SUMMARY
The continuously growing mouse incisor serves as a valuable model to study stem cell regulation during organ renewal. Epithelial stem cells are localized in the proximal end of the incisor in the labial cervical loop. Here, we show that the transcription factor Sox2 is a specific marker for these stem cells. Sox2+ cells became restricted to the labial cervical loop during tooth morphogenesis, and they contributed to the renewal of enamel-producing ameloblasts as well as all other epithelial cell lineages of the tooth. The early progeny of Sox2-positive stem cells transiently expressed the Wnt inhibitor Sfrp5. Sox2 expression was regulated by the tooth initiation marker FGF8 and specific miRNAs, suggesting a fine-tuning to maintain homeostasis of the dental epithelium. The identification of Sox2 as a marker for the dental epithelial stem cells will facilitate further studies on their lineage segregation and differentiation during tooth renewal.
INTRODUCTION
Continuously growing rodent incisors possess epithelial stem cell (SC) niches in contrast to most other mammalian teeth. As teeth share many developmental mechanisms with other ectodermal organs such as hair and mammary gland, the mouse incisor provides an excellent model to examine the characteristics and regulation of epithelial SCs in general. Incisors are covered on their labial side by enamel secreted by the ameloblasts, while the lingual side, lacking ameloblasts, is covered with softer dentin and cementum of mesenchymal origin (Figure 1A). Therefore, the softer, lingual side is more susceptible to abrasion leading to the formation of a cutting edge. Because of the enamel asymmetry, the labial side is often referred to as crown-analog (enamel) and the lingual side as root-analog (no enamel). The SCs are set apart in the proximal end of the incisor and provide a continuous supply of cells to counterbalance the constant abrasion (Harada et al., 1999).
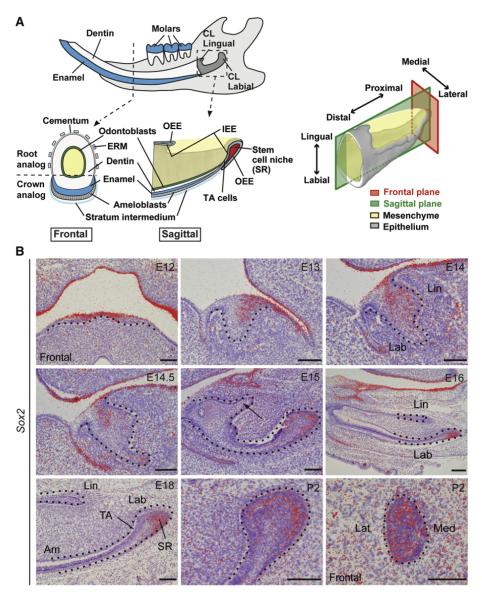
Figure 1. Sox2 Expression in the Oral Epithelium Is Progressively Restricted to the Labial CL during Incisor Development.
(A) Schematic illustrations of the mouse incisor. Left: Lower jaw and higher magnifications of the incisor in a frontal section, and a sagittal section from the proximal part illustrating labial and lingual CLs. Right: 3D reconstruction from histological sections of the proximal part of the incisor.
(B) In situ hybridization in the mouse lower incisor from E12 to P2 reveals gradual restriction of Sox2 mRNA expression to a subset of SR cells and adjacent enamel epithelium in the labial CL. The arrow at E15 indicates disappearance of Sox2 expression in the lingual CL. The dotted line marks the border between epithelium and mesenchyme. All sections are in the sagittal plane unless indicated otherwise.
Am, ameloblasts; CL, cervical loop; ERM, epithelial cell rests of Malassez; IEE, inner enamel epithelium; Lab, labial; Lat, lateral; Lin, lingual; Med, medial; OEE, outer enamel epithelium; SC, stem cell; SR, stellate reticulum; TA, transient amplifying cells. Scale bar, 100 μm.
See also Figure S1.
The epithelial SCs reside in structures called cervical loops (CLs) in the proximal end of the incisor. CLs are composed of inner and outer enamel epithelium (IEE and OEE, respectively) that surround the stellate reticulum (SR), a core of loosely arranged epithelial cells with mesenchymal appearance. Epithelial SCs have been localized to the SR and the adjacent enamel epithelium at the tip of the labial CL (Harada et al., 1999; Seidel et al., 2010). The progeny of the SCs proliferate in the transient amplifying (TA) zone of the IEE and differentiate into ameloblasts that form enamel on the labial crown-analog (Harada et al., 1999).
In contrast to the labial CL, the lingual CL is thin and largely depleted of SR cells. It does not generate ameloblasts, but it regulates dentin and cementum formation at the lingual surface of the incisor (Tummers et al., 2007). Later, the lingual epithelium, analogous to the epithelial cell rests of Malassez (ERMs) covering roots in molars, gives rise to the ERMs, which eventually remain as a network covering the lingual side of the incisor (Ten Cate, 1996; Tummers and Thesleff, 2008). Some SR cells and label-retaining cells (LRCs) have been localized in the tip of the lingual CL (Tummers and Thesleff, 2009; Seidel et al., 2010), but the identity and exact location of these putative SCs remain unknown.
Conserved signaling pathways such as fibroblast growth factor (FGFs), bone morphogenetic protein (BMP), transforming growth factor-β (TGF-β), and sonic hedgehog (Shh) regulate the maintenance and proliferation of the epithelial SCs and their progeny in the labial CL (Tummers and Thesleff, 2009; Seidel et al., 2010; Zhao et al., 2011). Fine-tuning of these signaling pathways affects the size, hard-tissue formation, and symmetry of the incisor (Wang et al., 2004; Plikus et al., 2005). Although some general SC marker genes such as Lgr5 (Suomalainen and Thesleff, 2010), ABCG2, Bmi-1, Oct-3/4, and Yap (Li et al., 2011) were recently detected in the SR of the labial CL, no specific marker for the epithelial SCs in incisors is known.
In this study, we characterized the gene-expression profile of the labial CL of the mouse incisor and assessed the expression patterns of selected candidate genes to find a specifically expressed marker for the epithelial SCs. We localized Sox2 expression in the incisor SCs, demonstrated that Sox2+ SCs contribute to all epithelial lineages of the incisor, and showed that Sox2 expression is regulated by FGF8 and miRNAs. Additionally, we identified the noncanonical Wnt inhibitor Sfrp5 as a marker for dental epithelial progenitors and showed that some of these progenitors derive from Sox2+ SCs.
RESULTS
Gene-Expression Profile of the Labial Cervical Loop of the Incisor
To find markers for epithelial SCs in the mouse incisor, we compared the gene-expression profiles and gene ontology processes (GOPs) between samples from postnatal day (P) 2 mouse incisors containing only labial CL to samples containing the entire proximal area of the incisor (Figure 1A). The microarray gene-expression profiling revealed that 1283 genes were enriched (p value < 0.005; fold > 1.5) in the labial CL and that 515 of these had at least 50% higher level of expression in the labial CL compared to the entire proximal area of the incisor (Table S1A available online). GOP analysis indicated that 95 GOPs were enriched over 2.5 fold in the labial CL (Table S1B). Several of these GOPs were specific to ectodermal processes (e.g., ectodermal organ formation and epidermal cell proliferation), which indicated a negligible amount of mesenchymal cells in the labial CL samples. Among the 515 prominently expressed genes, and associated with 24 of the 95 enriched GOPs, was the transcription factor Sox2. Because Sox2 plays a critical role in the maintenance of SCs in various organs (Suh et al., 2007; Arnold et al., 2011) and is an iPS cell co-inducer (Takahashi and Yamanaka, 2006), we decided to analyze its expression pattern during mouse incisor development and renewal.
