Abstract
Although more than half of genomic loci are believed to have antisense transcription, whether antisense transcription is involved in cytokine expression has not been studied. Here we show that some loci of innate immunity related genes do have antisense transcripts. We investigated the effect of several antisense RNAs, including anti-4-1BBL, anti-p100 and anti-IL-1β, on their cognate sense gene’s expression in macrophages. We found that overexpression of antisense IL-1β transcript suppressed IL-1β expression. Anti-IL-1β is complementary to the sequence in the 5′ upstream region of the IL-1β promoter. Its mediated inhibition of IL-1β production occurred at the transcriptional level. Anti-IL-1β did not alter the methylation status of the IL-1β promoter. However, chromatin immunoprecipitation (ChIP) assays revealed that the anti-IL-1β transcript can change the chromatin structure of the IL-1β promoter by decreasing H3K4 trimethylation on the promoter, which is at least part of the mechanism underlying the reduced binding of RNA polymerase II to the IL-1β promoter upon anti-IL-1β expression. Our data suggest that some antisense-transcripts of innate immunity related genes play a role by regulating cytokine expression.
Introduction
A broad definition of antisense RNA is the transcript(s) derived from any part of a gene including intron, exon, and promoter in the opposite direction of sense mRNA. Genome-wide transcriptional analyses have revealed a large amount of antisense transcription not only in repetitive regions but also in protein-coding areas. It is estimated that up to 72% of murine genomic loci showed evidence of antisense transcription (1). Although some of the antisense transcripts may represent transcription noise, there is clear evidence that many natural antisense direction transcripts play a role in gene regulation (2, 3). Genome-wide analyses suggested that natural antisense transcripts are over represented in certain genes such as the genes related to catalytic functions, nucleotide binding, etc., and are underrepresented in some others. The processed antisense RNA is significantly underrepresented on the mammalian X chromosome. This X chromosome bias only applies to transcript sense/antisense pairs in which there is exonic complementarity (4). These findings argue against the idea that natural antisense RNA is merely transcriptional noise or experimental artifact. Studies on selected antisense RNAs have revealed their functions in both positively and negatively regulating gene expressions. There is evidence suggesting that natural antisense RNA might function by affecting transcription (5), RNA splicing (6), RNA editing (7), or activation of PKR (8–10). It is proposed that the formation of RNA-RNA hybrids by sense and natural antisense RNA can generate “endo-siRNAs” (1, 10, 11). The antisense transcripts that complement to non-coding regions also affect gene expression (12). Despite the extensive data that has been accumulated, the function of the majority of these natural antisense RNAs remains obscure. Although it is anticipated that there are natural antisense RNA of immune response genes, the role of natural antisense RNA in cytokine gene expression has not been studied.
Macrophages constitute the first line of host defense and participate in inflammation triggered by both exogenous and endogenous stimuli. Production of pro-inflammatory cytokines such as tumor necrosis factor-α (TNF) or interleukin-1β is one of the most important functions of macrophages in modulating inflammation since the inflammatory cytokines are known to be involved in various acute and chronic inflammatory diseases; these include inflammatory bowel disease (13, 14), rheumatoid arthritis (15–17) and sepsis (18) among others. Bacterial endotoxin (or lipopolysaccharides, LPS), an outer membrane component of Gram negative bacteria and a potent activator of macrophages, triggers the abundant secretion of many cytokines from macrophages including IL-1β and TNF (19). It is known that LPS-induced cytokine expression is subject to regulation at multiple levels including at transcription, post-transcription, and translation. The involvement of transcription factors, RNA binding proteins, and miRNA in cytokine expression has been demonstrated. However, to date there is no report regarding to whether there is an involvement of natural antisense transcripts in cytokine expression. In this paper, we show the presence of natural antisense RNA for some innate immunity related genes. By analyzing the effects of the expression of these antisense RNAs, we found that some of the antisense RNAs had no effect on their cognate gene’s expression while others, such as IL-1β antisense transcript, suppressed their cognate gene’s expression. We further show that the suppression of IL-1β expression by antisense IL-1β is at least partially due to anti-IL-1β RNA-mediated changes of chromatin structure.
Materials and Methods
Cells and Cell Culture
RAW 264.7 cells were maintained in DMEM supplemented with 10% FBS, nonessential amino acids and glutamine. After 48 hours of transfection with linearized antisense transcripts overexpression plasmids and reporter plasmids, cells were selected with 800 μg/ml G418 for 6 days to set up stable overexpression lines. Primary peritoneal macrophages were prepared as previous described. Briefly, mice received intraperitoneal injections 2 ml of 3% thioglycolate 3 days before the preparation of peritoneal macrophages. After anesthesia, peritoneal macrophages were harvested from the mice by lavage of the peritoneal cavity with 5 ml of saline. The cells were then washed with PBS and resuspended in DMEM supplemented with 10% FBS, glutamine and antibiotics. After 2 hours of incubation at 37 °C, the non-adherent cells were removed by washing three times with PBS and cultured overnight.
Quantitative Real-Time PCR
RNA was isolated from RAW 264.7 cells and primary peritoneal macrophages with TRIzol (Invitrogen). First-strand cDNA was synthesized with SuperScript III (Invitrogen). Relative transcript levels were measured in a duplicate set of reactions for each gene. GAPDH levels were used for normalization. Quantitative real-time PCR was performed with SYBR Green Master Mix (Applied Biosystems) on an ABI Prism 7900HT Sequence Detection System (Applied Biosystems). Data analysis was performed with the manufacturer’s software (SDS version 2.4).
Chromatin Immunoprecipitation
The chromatin immunoprecipitation (ChIP) kit was purchased from Cell Signaling Technology (Cell Signaling Technology, #9002). ChIP was performed following the manufacturer’s instructions with control and anti-IL-1β overexpressing RAW 264.7 cells with antibodies against RNA polymerase II (Millipore), H3K4Me3 (Cell Signaling Technology) and H3K27Me3 (Cell Signaling Technology). ChIP DNA was analysed with SYBR Green Master Mix (Applied Biosystems) on an ABI Prism 7900HT Sequence Detection System (Applied Biosystems). Data analysis was performed using the manufacturer’s software (SDS version 2.4).
DNA Methylation Assay
DNA methylation analysis was performed following the manufacturer’s instructions using the EZ DNA methylation-gold kit (ZYMO Research). 100 ng genomic DNA from control and anti-IL-1β overexpression cells was applied for the assay. Converted DNA was used for PCR amplification. PCR products were cloned and sequenced.
mRNA Stability Assay
RAW 264.7 cells were pretreated with 10 ng/ml LPS for 4 hours followed by treatment with 50 μg/ml Actinomycin D. Total RNA was extracted with TRIzol reagent and reverse transcripted into cDNA. IL-1β and TNF-α cDNAs were quantitated by real-time PCR.
