Abstract
Surface area of the cerebral cortex is a highly heritable trait, yet little is known about genetic influences on regional cortical differentiation in humans. Using a data-driven, fuzzy clustering technique with magnetic resonance imaging data from 406 twins, we parceled cortical surface area into genetic subdivisions, creating a human brain atlas based solely on genetically informative data. Boundaries of the genetic divisions corresponded largely to meaningful structural and functional regions; however, the divisions represented previously undescribed phenotypes different from conventional (non–genetically based) parcellation systems. The genetic organization of cortical area was hierarchical, modular, and predominantly bilaterally symmetric across hemispheres. We also found that the results were consistent with human-specific regions being subdivisions of previously described, genetically based lobar regionalization patterns.
As early as the 1950s, Bergquist and Kallen postulated that the entire embryonic brain is divisible into an anteroposterior series of segmented neuromeres, each forming a complete ring around the brain’s longitudinal axis (1). Almost 40 years later, experimental data showed that many gene expression domains respect segment boundaries in the embryonic vertebrate hindbrain, suggesting a role of genetic control in regional differentiation (2, 3). This important finding prompted a search for similar genetic regulatory organization in other regions of the developing vertebrate brain (4). In particular, in the past decade the cerebral cortex has received substantial attention. Studies have shown, for example, that several signaling molecules and transcription factors are involved in establishing boundaries between mouse cortical regions (5, 6). Animal data demonstrate that the regional or positional identity of cortical regions is defined by the combinatorial expression pattern of various genes controlling for regional differentiation, each of which is expressed in a graded and restricted pattern with distinct spatiotemporal characteristics (7). Little is known, however, about the genetic patterning underlying the human cortex. In our previous work (8), we showed that genetic patterning underlying the anteroposterior gradient and four basic cortical divisions of cortical surface area demonstrated in mouse models (7) also existed in the human cortex. Furthermore, region-specific cortical areal expansion in humans has been linked to specific genetic polymorphisms (9, 10). We sought to go beyond the fundamental commonalities that humans share with other species and to investigate the genetic patterning specific to the human cortex with its 1000-fold increase in surface area relative to the mouse brain (11). In effect, we sought to develop a brain atlas of human cortical surface area that was based entirely on genetic correlations, rather than a priori structural or functional information.
To delineate the genetic patterning of the cortical area, we measured relative surface areal expansion using cortical surface reconstruction and spherical atlas mapping developed by Dale and colleagues (12–14). We divided the area measured at each location by the total surface area in order to account for global effects. Using the twin design, which compares monozygotic and dizygotic twins, we then estimated genetic correlations between different points on the cortical surface. These genetic correlations represent shared genetic influences on relative areal expansion between cortical regions (15). Details of these methods have been previously described (8, 16). After computing pairwise genetic correlations, we used an unsupervised pattern recognition method—fuzzy cluster analysis (17)—to demarcate the genetic topography of cortical surface area based on the genetic correlations of relative surface area measures. To determine the appropriate number of clusters, we computed the widely used silhouette coefficient.
On the basis of the peak of the silhouette coefficients (fig. S1), we identified 12 natural clusters. These clusters correspond closely to meaningful structural and functional regions (Fig. 1), even though the registration procedure did not rely on prespecified anatomical landmarks; rather, it makes use of the continuous pattern of surface curvature (13). In describing the subdivisions, we use conventional labels, but these only approximate the observed clusters. Subdivisions of the frontal cortex include the motor-premotor, dorsolateral prefrontal cortex extending to the anterior and superior parts, dorsomedial frontal, and orbitofrontal (Fig. 1, clusters 1 to 4). Another cluster is found between the frontal and parietal cortices, extending from pars opercularis to the subcentral region, including the inferior pre- and post-central gyri (Fig. 1, cluster 5). The temporal cortex includes the superior temporal, posterolateral temporal cortex extending to temporal and parietal junction, and anteromedial temporal cortex (Fig. 1, clusters 6 to 8). The parietal cortex includes the inferior parietal cortex, superior parietal cortex, and precuneus (Fig. 1, clusters 9 to 11). The occipital cortex constitutes a single cluster (Fig. 1, cluster 12). Some anatomical boundaries of these clusters map onto traditionally parcellated regions, such as cytoarchitectural areas or gyrus patterns; however, others do not follow classically defined boundaries (such as Brodmann areas). For example, there is no natural sulcal-gyral boundary between our dorsomedial and orbitofrontal clusters, but they still correspond reasonably well to the division between Brodmann areas 10 and 11. Conversely, the well-defined cytoarchitectural differentiation between Brodmann areas 17 and 18 is not manifest as separate genetically based clusters in our analyses.