Sox2 Is Expressed in the Incisor Stem Cell Niche
We localized Sox2 expression by radioactive in situ hybridization in mouse incisors from embryonic day (E) 12 to P2 (Figure 1B). Throughout this period, Sox2 expression was intense in the oral epithelium. At E12, Sox2 was expressed widely in the oral surface epithelium but was absent from the basal epithelium of incisor placodes, while intense expression remained in the epithelium between the placodes. At E13, the lingual side of the forming bud showed Sox2 expression, and at E14 cap stage, expression appeared in both lingual and labial CLs protruding from the bud. At this stage, the labial CL starts to grow more rapidly and the labial-lingual asymmetry becomes morphologically visible. At E14.5, the proximal end of the labial CL showed distinct Sox2 expression, while the expression in the lingual CL had decreased. Expression disappeared from the lingual CL by E15, but strong expression remained in the labial CL. At E16 and E18, Sox2 became increasingly restricted to the tip of the labial CL and was confined to this region at P2. Expression was restricted to an area consisting of SR cells and cells of the adjacent enamel epithelium layer. The same expression pattern of Sox2 remained in the incisor at P30 (Figure S1). The location of the most intense Sox2 expression corresponded to that of the putative epithelial SC population (Harada et al., 1999; Seidel et al., 2010). We also addressed Sox2 expression in upper incisors and molars: Sox2 was expressed in the labial cervical loop of the upper incisors in a manner similar to that observed in the lower incisors, whereas in molars, Sox2 was expressed in the cervical loops only before the initiation of root development (data not shown).
To further follow the dynamics of Sox2 expression during incisor development, we dissected E14.5 incisors from the Sox2-GFP reporter mouse expressing GFP under control of the Sox2 promoter (D’Amour and Gage, 2003) and monitored GFP expression in vitro (Figure 2A). Initially, GFP was present throughout the oral epithelium, as well as in both lingual and labial CLs, and later, after 24 hr, it became gradually confined to the tip of the labial CL. Like Sox2 mRNA, Sox2-GFP was absent from the lingual CL (Figures 1B and 2A).
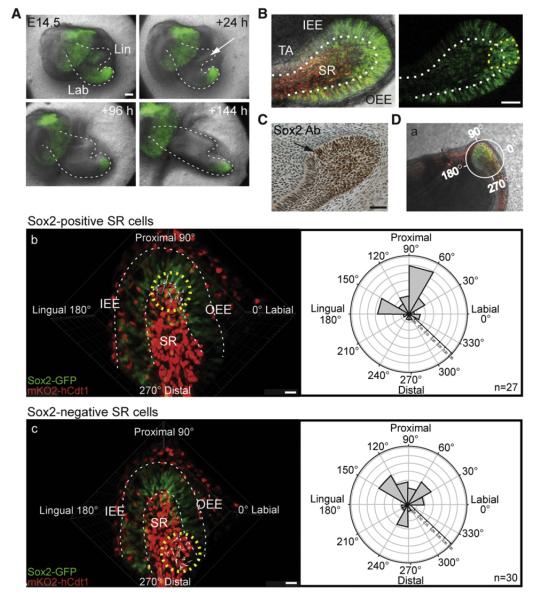
Figure 2. Culture of Sox2-GFP Mouse Incisor Demonstrates Restriction of Sox2+ Cells to Labial CL and Their Migration toward Enamel Epithelium.
(A) Expression of GFP in the Sox2-GFP reporter mouse incisor in tissue culture initiated at E14.5. The white arrow marks the disapperance of GFP signal from lingual CL.
(B) Confocal images of a living tissue slice of labial CL of Fucci-red;Sox2-GFP mouse at P2 with GFP (green) and mKO2 (red) expression on the left and only GFP expression on the right. GFP expression is intense in a subset of cells within the labial CL (yellow dotted line), whereas GFP gradually disappears from IEE when the cells reach the level of TA cells.
(C) Sox2 antibody staining in labial CL of P2 wild-type incisor. The arrow points to the border in the IEE where the Sox2 protein level decreases.
(Da–Dc) (Da) Living tissue slice of labial CL of P2 Fucci-red;Sox2-GFP mouse at P2 showing the imaged area and orientation for cell movement analysis. (Db and Dc) Still images of time-lapse movies show tracking (Movies S1 and S2) and polar plot analysis the orientation of the movement (small arrows point to the direction of overall movement of individual cells) of Sox2+ (Db) and Sox2-negative (Dc) cells (yellow circles).
Scale bars: 100 μm (A), 60 μm (B and C), 15 μm (D).
See also Figure S2.
To localize the Sox2-GFP-positive cells more precisely, we crossbred transgenic Fucci-red mice (Sakaue-Sawano et al., 2008), exhibiting G1 phase nuclei red, with Sox2-GFP mice. We used fluorescence confocal microscopy on 150-μm-thick sagittal slices of the labial CL at P2 (Figure 2B). For comparison, we detected the expression of Sox2 protein by antibody in the wild-type incisor. GFP (Figure 2B) and Sox2 (Figure 2C) were expressed in the proximal part of the labial CL in the same area where Sox2 mRNAs were located (Figure 1B). Additionally, expression was detected more widely in the IEE and OEE, where no Sox2 transcripts were detected. Thus, Sox2 protein seemed to perdure in the early progeny of Sox2+ cells.
To examine the movement of Sox2+ cells in the labial CL, we established a system that enables imaging of live tissue slices and 3D time-lapse tracking of individual cells. Transgenic Fucci-red crossbred with Sox2-GFP mice were used to detect nuclei and to follow cell viability during imaging. Live imaging revealed a preferential direction of cell movement of Sox2+ SR cells (n1 = 27) toward the tip of the loop where the cells reached the enamel epithelium (Figures 2Db and S2; Movie S1), whereas the movement of the Sox2-negative SR cells (n2 = 30) was more uniformly distributed (Figures 2Dc and S2; Movie S2). The track length and the direction of cell migration differed significantly between Sox2+ and Sox2-negative SR cells (Figure S2). The migration of the Sox2+ cells was also confirmed by the presence of cellular protrusions in the leading edge, and the cells appeared to migrate within the IEE (Movie S1). Collectively, these data indicate that during incisor morphogenesis, Sox2+ cells become enriched to the SR of the labial CL SC niche from where they migrate to the IEE and further to the TA region during ameloblast renewal.
Sox2+ Stem Cells Contribute to All Epithelial Cell Lineages of Incisor
To definitively verify that Sox2-expressing cells in the mouse incisor are SCs, we utilized genetic inducible fate mapping in the Sox2-CreER;R26R mouse line, in which tamoxifen transiently induces Cre-recombinase leading to permanent expression of lacZ in Sox2+ cells and their progeny (Arnold et al., 2011). We genetically labeled Sox2-expressing cells by administering tamoxifen in vivo at P2 and traced their descendants by detecting lacZ expression after 48 hr, 1 week, and 1 month (Figure 3A). After 48 hr, a small number of lacZ+ cells were detected in the epithelial compartments of the labial CL including SR, IEE, and OEE (Figure 3Ca) corresponding to the areas of Sox2-GFP expression (Figure 2B). After one week, more lacZ+ cells were located in these areas (Figure 3Cb). Additionally, lacZ+ cells were detected more distally in TA cells and ameloblasts as well as in the stratum intermedium (SI) cells that underlie ameloblasts and are derived from SR cells. This pattern is consistent with the progression of IEE cell differentiation into ameloblasts toward the incisor tip (Hwang and Tonna, 1965).
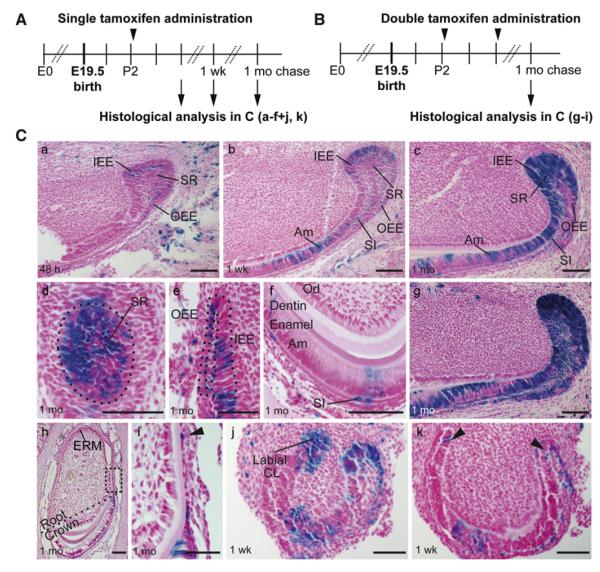
Figure 3. Genetic Inducible Fate Mapping Demonstrates that Sox2+ Cells in the Adult Incisor Are SCs.
(A) Timing of tamoxifen induction and histological analysis in Sox2Cre-ER;R26R mice.
(B) Timing of tamoxifen double induction and histological analysis.
