Abstract
Promiscuity of pattern recognition receptors, such as receptor for advanced glycation end products (RAGE), allows for a complex regulatory network controlling inflammation. Scavenging of RAGE ligands by soluble RAGE treatment is effective in reducing delayed-type hypersensitivity (DTH), even in RAGE−/− mice by 50% (p < 0.001). This has led to the hypothesis that molecules scavenged by soluble RAGE bind to receptors other than RAGE. This study identifies CD166/ALCAM (ALCAM) as a close structural and functional homolog of RAGE, and it shows that binding of S100B to CD166/ALCAM induces dose- and time-dependent expression of members of the NF-κB family in wild type (WT) and RAGE−/− mouse endothelial cells. Blocking CD166/ALCAM expression using small interfering RNA completely inhibited S100B-induced NF-κB activation in RAGE−/−, but not in WT cells. The in vivo significance of these observations was demonstrated by attenuation of DTH in WT and RAGE−/− animals pretreated with CD166/ALCAM small interfering RNA by 50% and 40%, respectively (p < 0.001). Experiments in ALCAM−/− animals displayed an only slight reduction of 16% in DTH, explained by compensatory reciprocal upregulation of RAGE in animals devoid of CD166/ALCAM, and vice versa. Consistently, ALCAM−/− mice, but not WT mice treated with RAGE small interfering RNA show a 35% reduction in DTH, and ALCAM−/− RAGE−/− double-knockout mice show a 27% reduction in DTH reaction. Thus, S100B is a proinflammatory cytokine bridging RAGE and CD166/ALCAM downstream effector mechanisms, both being compensatory upregulated after genetic deletion of its counterpart.
Introduction
Innate and adaptive immune responses underlying inflammation, though once thought of as segregated, are now considered overlapping, reinforcing, and closely linked on many levels. Consistent with this view, the innate immune system, protecting the organism against acute danger, and the adaptive immune system, initiating and exerting Ag-specific defense mechanisms, have multiple overlapping functions (1–4).
Various members of the pattern recognition receptor (PRR) family, such as TLRs, have been shown to provide a possible link between these two systems. For example, certain TLRs recognize bacterial cell wall components and viral RNA, thereby initiating a prompt inflammatory response (5, 6). Besides being crucial for an adequate innate host response to invading pathogens, TLRs also contribute to adaptive immunity by triggering the interaction between non-APCs and APCs (1). TLRs also bind danger-associated molecular patterns (DAMPs), such as S100 proteins and HMGB-1, released by cells upon cellular stress and necrosis (7–11).
These observations indicated that TLRs share a common feature with another PRR, the receptor for advanced glycation end products (RAGE), known to bind members of the S100 family (12, 13) and HMGB-1 (14–16). RAGE is a transmembrane receptor constitutively expressed at low levels in many tissues and is markedly upregulated by inflammatory mediators. RAGE engagement by ligands propagates inflammation associated with acute and chronic metabolic (17–19), inflammatory (20, 21), and malignant (22–24) processes. Activation of RAGE through its ligands leads to sustained activation of NF-κB (25, 26).
Although important in the innate immune system, RAGE was originally considered as noncontributory to delayed-type hypersensitivity and adaptive immunity (27). The role of RAGE in innate immunity (16, 27, 28) was associated with NF-κB activation via subunits p50/p65. Meanwhile, various aspects of a role of RAGE in adaptive immune processes have been shown (29, 30). The unlikely simplicity of this view was further underscored by the observation that although delayed-type hypersensitivity (DTH) is not impaired in RAGE−/− mice, soluble RAGE (sRAGE), functioning as a decoy receptor sequestering RAGE ligands and preventing them from interacting with cellular RAGE, ameliorates DTH in wild type (WT) mice as well as in RAGE−/− mice (27). These observations suggest that a receptor distinct from RAGE, but one that also recognizes DAMPs, presumably contributes to inflammation in DTH, the latter highly dependent on activation of the NF-κB subunit p50. Therefore, the current study was initiated to identify a RAGE ligand interacting with a receptor distinct from RAGE and relevant for DTH.
Materials and Methods
Mouse models
The RAGE targeting construct and the generation of RAGE−/− mice have been described in detail elsewhere (27). Animals used in the experiments described in this study were from F2 or F5 backcrosses onto the C57BL/6 genetic background. ALCAM−/− mice were purchased from the Jackson Laboratory in the C57BL/6 genetic background. ALCAM−/− RAGE−/− double knockout (DKO) mice were generated by cross-breeding ALCAM−/− and RAGE−/− mice (Supplemental Fig. 1). Mice were housed individually with a 12-h light-dark cycle and were given free access to food and water. All procedures in this study were approved by the Animal Care and Use Committees at the Regierungspräsidium Tübingen and Karlsruhe, Germany.
Model of DTH
Two- to three-month-old mice were sensitized by s.c. injection over the left inguinal lymph node with 100 μl of an emulsion containing 25 mg/ml methylated BSA (mBSA; Sigma-Aldrich), 0.9% NaCl, 50 mg/ml dextran (Sigma-Aldrich), and 50% IFA (Sigma-Aldrich). After 3 wk, the left plantar hind paw was injected s.c. with 50 μl of 0.4 mg/ml mBSA in 0.9% NaCl (control groups, 50 μl of 0.4 mg/ml OVA). In transfection experiments, small interfering RNA (siRNA) against CD166/ALCAM (ALCAM siRNA; Santa Cruz Biotechnology, cat. no. sc-43023) or RAGE (RAGE siRNA; Santa Cruz Biotechnology, cat. no sc-36375) dissolved in DOTAP (Roche, cat. no. 11811177001) to a final volume of 150 μl was used (1.5 μg siRNA per gram of body weight). The solution was incubated (15 min, room temperature [rt]) before injection. Control animals received scrambled siRNA devoid of function (Santa Cruz Biotechnology). siRNA was injected into the tail vein 3 d and 1 d prior to induction of DTH. Twenty-four hours after mBSA injection, mice were assigned clinical inflammation score by two investigators blinded to genotype of mouse as follows: (1) absence of inflammation, (2) slight rubor and edema, (3) moderate rubor and edema with skin wrinkles, (4) severe rubor and edema without skin wrinkles, and (5) severe rubor and edema with toe spreading (27). For intermediate conditions, half points were used when necessary. Mice not responding to DTH (clinical score ≤ 2; 10–15% of animals) were excluded from the analysis (data with no exclusion, see Supplemental Fig. 3 A–C). Mice were sacrificed, and feet were immediately quick-frozen in liquid nitrogen or fixed in 4% formalin and decalcified.
Identification of RAGE homologs
The area 1–250 of the RAGE protein was compared with the protein domain family (PRODOM) database using the BLAST algorithm (31). The alignment between RAGE_HUMAN 1-250 and CD166_HUMAN 95-221 was performed with the program GAP (32). Modeling and energy minimization of the RAGE protein domain using the CD166/ALCAM structure (PDB:1KJC) as template were performed as described previously (33). Because the CD166/ALCAM structure entry 1KJC comprises only the positions 28–133 of the protein, for the modeling a multiple alignment between RAGE_HUMAN 25-120, CD166_HUMAN 28-133, and 1KJC was generated using the program PRRN (34).
Cell culture
Mouse aortic endothelial cells (MAECs) or fibroblasts were maintained in RPMI 1640 medium containing 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (BioWhittaker, Walkersville, MD) and 20% FCS (Life Technologies, Dreieich, Germany) at 37°C and seeded 2–3 d before experiments.
Harvesting for cytoplasmic and nuclear proteins
Confluent MAECs or fibroblasts in T75 flasks (Nunc) were washed twice with cold PBS, harvested by scraping in ice-cold PBS, pelleted in 1.5-ml Eppendorf tubes (3000 rpm, 4°C, 5 min) and combined with 400 μl Buffer A (10 mM HEPES-KOH, pH 7.9, at 4°C, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.2 mM PMSF, and 0.6% NP-40). After vortexing (30 s), incubation on ice (30 min), and centrifugation (14,000 rpm, 4°C, 2 min), supernatant was quick-frozen in liquid nitrogen and stored at −80°C. The pellet was incubated with 110 μl Buffer B (25% glycerol, 20 mM HEPES-KOH, pH 7.9, at 4°C, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF, 2 mM benzamidine, and 5 mg/ml leupeptin), vortexed, incubated on ice (30 min) and centrifuged (14,000 rpm, 4°C, 5 min). The supernatant was stored as described above.
Harvesting for total protein lysates
Confluent MAEC or fibroblasts in T75 flasks were washed (cold PBS), harvested by scraping in RIPA buffer (150 mM NaCl, 50 mM Tris, 5 mM EDTA, 0.1% SDS, 1% Triton X-100, 0.5% sodium deoxycholate, 10 μg/ml leupeptin, 1 mM PMSF), incubated (30 min, 4°C, gentle shaking) and centrifuged (14,000 rpm, 15 min, 4°C). Supernatant was stored at −80°C. Frozen organs were crushed in liquid nitrogen, resuspended in 5× extraction buffer (20 mM HEPES buffer, pH7.9; 1 mM MgCl2; 5 mM EDTA; 1 mM PMSF; 1 μl per 100 μl HALT−proteinase-inhibitor cocktail; 0.5% TritonX100; 0.5% NP-40S 0.25% CHAPS, vortexed (30 s) and centrifuged (14,000 rpm, 30 min, 4°C), as described by Alhamdani et al. (35). The supernatant was stored at −80°C.