Fig. 1.
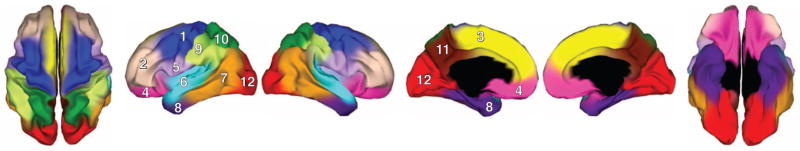
Genetic clustering map for 12-cluster solution. 1, motor-premotor cortex; 2, dorsolateral prefrontal cortex; 3, dorsomedial frontal cortex; 4, orbitofrontal cortex; 5, pars opercularis and subcentral region; 6, superior temporal cortex; 7, posterolateral temporal cortex; 8, anteromedial temporal cortex; 9, inferior parietal cortex; 10, superior parietal cortex; 11, precuneus; and 12, occipital cortex. Views shown from left to right are, respectively, superior, left hemisphere lateral, right hemisphere lateral, left hemisphere medial, right hemisphere medial, and inferior.
The genetically based clusters presented a spatially contiguous pattern within hemispheres. However, the cluster algorithm placed no constraint against noncontiguous clusters. Indeed, all 12 clusters were noncontiguous clusters bilaterally located in the homologous regions between hemispheres. There were some indications of surface-area asymmetry around perisylvian regions (8), but the patterns of the left and right hemispheres were almost mirror images of one another. Because the clustering was conducted on both hemispheres simultaneously with no constraint for hemispheric symmetry, the results clearly indicate a predominantly bilateral symmetric and within-hemisphere modular pattern.
In order to obtain reliable estimates of the genetic correlations, we applied spatial smoothing, which limited our ability to address the fine spatial structure of the genetic patterning (16). We focused on the large-scale, primary structure of genetic patterning. Other techniques, such as gene expression analysis of brain tissue, could reveal finer-scale genetic patterning that may show more asymmetrical features or more subdivisions (18, 19) than can our magnetic resonance imaging (MRI)–based approach.
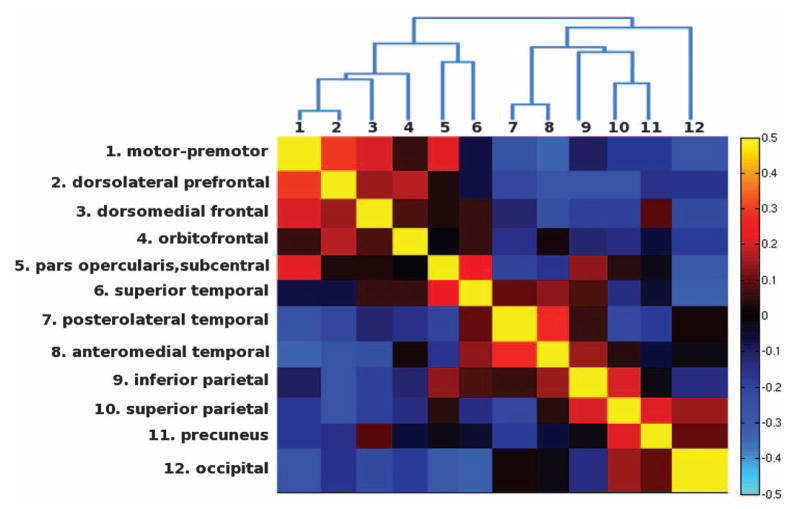
After identifying the boundaries of the genetically based parcellation, we next sought to examine the genetic relations between the 12 clusters; in particular, we searched for underlying organizational principles among these genetic subdivisions. We calculated the genetic similarity matrix to determine the genetic relatedness between clusters (Fig. 2). We found that genetic correlations are higher between clusters within the same lobe than between clusters in different lobes. Also included in Fig. 2 is a dendrogram derived from hierarchical clustering that summarizes the genetic relations between clusters. The dendrogram depicts a hierarchical structure of genetic patterning. The most distinct genetic partitions located at the highest level of the hierarchy correspond to the basic anteroposterior division between motor and sensory cortices; below that are the functionally specialized subdivisions generally nested within lobes. Similarly, a clear basic frontal/nonfrontal division and lobar-like clusters have been revealed by hierarchical clustering derived from transcriptome analyses of the fetal human brain (20, 21). One exception to these general patterns is that clusters belonging to the perisylvian region have relatively high correlations with one another, even though they are in different lobes. Cross-lobe clustering in these regions is consistent with a human-specific subdivision specialized for language.