(Ca–Ck) LacZ expression 48 hr (Ca), one week (Cb, Cj, and Ck), and one month (Cc–Cf) after single tamoxifen induction. Frontal sections (Cd–Cf) show the distinct lacZ+ epithelial cell types derived from Sox2+ cells: SR (Cd), IEE and OEE (Ce), ameloblasts and SI cells (Cf), ERMs (Ch and Ci), and IEE/OEE ridge (Cj and Ck). (Cg–Ci) LacZ expression one month after double induction. The region marked in (Ch) is shown in higher magnification in (Ci). The arrowhead in (Ci) points to ERM cells on the lateral side of the incisor.
CL, cervical loop; ERM, epithelial cell rests of Malassez; IEE, inner enamel epithelium; OEE, outer enamel epithelium; SR, stellate reticulum; TA, transient amplifying cells; Am, ameloblasts. Scale bar: 100 μm, except in (Ci), where it is 50 μm.
See also Figure S3.
One month after induction, the amount of lacZ+ cells had further increased, covering almost the whole SR and the IEE compartment (Figures 3Cc and 3Cd), demonstrating the self-renewing capacity of Sox2+ cells. LacZ+ cells were apparent in the OEE and IEE layers, as well as in ameloblasts and SI cells (Figures 3Ce and 3Cf). No lacZ+ cells were detected at any time point in the lingual cervical loop or elsewhere in the lingual side of the incisor. A Sox2-wt;R26R mouse line was used as a control; it showed no lacZ-stained cells in the dental epithelium (data not shown).
The absence of lacZ+ cells on the lingual side could be due to a low participation of the labial CL in the renewal of the lingual side of the incisor. To increase the labeling efficiency and to decrease mosaicism, we administered tamoxifen at P2 and P4 (Figure 3B). More lacZ+ cells were detected in the epithelial compartments of the labial side after double induction, as compared to single induction, and the SR of the labial CL was totally covered with lacZ+ cells (Figure 3Cg). Additionally, we detected some lacZ+ cells in the ERM cell layer close to the crown-root junction (Figures 3Ch and 3Ci).
To confirm that Sox2+ SCs are present in adult mice, we applied tamoxifen at 5 weeks (Figure S3A). After 3 days, the lacZ expression was similar to that at 48 hr after induction at P2 (Figure S3Cb).
These results conclusively demonstrate that adult SCs in the labial CL express Sox2, and that these Sox2+ SCs contribute to all epithelial cell lineages in mouse incisor.
The Progeny of Sox2+ Stem Cells Express Sfrp5 Prior to Differentiation
Canonical Wnt signaling is absent from the SC niche in embryonic incisors (Suomalainen and Thesleff, 2010; Liu et al., 2010). We did not detect canonical Wnt activity in the SC niche either in postnatal incisors using Axin2 expression (Lustig et al., 2002) or in TOPGAL (DasGupta and Fuchs, 1999) and BATGAL (Maretto et al., 2003) mice as reporters (Figure S4A). Sox2 is known to downregulate canonical Wnt signaling (Mansukhani et al., 2005), but our microarray showed that Wnt inhibitors CD9 (Huang et al., 2004), Ldb1 (Dey-Guha et al., 2009), and Sfrp5 (Li and Clevers, 2010) were also enriched in the labial CL (Table S1A). In situ hybridization indicated that CD9 and Ldb1 were indeed expressed in the SC area (Figure S4B). In addition, CD9 was found in one GOP and Ldb1 in two GOPs enriched in the labial CL (Table S1B). These data suggest that these Wnt inhibitors may contribute to the suppression of canonical Wnt signaling in the labial CL together with Sox2.
Secreted frizzled-related protein 5 (Sfrp5) is an antagonist of the noncanonical Wnt signaling that is associated with the regulation of SC niche activity in the intestine (Li and Clevers, 2010). It was not expressed in the labial CL SC area. Instead, it showed an intense, strictly localized expression in the junction between IEE and OEE surrounding the open proximal end of the incisor and was completely absent from the SR compartment at P2 (Figures 4Aa–4Ac). Sfrp5 was also expressed in the IEE/OEE junction in embryonic incisors at E16 and E18 (Figure S4C). Interestingly, in the labial CL, the expression pattern of Sfrp5 was complementary to that of Shh, which marks proliferating and differentiating ameloblast precursors (Gritli-Linde et al., 2002): in contrast to Sfrp5, Shh was not expressed in the IEE/OEE junction (Figures 4Ad and 4Ae).
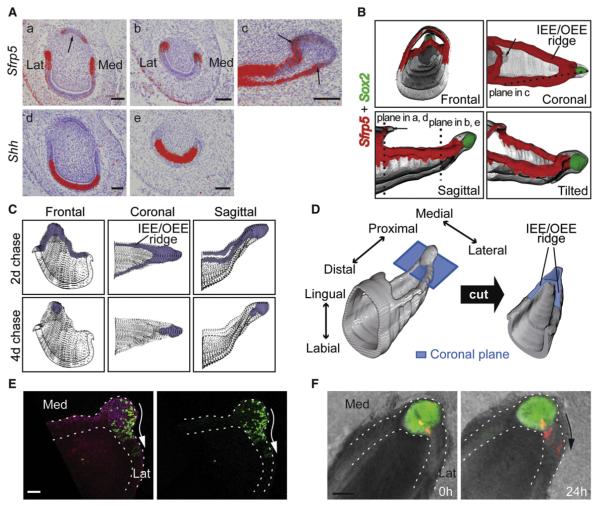
Figure 4. The Early Progeny of Sox2+ SCs Express Sfrp5.
(Aa–Ae) In situ hybridization of Sfrp5 (Aa–Ac) and Shh (Ad and Ae) in frontal and sagittal (Ac) sections of P2 incisor. Notice that Sfrp5 is absent from part of the lingual CL, as indicated by the arrow in (Aa). The planes of the frontal and sagittal sections are shown in the sagittal and coronal views of the proximal part of the incisor in (B). (B) 3D reconstruction of Sfrp5 (red) and Sox2 (green) expression patterns from in situ hybridization of serial sections. Notice that in the coronal view, Sfrp5 expression forms a circle marking the cells of the IEE/OEE ridge in the proximal opening of the incisor. Sox2 expression is restricted to the tip of the labial CL and does not overlap with Sfrp5 expression in either coronal or sagittal views. Arrows in coronal and sagittal views point out Sfrp5-negative cells.
(C) Schematic 3D representation of the LRCs of rat incisor redrawn from Smith (1980). 3H-Thymidine incorporation in rat incisor was monitored 2 and 4 days after injection. After 2 days, LRCs were localized in both the labial CL and IEE/OEE ridge. After 4 days, only the cell population within the labial CL exhibited labeling.
(D) 3D illustration of the proximal area of the P2 incisor shows the coronal section plane of thick slices from the Sox2-GFP incisor in (E) and (F).
(E) GFP expression in a thick coronal slice with BodipyTR and Draq5 counterstaining (left), and alone (right). The arrow shows the proposed direction of cell movement.
(F) DiI-labeled labial CL cells are detected in the IEE/OEE ridge 24 hr after injection. The arrow shows the direction of cell migration.
Lat, lateral; Med, medial. Scale bars: 60 μm in (E), 100 μm in (A) and (F). See also Figure S4.
To examine the relationship of the Sox2+ SCs and Sfrp5+ progenitor cells in the incisor, we made 3D reconstructions of Sfrp5 and Sox2 expression patterns from identical serial sections of the proximal part of the incisor. The coronal view revealed that Sfrp5 expression was precisely located in the IEE/OEE junction between the tips of labial and lingual CLs, and surrounded the open proximal end of the incisor, which formed a morphologically detectable ridge (Figure 4B). The Sfrp5+ IEE/OEE ridge was morphologically visible also in the sagittal view, which in addition revealed a restricted population of Sfrp5-negative cells at the tip of the lingual CL (Figures 4B and S4C). The expression patterns of Sfrp5 and Sox2 in the labial CL were complementary, with no overlap, as the two genes were expressed in two adjacent cell populations (Figure 4B). In addition, the Sfrp5 expression surrounded the Sox2+ cells in the labial CL (Figures 4Ac and 4B).
Strikingly, the expression patterns of Sox2 and Sfrp5 closely resemble the 3H-Thymidine incorporating LRC populations in the adult rat incisor reported by Smith (1980). We adapted these LRC patterns to our 3D reconstructions and observed that Sox2+ cells corresponded with the LRCs after 4 days chase and Sfrp5+ cells with the LRCs after 2 days chase (Figure 4C). Interestingly, Smith (1980) suggested that the labeling dynamics of the LRCs after 2 days chase reflected their derivation from the labial CL SCs.
