: a regulatory response to injury. Immunol Today. 1997;18:483–486. doi: 10.1016/s0167-5699(97)01129-8. [DOI] [PubMed] [Google Scholar]
- 11.Brien G, Debaud AL, Robert X, Oliver L, Trescol-Biemont MC, Cauquil N, Geneste O, Aghajari N, Vallette FM, Haser R, Bonnefoy-Berard N. C-terminal residues regulate localization and function of the antiapoptotic protein Bfl-1. J Biol Chem. 2009;284:30257–30263. doi: 10.1074/jbc.M109.040824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Sa-Eipper C, Chinnadurai G. Functional dissection of Bfl-1, a Bcl-2 homolog: anti-apoptosis, oncogene-cooperation and cell proliferation activities. Oncogene. 1998;16:3105–3114. doi: 10.1038/sj.onc.1201851. [DOI] [PubMed] [Google Scholar]
- 13.Herman M, Nyman T, Welin M, Lehtio L, Flodin S, Tresaugues L, Kotenyova T, Flores A, Nordlund P. Completing the family portrait of the anti-apoptotic Bcl-2 proteins: Crystal structure of human Bfl-1 in complex with Bim. FEBS Letters. 2008;582:3590–3594. doi: 10.1016/j.febslet.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 14.Hirotani M, Zhang Y, Fujita N, Naito M, Tsuruo T. NH2-terminal BH4 domain of Bcl-2 is functional for heterodimerization with Bax and inhibition of apoptosis. J. Biol. Chem. 1999;274:20415–20420. doi: 10.1074/jbc.274.29.20415. [DOI] [PubMed] [Google Scholar]
- 15.Huang DCS, Adams JM, Cory S. The conserved N-terminal BH4 domain of BCl-2 homologues is essential for inhibition of apoptosis and interaction with CED-4. EMBO J. 1998;17:1029–1039. doi: 10.1093/emboj/17.4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindsay J, Esposti MD, Gilmore AP. Bcl-2 proteins and mitochondria—Specificity in membrane targeting for death. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2010 doi: 10.1016/j.bbamcr.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 17.Cooper JT, Stroka DM, Brostjan C, Palmetshofer A, Bach FH, Ferran C. A20 blocks endothelial cell activation through a NF-κB-dependent mechanism. J. Biol. Chem. 1996;271:18068–18073. doi: 10.1074/jbc.271.30.18068. [DOI] [PubMed] [Google Scholar]
- 18.Richard L, Velasco P, Detmar M. A simple immunomagnetic protocol for the selective isolation and long-term culture of human dermal microvascular endothelial cells. Exp Cell Res. 1998;240:1–6. doi: 10.1006/excr.1998.3936. [DOI] [PubMed] [Google Scholar]
- 19.Ferran C, Stroka DM, Badrichani AZ, Cooper JT, Wrighton CJ, Soares M, Grey ST, Bach FH. A20 inhibits NF-kappaB activation in endothelial cells without sensitizing to tumor necrosis factor-mediated apoptosis. Blood. 1998;91:2249–2258. [PubMed] [Google Scholar]
- 20.Aiyar A, Xiang Y, Leis J. Site-directed mutagenesis using overlap extension PCR. Methods Mol Biol. 1996;57:177–191. doi: 10.1385/0-89603-332-5:177. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Duriez PJ, Wong F, Dorovini-Zis K, Shahidi R, Karsan A. A1 functions at the mitochondria to delay endothelial apoptosis in response to tumor necrosis factor. J Biol Chem. 2000;275:18099–18107. doi: 10.1074/jbc.M908925199. [DOI] [PubMed] [Google Scholar]
- 23.Werner AB. Bcl-2 Family Member Bfl-1/A1 Sequesters Truncated Bid to Inhibit Its Collaboration with Pro-apoptotic Bak or Bax. Journal of Biological Chemistry. 2002;277:22781–22788. doi: 10.1074/jbc.M201469200. [DOI] [PubMed] [Google Scholar]
- 24.Herold MJ. The Stability and Anti-apoptotic Function of A1 Are Controlled by Its C Terminus. Journal of Biological Chemistry. 2006;281:13663–13671. doi: 10.1074/jbc.M600266200. [DOI] [PubMed] [Google Scholar]