In vitro translation Analysis
Anti-IL-1β was cloned into pT7CFE1-CHis vector. The in vitro translation assay was performed following the manufacturer’s instructions (Pierce, #88855). Briefly, 1 μg linearized plasmid was used as the template to transcribe mRNA with T7 RNA polymerase. Transcription products were translated with [35S] methionine containing amino acid mixture for protein expression at 30 °C for 90 min. Translation products were analyzed by SDS-PAGE.
Strand-specific RT-PCR
According to the manufacturer’s protocol, 200ng of total RNA was used for reverse transcription. A final concentration of 2.5μM Oligo (dT)16 primer, 200 nM forward or reverse strand primer were used to synthesize the first-strand cDNA with the SuperScript III (Invitrogen). Relative transcript levels were measured in a duplicate set of reactions for each gene.
Web sites references
http://genome.gsc.riken.go.jp/m/antisense/viewer/ List of antisense transcripts. http://genome.ucsc.edu/ Genome Browser.
RESULTS
Expression of innate immunity related antisense RNAs
To get an overall idea of the expression of antisense RNAs that relate to innate immune responses, we searched the FANTOM2 database (4) for 216 genes that are involved in innate immune reactions; we found 27 natural antisense transcripts (Table 1). The genes that have antisense RNA include cytokines (IL-1α, IL-1β, IL-18, IFNγ, etc.), receptors (IL6rα, IL1r2, IL20rα, etc.), and signal transduction genes (IRF1, Traf5, Tirap, p100, p105, etc.). We did not find antisense transcripts of MyD88, IRAK1/4, CD14 and MD2, which are known to be important in innate immunity. Although we cannot exclude the possibility that their antisense transcripts have not yet been identified, based on current studies in natural antisense RNA, we believe that as with the genes involved in other biological processes, only a fraction of the innate immunity related genes have antisense transcripts.
Table 1.
Antisense Transcripts to Innate Immune Response Related Genes
| Sense Gene | Antisense EST/Gene | Sense Gene | Antisense EST/Gene |
|---|---|---|---|
| Ifng | AK089852 | Ilf2 | Snapap |
| Ifrd1 | AK004585 | Ilf3 | AK076941 |
| Il12rb2 | AK046984 | Irf1 | AK053162 |
| Il16 | AK052238 | Irf2 | AK082031 |
| Il17b | AK005683 | Isg20 | AF159455 |
| Il18 | AK014896 | Nf3b1 | AK090099 |
| Il1a | AK042010 | Nf3b2 | AK029443 |
| Il1b | AK076405 | Triap | AK084664 |
| Il1r2 | AK089728 | Tnfaip8 | AK043286 |
| Il20ra | AK039957 | Tnfsf9 | AK154177 |
| Il2r | Zfp281 | Traf5 | AK016485 |
| Il4i1 | AK087754 | Trap1 | Dnase1 |
| Il6ra | AK054125 | Peli2 | AK020658 |
| Cradd | AK087886 |
Antisense transcripts were chosen from FANTOM2 antisense database.
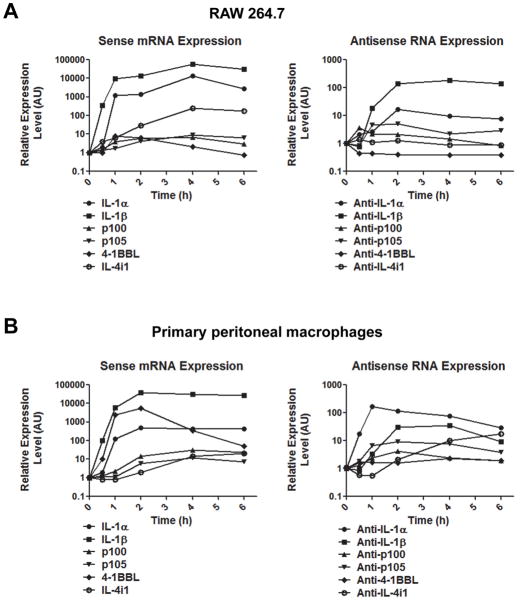
To gain a better understanding of antisense RNA in innate immunity, we chose six antisense transcripts, anti-IL-1α, anti-IL-1β, anti-4-1BBL, anti-IL-4i1, anti-p100 and anti-p105, for further analysis. The UCSC Genome Browser was used to plot the loci of these sense-antisense pairs of genes and revealed that these antisense transcripts can be divided into two groups according to the classification by Werner et al (1) (Fig. S1). One is head-to-head (Fig. S1A); their overlap is in the 5′ region of the genes. The other is tail-to-tail (Fig. S1B); they overlap in the 3′ region of the genes. Primers were designed based on this information to specifically detect expression of these antisense RNAs in macrophage cell line RAW264.7 and peritoneal macrophages. Since LPS activation of macrophages is a model cell system to innate immune responses, we treated RAW264.7 cells with LPS for different periods of time and measured the above mentioned six antisense RNAs by real-time PCR. As shown in Figure 1A, LPS stimulation reduced the level of anti-4-1BBL, but increased the expression of the other five antisense transcripts to different extents. As previously reported, the expression of all of the six sense genes were induced to different levels by LPS stimulation (20–24). Similar results were obtained when the expression of these antisense RNAs were analyzed using murine primary peritoneal macrophages (Fig. 1B). It needs to be noted that the expression levels of the sense transcripts are much higher than the antisense transcripts. The expression of sense and antisense transcripts decreased in similarly at a later time (6 to 24 hours) of LPS stimulation (data not shown). In short, we confirmed the presence of anti-IL-1α, anti-IL-1β, anti-4-1BBL, anti-IL-4i1, anti-p100 and anti-p105 transcripts in macrophages and found that their expression is dynamically regulated during LPS-induced macrophage activation.
Figure 1. Antisense transcripts were induced by LPS in both RAW 264.7 cells and primary peritoneal macrophages.
(A) Sense and antisense transcript expression in LPS stimulated RAW 264.7 cells. RAW 264.7 cells were treated with 100ng/ml LPS from 0 to 6 hours. Total RNA was extracted for real-time qPCR. (B) Sense and antisense transcript expression in LPS stimulated primary peritoneal macrophages. Cells were treated as in (A). All values were represented as mean from triplicated samples (n=3) and all samples at time 0 are set to relative abundance of 1. One representative experiment out of three is shown.
