Abstract
Autophagy is a lysosomal pathway that degrades and recycles intracellular organelles and proteins to maintain energy homeostasis during times of nutrient deprivation and to remove damaged cell components. Recent studies have identified new functions for autophagy under basal and stressed conditions. In the liver and pancreas, autophagy performs the standard functions of degrading mitochondria and aggregated proteins and regulating cell death. In addition, autophagy functions in these organs to regulate lipid accumulation in hepatic steatosis, trypsinogen activation in pancreatitis, and hepatitis virus replication. This review discusses the effects of autophagy on hepatic and pancreatic physiology and the contribution of this degradative process to diseases of these organs. The discovery of novel functions for this lysosomal pathway has increased our understanding of the pathophysiology of diseases in the liver and pancreas and suggested new possibilities for their treatment.
Keywords: Steatosis, Cell Death, Pancreatitis, Hepatitis Virus
Autophagy is an intracellular pathway by which lysosomes degrade and recycle long-lived proteins and cellular organelles. This pathway degrades cellular components that are worn out or damaged or are needed to generate substrates that maintain cellular energy homeostasis under conditions of limited nutrients or stress.1–3 Studies of the effects of altered autophagy in the liver have demonstrated the importance of the function of this pathway. In rats, a starvation-induced increase in autophagy led to the degradation of 35% of total liver protein within 24 hours.4 Conversely, inhibition of macroautophagy, by knockout of the autophagy gene atg7 in hepatocytes, led to a 4-fold increase in liver mass because of failure to degrade a variety of cellular components.5 Individual organs have specific and selective functions for this lysosomal pathway. In liver and pancreas, autophagy can regulate levels of lipids and contribute to pancreatitis and viral hepatitis. Although 3 distinct autophagic pathways have been described, most studies have been on macroautophagy, and this review summarizes the basic physiologic functions of macroautophagy and their role in the pathophysiology of hepatic and pancreatic diseases.
Autophagic Pathways
Macroautophagy
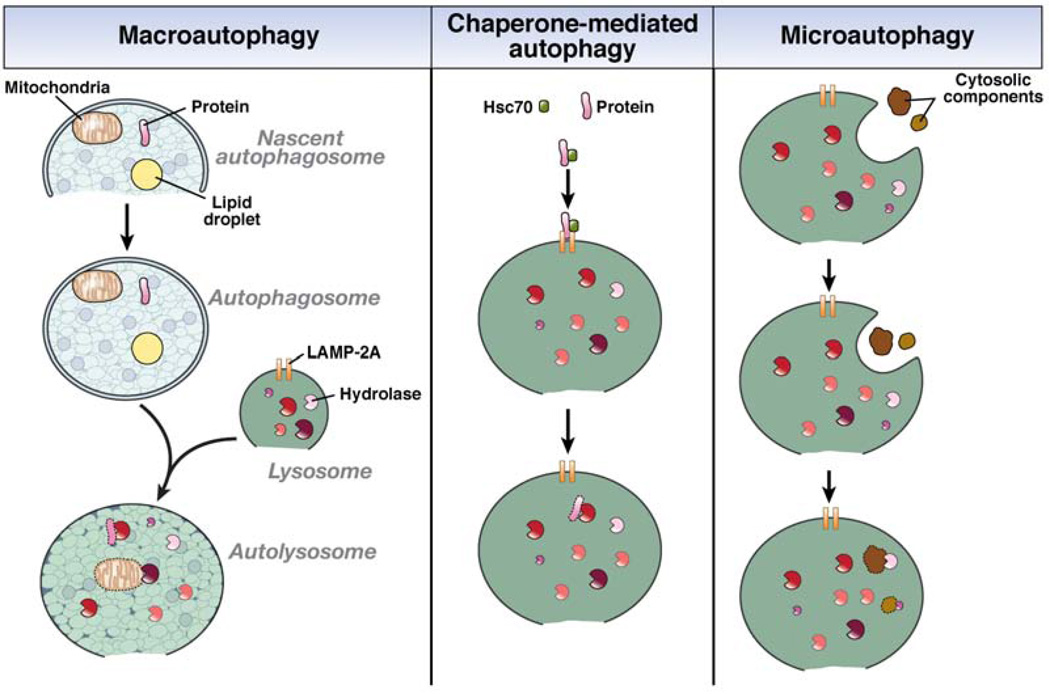