To see if the Sfrp5+ cells arose from the Sox2+ SCs, we made 150-mm-thick living coronal slices of the labial CL of Sox2-GFP incisor (Figure 4D) and checked the pattern of GFP-expressing cells with confocal imaging (Figure 4E). GFP expression was intense in SR and relatively faint in IEE/OEE ridge. This would be accountable for by the persistence of GFP in the early progeny of Sox2+ cells in the IEE/OEE and would reconcile with the persistence of Sox2 itself (compare to Figures 2B and 2C). By injecting DiI to the SR compartment of the labial CL slice in vitro cultures, we observed that cells advanced from the SR of the labial CL to the IEE/OEE ridge within two days (Figure 4F). These results were consistent with the localization of lacZ+ cells to the Sfrp5+ area in the IEE/OEE ridge by the genetic fate mapping (Figures 3Cj and 3Ck). Together, the findings above suggest that Sfrp5+ cells represent the progeny of Sox2+ cells in the labial CL.
Sox2 Expression in Incisor SCs Is Induced by FGF8 and Fine-Tuned by MicroRNAs
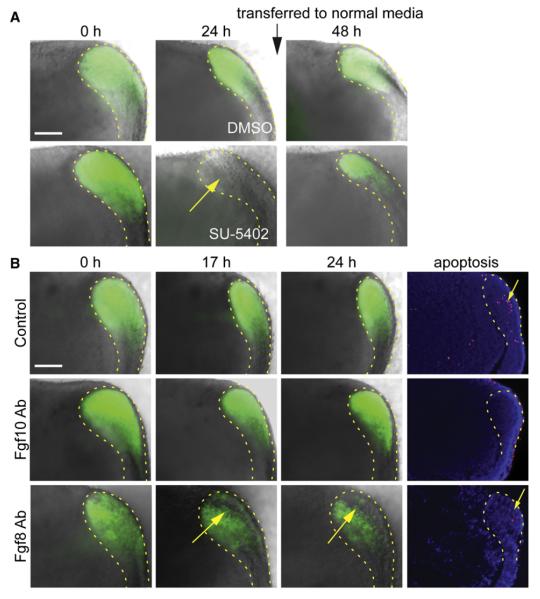
Because FGFs have been repeatedly associated with the regulation of incisor epithelial SCs (for review, see Tummers and Thesleff, 2009), we examined the consequences of inhibition of Fgf signaling in cultured CLs of P2 Sox2-GFP mice (Figure 5A). After exposure to the global FGF inhibitor SU-5402 for 24 hr, GFP expression disappeared (Figure 5A). This effect was reversible, such that after 24 hr culture in control medium, the GFP expression reappeared (Figure 5A). Thus, FGF signaling is required for Sox2 expression.
Figure 5. FGF8, but Not FGF10, Is Required for Sox2 Expression in the CL.
(A) Sox2-driven GFP expression in a cultured labial CL of P2 incisor under global inhibition of FGFs by SU-5402 (bottom) and in control with DMSO (top). Note the suppression of GFP expression after 24 hr with SU-5402 (yellow arrow) and the reappearance of expression within 24 hr after removal of inhibition.
(B) Sox2-driven GFP expression (green) and apoptotic cells (red) in a cultured labial CL of P2 incisor in control (top row) and under inhibition of FGF10 and FGF8 (middle and bottom rows, respectively) by blocking antibodies. Note the suppression of GFP expression and unaffected apoptosis by FGF8 inhibition (yellow arrows) and increased apoptosis in the periphery of the explants under FGF10 inhibition.
Scale bars, 100 μm. See also Figure S5A.
Since global FGF inhibition does not differentiate between individual Fgfs, we next attempted to identify the FGF(s) inducing Sox2 expression. FGF10 is generally considered as the key signal in the maintenance of SR cells (Harada et al., 2002). In accordance with previous studies (Harada et al., 1999; Yokohama-Tamaki et al., 2008), FGF10 was expressed in the mesenchyme close to the Sox2+ cells in the CL, and the expression of its receptors Fgfr2b and Fgfr1b overlapped with the Sox2+ cells (Figure S5). To test the involvement of FGF10 in Sox2 expression, we introduced blocking antibodies against FGF10 to P2 CL cultures (Harada et al., 2002). However, the pattern and intensity of GFP expression remained similar to the control after 17 and 24 hr, and no increase of apoptosis was detected (Figure 5B). FGF8, necessary for early tooth development (Trumpp et al., 1999), is expressed in the adult incisor (Wang et al., 2009) and induces Sox2 expression in the olfactory epithelium (Tucker et al., 2010). We found that Fgf8 and its receptor Fgfr1c (Zhang et al., 2006) were expressed in the SR and colocalized with Sox2 (Figures 1 and 6A). Moreover, the addition of FGF8 blocking antibodies (Jacques et al., 2007) to organ culture resulted in a progressive reduction of Sox2-GFP during 24 hr of culture in a restricted cell population overlapping with Fgf8 expressing cells. Increased apoptosis was not detected (Figure 5B). These results indicate that FGF8, but not FGF10, is required for expression of Sox2.
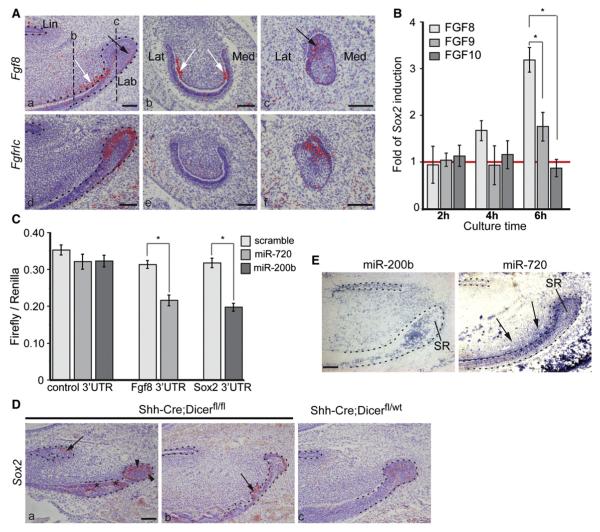
Figure 6. Sox2 Expression Is Induced by FGF8, and Expression of Both Sox2 and Fgf8 Is Regulated by miRNAs.
(Aa–Af) In situ hybridization of Fgf8 and FgfrIc in sagittal (Aa and Ad) and frontal (Ab, Ac, Ae, and Af) sections of the P2 incisor. Fgf8 is expressed in the labial CL in a restricted cell population in the distal part of the SR, as indicated by black arrows (Aa and Ac), and in preodontoblasts, as indicated by white arrows (Aa and Ab). Fgfr1c expression is restricted to the CL epithelium and localizes to the same area as Fgf8 in the distal part of the CL seen in frontal section (Af). Dashed lines in (Aa) show the positions of frontal sections depicted in (Ab) and (Ac).
(B) Effects of FGF8, FGF9, and FGF10 proteins on Sox2 expression in E14 incisor as measured by qRT-PCR.
(C) Effects of miRNAs on Fgf8 and Sox2 expression in a Luciferase assay.
(Da–Dc) Sox2 expression in sagittal sections from P5 Shh-Cre;Dicerfl/fl (Da and Db) and Shh-Cre;Dicerfl/wt incisors (Dc). (Da) Note ectopic Sox2 expression in the lingual CL (arrow) and in the preameloblasts (asterisks). Expression seems to be more intense in the IEE and OEE of labial CL (arrowheads) (Da) compared to Sox2 expression in the control (Dc). In the mutant, Sox2 expression is found also in the tips of the extra foldings of epithelium (arrow) (Db).
(E) Expression of miR-200b and miR-720 in sagittal sections of P2 wild-type incisor. MiR-200b expression is located in the SR of the labial CL, whereas in the lingual CL, expression is seen in all epithelial cells. MiR-720 is located in the enamel epithelium and in the mesenchyme next to the TA cells and in the preodontoblasts (arrows). No expression is detected in the SR.
Bars in (B) and (C) correspond to average ± SD, n = 3. Asterisk indicates p < 0.01. Lab, labial; Lat, lateral; Lin, lingual; Med, medial. Scale bars: 100 mm. See also Figure S5B.