Overexpression of antisense IL-1β transcript suppresses the induction of IL-1β
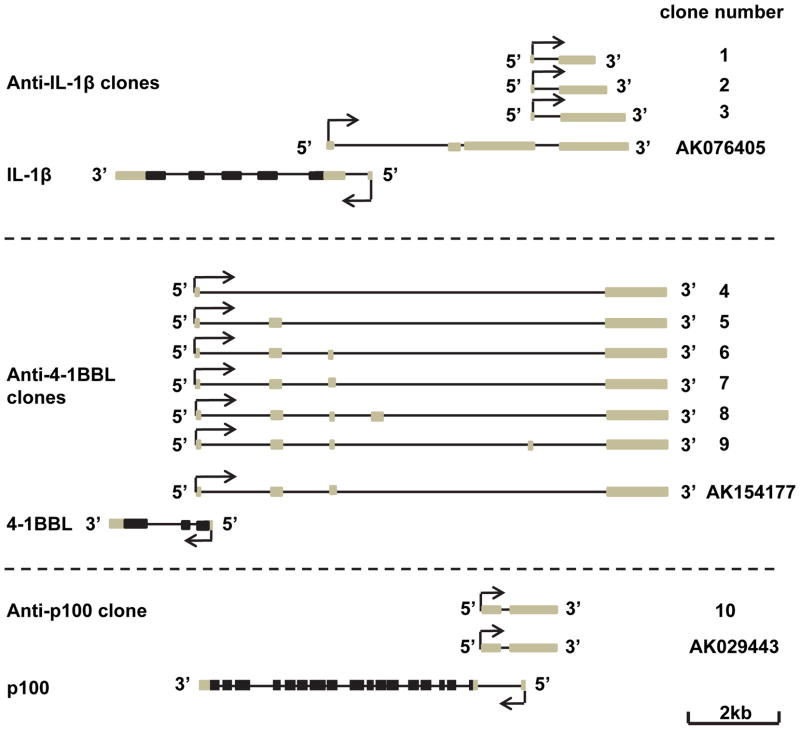
To further study the function of antisense RNA in LPS-induced macrophage activation, we chose highly induced anti-IL-1β, moderately induced anti-p100, and suppressed anti-4-1BBL for further study. All of these three antisense RNAs are head to head type (Fig. S1). To specifically study these antisense transcripts in RAW 264.7 cells, we cloned anti-IL-1β, anti-p100, and anti-4-1BBL from this cell line. The anti-IL-1β transcripts we cloned are varied in length and shorter than the sequence in the database (named AK076405) (Fig. 2). Multiple attempts did not add more antisense transcripts than those shown in Figure 2. Northern blot analysis only detected smeared signals but not bands (data not shown), suggesting that the antisense transcripts might be heterogeneous in length. The sequence alignment of our longest anti-IL-1β cDNA (Clone 3, Fig. 2) with AK076405 and IL-1β genome is shown in Figure S2. We used our longest anti-IL-1β transcript for further study. The anti-4-1BBL transcripts we cloned are similar to the sequence in the data base except that six different splicing forms were isolated in our experiments. We chose to use the transcript that has the same sequence as that in the database for further study (Clone 7, Fig. 2). The anti-p100 we cloned is the same as that in the database (Clone 10, Fig. 2).
Figure 2. Schematic of anti-IL-1β, anti-4-1BBL and anti-p100 clones and their annotated EST in GenBank.
Lines represent gene loci; black boxes represent coding region; grey boxes represent non-coding region.
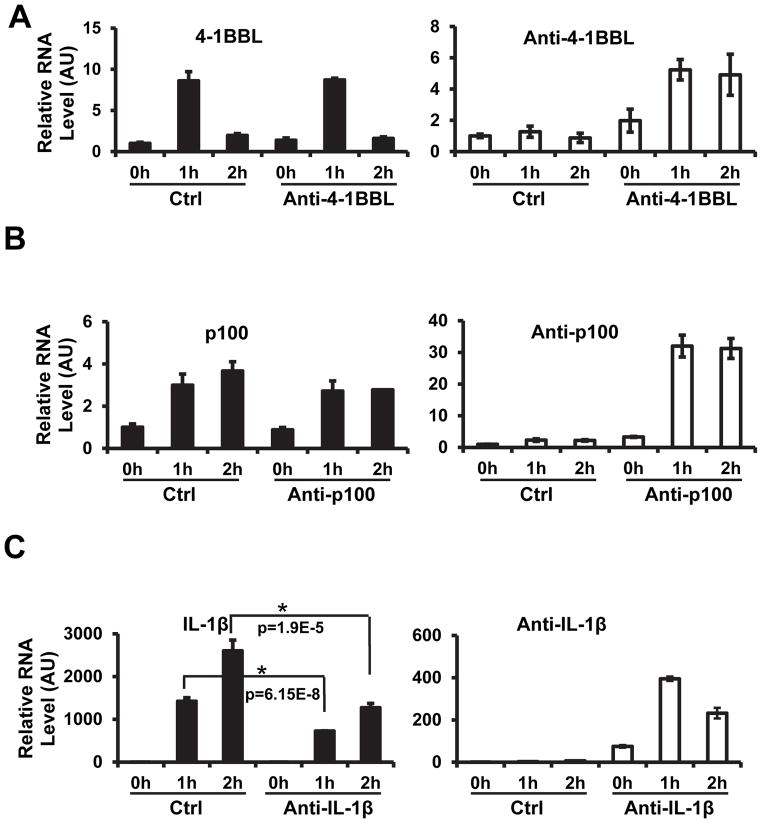
We subcloned the above mentioned three antisense transcripts (cDNA) under the control of EF1α promoter and transfected them into RAW264.7 cells to establish stable cell lines. The EF1α promoter was chosen because its responsiveness to LPS is less than the more commonly used CMV and SV40 promoters in RAW264.7 cells (data not shown). The pool and individual cell lines were isolated and characterized for their expression of the transfected antisense RNA. Since the expression levels of these antisense RNAs in the individual clones and the pools are similar, we used pools to conduct experiments. It was reported that antisense transcripts can affect the expression of sense genes. So we determined whether the overexpression of antisense transcripts influenced LPS-induced induction of endogenous sense genes. Despite of the use of EF1α promoter, the exogenously introduced antisense transcripts were still induced to various extents by LPS-treatment (right panels of Fig. 3A–C). These exogenous antisense transcripts were expressed at much higher levels than the endogenous antisense transcripts and thus it is the result of overexpression that should be observed in these experiments. Figure 3A shows that RAW264.7 cells transfected with empty vector or anti-4-1BBL expressing vector had comparable expression of 4-1BBL sense RNA, although much more anti-4-1BBL RNA was detected in the cells transfected with the anti-4-1BBL expressing vector. Similarly, expression of anti-p100 had no significant effect on the induction of p100 after LPS treatment (Fig. 3B). In contrast, expression of anti-IL-1β significantly reduced LPS-induced IL-1β expression at both time points (Fig. 3C). As controls, overexpression of anti-4-1BBL or complementary strand of anti-IL-1β did not affect IL-1β expression (Figure S3). These data indicate that different antisense RNAs had different effects on the expression of their sense transcripts. It needs to be noted that the anti-IL-1β we overexpressed is situated about 3 kb away from the sense transcription unit. Its ability to regulate sense transcription from such a long distance is not a surprise since regulatory elements of a given gene can be dispersed far away, even hundreds of kb, from the transcriptional initiation site(25, 26).