The 3 known types of autophagy are macroautophagy, chaperone-mediated autophagy (CMA), and microautophagy (Figure 1).6,7 In macroautophagy, a portion of cytosol is engulfed by a double-membrane structure, termed an autophagosome, that fuses with a lysosome, whose enzymes degrade the cellular constituents sequestered in the autophagosome.6 The regulation of this process is complex and controlled by the coordinated actions of autophagy-related genes (Atgs), over 30 of which have been identified in yeast and humans.8 Studies in yeast indicated that an initial structure, called an isolation membrane or phagophore, becomes a nascent autophagosome, whose ends elongate until they form the completely enclosed autophagosome. The source of the double membrane is controversial, but it might be derived from the endoplasmic reticulum (ER), mitochondria, or plasma membrane.9 The double membrane of the autophagosome is formed and elongated by unclear mechanisms, but a number of multiprotein complexes are known to mediate these processes.
Figure 1.
The 3 pathways of autophagy .In macroautophagy, doubie membrane of unclear origin forms around cytosolic components such as mitochondria, lipid droplets, and proteins. The membrane elongates to completely enclose the cellular elements within an autophagosome that translocates to a lysosome containing degradative hydrolases. The 2 structures fuse into an autolysosome, in which the cellular components are degraded by the lysosomal hydrolases. In CMA, cytosolic proteins with a specific pentapeptide motif are recognized by the chaperone Hsc70. This complex binds to the lysosomal LAMP-2A receptor for protein internalization and proteolytic degradation. Microautophagy involves the uptake of cellular components, both organelles and proteins, within an invagination of the lysosomal membrane for enzymatic degradation in the lysosome.
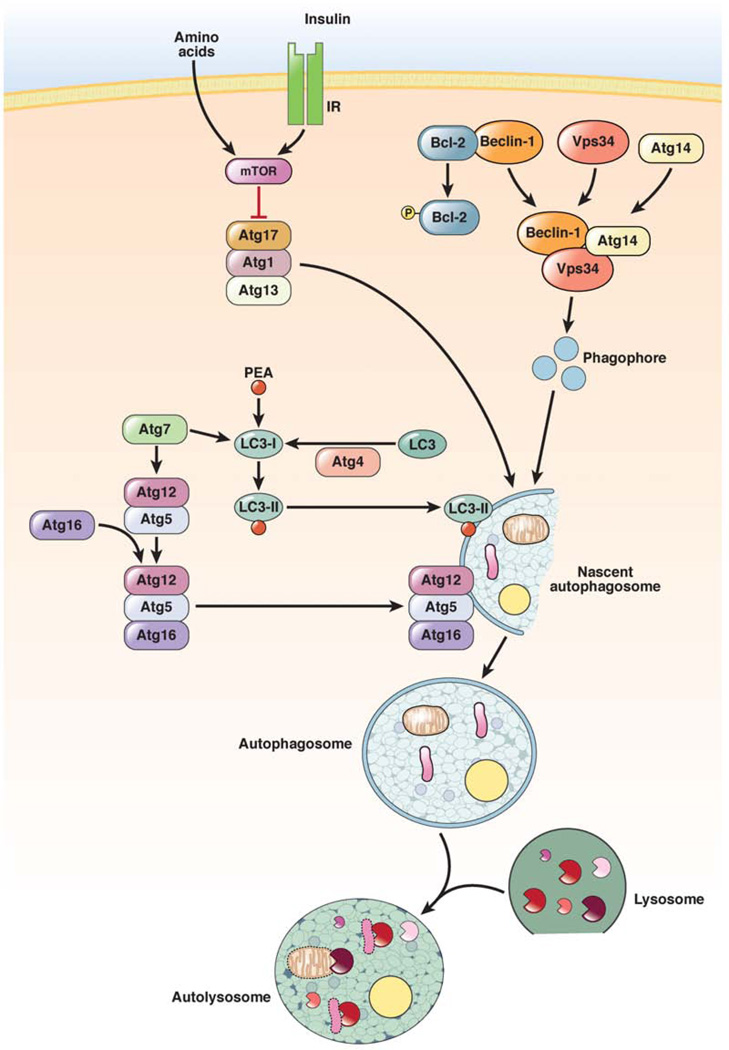
There are 3 major pathways that regulate macroautophagy (Figure 2). The first is the inhibitory mammalian target of rapamycin (mTOR) pathway. In direct response to nutrients (particularly amino acids), or through nutrient-induced insulin, class I phosphatidylinositol 3-kinase (PI3K) activates Akt and mTOR. This signaling pathway blocks macroautophagy through the ability of mTOR to inhibit Atg1 from recruiting its partners Atg13 and Atg17.10 The Atg1–Atg13–Atg17 complex recruits and organizes other proteins for the developing autophagosome.11,12 The mTOR inhibitor rapamycin is the most commonly used agent to increase autophagy; however, findings from studies with this agent cannot always be ascribed to its effects on autophagy, because mTOR regulates many other cellular pathways.13 Another pathway that regulates autophagy is mediated by Atg6/beclin-1, which forms a complex with the class III PI3K Vps34. Activation of the Atg1–Atg13–Atg17 complex leads