Western blotting
Fifteen micrograms of total protein extract, 20 μg cytoplasmic, or 10 μg nuclear extracts were separated on an 8% Tris gel (CD166/ALCAM) respectively on a 10% Nu-Page gel or 12% Tris gel (RAGE), electroblotted onto nitrocellulose-ECL membranes, and membranes were blocked with 5% milk solution for 1 h. Membranes were incubated with a primary Ab (anti-ALCAM [anti-mouse ALCAM Ab, AF1172, dilution 1:800 (R&D Systems)], anti-NF-κBp50 [Epitomics, Burlingame, CA], anti–NF-κBp65 and anti–NF-κBcRel [Rockland Immunochemicals, Gilbertsville, PA], anti–NF-κBRelB [Santa Cruz Biotechnology, cat. no. sc-28689; 1:1000 dilution], anti-RAGE [BioLogo, Kronshagen, Germany, cat. no. AGE001], anti-CD6 [R&D Systems, cat. no. AF750], or anti-S100B [Santa Cruz Biotechnology, cat. no. sc-28533]) for 1 h, washed in PBS with 0.05% Tween three times, and incubated with HRP-coupled secondary IgGs (Santa Cruz Biotechnology or Cell Signaling Technology, Beverly, MA) at a dilution of 1:2000 for 45 min at room temperature. Immunoreactive proteins were visualized on x-ray film using ECL-detection reagents (Amersham Bioscience, Little Chalfont, U.K.) and an exposure time of 3 min.
Immunoprecipitation
Two hundred micrograms of protein (RIPA buffer) in 1.5-ml Eppendorf tubes were increased in volume to 1 ml by adding immunoprecipitation (IP) buffer, incubated with 30 μl agarose beads (Protein A/G PLUS-Agarose; Santa Cruz Biotechnology, cat. no. sc-2003) for 30 min at 4°C, gently shaken, and centrifuged at 10,000 × g for 5 min at 4°C. The supernatant was incubated at 4°C for 1 h with 3 μg of the respective Ab against CD166/ALCAM (goat anti-mouse ALCAM; R&D Research, cat. no. AF 1172), RAGE (goat anti-human RAGE Ab; BioLogo, cat. no. AGE001) or S100B (goat anti-human S100B; R&D Research, cat. no. AF1820) gently shaken. Thirty microliters of protein A/G PLUS-Agarose was added, and probes were incubated for 16 h overnight at 4°C, gently shaken, and then centrifuged (10,000 × g, 10 min, 4°C). Pellets were washed three times with IP buffer (1200 rpm, 5 min, 4°C for each wash step) and once with final wash buffer, resuspended in 40 μl denaturing buffer, and incubated at 97°C for 5 min. After centrifugation (4000 rpm, 1 min, room temperature), 10 μl supernatant was separated on 10% SDS gel. After blotting onto a membrane, immune visualization was performed as described for Western blotting.
Far-Western blotting
Far-Western blotting was performed as described previously (36). Briefly, 20 μg cell lysate from RAGE−/− MAEC or 1.5 μg recombinant CD166/ALCAM protein (Recombinant Mouse ALCAM/CD166 Fc Chimera; R&D Systems, cat. no. 1172-AL) were separated on SDS gels and transferred to membranes. Proteins on membrane were denatured with AC buffer (100 mM NaCl, 20 mM Tris [pH 7.6], 0.5 mM EDTA, 10% glycerol, 0.1% Tween-20, 2% skim milk powder, and 1 mM DTT) containing guanidine-HCl 6M, renatured in AC-buffer (guanidine-HCl 3, 1, and 0.1 M), and incubated in only AC buffer overnight at 4°C. Membranes were blocked (5% milk solution, 1 h, room temperature). Incubation with 100 μg of S100B (S100B protein, bovine brain; Calbiochem, cat. no. 559290) in protein-binding buffer (100 mM NaCl, 20 mM Tris [pH 7.6], 10% glycerol, 0.1% Tween-20, 2% skim milk powder, 1 mM DTT, 4 mM CaCl2) was performed for 3 h at room temperature. EDTA was excluded because of calcium dependency of S100B binding, and 4 mM CaCl2 was included in all subsequent buffers. After washing four times with PBS with Tween 20, the membrane was probed with anti-S100B Ab (goat anti-human S100B; R&D Research, cat. no. AF1820) diluted 1:5000 in 3% milk overnight at 4°C. After washing in PBS with Tween 20, the membrane was probed with donkey–anti-goat secondary Ab (Santa Cruz Biotechnology, cat. no. SC-2020) diluted 1:2000 in 3% milk for 1 h. Immunoreactive proteins were visualized as described for Western blotting.
siRNA experiments in cell culture
For silencing or suppressing CD166/ALCAM expression in MAEC, predesigned siRNA for ALCAM (Santa Cruz Biotechnology) was transfected by electroporation (Microporator; Peqlab, Erlangen, Germany). Nonspecific siRNA served as a control. The efficiency of silencing was confirmed by Western blot and RT-PCR. Confluent cells were used for experiments 2 d after transfection.
EMSA
Nuclear extracts from MAEC or fibroblasts were assayed for transcription factor–binding activity using the NF-κB consensus sequence 5′-AGTTGAGGGGACTTTCCCAGGC-3′. Specificity of binding was ascertained by competition with a 160-fold molar excess of unlabeled consensus oligonucleotides, and protein-DNA complexes were separated from unbound DNA probe by electrophoresis through 5% native polyacrylamide gels containing 2.5% glycerol and 0.5× TBE. Gels were dried and exposed to x-ray film (Amersham Pharmacia, Freiburg, Germany) for 28–48 h at −80°C with intensifying screens. For supershift assays, appropriate Abs (same Abs used in Western blots) against NF-κB subunits were added as indicated in the figure and incubated on ice for 40 min. Next, they were separated on a 5% polyacrylamide gel for 6 h and processed as described above.
RT-PCR
RT-PCR was performed after transcription of 2 μg RNA into cDNA (3 μl oligo-dTs [Promega], 2 μl RNasin, 3 μl dNTPmix, 7 μl 5xAMV buffer, 1 μl reverse transcriptase); 1.5 μg cDNA was used as starting material in RT-PCR, and the signal was normalized using the house-keeping gene β-actin with the following conditions: murine β-actin, forward 5′- GCA GCT CCT TCG TTG CCG GT-3′; reverse 5′- GGG GCC ACA CGC AGC TCA TT-3′; 95°C, 180 s, 26 x (95°C, 30 s, 58°C [35S], 72°C 40 s), 72°C 180s, 4°C pause. Primers and conditions for CD166/ALCAM and RAGE were as follows: murine ALCAM, forward 5′-AGT CCA AGA GGA CCC AAG GT -3′; reverse 5′-GAC CAA GTC GGA GAC TGA GC -3′; 95°C 180s, 31 x (95°C 30s, 56°C 30s, 72°C 30s), 72°C 180s, 4°C pause; murine RAGE TR GFP forward 5′-ATG CTG GCC TTG GGG ATC CTG GGA GGC -3′; FL GFP reverse 5′-CGG TCC CCC GGC ACC ATT CTC TG-3′; 94°C 180s, 30 x (94°C 60s, 70°C 45s, 72°C 30s), 72°C 240s, 4°C pause. The PCR products were separated onto 1.7% agarose gels and visualized by ethidium bromide staining. Amplification of β-actin served as a control for sample loading and integrity. Reactions lacking cDNA and polymerase served as internal controls.
Histology
Feet were fixed in 4% formalin and decalcified, and 5-μm–thick sections were cut and stained with H&E according to standard protocols.
Bio-Plex assay
Snap-frozen feet samples were mechanically disrupted and treated with lysis solution (Bio-Rad). Protein concentration in lysates was determined using Bradford assay (Bio-Rad). Concentrations of the indicated chemokines were determined using bead-based multiplex protein array technology (Luminex) according to manufacturer’s instructions for protein multiplexing (Panomics Affymetrix). Concentrations for a particular sample were calculated by the Bio-Plex Manager 4.1.1 software based on the five-parameter logistic plot regression formula. The detection sensitivity of all analyses was between 2 pg/ml and 40 ng/ml.
Statistical analysis
Where indicated, values of experimental groups are given as means, with bars showing the SEM. The means of groups were compared by ANOVA using Student t test. A probability of p < 0.05 was considered statistically significant.
Results
An identical degree and time course of the inflammatory response after induction of DTH was observed in WT and RAGE−/− mice (Fig. 1A), as has been noted previously (27). However, sRAGE, which scavenges all ligands binding to the promiscuous pattern recognition receptor RAGE, reduced the adaptive immune response and foot pad swelling in WT and RAGE knockout (RAGE−/−) mice to the same degree (27). These data suggested the existence of a cell surface recognition site distinct from RAGE (but capable of binding RAGE ligands, as sRAGE served as an effective soluble decoy receptor), contributing to the adaptive immune response.