Fig. 2.
Genetic similarity matrix and dendrogram. The color scale represents the weighted mean genetic correlations within and between clusters. Negative genetic correlations indicate that the genes that cause areal expansion in anterior regions also cause relative areal contraction in posterior regions and vice versa (8).
We also examined the progression of cluster solutions, from 2 to 12 clusters, using fuzzy clustering (fig. S2). If the structure of the data are hierarchical, then successive clusters will tend to be subdivisions of previous clusters (22). In contrast to hierarchical clustering, our approach imposed no constraint for hierarchical organization; each level of the fuzzy clustering analysis was performed independently. Yet, the sequentially unfolding pattern revealed that the emerging clusters tended to respect the boundaries of preceding clusters and appeared to be nested subdivisions. The convergence of results of this analysis and the dendogram method thus provide further evidence for a hierarchical structure of genetic patterning that is intrinsic to the data.
The organization of genetic patterning is consistent with a ubiquitous pattern in the development of biological forms—increasing differentiation along with what appeared to be increasing hierarchical integration, as reflected by functionally specialized subdivisions (23). We previously showed that the four-cluster solution revealed fundamental genetic divisions comprising primary functional regions largely corresponding to the lobar divisions in all mammalian species (8). Our current results demonstrate further differentiation of each of the lobes into several nested subdivisions that correspond specifically to human functional specialization, such as the lateral or granular prefrontal cortex, and regions around Broca’s area and the subcentral region associated with vocalization essential for human language (24). Our results suggest that human specialization regions are not genetically more distinct than primary functional lobar regions. However, small genetic differences resulting in functional importance have become increasingly recognized (11, 25). These findings support the notion that the human cortex is built on the foundation of the primary functional divisions, which are shared among mammals (7). Without any incorporation of prior anatomical knowledge, this statistically constructed hierarchy demonstrated a biologically sensible organizational structure of the human brain.
We described a previously unidentified parcellation system for the human cortex that reflects shared genetic influences on cortical areal expansion. This system constitutes the first human brain atlas based solely on genetically informative data, which may provide presently undescribed phenotypes that will have greater statistical power for genome-wide genetic association studies in comparison with traditional cortical parcellations. We found evidence for a hierarchical, modular, and bilaterally symmetric genetic architecture. Genetically based lobar regions have been demonstrated across mammalian species (7, 8), and our results are consistent with genetically based regions of human specialization being increasingly differentiated subdivisions of these lobar regions. Our findings may thus be useful for translating results from model organisms into functional and clinical insights about human specializations, so as to understand both order and disorder in the human brain.