To examine whether the FGFs could directly activate Sox2 expression, we quantified Sox2 mRNA levels in E14.5 incisor tooth germs cultured in hanging drops with FGF8, FGF10, or FGF9, another potential regulator of Sox2 in the Fgf family, which is expressed at a close range to the Sox2+ cells in the TA cells (Yokohama-Tamaki et al., 2008). FGF8 increased Sox2 expression after 6 hr by 318% and FGF9 by 177% (Figure 6B). In comparison, FGF10 did not affect Sox2 expression (Figure 6B). These results further support the specific requirement of FGF8 for Sox2 expression in epithelial SCs and also suggest a minor role for FGF9.
MicroRNAs are required for proper incisor development, and several miRNAs are specifically expressed in the labial CL (Michon et al., 2010). Fgf8 is a predicted target of miR-720 and Sox2 a target of the miR-200 family, both of which are expressed in the CL (Michon et al., 2010). We used an in vitro luciferase assay to assess the possible regulation of Sox2 and Fgf8 expression by these miRNAs. MiR-720 decreased luciferase activity linked to Fgf8 3′UTR by 31.8% and MiR-200b activity linked to Sox2 3′UTR by 37.6% compared to scramble miRNA (Figure 6C). These results indicate that Sox2 is regulated by miR-200b and Fgf8 is regulated by miR-720 posttranscriptionally.
To validate the involvement of miRNA regulation in Sox2 and Fgf8 expression, we analyzed the expression of both genes in the incisor of P5 Shh-Cre;Dicer-1fl/fl mice in which the Dicer-1 enzyme required for miRNA processing is conditionally deleted in the epithelium. The Cre recombination efficiency was validated after LacZ staining of P5 Shh-Cre;Rosa26R incisor (Figure S5B). Extra foldings appeared in the epithelium of the incisor, as previously reported for K14-Cre;Dicer-1fl/fl mice (Michon et al., 2010). The domain of Sox2 expression was clearly extended toward the distal tip of the P5 Shh-Cre;Dicer-1fl/fl incisor and covered the TA cells and ameloblasts (Figure 6D). Moreover, Sox2 was ectopically expressed in the lingual cervical loop. No clear modification of Fgf8 expression was visible in the mutant incisor (data not shown).
In situ hybridization in P2 mouse incisor revealed that MiR-200b was expressed in the SR, preameloblasts, and ameloblasts on the labial side, and in the lingual CL (Figure 6E). Similar to E18 mouse incisor (Michon et al., 2010), miR-720 was expressed in IEE and OEE surrounding the SC niche, as well as in preameloblasts and preodontoblasts (Figure 6E). This expression pattern correlated with the ectopic Sox2 expression observed in the Shh-Cre;Dicer-1fl/fl mutant (Figure 6D).
These results confirm that Sox2 expression is regulated by miRNA. Additionally, the ectopic Sox2 expression in Shh-Cre; Dicer-1fl/fl might explain the defects in ameloblast differentiation previously reported with conditional deletion of Dicer-1 (Michon et al., 2010; Cao et al., 2010).
DISCUSSION
In this study, we show that (1) Sox2 is a specific marker for the epithelial SCs in the labial CL of the continuously growing incisor; (2) the Sox2+ SCs contribute to all cell lineages of dental epithelium: the ameloblasts, SI, SR, OEE, and ERM cells; (3) Sfrp5 is a specific marker for the early progeny of the CL SCs; and (4) Sox2 expression is induced by FGF8 and fine-tuned by microRNAs.
Sox2 is an SC marker in various contexts and a cofactor in iPS cell reprogramming (Arnold et al., 2011; Takahashi and Yamanaka, 2006). Our findings indicate that Sox2 expression also marks the epithelial SCs in the incisor. Sox2 expression appeared during early phases of tooth morphogenesis and was progressively restricted to the SC niche. This suggests an early function for Sox2 in the specification of SCs in the dental epithelium, and it is supported by recent findings in other tissues where developmental fate mapping of Sox2 and Sox9 expressing cells revealed that adult SCs originate from embryonic tissue progenitors (Nowak et al., 2008; Arnold et al., 2011). Sox2 was expressed in postnatal incisor at the tip of the labial CL in the restricted population of SR cells and adjacent enamel epithelium. This expression pattern is in line with the localization of SCs previously defined by the presence of LRCs and lineage tracing (Smith, 1980; Harada et al., 1999; Seidel et al., 2010). Moreover, the observed pattern of the Sox2+ genetic inducible fate mapping demonstrated conclusively that Sox2+ cells are adult SCs in the mouse incisor and that they replenish all dental epithelial cell lineages during incisor renewal (Figure 7B). However, our results from genetic inducible fate mapping do not exclude the existence of a Sox2-negative SC population in addition to the Sox2+ SCs in the labial CL. Additional experiments are also required to test whether all Sox2+ cells in the labial CL harbor stem cell activity.
Figure 7. Schematic Presentation of the Movement of Sox2+ SC Progeny and the Generation of Distinct Epithelial Cell Types.
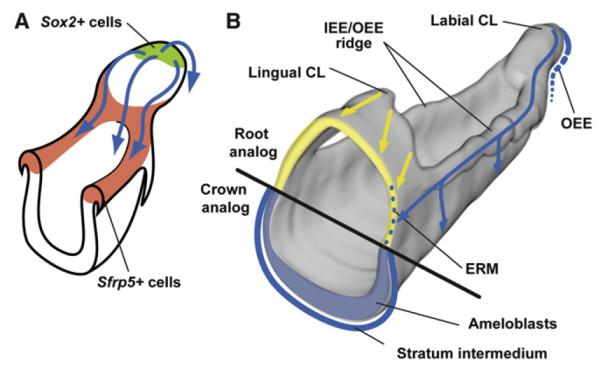
(A) Sox2+ SCs (green) in the distal tip of the labial CL give rise to progeny that move in four main directions (blue arrows), all going through the Sfrp5+ area (red): the ameloblast lineage along the labial surface of the incisor, the IEE/OEE ridge on both sides, and the OEE in the posterior part of the labial CL.
(B) The progeny from Sox2+ SCs (blue) form ameloblasts and SI, SR, and OEE cells and take part in the renewal of the ERM (yellow).
CL, cervical loop; ERM, epithelial rests of Malassez.
Sfrp5+ cells surrounded the Sox2+ SC niche and these populations appeared to correspond with the two LRC populations described by Smith (1980) in the labial CL of rat incisor. Smith suggested that the LRCs in the IEE/OEE ridge are derived from the SCs in the tip of the CL, which is supported by our analysis of the movement of cells from the Sox2+ SC niche to the Sfrp5+ IEE/OEE ridge. We propose that Sfrp5 expression marks the early progeny of Sox2+ SCs and suggest that this stage of Sfrp5 expression precedes their differentiation toward distinct cell fates (Figure 7A). It is intriguing that Sfrp5 has been associated with the regulation of the SC niche in the intestine and that it has been proposed to play a role in separating quiescent from active SCs through inhibition of the Wnt pathway (Li and Clevers, 2010).
Interestingly, Sox2 and Sfrp5 are both involved in Wnt pathway inhibition. Sox2 encodes an inhibitor of the canonical Wnt pathway (Kelberman et al., 2008) and Sfrp5 encodes a specific inhibitor of Wnt5a and Wnt11, which activate the canonical and noncanonical Wnt pathways (Li et al., 2008) and are expressed in the incisor (Suomalainen and Thesleff, 2010). Canonical Wnt signaling is inhibited in the labial CL epithelium including the areas of Sox2 and Sfrp5 expression (Suomalainen and Thesleff, 2010; this study). We identified two additional Wnt inhibitors, Ldb1 and CD9, in the CL, suggesting the importance of Wnt signaling inhibition for the epithelial SCs and their early progeny. Indeed, ectopic activation of Wnt signaling leads to the loss of differentiated ameloblasts and disrupts enamel formation (Millar et al., 2003). In addition to Wnt inhibition, Sox2 and Sfrp5 may have other roles during incisor renewal, as Sox2 is known to induce the expression of hundreds of genes, including some necessary for stemness (Kim et al., 2008).