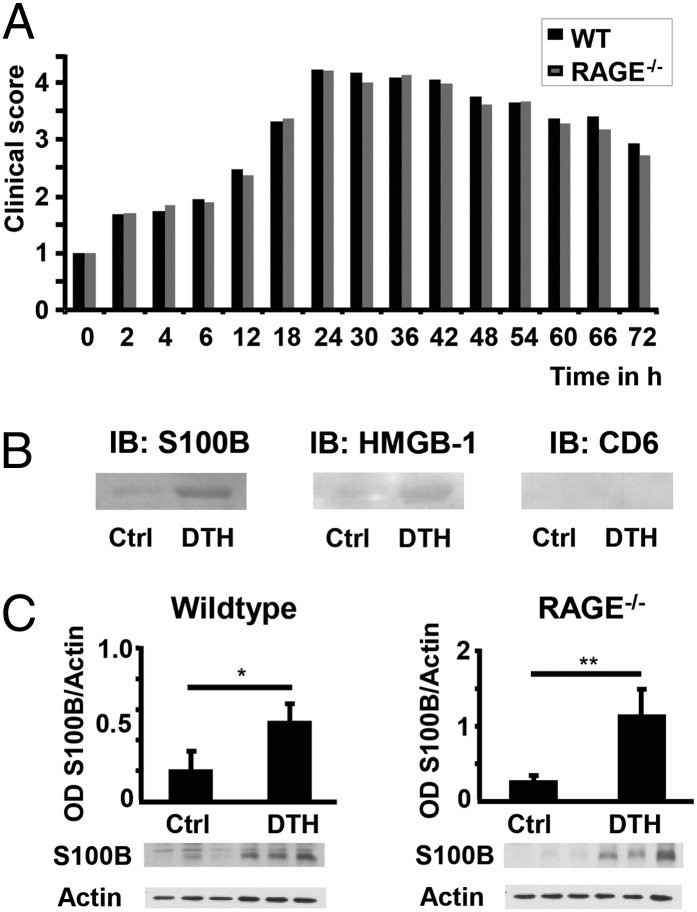
FIGURE 1.
Clinical grade of inflammation and upregulation of RAGE ligands in DTH. (A) DTH time course in WT (n = 20) and RAGE−/− (n = 19) mice; data from three independent experiments. The grading of the clinical score was performed as described in Materials and Methods. (B) Protein extract from footpads of WT mice (one control, one DTH) was passed along Ni-TED-Resin with attached soluble RAGE, and eluted proteins were immunoblotted with anti-S100B, anti-CD6, and anti-HMGB-1 as described in Materials and Methods. (C) Western blot analysis of total protein extract of WT and RAGE−/− footpads (Control = Ctrl and DTH) using anti-S100B (30 μg of total protein extract), n = 3 animals per group. Densitometry is normalized to actin, and data are the mean percentage ± SE from three different animals per group. *p < 0.03, **p < 0.013.
To identify ligands bound to sRAGE, proteins extracted from foot pads after induction of DTH were extracted and loaded onto a column to which sRAGE was immobilized. Elution of the columns revealed several proteins, among them HMGB-1 and S100B, but not CD6 (Fig. 1B). Western blot analysis of protein extract from control and DTH foot pads showed a significant upregulation of S100B Ag in WT and RAGE−/− mice (Fig. 1C). S100 proteins are known to elicit immune responses in diseases characterized by cross talk between adaptive and innate immune responses (37–41). Therefore, a search for structural homologs of RAGE was initiated by searching the PRODOM database, yielding a significant hit comparing the range from amino acid residues 1–250 of the mature RAGE protein with the PRODOM database entry PRODOM:3993. Among the 51 sequences of this protein domain, CD166/ALCAM was chosen as a potential RAGE homolog because of the similarity of 35% seen by an alignment between RAGE_HUMAN 1-250 and CD166_HUMAN 95-221 (Fig. 2A). Modeling and energy minimization showed a structural homology between CD166/ALCAM (position 28–133) and RAGE (position 25–120; Fig. 2B). Meanwhile, these findings have been reported in a recent publication by Koch et al. (42), who proved CD166/ALCAM to be the closest structural homolog to RAGE.
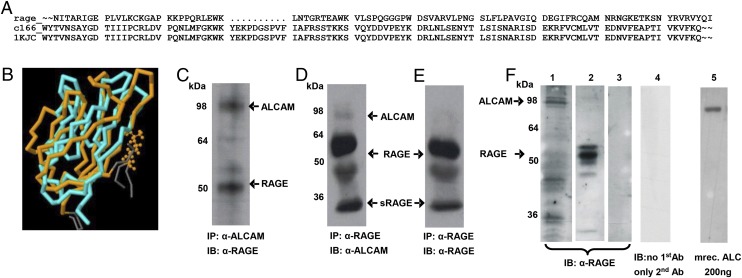
FIGURE 2.
RAGE and ALCAM show similarity in quaternary structure and Ab cross-reaction. (A) Sequence alignment between RAGE_HUMAN 1-250 and CD166_HUMAN 95-221 was performed with the program GAP. (B) Superposition of ALCAM (blue) and RAGE (orange; JCE V.1.1). (C) Total cell lysate from WT MAEC was subjected to IP with anti-ALCAM, separated, and stained with anti-RAGE (1:200). (D) IP with anti-RAGE Ab was separated and stained anti-ALCAM (1:200). (E) IP material as in (D) with immunoblot anti-RAGE. (F) Immunoblot anti-RAGE (1:1250) with different murine tissue protein extract and recombinant ALCAM protein: 1, 35 μg RAGE−/− liver extract; 2, 30 μg ALCAM−/− lung extract; 3, 15 μg bovine serum albumin; 4, 35 μg RAGE−/− liver extract, no first Ab, only second Ab; 5, 200 ng recombinant mouse ALCAM.
To confirm the results of our modeling analysis through functional studies in vivo, additional studies were performed. First, WT MAECs were harvested, and IP was performed with an Ab directed against either CD166/ALCAM or RAGE. Material immunoprecipitated with CD166/ALCAM Ab was separated on a gel and immunoblotted with anti-RAGE Ab, showing two bands of equal intensity at the expected size of RAGE (∼50 kDa) and CD166/ALCAM (∼100 kDa; Fig. 2C). Controls stained with either first or second Ab alone showed no signal (Supplemental Fig. 2A). When material was subjected to IP with RAGE Ab and separated on a gel, immunoblotting with anti-ALCAM Ab showed two bands (55 and 100 kDa) in which the RAGE signal at 55 kDa was significantly stronger (Fig. 2D). For further verification, material immunoprecipitated with RAGE Ab was subjected to immunoblotting with anti-RAGE Ab, revealing only a band corresponding to what would be expected for the size of RAGE (and possibly sRAGE at ∼30 kDa), but no CD166/ALCAM signal (Fig. 2E). These data suggested that the anti-RAGE Ab might be relatively selective for RAGE, and failure to visualize small amounts of CD166/ALCAM in Fig. 2E was likely due to the lesser cross-reactivity of anti-RAGE with CD166/ALCAM). Consistently, anti-RAGE immunoblotting of total protein lysate from RAGE−/− mouse liver rich in ALCAM protein (Supplemental Fig. 2B) revealed an CD166/ALCAM signal at 98 kDa, a clear RAGE signal at 55 kDa in CD166/ALCAM−/− lung protein lysate and no unspecific band either with BSA or secondary Ab treatment only (Fig. 2F). Commercially available recombinant CD166/ALCAM protein revealed a dose-dependent signal that disappeared at CD166/ALCAM concentrations <150 ng of protein (Supplemental Fig. 2C) and was clearly visible at concentrations of 200 ng recombinant ALCAM protein (Fig. 2F).
To test the functional role of CD166/ALCAM, studies were performed in MAEC. CD166/ALCAM Ag expression in MAEC was confirmed by Western blot, and CD166/ALCAM mRNA was confirmed by PCR analysis (Fig. 3A). WT MAECs were cultured to confluence and were treated with S100B (800 nM, 6 h), followed by cross-linking with 1.2% formaldehyde and IP with anti-S100B Ab. After gel separation, immunostaining with anti-ALCAM showed a band at 120 kDa, the expected size of a CD166/ALCAM-S100B complex (Fig. 3B). To verify the results and prove binding, Far-Western blotting was performed. Recombinant full-length CD166/ALCAM protein was separated, blotted onto a PVDF membrane, renaturated, and incubated with S100B. After washing, the membrane was probed with anti-S100B Ab, yielding a signal at the expected size of the CD166/ALCAM-S100B complex (Fig. 3C). Because S100B is known to bind RAGE, MAECs or mouse fibroblasts (where indicated) from RAGE−/− mice were used for additional functional studies to rule out confounding interactions in S100B stimulation experiments.