Figure 3. Over-expression of anti-IL-1β suppressed the induction of IL-1β triggered by LPS.
(A) Effect of anti-4-1BBL on LPS induced 4-1BBL expression. RAW 264.7 cells were transfected with anti-4-1BBL over-expression vector and selected as a stable line. After the treatment of LPS, total RNA was extracted, reverse transcribed to cDNA and applied to real-time qPCR. The expression levels of 4-1BBL and total anti-4-1BBL were converted to absolute units and normalized to GAPDH expression. The relative RNA levels were obtained by setting the control samples at time 0 as abundance of 1. Data are the mean ± SEM of triplicates. (B) Effect of anti-p100 on LPS induced p100 expression. Samples collected and data presented as in (A). (C) Effect of anti-IL-1β on LPS induced IL-1β expression. Samples collected and data presented as in (A).
We then characterized anti-IL-1β RNA in RAW264.7 cells. We found that only oligo d(T) primer and reverse primer, but not the forward primer of anti-IL-1β can reverse transcribe anti-IL-1β RNA, indicating it is a poly-A RNA (Fig. S4A). By extracting cytosolic and nuclear RNA we found that anti-IL-1β RNA was primarily located in the nuclei of RAW264.7 cells (Fig. S4B). Furthermore, by using an in vitro translation system we determined that anti-IL-1β RNA cannot translate into protein (Fig. S4C). Thus, anti-IL-1β RNA is a non-coding nuclear poly-A RNA.
The two directional transcriptions at IL-1β locus is affected differently by anti-IL-1β RNA
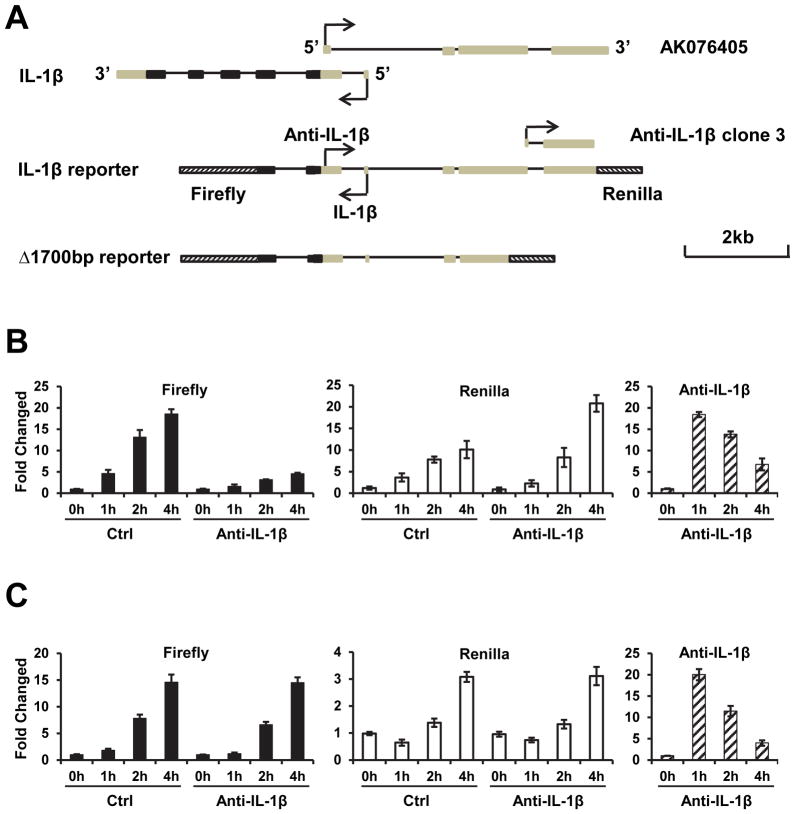
The anti-IL-1β and IL-1β mRNA are in head to head position (Fig. 2). The anti-IL-1β overlaps with 5′ upstream promoter region of IL-1β gene, suggesting that the promoters of the two directional transcriptions at IL-1β locus are located at the same or nearby genomic fragments. We constructed a reporter gene termed IL-1β reporter using ~8 kb IL-1β genome fragment containing part of IL-1β coding region, promoter, and the 5′ upstream region corresponding to anti-sense transcript (Fig. 4A). Firefly luciferase gene with a poly A signal sequence was linked to the second exon of IL-1β gene in the direction of IL-1β transcription. Renilla luciferase gene with a poly A signal sequence was added after anti-IL-1β in anti-IL-1β direction (Fig. 4A). The reporter was stably transfected into RAW264.7 cells with the anti-IL-1β expression plasmid or empty vector (control). The cells were treated with LPS for different periods of time and the luciferase activities were determined. As shown in Figure 4B, both firefly and renilla luciferase activities were induced by LPS stimulation, indicating that the reporter mimicked endogenous IL-1β gene in the bi-directional transcription. We estimated by qRT-PCR that the antisense transcription level is about one tenth of sense transcription (data not shown), which is a ratio similar to the ratio of the expression of endogenous antisense IL-1β RNA to that of antisense IL-1β mRNA. (Fig. 1). As with the endogenous IL-1β (Fig. 3C), the LPS-induced firefly luciferase expression was inhibited by anti-IL-1β expression (Fig. 4B, left panel). In contrast, the anti-IL-1β expression did not inhibit but even enhanced LPS-induced renilla luciferase expression (Fig. 4B, central panel). These results were reproducible when independently generated stable reporter cell lines were used. This different effect of anti-IL-1β on the two directions of gene transcription is unlikely to result from a particular locus of reporter gene insertion site because we used pools of the stably transfected cells.
Figure 4. Different effects of anti-IL-1β overexpression on sense and antisense transcription in IL-1β locus.