- 25.Lee EG, Boone DL, Chai S, Libby SL, Chein J, Lodolce JP, Ma A. Failure to regulate TNF-induced NF-κB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kucharczak JF, Simmons MJ, Duckett CS, Gelinas C. Constitutive proteasomemediated turnover of Bfl-1/A1 and its processing in response to TNF receptor activation in FL5.12 pro-B cells convert it into a prodeath factor. Cell Death Differ. 2005;12:1225–1239. doi: 10.1038/sj.cdd.4401684. [DOI] [PubMed] [Google Scholar]
- 27.Yang WS, Ko JK, Park SO, Choi HY, Kim YN, Kim CW. C-terminal region of Bfl-1 induces cell death that accompanies caspase activation when fused with GFP. J Cell Biochem. 2005;94:1234–1247. doi: 10.1002/jcb.20381. [DOI] [PubMed] [Google Scholar]
- 28.Crosio C, Casciati A, Iaccarino C, Rotilio G, Carrì MT. Bcl2a1 serves as a switch in death of motor neurons in amyotrophic lateral sclerosis. Cell Death and Differentiation. 2006;13:2150–2153. doi: 10.1038/sj.cdd.4401943. [DOI] [PubMed] [Google Scholar]
- 29.Simmons MJ, Fan G, Zong WX, Degenhardt K, White E, Gélinas C. Bfl-1/A1 functions, similar to Mcl-1, as a selective tBid and Bak antagonist. Oncogene. 2007;27:1421–1428. doi: 10.1038/sj.onc.1210771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antonsson B, Conti F, Ciavatta AM, Montessuit S, Lewis S, Martinou I, Bernasconi L, Bernard A, Mermod JJ, Mazzei G, Maundrell K, Gambale F, Sadoul R, Martinou JC. Inhibition of Bax channel-forming activity by Bcl-2. Science. 1997:370. doi: 10.1126/science.277.5324.370. [DOI] [PubMed] [Google Scholar]
- 31.Boise LH, Gonzalez-Garcia M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nunez G, Thompson CB. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 32.Ko JK, Lee MJ, Cho SH, Cho JA, Lee BY, Koh JS, Lee SS, Shim YH, Kim CW. Bfl-1S, a novel alternative splice variant of Bfl-1, localizes in the nucleus via its C-terminus and prevents cell death. Oncogene. 2003;22:2457–2465. doi: 10.1038/sj.onc.1206274. [DOI] [PubMed] [Google Scholar]
- 33.Hamasaki A, Sendo F, Nakayama K, Ishida N, Negishi I, Hatakeyama S. Accelerated neutrophil apoptosis in mice lacking A1-a, a subtype of the bcl-2-related A1 gene. The Journal of experimental medicine. 1998;188:1985–1992. doi: 10.1084/jem.188.11.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bilbao G, Contreras JL, Mikheeva G, Krasnykh V, Eckhoff DE, Thomas FT, Thomas J, Curiel DT. Genetic cytoprotection of human endothelial cells during preservation time with an adenoviral vector encoding the anti-apoptotic human Bcl-2 gene. Transplantation proceedings. 1999;31:1012–1015. doi: 10.1016/s0041-1345(98)01880-6. [DOI] [PubMed] [Google Scholar]
- 35.Karsan A, Yee E, Harlan JM. Endothelial cell death induced by tumor necrosis factor-alpha is inhibited by the Bcl-2 family member, A1. J Biol Chem. 1996;271:27201–27204. doi: 10.1074/jbc.271.44.27201. [DOI] [PubMed] [Google Scholar]
- 36.Iwata A, Morgan-Stevenson V, Schwartz B, Liu L, Tupper J, Zhu X, Harlan J, Winn R. Extracellular BCL2 proteins are danger-associated molecular patterns that reduce tissue damage in murine models of ischemia-reperfusion injury. PLoS One. 2010;5:e9103. doi: 10.1371/journal.pone.0009103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jenal M, Batliner J, Reddy VA, Haferlach T, Tobler A, Fey MF, Torbett BE, Tschan MP. The anti-apoptotic gene BCL2A1 is a novel transcriptional target of PU.1. Leukemia. 2010;24:1073–1076. doi: 10.1038/leu.2010.26. [DOI] [PubMed] [Google Scholar]
- 38.Olsson A, Norberg M, ökvist A, Derkow K, Choudhury A, Tobin G, Celsing F, österborg A, Rosenquist R, Jondal M, Osorio LM. Upregulation of bfl-1 is a potential mechanism of chemoresistance in B-cell chronic lymphocytic leukaemia. British Journal of Cancer. 2007;97:769–777. doi: 10.1038/sj.bjc.6603951. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.