Supplementary Material
Acknowledgments
This work was funded by the National Institute on Aging (AG022381, AG018386, AG018384, AG022982, and AG031224), National Institute of Drug Abuse (DA029475), National Institute of Neurological Disorders and Stroke (NS056883), National Center for Research Resources (P41-RR14075, BIRN002, and U24 RR021382), National Institute for Biomedical Imaging and Bioengineering (EB006758), National Center for Alternative Medicine (RC1 AT005728-01), National Institute for Neurological Disorders and Stroke (NS052585-01, 1R21NS072652-01, and 1R01NS070963), National Institutes of Health (T32DC000041), and the Ellison Medical Foundation. This material is also partly the result of work supported with resources of the VA San Diego Center of Excellence for Stress and Mental Health. The Cooperative Studies Program, Office of Research and Development, U.S. Department of Veterans Affairs has provided financial support for the development and maintenance of the Vietnam Era Twin (VET) Registry. A.M.D. is a founder and holds equity in CorTechs Laboratories and also serves on its Scientific Advisory Board.
Footnotes
References and Notes
- 1.Bergquist H, Kallen B. Acta Anat (Basel) 1953;18:65. doi: 10.1159/000140825. [DOI] [PubMed] [Google Scholar]
- 2.Fraser S, Keynes R, Lumsden A. Nature. 1990;344:431. doi: 10.1038/344431a0. [DOI] [PubMed] [Google Scholar]
- 3.Wilkinson DG, Bhatt S, Cook M, Boncinelli E, Krumlauf R. Nature. 1989;341:405. doi: 10.1038/341405a0. [DOI] [PubMed] [Google Scholar]
- 4.Puelles L, Rubenstein JL. Trends Neurosci. 2003;26:469. doi: 10.1016/S0166-2236(03)00234-0. [DOI] [PubMed] [Google Scholar]
- 5.Bishop KM, Goudreau G, O’Leary DD. Science. 2000;288:344. doi: 10.1126/science.288.5464.344. [DOI] [PubMed] [Google Scholar]
- 6.Fukuchi-Shimogori T, Grove EA. Science. 2001;294:1071. doi: 10.1126/science.1064252. [DOI] [PubMed] [Google Scholar]
- 7.O’Leary DD, Chou SJ, Sahara S. Neuron. 2007;56:252. doi: 10.1016/j.neuron.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 8.Chen CH, et al. Neuron. 2011;72:537. doi: 10.1016/j.neuron.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rimol LM, et al. Proc Natl Acad Sci USA. 2010;107:384. [Google Scholar]
- 10.Joyner AH, et al. Proc Natl Acad Sci USA. 2009;106:15483. doi: 10.1073/pnas.0901866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rakic P. Nat Rev Neurosci. 2009;10:724. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dale AM, Fischl B, Sereno MI. Neuroimage. 1999;9:179. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 13.Fischl B, Sereno MI, Tootell RBH, Dale AM. Hum Brain Mapp. 1999;8:272. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dale AM, Sereno MI. J Cogn Neurosci. 1993;5:162. doi: 10.1162/jocn.1993.5.2.162. [DOI] [PubMed] [Google Scholar]
- 15.Eaves LJ, Last KA, Young PA, Martin NG. Heredity. 1978;41:249. doi: 10.1038/hdy.1978.101. [DOI] [PubMed] [Google Scholar]
- 16.Materials and methods are available as supporting material on Science Online.
- 17.Kaufman L, Rousseeuw P. Finding Groups in Data: An Introduction to Cluster Analysis. Wiley; New York: 1990. [Google Scholar]
- 18.Sun T, et al. Science. 2005;308:1794. doi: 10.1126/science.1110324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abrahams BS, et al. Proc Natl Acad Sci USA. 2007;104:17849. doi: 10.1073/pnas.0706128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson MB, et al. Neuron. 2009;62:494. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang HJ, et al. Nature. 2011;478:483. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan P-N, Steinbach M, Kumar V. Introduction to Data Mining. 1. Pearson Education; UK: 2006. [Google Scholar]
- 23.Werner H. Comparative Psychology of Mental Development. International Universities Press; New York: 1948. [Google Scholar]
- 24.Jürgens U. Neurosci Biobehav Rev. 2002;26:235. doi: 10.1016/s0149-7634(01)00068-9. [DOI] [PubMed] [Google Scholar]
- 25.Konopka G, Geschwind DH. Neuron. 2010;68:231. doi: 10.1016/j.neuron.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.