The lingual CL has been suggested to house SCs based on the localization of a small population of SR cells including LRCs (Tummers and Thesleff, 2009; Seidel et al., 2010). These may correspond to the small area of Sox2- and Sfrp5-negative cells in the lingual CL observed in this study. We suggest that this small SC population in the lingual CL may replenish the Sfrp5+ IEE/OEE ridge forming the root sheath, which generates the most lingual part of the root analog. Additionally, Sox2+ SCs from the labial CL may replenish the part of the root analog closer to the crown analog where we detected lacZ+ ERM cells (Figures 1A, 3Ci, and 7B).
As previously demonstrated, FGFs are important regulators of the maintenance of the incisor SC niche (Harada et al., 2002;Klein et al., 2008) as well as of the transition of crown to root during molar development (Yokohama-Tamaki et al., 2006). Our findings uncovered a role for FGF8 as an autocrine regulator of the epithelial SCs in the incisor. This finding was unexpected, as mesenchymal FGFs, in particular FGF10, are currently regarded as the key regulators of epithelial SCs in the incisor (Harada et al., 2002; Parsa et al., 2010). However, FGF10 did not influence Sox2 expression, although its receptors Fgfr1b and Fgfr2b are expressed in the SC area. We showed that Fgf8, its receptor Fgfr1c, and Sox2 were coexpressed in the tip of the labial CL, and that Sox2 expression was induced by FGF8 and inhibited by FGF8 blocking antibodies in cultured CLs. Our results suggest a direct role for FGF8 in the maintenance of stemness in the Sox2+ cells. The posttranscriptional regulation of Fgf8 and Sox2 by miRNAs is in line with the loss of epithelial homeostasis observed in the Shh-Cre;Dicer-1 cKO and suggests that fine-tuning of Fgf8 and Sox2 expression levels might be necessary to maintain the homeostasis of the dental epithelium.
The availability of new markers for dental epithelial SCs and progenitors, together with an increased understanding of mechanisms of SC maintenance and differentiation in the tooth, will be important assets in advancing our knowledge about tooth renewal. Furthermore, as the SC niche in the incisor resembles the niches in other organs, such as the hair bulge and the intestinal crypt, our results will benefit studies on multipotent SCs in the renewal of epithelial organs.
EXPERIMENTAL PROCEDURES
Animals and Tissue Preparation
NMRI mice were used at various embryonic and early postnatal stages. Plug day was taken as E0 and embryos were staged according to morphological criteria. Sox2-GFP mice, in which eGFP expression is under the control of a 5.5 kb fragment of the upstream regulatory element of the Sox2 promoter (D’Amour and Gage, 2003), were a kind gift from Fred H. Gage (Salk Institute). Fucci-red (mKO2-hCdt1) mice were from RIKEN (Sakaue-Sawano et al., 2008). Mice carrying the Shh-GFPCre (Harfe et al., 2004), Dicer-1fl/fl (Harfe et al., 2005), R26R (Soriano, 1999), or Sox2CreERT2 (Arnold et al., 2011) alleles or transgenes were maintained and genotyped as previously described. Incisors were dissected in Dulbecco’s PBS, pH 7.4. For histology, tissues were fixed, decalcified, dehydrated, and embedded in paraffin. For culture, the incisors from lower jaws at E14.5 or P2 stages were dissected and cultured as previously described (Harada et al., 1999). The preparation of living tissue slices is described in the Supplemental Information.
Transcriptome Microarray Analysis
Dissected P2 labial CLs from NMRI mice composed the first sample, and the total proximal area of the incisor, containing both lingual and labial CLs, composed the second sample. Most mesenchymal tissue was mechanically removed from the first sample. Biological triplicates for each sample were analyzed. RNA quality and concentration were monitored using a 2100 Bioanalyzer (Agilent Technologies). RNAs were processed and hybridized by Biomedicum Genomics (Helsinki, Finland) on Agilent mouse genome 44K array and extensive data analysis was performed using GeneSpring GX11.0 software (Agilent). The genes exhibiting a p value below 0.005 were chosen for further analysis. The functional enrichment of gene ontologies was sorted out using Limma software (t-values).
In Situ Hybridization and Immunostaining
Radioactive in situ hybridization on 7 μm paraffin frontal and sagittal sections was carried out according to standard protocols (Suomalainen and Thesleff, 2010). 35S (Amersham)-labeled RNA probes were used to detect the expression of Sox2 (Ferri et al., 2004), Sfrp5 (Witte et al., 2009), Fgf8 (Kettunen and Thesleff, 1998), and FgfrIc (Kettunen et al., 1998).
MicroRNA in situ hybridization on 7 μm sagittal sections was carried out as previously described (Michon et al., 2010) using double DIG labeled LNA probes for U6 (positive control), scramble (negative control), mmu-miR-200b, and mmu-miR-720 (Exiqon).
For Sox2 immunostaining, paraffin sections were rehydrated and heated in a microwave in 10 mM Na-citrate buffer (pH 6.0). Staining was performed using rabbit anti-Sox2 antibody (Millipore), Ultravision Large Volume Detection System Anti-Rabbit, HRP kit (Thermo Scientific), and DAB Peroxidase Substrate Kit SK4100 (Vector Laboratories).
3D Reconstruction of Gene-Expression Patterns from In Situ Hybridization Analyses
Three-dimensional (3D) reconstructions of Sox2 and Sfrp5 expression in P2 incisor were made from serial frontal 7 mm identical sections. To sustain the original size of the incisor, the distance between the identical sections was doubled (14 mm). The pictures were imported into a stack with Photoshop CS4 software (PS), and individual pictures were aligned using the midline of the jaw as reference. The shape of the epithelium was manually traced (pen tool, PS). This subpath was filled with white color on a black layer. Aligned pictures were resliced as sagittal sections by ImageJ software (NCBI). The jagged epithelial shape in the resulting slices was smoothened in PS with a batch command: select white, expand two pixels, smooth 10 pixels. The filled epithelial shape was chosen with white color range and transferred into the path (tolerance 2 pixels). The contour line was traced by pencil tool as white dotted lines (brush tip size 1–2 pixels, spacing 300%–600%) on a new black layer. Subsequently, the expression patterns of Sox2 and Sfrp5 were isolated and treated in the same sequence of steps as the epithelial shape and projected on the corresponding slice of the shape projection. The 3D image of the epithelium and Sox2 and Sfrp5 expression were superimposed, and frontal, sagittal, and tilted projections were selected for presentation.
Confocal Microscopy and Migration Analysis
CLs of P2 Sox2-GFP and Fucci-red (mKO2-hCdt1);Sox2-GFP mice were dissected in 2% glucose/PBS containing 100 U/ml penicillin and 100 mg/ml streptomycin, embedded in 5% low-melting point agarose (TopVision, Fermentas) and cut in sagittal orientation to 150 mm slices with a vibratome. The slices from Sox2-GFP mice were counterstained with BodipyTR (Invitrogen) and Draq5 (Biostatus).
For 3D time lapse imaging studies tissue slices were allowed to recover from sectioning for a minimum of 2 hr and maintained in DMEM/F12 without phenol red and supplemented with 5% serum, antibiotics, and 15 mM HEPES (GIBCO). Sections were staged on a filter on top of a grid in the air-liquid interface for imaging with an upright Leica SP5 laser scanning confocal microscope equipped with an environmental chamber (5% CO2 humidified and maintained at 37°C) and HC PL APO 10×/0,4 air objective. Confocal images were acquired as z stacks at 2 μm intervals (50 μm in total) and tissues were imaged every 20 min for up to 20 hr with low laser power (<10%), 600 Hz scanning speed, and suboptimal sampling by averaging of two scans to reduce extended-illumination-induced cell damage. The cells were followed for 8 hr to ensure good condition of the tissue. A lack of pyknotic nuclei and frequency of mitoses in every acquired z-stack, as visualized directly with the aid of the Fucci-red reporter, confirmed good tissue health.
Images were deconvoluted with AutoQuant software (Media Cybernetics), and analysis and quantitative measurements were performed with Imaris 7.2.1 software (Bitplane). Data were further statistically analyzed and processed for graphing with Prism 5 (GraphPad) and Sigmaplot 11.0 (Sigmaplot) softwares. Image panels were processed for presentation with Adobe Photoshop CS5 and Illustrator CS5 software (Adobe Systems, San Jose, CA).