(A) Schematic of the reporters of bi-directional transcription. (B) The RAW264.7 cells, stably transfected with a bi-direction reporter (IL-1β reporter) together with empty vector (Ctrl) or anti-IL-1β overexpression vector, were treated with LPS for different times as indicated. The sense expressions were determined by measuring firefly luciferase activity and the antisense expressions were determined by measuring renilla luciferase activity. The expression levels of exogenously introduced anti-IL-1β were determined by qPCR. (C) The same as in B except that Δ1700 reporter was used.
Because the reporter contains the anti-IL-1β sequence, we used one primer located in the flanking sequence derived from the anti-IL-1β expression vector to specifically measure exogenously introduced anti-IL-1β transcript. We did not detect exogenous anti-IL-1β in control cells and detected anti-IL-1β in anti-IL-1β transfected cells (Fig. 4B, right panel). Similar to the data shown in Figure 3C, EF1α driven anti-IL-1β expression was induced by LPS stimulation.
We also constructed an IL-1β reporter termed Δ1700 reporter, in which the 1700 bp sequence corresponding to anti-IL-1β was deleted (Fig. 4A). Δ1700 reporter was stably transfected into RAW264.7 cells with the anti-IL-1β expression plasmid or empty vector (control). The expression of anti-IL-1β did not inhibit LPS-induced firefly and renilla luciferase expression (Fig. 4C), suggesting that the suppression of IL-1β expression by anti-IL-1β depends on its homologous sequence in the IL-1β promoter.
The suppressive effect of anti-IL-1β RNA on IL-1β expression derives from multiple regions of this RNA
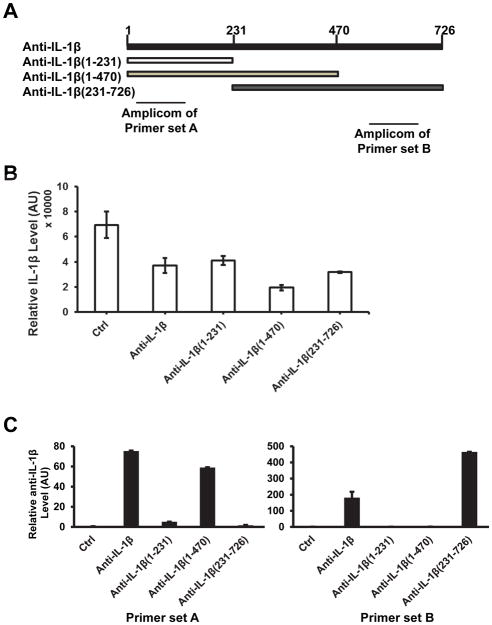
As noted earlier the anti-IL-1β RNA derives from the promoter region of IL-1β (Fig. 2). Thus, it cannot form RNA-RNA hybrids with IL-1β mRNA, and therefore, generating “endo-siRNA” is clearly not the mechanism by which anti-IL-1β functions. We constructed vectors to express three different truncations of anti-IL-1β to determine whether any subregion of this RNA plays a key role in suppression of IL-1β expression. The expression of these deletion mutants was measured using two sets of primers as shown in Figure 5A. Inhibition of IL-1β expression was observed in the cells expressing any one of these three deletions (Fig. 5B), suggesting that all sequences in the anti-IL-1β RNA can affect IL-1β expression. However, the levels of inhibition by anti-IL-1β and its deletions were not correlated with their expression level (Fig. 5B, 5C), suggesting that some sequences in the anti-IL-1β RNA may have stronger inhibitory effects than others.
Figure 5. All parts of anti-IL-1β are important for its suppression effect on IL-1β expression.
(A) Schematic of anti-IL-1β mutants and primer sets. Anti-IL-1β (1–231) has the sequence from 1 to 231bp of anti-IL-1β. Anti-IL-1β (1–470) has the sequence from 1 to 470bp of anti-IL-1β. Anti-IL-1β (231–726) has the sequence from 231 to 726bp of anti-IL-1β. Primer set A was designed for anti-IL-1β (1–231)/ (1–470), set B was designed for anti-IL-1β (231–726) only. (B) Effect of anti-IL-1β mutants on IL-1β induction. Stable cell lines of control, anti-IL-1β and the three mutants were treated with LPS for 6 hours. IL-1β mRNA expression was determined by real-time qPCR. (C) Expression of the three anti-IL-1β mutants. Primer sets A and B were used to determine the expression of the mutants. All values were represented as mean±SEM from triplicated samples (n=3). One representative experiment out of three is shown.
Anti-IL-1β has no effect on IL-1β mRNA stability
Antisense transcripts has been reported to influence sense mRNA stability. Inducible nitric oxide synthase (iNOS) mRNA can be stabilized by its antisense RNA (27). Although anti-IL-1β is unlikely to pair with IL-1β mRNA since it is located in the promoter region of IL-1β (Fig. 2), whether it has an effect on the stability of IL-1β still needed to be determined. We pretreated RAW264.7 cells with 10ng/ml of LPS for 4h, followed by 50μg/ml Actinomycin D treatment. Total RNA was collected from 0 to 3 h after Actinomycin D treatment. The stability of IL-1β mRNA was compared between control and anti-IL-1β overexpressing RAW 264.7 cells. The stability of TNF mRNA was also measured and included as a control. The stability of IL-1β mRNA is very similar in vector and anti-IL-1β expressing lines (Fig. 6A). As expected, TNF mRNA stability was not affected by anti-IL-1β expression (Fig. 6B). The higher anti-IL-1β level in the overexpressing cell line was confirmed (Fig. 6C). These results demonstrated that anti-IL-1β has no effect on IL-1β mRNA stability.
Figure 6. Overexpression of anti-IL-1β has no effect on IL-1β mRNA stability.
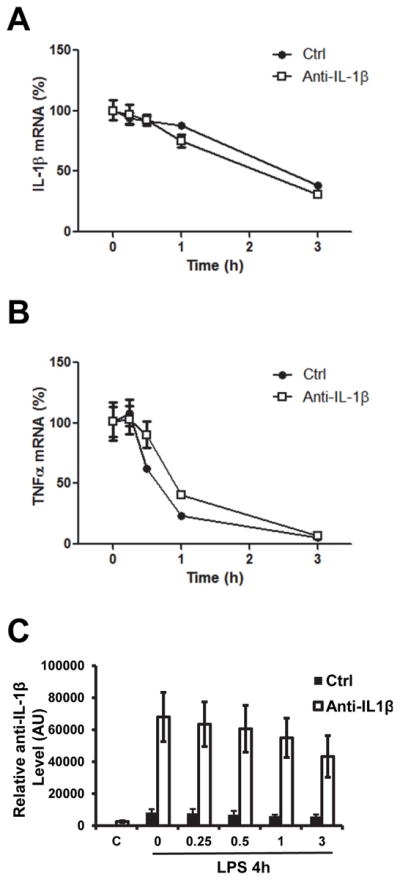
RAW 264.7 cells were stimulated with LPS for 4 hours, followed by 50μg/ml Actinomycin D treatment. Total RNA was extracted for real-time PCR. IL-1β (A) and TNFα (B) degradation, and anti-IL-1β (C) overexpression were determined by real-time qPCR.