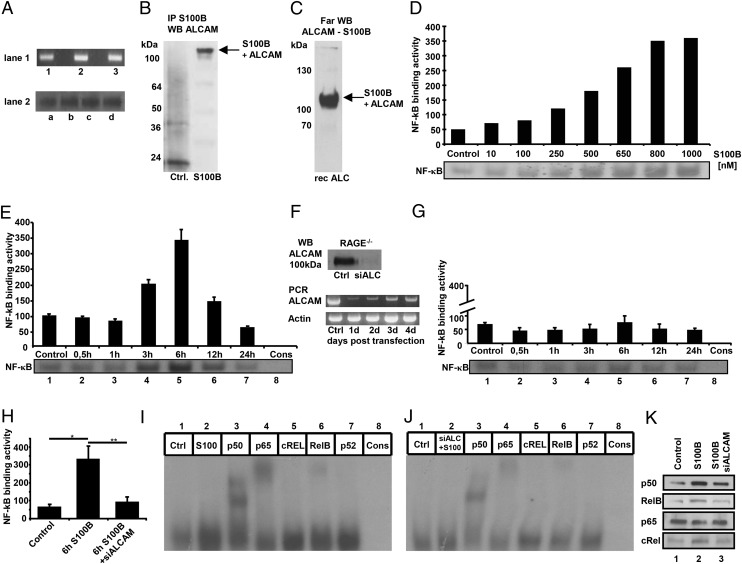
FIGURE 3.
ALCAM is expressed on MAECs and binds S100B, leading to an ALCAM-dependent NF-κB activation in RAGE−/− MAECs. (A) RAGE−/− MAECs express ALCAM mRNA as shown by PCR in lane 1 (results of three different cell populations 1–3 are shown) and ALCAM Ag as shown by Western blot in lane 2 (results of four different cell populations a–d are shown). (B) RAGE−/− MAECs incubated with 800 nM S100B for 6 h were cross-linked (control: no cross-linking) and harvested. Next, immunoprecipitation of protein lysate was performed with anti-S100B as described in Materials and Methods. The IP material was separated and stained with anti-ALCAM Ab (1:600). (C) Recombinant ALCAM was separated using electrophoresis, transferred to a membrane, and renatured. Incubation with S100B overnight was followed by washing and anti-S100B staining as described in Materials and Methods. (D) S100B treatment for 6 h leads to a dose-dependent increase of NF-κB–binding activity in RAGE−/− MAECs, determined by EMSA. Densitometry normalized to consensus (background). (E) Time course of NF-κB–binding activity from MAECs incubated with 800 nM S100B. Time points are indicated on the x-axis. Lane 8 contains specificity control with unlabeled consensus NF-κB oligonucleotide. The experiment was repeated two times, and EMSA of one representative experiment shown. Densitometry of two experiments, normalized to consensus (background). (F) ALCAM silencing in RAGE−/− MAECs leads to highly efficient suppression of ALCAM protein and mRNA expression 2 d after transfection, as shown in Western blot and PCR. (G) ALCAM silencing in RAGE−/− MAEC 2 d prior to treatment with 800 nM S100B (for time points as indicated on x-axis) leads to 90% inhibition of NF-κB–binding activity. The experiment was repeated two times, and EMSA of one representative experiment is shown. Densitometry of two experiments, normalized to consensus (background). (H) S100B treatment (800 nM) at optimal time point of 6 h on NF-κB–binding activity of MAEC RAGE−/− with and without ALCAM silencing. Data are the mean percentage ± SE from three independent experiments. *p < 0.01, **p < 0.009. (I) Supershift (anti-p50, anti-p65, anti-cREL, anti-RelB, or anti-p52 Abs) from RAGE−/− MAECs treated with 800 nM S100B for 6 h. Lanes: 1, control; 2, S100B treated; 3–7, S100B treated (Abs as specified); 8, consensus oligo. (J) Supershift as in (I), but with prior ALCAM silencing. (K) Western blot for different NF-κB subunits with 10 μg of nuclear extract from RAGE−/− MAECs after 6 h of 800 nM S100B with (2) or without (3) prior silencing of ALCAM. Lane 1, control without S100B.
Limited data are available regarding concentrations of S100B in inflammatory settings in vivo, with reported ranges from high nanomolar to micromolar concentrations (43–45). We treated MAECs with increasing concentrations of S100B and found a dose-dependent activation of NF-κB, with activation being half maximal at a concentration of ∼400 nM and reaching a plateau at 800–1000 nM (Fig. 3D). This corresponds well to the micromolar concentrations of S100B found in vivo (42–44). Therefore, an S100B concentration of 800 nM was used in our additional studies. Treatment of MAECs with 800 nM S100B led to a time-dependent activation of NF-κB, with an apparent maximum at 6 h (Fig. 3E). Next, silencing of CD166/ALCAM expression in RAGE−/− MAECs was performed 2 d prior to treatment with S100B using siRNA against CD166/ALCAM. Transfection resulted in virtually complete suppression of CD166/ALCAM Ag expression as shown by PCR and Western blotting (Fig. 3F) without affecting cell viability or attachment of cells to the growth substrate. NF-κB activation upon stimulation with S100B was completely abolished when RAGE−/− MAECs were pretreated with siRNA against CD166/ALCAM (Fig. 3G). Thus, NF-κB activation by S100B is dependent on the ALCAM-S100B interaction in the absence of RAGE (Fig. 3H). The same results were seen with RAGE−/− fibroblasts (data not shown).
To determine which specific subunits of NF-κB were induced or activated by CD166/ALCAM-mediated S100B-induced signaling, a super shift was performed either without (Fig. 3I) or with (Fig. 3J) prior silencing of CD166/ALCAM expression in MAECs from RAGE null mice. S100B induced a strong signal for p50 and a weaker signal for p65, both of which were significantly attenuated by siRNA against CD166/ALCAM. In addition, a weak RelB signal was attenuated by siRNA against CD166/ALCAM. Western blot analysis of nuclear cell extracts in part confirmed these results, showing suppression of p50 and RelB, but no significant difference in the p65 signal after siRNA against CD166/ALCAM (Fig. 3K).
To understand the potential significance of our findings in the in vivo milieu, a model of DTH was used. WT and RAGE−/− mice sensitized to mBSA either were pretreated with siRNA against CD166/ALCAM before injection of mBSA or received control injections before induction of inflammation. Animals were subsequently studied at the peak of inflammation, 24 h after mBSA injection. Pretreatment with siRNA against CD166/ALCAM significantly decreased the clinical inflammation score in the footpad of WT by 50% (Fig. 4A) and in RAGE−/− mice by 40% (Fig. 4B), and PCR from foot material confirmed significant suppression of CD166/ALCAM expression (Fig. 4C). Consistent with these results, siRNA against CD166/ALCAM decreased DTH-mediated induction of proinflammatory cytokines IL-1β, IL-17, CCL-3, CCL-4, CCL-11, and KC by >60% in WT mice (Fig. 4D–I) as well as in RAGE−/− mice (not shown). At the same time, expression of GM-CSF and G-CSF remained unchanged (Fig. 4J, 4K), and expression of anti-inflammatory cytokines IL-10 and IL-13 was even enhanced in the CD166/ALCAM-devoid inflammatory setting (Fig. 4L, 4M), and Sham-transfected mice showed no diminished inflammation. Therefore, siRNA against CD166/ALCAM has a strong inhibitory effect on inflammation in this setting.
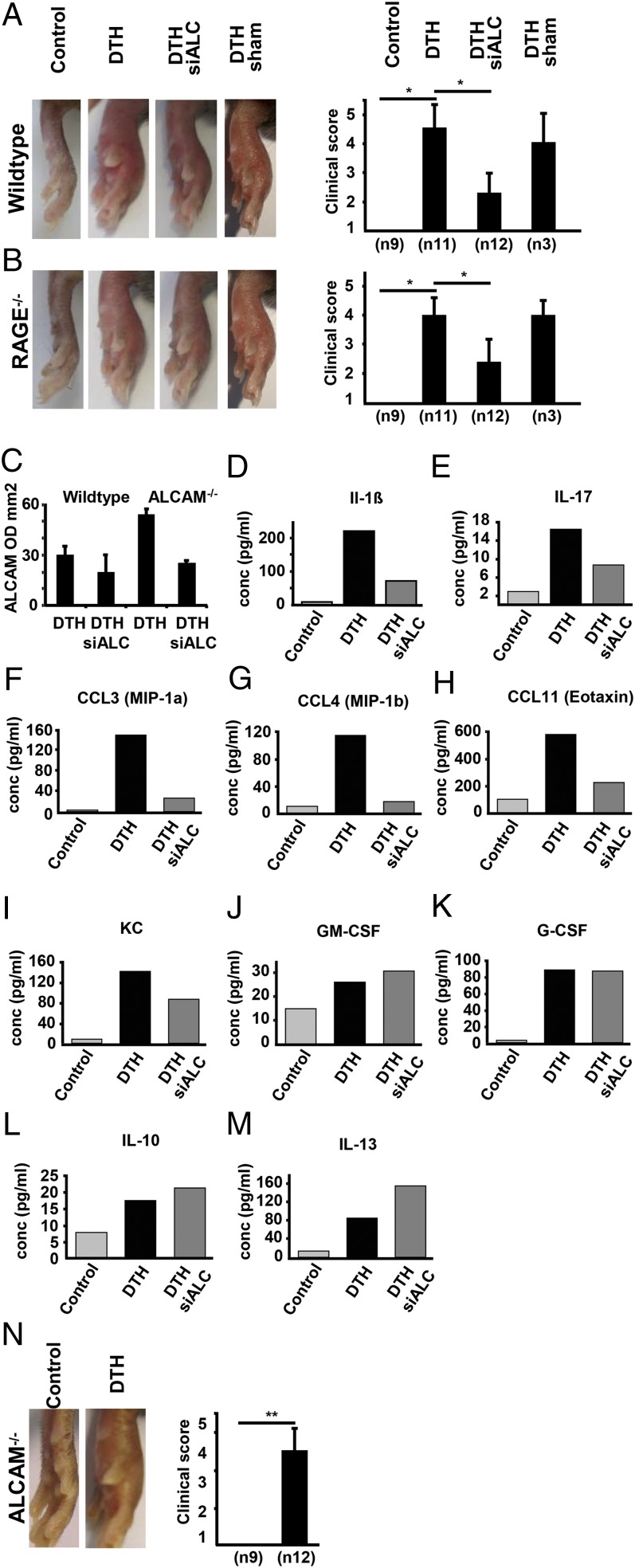
FIGURE 4.