The angle (from 0° to 359°) at which the cell moved with respect to the labiallingual axis (used as the 0°/180° line of reference) was measured for individual cells. Measurements were grouped into 12 segments of 30° and were represented on a polar plot. For 4D cell migration tracking, track length, duration, cell net displacement distance, mean speed, and track straightness as the ratio of the displacement to total distance traveled (1 = straight line) were analyzed. A box-and-whiskers plot represented the minimum (25th percentile), median (75th percentile), and maximum values for the data set. Statistical analysis used was the nonparametric Mann-Whitney U-test.
Genetic Inducible Fate Mapping
For genetic fate mapping of Sox2+ cells, 0.3 mg Tamoxifen (Sigma T-5648) in corn oil/pup was given by injection either once (P2) or twice (P2 and P4) to Sox2CreERT2;R26R mice and control animals lacking the Cre-driver. For lineage tracing in adult animals, 10 mg Tamoxifen/mouse was given by oral gavage at 5 weeks of age. Induced R26R mice lacking the Sox2CreERT2 allele or double heterozygote animals gavaged with corn oil were used as controls. Whole-mount X-Gal staining, as well as subsequent processing of jaws, was performed as previously described (Seidel et al., 2010). Sections were counterstained with Fast Red (Sigma-Aldrich). Animal experiments involving living animals were performed under the research protocol approved by the Animal Research Committee at Children’s Hospital Los Angeles.
Organ Culture and Apoptosis Detection
The FGF inhibition assays were performed on dissected CLs of P2 Sox2-GFP mice. Tissues were cultured as previously described (Harada et al., 1999). For global FGF inhibition, 25 μM InSolution SU-5402 (Calbiochem) was added in standard medium (DMEM/F12 + 10% FCS). To block specific FGFs, monoclonal antibody Anti-FGF8 (R&D) (200 μg/μl) with heparin (0.1 μg/μl) or anti-FGF10 (C-17): sc-7375 (50 μg/μl; Santa Cruz Biotechnology, Inc.) was included in the medium without serum. Apoptosis was detected from methanol and 4% PFA fixed paraffin sections (7 μm) with ApopTag Red In Situ Apoptosis Detection Kit (CHEMICON).
Hanging Drop Cultures and Quantitative RT-PCR
To analyze the induction of Sox2 expression by FGF8, FGF9, and FGF10, tissues were cultured in hanging drops. E14.5 wild-type incisors were dissected and allowed to recover for 30 min before placing them in hanging drops. A minimum of triplicate samples was assayed each time. Tissues were collected after 2, 4, and 6 hr and placed in 700 μl TriReagent. Total RNA was isolated according to the manufacturer’s instructions using RNeasy Mini Kit (QIAGEN, DE) and quantified by Nanodrop spectrophotometer. cDNA synthesis and quantitative PCR (qRT-PCR) were performed as described previously (Fliniaux et al., 2008), and the data were normalized against Actin expression. The primer sequences are available upon request. For each triplicate, the SD was calculated and Student’s t test was used to determine p values, with p < 0.01 deemed to be significant.
DiI Labeling
Microinjections were performed by injecting fluorescent DiI [1,1′-di-octadecyl-6,6-di(4-sulfophenyl)-3,3,3′,3′-tetramethylindocarbocyanine] (Invitrogen) to coronal slices of the labial CLs of P2 Sox2-GFP mouse. The slices were counterstained with BodipyTR (Invitrogen) and Draq5 (Abcam) according to the manufacturer’s protocol.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Finnish Doctoral Program in Oral Sciences (E.J.), Grant-in-Aid for Scientific Research (C) 21592605 (K. Saito), Sigrid Juselius Foundation (I.T.), National Institutes of Health and California Institute of Regenerative Medicine (O.D.K.), NIH SysCODE training fellowship (K. Seidel), and Academy of Finland (M.T., F.M., and I.T.). We thank Fred Gage for the Sox2-GFP mouse strain. We also thank Merja Mäkinen, Riikka Santalahti, and Raija Savolainen for excellent technical help and Maria Jussila and Juuso Juuri for critical reading of the manuscript.
Footnotes
SUPPLEMENTAL INFORMATION Supplemental Information includes five figures, one table, Supplemental Experimental Procedures, and two movies and can be found with this article online at http://dx.doi.org/10.1016/j.devcel.2012.05.012.
REFERENCES
- Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R, Sengupta S, Seandel M, Geijsen N, Hochedlinger K. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell. 2011;9:317–329. doi: 10.1016/j.stem.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Wang J, Li X, Florez S, Huang Z, Venugopalan SR, Elangovan S, Skobe Z, Margolis HC, Martin JF, Amendt BA. MicroRNAs play a critical role in tooth development. J. Dent. Res. 2010;89:779–784. doi: 10.1177/0022034510369304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amour KA, Gage FH. Genetic and functional differences between multipotent neural and pluripotent embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2003;100(Suppl 1):11866–11872. doi: 10.1073/pnas.1834200100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- Dey-Guha I, Mukhopadhyay M, Phillips M, Westphal H. Role of ldb1 in adult intestinal homeostasis. Int. J. Biol. Sci. 2009;5:686–694. doi: 10.7150/ijbs.5.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri AL, Cavallaro M, Braida D, Di Cristofano A, Canta A, Vezzani A, Ottolenghi S, Pandolfi PP, Sala M, DeBiasi S, Nicolis SK. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development. 2004;131:3805–3819. doi: 10.1242/dev.01204. [DOI] [PubMed] [Google Scholar]
- Fliniaux I, Mikkola ML, Lefebvre S, Thesleff I. Identification of dkk4 as a target of Eda-A1/Edar pathway reveals an unexpected role of ectodysplasin as inhibitor of Wnt signalling in ectodermal placodes. Dev. Biol. 2008;320:60–71. doi: 10.1016/j.ydbio.2008.04.023. [DOI] [PubMed] [Google Scholar]
- Gritli-Linde A, Bei M, Maas R, Zhang XM, Linde A, McMahon AP. Shh signaling within the dental epithelium is necessary for cell proliferation, growth and polarization. Development. 2002;129:5323–5337. doi: 10.1242/dev.00100. [DOI] [PubMed] [Google Scholar]
- Harada H, Kettunen P, Jung HS, Mustonen T, Wang YA, Thesleff I. Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J. Cell Biol. 1999;147:105–120. doi: 10.1083/jcb.147.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Toyono T, Toyoshima K, Yamasaki M, Itoh N, Kato S, Sekine K, Ohuchi H. FGF10 maintains stem cell compartment in developing mouse incisors. Development. 2002;129:1533–1541. doi: 10.1242/dev.129.6.1533. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, Tabin CJ. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc. Natl. Acad. Sci. USA. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CL, Liu D, Masuya D, Kameyama K, Nakashima T, Yokomise H, Ueno M, Miyake M. MRP-1/CD9 gene transduction downregulates Wnt signal pathways. Oncogene. 2004;23:7475–7483. doi: 10.1038/sj.onc.1208063. [DOI] [PubMed] [Google Scholar]
- Hwang WS, Tonna EA. Autoradiographic analysis of labeling indices and migration rates of cellular component of mouse incisors using tritiated thymidine (H3TDR) J. Dent. Res. 1965;44:42–53. doi: 10.1177/00220345650440012901. [DOI] [PubMed] [Google Scholar]
- Jacques BE, Montcouquiol ME, Layman EM, Lewandoski M, Kelley MW. Fgf8 induces pillar cell fate and regulates cellular patterning in the mammalian cochlea. Development. 2007;134:3021–3029. doi: 10.1242/dev.02874. [DOI] [PubMed] [Google Scholar]