Anti-IL-1β inhibits IL-1β mRNA transcription
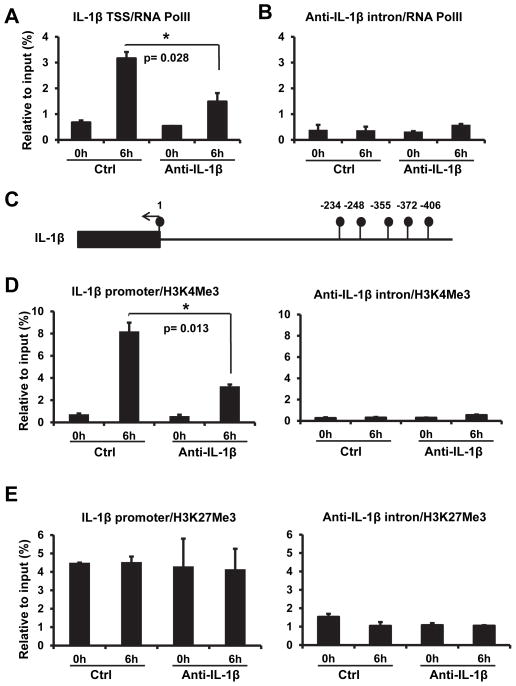
RNA polymerase II is the enzyme responsible for the transcription of protein-coding genes. The binding ability of this enzyme to the promoter would be increased when gene expression is activated. To determine whether anti-IL-1β mediated inhibition of IL-1β mRNA expression occurred at transcriptional level, we measured binding of RNA polymerase II to the transcription start site of IL-1β. Chromatin immunoprecipitation (ChIP) assay showed that the binding of RNA polymerase II at the IL-1β transcription start site was decreased in anti-IL-1β overexpressing cells (Fig. 7A). ChIP assay with anti-IL-1β intron region showed no difference between vector and anti-IL-1β transfected cells (Fig. 7B), confirming a specific inhibitory effect of anti-IL-1β RNA on the transcription of IL-1β.
Figure 7. Anti-IL-1β leads to modification IL-1 β chromatin structure.
(A) Over-expression of anti-IL-1β decreased recruitment of RNA polymerase II to the IL-1β transcription start region. Genomic DNA was pulled down by anti-RNA polymerase II antibody and quantitated by real-time qPCR with the primers specific to the IL-1β transcription start region. The amount of DNA determined by qPCR is plotted. (B) Anti-IL-1β has no effect on the binding of RNA polymerase II to the anti-IL-1β intronic region. Samples were collected as in (A) and quantitated by qPCR with anti-IL-1β intron specific primers. All values were represented as mean±SEM from triplicated samples (n=3). (C) Schematic of IL-1β promoter region. Filled circles represent CpG sites and the filled box represents the start of the IL-1β mRNA. Anti-IL-1β overexpression did not lead to CpG methylation change at these sites. (D) Anti-IL-1β reduced H3K4Me3 bound to the IL-1β promoter. Genomic DNA, pulled down with anti-H3K4Me3 antibody from control and anti-IL-1β overexpression cell lines, were analysed by real-time PCR analysis to quantitate the IL-1β promoter (-1 to -150) and intronic DNA. (E) H3K27Me3 levels were not affected by anti-IL-1β overexpression. Genomic DNA was pull down as in (D) with H3K27Me3 antibody. All values were represented as mean±SEM from triplicated samples (n=3). One representative experiment out of three is shown. The symbol * marks a significantly different result (p<0.05) compared to respective control value.
Anti-IL-1β does not affect methylation modification of IL-1β promoter
Since DNA methylation plays an important role in gene expression, we examined whether anti-IL-1β affected the methylation status of the IL-1β promoter. To investigate this possibility we analyzed the methylation of the IL-1β promoter. There are five CpG sites in the −1 to −500 region of IL-1β promoter, and one at the transcription start site (Circles, Fig. 7C). Sodium bisulfite converts cytosines in RNA to uracil while methylation on cytosines prevents this conversion (28). By treating the DNA sample with sodium bisulfite, amplifying with PCR and sequencing, the methylation of CpG was determined. We found that CpG sites at −406, −372 and −355 were always methylated, but the sites at −248, −234 and +1 were not methylated (Fig. 7C). No difference of the methylation was found between control and anti-IL-1β expressing RAW 264.7 cells. In LPS treated samples, we found that 17% of the CpG at −372 was unmethylated in control cells and 10% of the CpG site at +1 was methylated in anti-IL-1β overexpression cells (Fig. 7C). Since there was only a very slight difference of CpG methylation between control and anti-IL-1β overexpression cells, we conclude that DNA methylation is unlikely to play a key role in anti-IL-1β mediated suppression of IL-1β expression.
Anti-IL-1β expression affects chromatin structure of IL-1β promoter
Histone H3 is a component of nucleosomes. The amino acid tail of histone H3 can be methylated, acetylated, phosphorylated and ubiquitinated. These modifications affect chromatin structure and thus regulate gene expression (29). It is known that H3K4Me3 is enriched on highly activated promoters (30), and H3K27Me3 is associated with low expression promoters (31). We used ChIP assay to investigate histone 3 modifications in the IL-1β promoter region (−1 to −150). IL-1β intron was included as a control. As shown in Figure 7D, LPS stimulation dramatically enriched H3K4Me3 (from 0.73% to 8.2%) at IL-1β promoter region, which is consistent with IL-1β mRNA induction (Fig. 3C). In the resting stage (no LPS stimulation), H3K4Me3 at IL-1β promoter was not different between control and anti-IL-1β overexpression cells. In contrast, following LPS stimulation anti-IL-1β overexpression significantly reduced H3K4Me3 at IL-1β promoter. The anti-IL-1β mediated reduction of H3K4Me3 at IL-1β promoter correlated with inhibition of IL-1β mRNA expression (Fig. 7D and 3C). We also measured H3K27Me3 on IL-1β promoter and found that neither LPS treatment nor anti-IL-1β overexpression had an effect on the level of H3K27Me3 at IL-1β promoter (Fig. 7E). These results indicate that inhibition of H3K4 trimethylation on IL-1β promoter is at least part of the mechanism by which anti-IL-1β mediated suppression of IL-1β gene expression.