DTH clinical score and cytokine expression in WT and RAGE−/− mice with and without prior siRNA ALCAM and DTH in ALCAM−/− mice. Time point is 24 h after DTH induction. (A) Representative pictures of DTH in left footpads of WT mice, compared with control, siALCAM-treated, and sham-transfected animals. Data of clinical inflammation scores are the mean ± SE from three independent experiments, with a total number of animals per group as indicated (n). *p < 0.01. (B) DTH in RAGE−/− mice with and without prior treatment with siRNA ALCAM or sham transfection. Data for clinical inflammation score are the mean ± SE from three independent experiments with a total number of animals per group as indicated (n). *p < 0.01. (C) Densitometry of ALCAM PCR from footpads of WT and RAGE−/− mice, with and without prior ALCAM siRNA transfection; n = 2 animals per group. (D–M) Expression of inflammation-related cytokines determined by Plex analysis of footpad lysate 24 h after DTH induction in WT mice. Footpad material was processed for Plex analysis as described in Materials and Methods. (D–I) Expression of proinflammatory cytokines is augmented in DTH footpads of WT mice treated with siRNA ALCAM before induction of inflammation. (J and K) Expression of GM-CSF and G-CSF is not changed. (L and M) IL-10 and IL-13 expression is elevated. Results of multiplex analysis from one experiment. (N) Control and DTH in ALCAM−/− mice. Data for clinical inflammation score are the mean ± SE from three independent experiments, with a total number of animals per group as indicated (n). **p < 0.001.
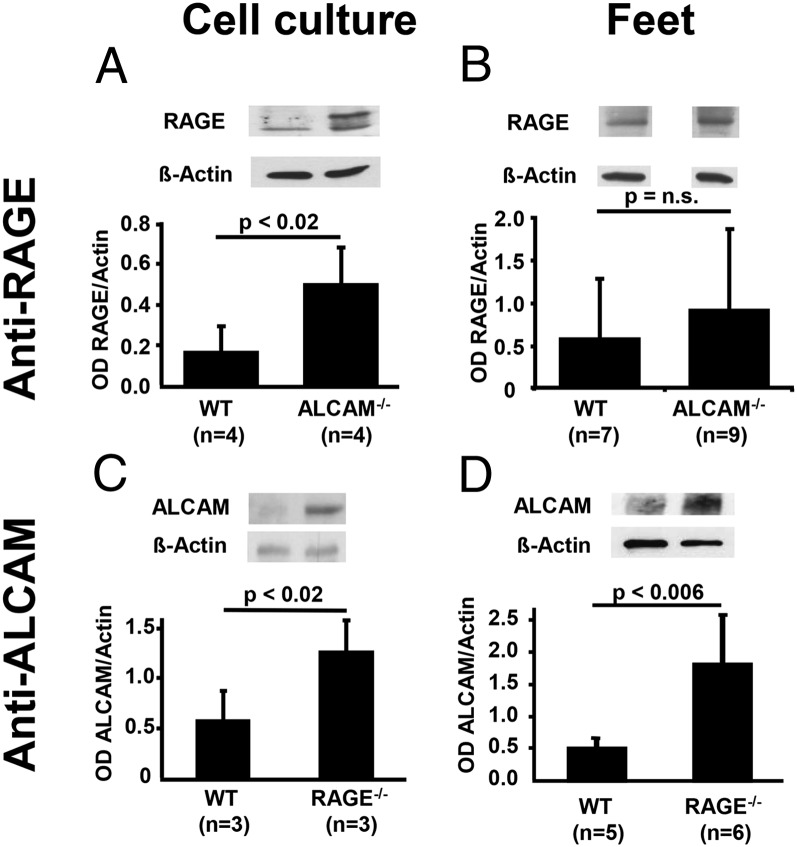
For further investigation of these findings, CD166/ALCAM−/− mice were studied in more detail. Despite a complete absence of detectable ALCAM mRNA or protein in homozygous null ALCAM mice, these animals displayed only a tendency toward less inflammation, compared with WT animals (Fig. 4A), but no significant difference in the intensity (Fig. 4N) or time-course of inflammation (data not shown). At first, these results seemed contradictory to the findings described above, regarding the role of CD166/ALCAM in DTH, unless a compensatory response in another limb of the inflammatory response accompanied the loss of CD166/ALCAM expression. These results led us to consider whether loss of CD166/ALCAM could be accompanied by upregulation of RAGE in light of their cooperation in the context of S100B-mediated inflammation. To examine this hypothesis, total protein extract from WT, RAGE−/−, or CD166/ALCAM−/− MAEC or total protein extract from WT, RAGE−/−, or CD166/ALCAM−/− feet was subjected to Western blotting. Indeed, significant upregulation of RAGE was found in the CD166/ALCAM−/− MAEC (Fig. 5A), and RAGE showed a tendency toward upregulation in the feet of ALCAM−/− mice, although it was not statistically significant (Fig. 5B). We also observed a statistically significant upregulation of CD166/ALCAM in RAGE−/− MAEC (Fig. 5C) compared with WT MAEC, and upregulation of CD166/ALCAM was regulated statistically significant in the feet of RAGE−/− mice (Fig. 5D). The same upregulatory mechanisms were seen in feet after induction of DTH. Therefore, the loss of CD166/ALCAM is accompanied by reciprocal upregulation of RAGE in baseline cultured cells and animals with and without DTH, and vice versa.
FIGURE 5.
Compensatory upregulation of RAGE in ALCAM−/− and of ALCAM in RAGE−/− mice. (A) Western blot using anti-RAGE in WT and ALCAM−/− MAECs. (B) Western blot using anti-RAGE in WT and ALCAM−/− feet. (C) Western blot using anti-ALCAM in WT and RAGE−/− MAECs. (D) Western blot using anti-ALCAM in WT and RAGE−/− feet. The bar graphs show densitometry normalized to actin, number of different cell populations, or feet from separate animals as indicated. Data are the mean percentage ± SE. p < 0.05 was considered statistically significant.
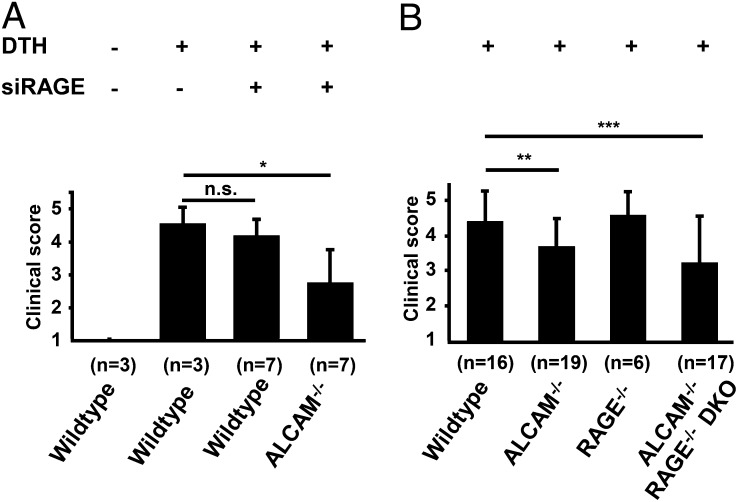
To further test the hypothesis of compensatory upregulation of RAGE in CD166/ALCAM−/− animals, DTH experiments were initiated in WT and ALCAM−/− animals with siRNA against RAGE as indicated (Fig. 6A). In WT animals, siRNA against RAGE did not significantly influence the degree of inflammation, in contrast to siRNA against CD166/ALCAM (Fig. 4A). These results are consistent with primacy of CD166/ALCAM-dependent mechanisms in DTH in WT animals. In contrast, in ALCAM−/− animals, pretreatment with siRNA against RAGE led to a significant decrease in the DTH response by 35% (Fig. 6A), confirming the functional substitution of RAGE for CD166/ALCAM in ALCAM−/− animals. To verify and confirm these results, ALCAM−/− RAGE−/− DKO mice were produced. DTH was induced in WT, ALCAM−/−, RAGE−/−, and ALCAM−/− RAGE−/− DKO mice (Fig. 6B) in two separate experiments with a large number of animals. Experiments revealed a statistically significant reduction in DTH by 15% in the ALCAM−/− animals (p < 0.015), compared with WT animals. This finding does not contradict the results shown in Fig. 4N; rather, it reinforces that the RAGE-dependent compensation in ALCAM−/− animals does not fully restore the inflammatory response to the normal level and that large group sizes are crucial to reveal this.