- Kelberman D, de Castro SC, Huang S, Crolla JA, Palmer R, Gregory JW, Taylor D, Cavallo L, Faienza MF, Fischetto R, et al. SOX2 plays a critical role in the pituitary, forebrain, and eye during human embryonic development. J. Clin. Endocrinol. Metab. 2008;93:1865–1873. doi: 10.1210/jc.2007-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettunen P, Thesleff I. Expression and function of FGFs-4, -8, and -9 suggest functional redundancy and repetitive use as epithelial signals during tooth morphogenesis. Dev. Dyn. 1998;211:256–268. doi: 10.1002/(SICI)1097-0177(199803)211:3<256::AID-AJA7>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Kettunen P, Karavanova I, Thesleff I. Responsiveness of developing dental tissues to fibroblast growth factors: expression of splicing alternatives of FGFR1, -2, -3, and of FGFR4; and stimulation of cell proliferation by FGF-2, -4, -8, and -9. Dev. Genet. 1998;22:374–385. doi: 10.1002/(SICI)1520-6408(1998)22:4<374::AID-DVG7>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein OD, Lyons DB, Balooch G, Marshall GW, Basson MA, Peterka M, Boran T, Peterkova R, Martin GR. An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors. Development. 2008;135:377–385. doi: 10.1242/dev.015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Kwon HJ, Harada H, Ohshima H, Cho SW, Jung HS. Expression patterns of ABCG2, Bmi-1, Oct-3/4, and Yap in the developing mouse incisor. Gene Expr. Patterns. 2011;11:163–170. doi: 10.1016/j.gep.2010.11.001. Published online November 10, 2010. http://dx.doi.org/10.1016/j.gep.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Li Y, Rankin SA, Sinner D, Kenny AP, Krieg PA, Zorn AM. Sfrp5 coordinates foregut specification and morphogenesis by antagonizing both canonical and noncanonical Wnt11 signaling. Genes Dev. 2008;22:3050–3063. doi: 10.1101/gad.1687308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Dangaria S, Andl T, Zhang Y, Wright AC, Damek-Poprawa M, Piccolo S, Nagy A, Taketo MM, Diekwisch TG, et al. beta-Catenin initiates tooth neogenesis in adult rodent incisors. J. Dent. Res. 2010;89:909–914. doi: 10.1177/0022034510370090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig B, Jerchow B, Sachs M, Weiler S, Pietsch T, Karsten U, van de Wetering M, Clevers H, Schlag PM, Birchmeier W, Behrens J. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol. Cell. Biol. 2002;22:1184–1193. doi: 10.1128/MCB.22.4.1184-1193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansukhani A, Ambrosetti D, Holmes G, Cornivelli L, Basilico C. Sox2 induction by FGF and FGFR2 activating mutations inhibits Wnt signaling and osteoblast differentiation. J. Cell Biol. 2005;168:1065–1076. doi: 10.1083/jcb.200409182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretto S, Cordenonsi M, Dupont S, Braghetta P, Broccoli V, Hassan AB, Volpin D, Bressan GM, Piccolo S. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc. Natl. Acad. Sci. USA. 2003;100:3299–3304. doi: 10.1073/pnas.0434590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michon F, Tummers M, Kyyrönen M, Frilander MJ, Thesleff I. Tooth morphogenesis and ameloblast differentiation are regulated by micro-RNAs. Dev. Biol. 2010;340:355–368. doi: 10.1016/j.ydbio.2010.01.019. [DOI] [PubMed] [Google Scholar]
- Millar SE, Koyama E, Reddy ST, Andl T, Gaddapara T, Piddington R, Gibson CW. Over- and ectopic expression of Wnt3 causes progressive loss of ameloblasts in postnatal mouse incisor teeth. Connect. Tissue Res. 2003;44(Suppl 1):124–129. [PubMed] [Google Scholar]
- Nowak JA, Polak L, Pasolli HA, Fuchs E. Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell. 2008;3:33–43. doi: 10.1016/j.stem.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsa S, Kuremoto K, Seidel K, Tabatabai R, Mackenzie B, Yamaza T, Akiyama K, Branch J, Koh CJ, Al Alam D, et al. Signaling by FGFR2b controls the regenerative capacity of adult mouse incisors. Development. 2010;137:3743–3752. doi: 10.1242/dev.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plikus MV, Zeichner-David M, Mayer JA, Reyna J, Bringas P, Thewissen JG, Snead ML, Chai Y, Chuong CM. Morphoregulation of teeth: modulating the number, size, shape and differentiation by tuning Bmp activity. Evol. Dev. 2005;7:440–457. doi: 10.1111/j.1525-142X.2005.05048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Seidel K, Ahn CP, Lyons D, Nee A, Ting K, Brownell I, Cao T, Carano RA, Curran T, Schober M, et al. Hedgehog signaling regulates the generation of ameloblast progenitors in the continuously growing mouse incisor. Development. 2010;137:3753–3761. doi: 10.1242/dev.056358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE. Cell turnover in the odontogenic organ of the rat incisor as visualized by graphic reconstructions following a single injection of 3H-thymidine. Am. J. Anat. 1980;158:321–343. doi: 10.1002/aja.1001580307. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Suh H, Consiglio A, Ray J, Sawai T, D’Amour KA, Gage FH. In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell. 2007;1:515–528. doi: 10.1016/j.stem.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen M, Thesleff I. Patterns of Wnt pathway activity in the mouse incisor indicate absence of Wnt/beta-catenin signaling in the epithelial stem cells. Dev. Dyn. 2010;239:364–372. doi: 10.1002/dvdy.22106. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Ten Cate AR. The role of epithelium in the development, structure and function of the tissues of tooth support. Oral Dis. 1996;2:55–62. doi: 10.1111/j.1601-0825.1996.tb00204.x. [DOI] [PubMed] [Google Scholar]
- Trumpp A, Depew MJ, Rubenstein JL, Bishop JM, Martin GR. Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes Dev. 1999;13:3136–3148. doi: 10.1101/gad.13.23.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker ES, Lehtinen MK, Maynard T, Zirlinger M, Dulac C, Rawson N, Pevny L, Lamantia AS. Proliferative and transcriptional identity of distinct classes of neural precursors in the mammalian olfactory epithelium. Development. 2010;137:2471–2481. doi: 10.1242/dev.049718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummers M, Thesleff I. Observations on continuously growing roots of the sloth and the K14-Eda transgenic mice indicate that epithelial stem cells can give rise to both the ameloblast and root epithelium cell lineage creating distinct tooth patterns. Evol. Dev. 2008;10:187–195. doi: 10.1111/j.1525-142X.2008.00226.x. [DOI] [PubMed] [Google Scholar]
- Tummers M, Thesleff I. The importance of signal pathway modulation in all aspects of tooth development. J. Exp. Zoolog. B Mol. Dev. Evol. 2009;312B:309–319. doi: 10.1002/jez.b.21280. [DOI] [PubMed] [Google Scholar]
- Tummers M, Yamashiro T, Thesleff I. Modulation of epithelial cell fate of the root in vitro. J. Dent. Res. 2007;86:1063–1067. doi: 10.1177/154405910708601108. [DOI] [PubMed] [Google Scholar]
- Wang XP, Suomalainen M, Jorgez CJ, Matzuk MM, Werner S, Thesleff I. Follistatin regulates enamel patterning in mouse incisors by asymmetrically inhibiting BMP signaling and ameloblast differentiation. Dev. Cell. 2004;7:719–730. doi: 10.1016/j.devcel.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Wang XP, O’Connell DJ, Lund JJ, Saadi I, Kuraguchi M, Turbe-Doan A, Cavallesco R, Kim H, Park PJ, Harada H, et al. Apc inhibition of Wnt signaling regulates supernumerary tooth formation during embryogenesis and throughout adulthood. Development. 2009;136:1939–1949. doi: 10.1242/dev.033803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte F, Dokas J, Neuendorf F, Mundlos S, Stricker S. Comprehensive expression analysis of all Wnt genes and their major secreted antagonists during mouse limb development and cartilage differentiation. Gene Expr. Patterns. 2009;9:215–223. doi: 10.1016/j.gep.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Yokohama-Tamaki T, Ohshima H, Fujiwara N, Takada Y, Ichimori Y, Wakisaka S, Ohuchi H, Harada H. Cessation of Fgf10 signaling, resulting in a defective dental epithelial stem cell compartment, leads to the transition from crown to root formation. Development. 2006;133:1359–1366. doi: 10.1242/dev.02307. [DOI] [PubMed] [Google Scholar]
- Yokohama-Tamaki T, Fujiwara N, Shibata S, Wakisaka S, Harada H. The epithelial-mesenchymal interaction plays a role in the maintenance of the stem cell niche of mouse incisors via fgf10 and fgf9 signaling. Open Biotechnol. J. 2008;2:111–115. [Google Scholar]
- Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J. Biol. Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Li S, Han D, Kaartinen V, Chai Y. Alk5-mediated transforming growth factor β signaling acts upstream of fibroblast growth factor 10 to regulate the proliferation and maintenance of dental epithelial stem cells. Mol. Cell. Biol. 2011;31:2079–2089. doi: 10.1128/MCB.01439-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.