DISCUSSION
Inflammatory cytokine expression is known to be controlled at multiple levels. We show in this report that the antisense transcript of IL-1β may add one more layer of control to IL-1β expression. Overexpressed anti-IL-1β has an inhibitory effect on LPS-induced IL-1β expression. We found that at least one of the effects of the anti-IL-1β is to inhibit LPS-induced trimethylation of H3K4 bound to the IL-1β promoter, thus reducing LPS-induced IL-1β transcription.
Anti-IL-1β RNA appears to have splicing variants since the anti-IL-1β RNAs we cloned have different lengths and are also different from the anti-IL-1β sequence in the database. Since we were unable to clone the same anti-IL-1β as that in the database, it is possible that different anti-IL-1β RNAs are transcribed in different types of cells. Although we show in this report that anti-IL-1β can alter H3K4 trimethylation in the IL-1β promoter, we cannot exclude the possibility that an anti-IL-1β with a different length, such as that deposited in the database, might have a different function or functions.
Because anti-IL-1β was expressed at a much lower level than IL-1β mRNA, whether it would have measurable effect on sense transcription in vivo cannot be assessed by current techniques. However, the low level of anti-IL-1β might not limit its function in modulating H3K4 trimethylation because targeting genomic DNA may not need many RNA molecules. For example, if anti-IL-1β activates demethylation of H3K4Me3, its effect could be catalytic rather than stoichiometric. The expression of anti-IL-1β RNA is associated with the activation of sense transcription because the induction of anti-IL-1β by LPS stimulation has similar kinetics to that of IL-1β sense transcription (Fig. 1). However, the promoter elements that control the transcriptions in the two directions should not all be the same because the anti-IL-1β has different effects on sense and antisense transcriptions (Fig. 4B). Probably H3K4Me3 only affects nearby transcriptional initiation and H3K4 trimethylation near the initiation site of IL-1β mRNA only reduces IL-1β transcription but not anti-IL-1β transcription.
Our data presented in this manuscript demonstrate that overexpressed anti-IL-1β can inhibit H3K4 trimethylation in the IL-1β promoter. The chromatin modification suggests that anti-IL-1β functions by targeting the DNA of the IL-1β promoter. This notion is supported by the fact that the sequence of anti-IL-1β is derived from the 5′ upstream sequence of the IL-1β promoter and anti-IL-1β transcripts are located in the nucleus. Small interfering RNA or antisense non-coding RNA mediated chromatin remodeling has been reported (32, 33). Here we provide another example where antisense RNA plays a role in modifying chromatin. Because pairing with the sequence of the IL-1β 5′ upstream region of its promoter should be the mechanism for anti-IL-1β to specifically regulate IL-1β promoter, how RNA interacts with genomic DNA is still an open question. It is highly possible that anti-IL-1β interacts with the sense strand of the IL-1β promoter immediately after it is transcribed since unwinding double stranded DNA in the IL-1β promoter region is required for it to pair with anti-IL-1β RNA. Nuclear RNA and promoter interaction could be a very important regulatory mechanism of gene expression.
Because of the essential role of IL-1β in inflammation, we expect that suppression of IL-1β by anti-IL-1β should affect inflammatory responses in vivo. Although no data is available regarding to the involvement of natural antisense RNA in inflammation, the involvement of antisense RNA in other diseases have been reported. Span et al reported recently that overexpression of a natural antisense transcript of hypoxia-inducible factor 1 (αHIF) is associated with malignant pheochromocytoma/paraganglioma (34). Differential expression of hundreds of antisense RNAs between colorectal cancer tissues and non-cancerous tissues was also reported recently (35). It will not be a surprise if alteration of antisense expression affects inflammatory responses.
In summary, we have analyzed a number of antisense transcripts of innate immune response genes. Diversities can be found in these transcripts in terms of complementarity regions (coding, non-coding, promoter, intron/exon), and effects on sense transcripts. We show that the anti-IL-1β transcript, which should target the promoter region of IL-1β, can suppress IL-1β transcription by modifying the chromatin structure of the IL-1β promoter. Our data suggest that interaction of a nuclear RNA with a promoter is a mechanism that regulates gene expression.
Supplementary Material
Acknowledgments
This work was supported by grants from NSF China 91029304, 81061160512, 31221065, 91229201, National Institutes of Health AI41637 and AI68896.
Abbreviations used in this article
- ChIP
chromatin immunoprecipitation
- H3K4Me3
tri-methylated Histone H3 at lysine 4
- H3K27Me3
tri-methylated Histone H3 at lysine 27
- IL-1β
interleukin-1β
- LPS
lipopolysaccharides
- NF-κB
nuclear factor-κB
- TNFα
tumor necrosis factor-α
- TRAF6
TNF receptor-associated factor 6
References
- 1.Werner A, Carlile M, Swan D. What do natural antisense transcripts regulate? RNA Biol. 2009;6:43–48. doi: 10.4161/rna.6.1.7568. [DOI] [PubMed] [Google Scholar]
- 2.Mahmoudi S, Henriksson S, Corcoran M, Mendez-Vidal C, Wiman KG, Farnebo M. Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Mol Cell. 2009;33:462–471. doi: 10.1016/j.molcel.2009.01.028. [DOI] [PubMed] [Google Scholar]
- 3.Fish JE, Matouk CC, Yeboah E, Bevan SC, Khan M, Patil K, Ohh M, Marsden PA. Hypoxia-inducible expression of a natural cis-antisense transcript inhibits endothelial nitric-oxide synthase. J Biol Chem. 2007;282:15652–15666. doi: 10.1074/jbc.M608318200. [DOI] [PubMed] [Google Scholar]