FIGURE 6.
Effects of siRNA RAGE on DTH clinical score in WT and ALCAM−/− mice, and effect of ALCAM−/− RAGE−/− double knockout on DTH clinical score. The time point is 24 h after DTH induction. (A) Mean clinical inflammation score in left footpads of WT mice with and without prior siRAGE treatment and of ALCAM−/− mice with prior siRAGE treatment. Number of animals per group is indicated (n). (B) Mean clinical inflammation score in WT, ALCAM−/−, RAGE−/−, and ALCAM−/− RAGE−/− DKO mice with number of animals per group as indicated (n). Control animals of all groups without DTH showed no inflammation (clinical score = 1). Data for clinical inflammation score are the mean ± SE from two independent experiments in (A) and (B) each, with a total number of animals per group as indicated (n). *p < 0.02, **p = 0.015, ***p < 0.045.
Compared with WT animals, the ALCAM−/− RAGE−/− DKO mice showed a significant reduction by 27% in clinical inflammation score (p < 0.045). This reduction is smaller compared with that achieved by small inhibitory RNA against CD166/ALCAM (siALCAM) treatment in WT, because of the lack of compensatory RAGE upregulation at this early time point. The 27% reduction in inflammation in the ALCAM−/− RAGE−/− DKO mice is more than that seen in CD166/ALCAM−/− alone, but not as dramatic as expected. This result is probably due to several mechanisms that are under active investigation, involving significant genotype specific variations in the expression of TLRs with, for example, TLR2 being upregulated 2.5-fold in ALCAM−/− RAGE−/− DKO, compared with WT, and even downregulated 0.5-fold in ALCAM−/−. In addition, RAGE−/− mice show an increased leukocyte adherence and infiltration at the site of inflammation (R. von Bauer and A. Sulaj, unpublished observations). Taken together, the findings are consistent with a compensatory pathway by which S100B can bridge the RAGE- and CD166/ALCAM-mediated immune response mechanisms, with a more dominant role of CD166/ALCAM, compared with RAGE, in DTH.
Discussion
The data in this study show that CD166/ALCAM and RAGE can functionally substitute for each other in the context of S100B-induced inflammatory responses (in this context, DTH). Whereas RAGE and S100B interaction has been described in detail (12, 46), CD166/ALCAM-S100B interaction and consequent activation of NF-κB have not been described. The results of our cross-linking data demonstrate CD166/ALCAM – S100B complex (Fig. 3B), and Far-Western blotting shows CD166/ALCAM–S100B binding (Fig. 3C). Further characterization of the binding properties is currently under active study in our laboratory.
The functional data show that S100B causes, via interaction with CD166/ALCAM, a dose- and time-dependent activation of NF-κB, reaching a maximum at an S100B concentration of ∼800 nM. This concentration is consistent with several published in vitro and in vivo data. In vitro, S100B binding to the tumor suppressor protein p53 has been shown to be saturated at ∼800–900 nM (47). In inflammatory diseases of the gut, S100B expression is strongly increased (41) and dose-dependently induces the expression of iNOS, with optimal NO production at 500–5000 nM S100B (44). In vivo, patients with rheumatoid arthritis patients show micromolar S100B serum concentrations (48), and patients with Alzheimer disease or HIV infection show micromolar concentrations of S100B in the cerebrospinal fluid (49). Lymphocytes as well as glioma cells and hippocampal tissue can produce and secrete S100B upon stimulation (50–53) with IL-1β and other proinflammatory cytokines. Despite the rather short plasma half-life time of S100B (30–120 min) (54, 55), local accumulation of S100B at sites of inflammation because of rapid and massive secretion by proinflammatory cells probably leads to the nanomolar concentrations needed for optimal interaction with ALCAM. Furthermore, posttranslational S100B modification can enhance its proinflammatory capacity (21).
The NF-κB subunits activated by S100B-ALCAM interaction, with p50 and p65 as the most prominent members, are of special interest. Until now, perpetuation of inflammation via NF-κB has been a hallmark of RAGE, based on its ability to induce de novo synthesis of p65 (25) and induction of epigenetic modulations through diabetes mellitus (56, 57). In the CD166/ALCAM-S100B axis, perpetuation of inflammation is more likely to depend on cell-cell interaction following the initial signaling event, because in vitro induction of NF-κB persisted for not more than 16 h. Accordingly, a recent work identified an innate immune gene response upon modulation of CD166/ALCAM in melanoma (35), a cancer also characterized by expression of S100B.
The data presented in this study show upregulation of CD166/ALCAM in the RAGE−/− mice and vice versa (to a lesser degree). In knockout animals, compensatory upregulation (58–60), as well as assumption of function by other protein family members, shown for the NF-κB family (61) and others (62), are mechanisms to cope with the loss of function. Prior treatment with siRNA against RAGE diminished the degree of DTH only in ALCAM−/− mice, but not in WT mice, supporting our findings that RAGE is of importance in DTH only when CD166/ALCAM expression is deleted. RAGE belongs to the family of innate immune receptors, sharing properties with TLRs and interacting in a complex network (16). The data presented in this study establish CD166/ALCAM as another member of this network and provide insight into a relationship between CD166/ALCAM and RAGE on both structural and functional levels. The only 27% reduction in clinical inflammation score in DKO mice compared with WT mice revealed additional proinflammatory changes in the regulatory network, involving TLR2 and others. These processes that also involve significant functional and expressional differences in IL-17 are now under active investigation (R. von Bauer and A. Sulaj, unpublished observations).
Although the biological and physiologic roles of potential CD166/ALCAM–RAGE compensation remain to be fully identified, the siRNA experiments revealed that this interaction is a complex spatiotemporal process. The close relationship of both receptors is important also in the context of experiments with RAGE−/− animals, where upregulation of CD166/ALCAM might contribute to the observed results. Furthermore, experimental results based on use of RAGE Abs or sRAGE might partly be CD166/ALCAM mediated instead. In experimental autoimmune encephalitis (EAE), anti-RAGE Ab, sRAGE, and anti-ALCAM Ab led to significantly reduced disease severity (63, 64), whereas animals with CD4+ T cells devoid of functional RAGE show diminished but still detectable EAE (63). In patients with multiple sclerosis, CD166/ALCAM is upregulated (64), and CD166/ALCAM polymorphism correlates with risk and progression of multiple sclerosis in humans (65). EAE in CD166/ALCAM−/−, RAGE−/−, and ALCAM−/− RAGE−/− DKO animals might reveal confounding effects of possible Ab cross-reactivity.
The orchestration of a specific CD166/ALCAM-dependent response to S100B in the presence of RAGE is not studied in detail, although we propose various mechanisms to explain the apparent primacy of the S100B-ALCAM axis: the receptor composition at the site of inflammation and its change by regulatory mediators is probably an important component. RAGE is constitutively expressed at high levels in the lung only (7), whereas CD166/ALCAM baseline expression in the footpad during DTH is higher and becomes enhanced by the rapid increase of TNF-α, G-CSF, and GM-CSF in DTH, factors known to upregulate CD166/ALCAM (66, 67). This potentially results in preferential binding of S100B to CD166/ALCAM. A positive feedback loop with involvement of the known p65 binding site in the ALCAM promoter (68) could also be operative, further increasing CD166/ALCAM expression. Furthermore, the increase of anti-inflammatory cytokines following siRNA-mediated CD166/ALCAM suppression (Fig. 4N, 4O) suggests that CD166/ALCAM leads to suppression of anti-inflammatory cytokines. In addition, metabolic acidosis at the site of inflammation (69) impairs the formation of RAGE homodimers needed for S100B-induced RAGE activation (42) and the formation of S100B oligomers with higher RAGE affinity (12, 47).
Our results show structural similarity and reciprocal complementary expression of CD166/ALCAM and RAGE. The data indicate possible time- and spatial-dependent contributions of CD166/ALCAM and RAGE in the context of S100B-driven initiation, perpetuation, and possibly termination of inflammation. S100B is therefore a ligand interacting with two pattern-recognition receptors, thereby bridging RAGE and CD166/ALCAM downstream effector mechanisms.
Acknowledgments
We thank Dr. Thomas Fleming and Claudia Schmidt for technical support during these studies.
Deceased.
This work was supported in part by Deutsche Forschungsgemeinschaft Grant SFB 938 and the Dietmar Hopp Foundation.
The online version of this article contains supplemental material.