- 4.Kiyosawa H, Yamanaka I, Osato N, Kondo S, Hayashizaki Y. Antisense transcripts with FANTOM2 clone set and their implications for gene regulation. Genome Res. 2003;13:1324–1334. doi: 10.1101/gr.982903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shearwin KE, Callen BP, Egan JB. Transcriptional interference--a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cayre A, Rossignol F, Clottes E, Penault-Llorca F. aHIF but not HIF-1alpha transcript is a poor prognostic marker in human breast cancer. Breast Cancer Res. 2003;5:R223–230. doi: 10.1186/bcr652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Z, Carmichael GG. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- 8.Adah SA, Bayly SF, Cramer H, Silverman RH, Torrence PF. Chemistry and biochemistry of 2′,5′-oligoadenylate-based antisense strategy. Curr Med Chem. 2001;8:1189–1212. doi: 10.2174/0929867013372445. [DOI] [PubMed] [Google Scholar]
- 9.Wang Q, Carmichael GG. Effects of length and location on the cellular response to double-stranded RNA. Microbiol Mol Biol Rev. 2004;68:432–452. doi: 10.1128/MMBR.68.3.432-452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlile M, Nalbant P, Preston-Fayers K, McHaffie GS, Werner A. Processing of naturally occurring sense/antisense transcripts of the vertebrate Slc34a gene into short RNAs. Physiol Genomics. 2008;34:95–100. doi: 10.1152/physiolgenomics.00004.2008. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, Surani MA, Sakaki Y, Sasaki H. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 12.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity. 1999;10:387–398. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- 14.Ligumsky M, Simon PL, Karmeli F, Rachmilewitz D. Role of interleukin 1 in inflammatory bowel disease--enhanced production during active disease. Gut. 1990;31:686–689. doi: 10.1136/gut.31.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feldmann M, Brennan FM, Maini RN. Role of cytokines in rheumatoid arthritis. Annu Rev Immunol. 1996;14:397–440. doi: 10.1146/annurev.immunol.14.1.397. [DOI] [PubMed] [Google Scholar]
- 16.Mori L, Iselin S, De Libero G, Lesslauer W. Attenuation of collagen-induced arthritis in 55-kDa TNF receptor type 1 (TNFR1)-IgG1-treated and TNFR1-deficient mice. J Immunol. 1996;157:3178–3182. [PubMed] [Google Scholar]
- 17.Kuiper S, Joosten LA, Bendele AM, Edwards CK, 3rd, Arntz OJ, Helsen MM, Van de Loo FA, Van den Berg WB. Different roles of tumour necrosis factor alpha and interleukin 1 in murine streptococcal cell wall arthritis. Cytokine. 1998;10:690–702. doi: 10.1006/cyto.1998.0372. [DOI] [PubMed] [Google Scholar]
- 18.Dinarello CA, Gelfand JA, Wolff SM. Anticytokine strategies in the treatment of the systemic inflammatory response syndrome. JAMA. 1993;269:1829–1835. [PubMed] [Google Scholar]
- 19.Guha M, Mackman N. LPS induction of gene expression in human monocytes. Cell Signal. 2001;13:85–94. doi: 10.1016/s0898-6568(00)00149-2. [DOI] [PubMed] [Google Scholar]
- 20.Kang YJ, Kim SO, Shimada S, Otsuka M, Seit-Nebi A, Kwon BS, Watts TH, Han J. Cell surface 4-1BBL mediates sequential signaling pathways ‘downstream’ of TLR and is required for sustained TNF production in macrophages. Nat Immunol. 2007;8:601–609. doi: 10.1038/ni1471. [DOI] [PubMed] [Google Scholar]
- 21.Pelegrin P, Barroso-Gutierrez C, Surprenant A. P2X7 receptor differentially couples to distinct release pathways for IL-1beta in mouse macrophage. J Immunol. 2008;180:7147–7157. doi: 10.4049/jimmunol.180.11.7147. [DOI] [PubMed] [Google Scholar]
- 22.de Wit H, Dokter WH, Koopmans SB, Lummen C, van der Leij M, Smit JW, Vellenga E. Regulation of p100 (NFKB2) expression in human monocytes in response to inflammatory mediators and lymphokines. Leukemia. 1998;12:363–370. doi: 10.1038/sj.leu.2400950. [DOI] [PubMed] [Google Scholar]
- 23.Saito T, Sasaki CY, Rezanka LJ, Ghosh P, Longo DL. p52-Independent nuclear translocation of RelB promotes LPS-induced attachment. Biochem Biophys Res Commun. 2010;391:235–241. doi: 10.1016/j.bbrc.2009.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patial S, Saini Y, Parvataneni S, Appledorn DM, Dorn GW, 2nd, Lapres JJ, Amalfitano A, Senagore P, Parameswaran N. Myeloid-specific GPCR kinase-2 negatively regulates NF-kappaB1p105-ERK pathway and limits endotoxemic shock in mice. J Cell Physiol. 2011;226:627–637. doi: 10.1002/jcp.22384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.West AG, Fraser P. Remote control of gene transcription. Hum Mol Genet. 2005;14(Spec 1):R101–111. doi: 10.1093/hmg/ddi104. [DOI] [PubMed] [Google Scholar]
- 26.Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsui K, Nishizawa M, Ozaki T, Kimura T, Hashimoto I, Yamada M, Kaibori M, Kamiyama Y, Ito S, Okumura T. Natural antisense transcript stabilizes inducible nitric oxide synthase messenger RNA in rat hepatocytes. Hepatology. 2008;47:686–697. doi: 10.1002/hep.22036. [DOI] [PubMed] [Google Scholar]
- 28.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wysocka J, Milne TA, Allis CD. Taking LSD 1 to a new high. Cell. 2005;122:654–658. doi: 10.1016/j.cell.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 30.Koch CM, Andrews RM, Flicek P, Dillon SC, Karaoz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, James KD, Lefebvre GC, Bruce AW, Dovey OM, Ellis PD, Dhami P, Langford CF, Weng Z, Birney E, Carter NP, Vetrie D, Dunham I. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 32.Malecova B, Morris KV. Transcriptional gene silencing through epigenetic changes mediated by non-coding RNAs. Current opinion in molecular therapeutics. 2010;12:214–222. [PMC free article] [PubMed] [Google Scholar]
- 33.Jamalkandi SA, Masoudi-Nejad A. Reconstruction of Arabidopsis thaliana fully integrated small RNA pathway. Functional & integrative genomics. 2009;9:419–432. doi: 10.1007/s10142-009-0141-z. [DOI] [PubMed] [Google Scholar]
- 34.Span PN, Rao JU, Oude Ophuis SB, Lenders JW, Sweep FC, Wesseling P, Kusters B, van Nederveen FH, de Krijger RR, Hermus AR, Timmers HJ. Overexpression of the natural antisense hypoxia-inducible factor-1alpha transcript is associated with malignant pheochromocytoma/paraganglioma. Endocr Relat Cancer. 2011;18:323–331. doi: 10.1530/ERC-10-0184. [DOI] [PubMed] [Google Scholar]
- 35.Kohno K, Chiba M, Murata S, Pak S, Nagai K, Yamamoto M, Yanagisawa K, Kobayashi A, Yasue H, Ohkohchi N. Identification of natural antisense transcripts involved in human colorectal cancer development. Int J Oncol. 2010;37:1425–1432. doi: 10.3892/ijo_00000794. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.