- DAMP
- danger-associated molecular pattern
- DKO
- double knockout
- DTH
- delayed-type hypersensitivity
- EAE
- experimental autoimmune encephalitis
- IP
- immunoprecipitation
- MAEC
- mouse aortic endothelial cell
- mBSA
- methylated BSA
- PRODOM
- protein domain family
- PRR
- pattern recognition receptor
- RAGE
- receptor for advanced glycation end products
- siALCAM
- small inhibitory RNA against CD166/ALCAM
- siRNA
- small interfering RNA
- sRAGE
- soluble RAGE
- WT
- wild type.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Kawai T., Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11: 373–384 [DOI] [PubMed] [Google Scholar]
- 2.Chin A. I., Dempsey P. W., Bruhn K., Miller J. F., Xu Y., Cheng G. 2002. Involvement of receptor-interacting protein 2 in innate and adaptive immune responses. Nature 416: 190–194 [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi K., Inohara N., Hernandez L. D., Galán J. E., Núñez G., Janeway C. A., Medzhitov R., Flavell R. A. 2002. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature 416: 194–199 [DOI] [PubMed] [Google Scholar]
- 4.Saiga H., Shimada Y., Takeda K. 2011. Innate immune effectors in mycobacterial infection. Clin. Dev. Immunol. 2011: 347594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akira S., Takeda K., Kaisho T. 2001. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2: 675–680 [DOI] [PubMed] [Google Scholar]
- 6.Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413: 732–738 [DOI] [PubMed] [Google Scholar]
- 7.Sorci G., Giovannini G., Riuzzi F., Bonifazi P., Zelante T., Zagarella S., Bistoni F., Donato R., Romani L. 2011. The danger signal S100B integrates pathogen- and danger-sensing pathways to restrain inflammation. PLoS Pathog. 7: e1001315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bianchi M. E. 2007. DAMPs, PAMPs and alarmins: all we need to know about danger. J. Leukoc. Biol. 81: 1–5 [DOI] [PubMed] [Google Scholar]
- 9.Lotze M. T., Zeh H. J., Rubartelli A., Sparvero L. J., Amoscato A. A., Washburn N. R., Devera M. E., Liang X., Tör M., Billiar T. 2007. The grateful dead: damage-associated molecular pattern molecules and reduction/oxidation regulate immunity. Immunol. Rev. 220: 60–81 [DOI] [PubMed] [Google Scholar]
- 10.Foell D., Wittkowski H., Vogl T., Roth J. 2007. S100 proteins expressed in phagocytes: a novel group of damage-associated molecular pattern molecules. J. Leukoc. Biol. 81: 28–37 [DOI] [PubMed] [Google Scholar]
- 11.Scaffidi P., Misteli T., Bianchi M. E. 2002. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 418: 191–195 [DOI] [PubMed] [Google Scholar]
- 12.Ostendorp T., Leclerc E., Galichet A., Koch M., Demling N., Weigle B., Heizmann C. W., Kroneck P. M., Fritz G. 2007. Structural and functional insights into RAGE activation by multimeric S100B. EMBO J. 26: 3868–3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donato R. 2007. RAGE: a single receptor for several ligands and different cellular responses: the case of certain S100 proteins. Curr. Mol. Med. 7: 711–724 [DOI] [PubMed] [Google Scholar]
- 14.Taguchi A., Blood D. C., del Toro G., Canet A., Lee D. C., Qu W., Tanji N., Lu Y., Lalla E., Fu C., et al. 2000. Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature 405: 354–360 [DOI] [PubMed] [Google Scholar]
- 15.Dumitriu I. E., Baruah P., Valentinis B., Voll R. E., Herrmann M., Nawroth P. P., Arnold B., Bianchi M. E., Manfredi A. A., Rovere-Querini P. 2005. Release of high mobility group box 1 by dendritic cells controls T cell activation via the receptor for advanced glycation end products. J. Immunol. 174: 7506–7515 [DOI] [PubMed] [Google Scholar]
- 16.van Zoelen M. A., Yang H., Florquin S., Meijers J. C., Akira S., Arnold B., Nawroth P. P., Bierhaus A., Tracey K. J., van der Poll T. 2009. Role of toll-like receptors 2 and 4, and the receptor for advanced glycation end products in high-mobility group box 1-induced inflammation in vivo. Shock 31: 280–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bucciarelli L. G., Wendt T., Rong L., Lalla E., Hofmann M. A., Goova M. T., Taguchi A., Yan S. F., Yan S. D., Stern D. M., Schmidt A. M. 2002. RAGE is a multiligand receptor of the immunoglobulin superfamily: implications for homeostasis and chronic disease. Cell. Mol. Life Sci. 59: 1117–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D’Agati V., Schmidt A. M. 2010. RAGE and the pathogenesis of chronic kidney disease. Nat. Rev. Nephrol. 6: 352–360 [DOI] [PubMed] [Google Scholar]
- 19.Yan S. F., Ramasamy R., Schmidt A. M. 2008. Mechanisms of disease: advanced glycation end-products and their receptor in inflammation and diabetes complications. Nat. Clin. Pract. Endocrinol. Metab. 4: 285–293 [DOI] [PubMed] [Google Scholar]
- 20.Ramasamy R., Yan S. F., Schmidt A. M. 2009. RAGE: therapeutic target and biomarker of the inflammatory response—the evidence mounts. J. Leukoc. Biol. 86: 505–512 [DOI] [PubMed] [Google Scholar]
- 21.Andrassy M., Igwe J., Autschbach F., Volz C., Remppis A., Neurath M. F., Schleicher E., Humpert P. M., Wendt T., Liliensiek B., et al. 2006. Posttranslationally modified proteins as mediators of sustained intestinal inflammation. Am. J. Pathol. 169: 1223–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sims G. P., Rowe D. C., Rietdijk S. T., Herbst R., Coyle A. J. 2010. HMGB1 and RAGE in inflammation and cancer. Annu. Rev. Immunol. 28: 367–388 [DOI] [PubMed] [Google Scholar]
- 23.Bierhaus A., Nawroth P. P. 2009. Multiple levels of regulation determine the role of the receptor for AGE (RAGE) as common soil in inflammation, immune responses and diabetes mellitus and its complications. Diabetologia 52: 2251–2263 [DOI] [PubMed] [Google Scholar]
- 24.Riehl A., Németh J., Angel P., Hess J. 2009. The receptor RAGE: Bridging inflammation and cancer. Cell Commun. Signal. 7: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bierhaus A., Schiekofer S., Schwaninger M., Andrassy M., Humpert P. M., Chen J., Hong M., Luther T., Henle T., Klöting I., et al. 2001. Diabetes-associated sustained activation of the transcription factor nuclear factor-kappaB. Diabetes 50: 2792–2808 [DOI] [PubMed] [Google Scholar]
- 26.Hermani A., De Servi B., Medunjanin S., Tessier P. A., Mayer D. 2006. S100A8 and S100A9 activate MAP kinase and NF-kappaB signaling pathways and trigger translocation of RAGE in human prostate cancer cells. Exp. Cell Res. 312: 184–197 [DOI] [PubMed] [Google Scholar]
- 27.Liliensiek B., Weigand M. A., Bierhaus A., Nicklas W., Kasper M., Hofer S., Plachky J., Gröne H. J., Kurschus F. C., Schmidt A. M., et al. 2004. Receptor for advanced glycation end products (RAGE) regulates sepsis but not the adaptive immune response. J. Clin. Invest. 113: 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Djuric Z., Kashif M., Fleming T., Muhammad S., Piel D., von Bauer R., Bea F., Herzig S., Zeier M., Pizzi M., et al. 2012. Targeting activation of specific NF-κB subunits prevents stress-dependent atherothrombotic gene expression. Mol. Med. 18: 1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y., Akirav E. M., Chen W., Henegariu O., Moser B., Desai D., Shen J. M., Webster J. C., Andrews R. C., Mjalli A. M., et al. 2008. RAGE ligation affects T cell activation and controls T cell differentiation. J. Immunol. 181: 4272–4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moser B., Desai D. D., Downie M. P., Chen Y., Yan S. F., Herold K., Schmidt A. M., Clynes R. 2007. Receptor for advanced glycation end products expression on T cells contributes to antigen-specific cellular expansion in vivo. J. Immunol. 179: 8051–8058 [DOI] [PubMed] [Google Scholar]
- 31.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- 32.Womble D. D. 2000. GCG: The Wisconsin Package of sequence analysis programs. Methods Mol. Biol. 132: 3–22 [DOI] [PubMed] [Google Scholar]
- 33.Hotz-Wagenblatt A., Dröge W. 2002. Redox-mediated functional and structural changes in insulin receptor kinase. Methods Enzymol. 348: 288–296 [DOI] [PubMed] [Google Scholar]
- 34.Gotoh O. 1996. Significant improvement in accuracy of multiple protein sequence alignments by iterative refinement as assessed by reference to structural alignments. J. Mol. Biol. 264: 823–838 [DOI] [PubMed] [Google Scholar]
- 35.van Kilsdonk J. W., Takahashi N., Weidle U., Burtscher H., Jarry J., Daha M. R., Swart G. W., van Kempen L. C. 2012. Modulation of activated leukocyte cell adhesion molecule-mediated invasion triggers an innate immune gene response in melanoma. J. Invest. Dermatol. 132: 1462–1470 [DOI] [PubMed] [Google Scholar]
- 36.Wu Y., Li Q., Chen X. Z. 2007. Detecting protein-protein interactions by Far western blotting. Nat. Protoc. 2: 3278–3284 [DOI] [PubMed] [Google Scholar]
- 37.Donato R. 2001. S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int. J. Biochem. Cell Biol. 33: 637–668 [DOI] [PubMed] [Google Scholar]
- 38.Foell D., Wittkowski H., Roth J. 2007. Mechanisms of disease: a ‘DAMP’ view of inflammatory arthritis. Nat. Clin. Pract. Rheumatol. 3: 382–390 [DOI] [PubMed] [Google Scholar]
- 39.Yan S. F., Ramasamy R., Naka Y., Schmidt A. M. 2003. Glycation, inflammation, and RAGE: a scaffold for the macrovascular complications of diabetes and beyond. Circ. Res. 93: 1159–1169 [DOI] [PubMed] [Google Scholar]
- 40.Prakken B., Albani S., Martini A. 2011. Juvenile idiopathic arthritis. Lancet 377: 2138–2149 [DOI] [PubMed] [Google Scholar]
- 41.Cirillo C., Sarnelli G., Esposito G., Turco F., Steardo L., Cuomo R. 2011. S100B protein in the gut: the evidence for enteroglial-sustained intestinal inflammation. World J. Gastroenterol. 17: 1261–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koch M., Chitayat S., Dattilo B. M., Schiefner A., Diez J., Chazin W. J., Fritz G. 2010. Structural basis for ligand recognition and activation of RAGE. Structure 18: 1342–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huttunen H. J., Kuja-Panula J., Sorci G., Agneletti A. L., Donato R., Rauvala H. 2000. Coregulation of neurite outgrowth and cell survival by amphoterin and S100 proteins through receptor for advanced glycation end products (RAGE) activation. J. Biol. Chem. 275: 40096–40105 [DOI] [PubMed] [Google Scholar]
- 44.Cirillo C, Sarnelli G., Esposito G., Grosso M., Petruzzelli R., Izzo P., Cali G., D'Armiento F. P., Rocco A., Nardone G., Iuvone T., Steardo L., Cuomo R. 2009. Increased mucosal nitric oxide production in ulcerative colitis is mediated in part by the enteroglial-derived s100b protein. Neurogastroenterol. Motil. 21: 1209–e1112 [DOI] [PubMed] [Google Scholar]
- 45.Esposito G., Cirillo C., Sarnelli G., De Filippis D., D’Armiento F. P., Rocco A., Nardone G., Petruzzelli R., Grosso M., Izzo P., et al. 2007. Enteric glial-derived S100B protein stimulates nitric oxide production in celiac disease. Gastroenterology 133: 918–925 [DOI] [PubMed] [Google Scholar]
- 46.Leclerc E., Fritz G., Weibel M., Heizmann C. W., Galichet A. 2007. S100B and S100A6 differentially modulate cell survival by interacting with distinct RAGE (receptor for advanced glycation end products) immunoglobulin domains. J. Biol. Chem. 282: 31317–31331 [DOI] [PubMed] [Google Scholar]
- 47.Thulin E., Kesvatera T., Linse S. 2011. Molecular determinants of S100B oligomer formation. PLoS ONE 6: e14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamed S. A., Selim Z. I., Elattar A. M., Elserogy Y. M., Ahmed E. A., Mohamed H. O. 2012. Assessment of biocorrelates for brain involvement in female patients with rheumatoid arthritis. Clin. Rheumatol. 31: 123–132 [DOI] [PubMed] [Google Scholar]
- 49.Du Pasquier R. A., Jilek S., Kalubi M., Yerly S., Fux C. A., Gutmann C., Cusini A., Günthard H. F., Cavassini M., Vernazza P. L., Swiss HIV Cohort Study 2013. Marked increase of the astrocytic marker S100B in the cerebrospinal fluid of HIV-infected patients on LPV/r-monotherapy. AIDS 27: 203–210 [DOI] [PubMed] [Google Scholar]
- 50.Steiner J., Westphal S., Schroeter M. L., Schiltz K., Jordan W., Müller U. J., Bernstein H. G., Bogerts B., Schmidt R. E., Jacobs R. 2012. Increased S100B+ NK cell counts in acutely ill schizophrenia patients are correlated with the free cortisol index, but not with S100B serum levels. Brain Behav. Immun. 26: 564–567 [DOI] [PubMed] [Google Scholar]
- 51.Miki Y., Gion Y., Mukae Y., Hayashi A., Sato H., Yoshino T., Takahashi K. 2013. Morphologic, flow cytometric, functional, and molecular analyses of S100B positive lymphocytes, unique cytotoxic lymphocytes containing S100B protein. Eur. J. Haematol. 90: 99–110 [DOI] [PubMed] [Google Scholar]
- 52.de Souza D. F., Wartchow K., Hansen F., Lunardi P., Guerra M. C., Nardin P., Gonçalves C. A. 2013. Interleukin-6-induced S100B secretion is inhibited by haloperidol and risperidone. Prog. Neuropsychopharmacol. Biol. Psychiatry 43: 14–22 [DOI] [PubMed] [Google Scholar]
- 53.de Souza D. F., Leite M. C., Quincozes-Santos A., Nardin P., Tortorelli L. S., Rigo M. M., Gottfried C., Leal R. B., Gonçalves C. A. 2009. S100B secretion is stimulated by IL-1beta in glial cultures and hippocampal slices of rats: Likely involvement of MAPK pathway. J. Neuroimmunol. 206: 52–57 [DOI] [PubMed] [Google Scholar]
- 54.Sedaghat F., Notopoulos A. 2008. S100 protein family and its application in clinical practice. Hippokratia 12: 198–204 [PMC free article] [PubMed] [Google Scholar]
- 55.Rothermundt M., Peters M., Prehn J. H., Arolt V. 2003. S100B in brain damage and neurodegeneration. Microsc. Res. Tech. 60: 614–632 [DOI] [PubMed] [Google Scholar]
- 56.El-Osta A., Brasacchio D., Yao D., Pocai A., Jones P. L., Roeder R. G., Cooper M. E., Brownlee M. 2008. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J. Exp. Med. 205: 2409–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nawroth P. 2012. Innovative models for investigation of pathomechanisms leading to late diabetic complications. Exp. Clin. Endocrinol. Diabetes. 120: 177–178 [DOI] [PubMed] [Google Scholar]
- 58.Rudnicki M. A., Schnegelsberg P. N., Stead R. H., Braun T., Arnold H. H., Jaenisch R. 1993. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 75: 1351–1359 [DOI] [PubMed] [Google Scholar]
- 59.Zeisberg M., Khurana M., Rao V. H., Cosgrove D., Rougier J. P., Werner M. C., Shield C. F., III, Werb Z., Kalluri R. 2006. Stage-specific action of matrix metalloproteinases influences progressive hereditary kidney disease. PLoS Med. 3: e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carracedo A., Ma L., Teruya-Feldstein J., Rojo F., Salmena L., Alimonti A., Egia A., Sasaki A. T., Thomas G., Kozma S. C., et al. 2008. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J. Clin. Invest. 118: 3065–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weih F., Warr G., Yang H., Bravo R. 1997. Multifocal defects in immune responses in RelB-deficient mice. J. Immunol. 158: 5211–5218 [PubMed] [Google Scholar]
- 62.Condie B. G., Capecchi M. R. 1994. Mice with targeted disruptions in the paralogous genes hoxa-3 and hoxd-3 reveal synergistic interactions. Nature 370: 304–307 [DOI] [PubMed] [Google Scholar]
- 63.Yan S. S., Wu Z. Y., Zhang H. P., Furtado G., Chen X., Yan S. F., Schmidt A. M., Brown C., Stern A., LaFaille J., et al. 2003. Suppression of experimental autoimmune encephalomyelitis by selective blockade of encephalitogenic T-cell infiltration of the central nervous system. Nat. Med. 9: 287–293 [DOI] [PubMed] [Google Scholar]
- 64.Cayrol R., Wosik K., Berard J. L., Dodelet-Devillers A., Ifergan I., Kebir H., Haqqani A. S., Kreymborg K., Krug S., Moumdjian R., et al. 2008. Activated leukocyte cell adhesion molecule promotes leukocyte trafficking into the central nervous system. Nat. Immunol. 9: 137–145 [DOI] [PubMed] [Google Scholar]
- 65.Wagner M., Wiśniewski A., Bilińska M., Pokryszko-Dragan A., Nowak I., Kuśnierczyk P., Jasek M. 2013. ALCAM - Novel multiple sclerosis locus interfering with HLA-DRB1*1501. J. Neuroimmunol. 258: 71–76 . [DOI] [PubMed] [Google Scholar]
- 66.Levesque M. C., Heinly C. S., Whichard L. P., Patel D. D. 1998. Cytokine-regulated expression of activated leukocyte cell adhesion molecule (CD166) on monocyte-lineage cells and in rheumatoid arthritis synovium. Arthritis Rheum. 41: 2221–2229 [DOI] [PubMed] [Google Scholar]
- 67.Ikeda K., Quertermous T. 2004. Molecular isolation and characterization of a soluble isoform of activated leukocyte cell adhesion molecule that modulates endothelial cell function. J. Biol. Chem. 279: 55315–55323 [DOI] [PubMed] [Google Scholar]
- 68.King J. A., Tan F., Mbeunkui F., Chambers Z., Cantrell S., Chen H., Alvarez D., Shevde L. A., Ofori-Acquah S. F. 2010. Mechanisms of transcriptional regulation and prognostic significance of activated leukocyte cell adhesion molecule in cancer. Mol. Cancer 9: 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harrison D. K., Spence V. A., Beck J. S., Lowe J. G., Walker W. F. 1986. pH changes in the dermis during the course of the tuberculin skin test. Immunology 59: 497–501 [PMC free article] [PubMed] [Google Scholar]