to organization of the beclin-1–Vps34 complex on the lipid membrane. Vps34 produces phosphatidylinositol 3-phosphate, which can recruit other proteins to the complex.14,15 It is important to distinguish this PI3K from the insulin-activated, class I PI3K, which activates mTOR. Vps34 is the target of the widely used pharmacologic inhibitor of autophagy 3-methyladenine.16 Beclin-1 is an important interface between the autophagic and cell death pathways, because the antiapoptotic proteins Bcl-2 and Bcl-XL bind beclin-1 to inhibit autophagy.17 The regulation of this interaction is complex but includes its disruption by c-Jun N-terminal kinase 1–mediated phosphorylation of Bcl-2.18
Figure 2.
Pathways that control levels of macroautophagy. Macroautophagy is regulated by 3 major pathways. The first is an inhibitory pathway in which nutrient or insulin stimulation of the mTOR signaling pathway blocks autophagosome formation (red line). Two other pathways are stimulatory. In one, phosphorylation of Bcl-2 dissociates it from beclin-1, which allows beclin-1 to form a complex with the PI3K Vps34 that produces phosphatidylinositol 3-phosphate, which is required for induction of autophagy. The other pathway involves a series of conjugation steps that generate LC3-II and the Atg5–Atg12–Atg16 protein complex, which are both necessary for autophagosome formation. IR, insulin receptor; PEA, phosphatidylethanolamine.
The third major pathway that mediates autophagosome formation and elongation involves 2 ubiquitin-like conjugation processes that generate membrane-bound protein complexes. In the first, Atg7 and Atg10 mediate the conjugation of Atg12 to Atg5,19 which subsequently interact with Atg16.20 The Atg12–Atg5 complex associates with the membrane and then dissociates upon completion of the autophagosome. The second critical conjugation reaction involves Atg8 or microtubule-associated protein 1 light chain 3 (LC3). LC3 is constitutively cleaved by Atg4 to produce LC3-I. With a signal to induce autophagy, Atg7 and Atg3 mediate the conjugation of LC3-I to the membrane lipid phosphatidylethanolamine to form LC3-II.21 LC3-II associates with the autophagosomal membrane, where the lipidated protein can mediate membrane elongation and closure. LC3-II is degraded late in the autophagic pathway, after autophagosome fusion with a lysosome. In immunoblot analyses, increased levels of LC3-II are often misinterpreted as conclusive evidence of increased autophagic function. Although increased levels of LC3-II can indicate an increase in autophagy, they can also indicate a decrease in autophagic function due to a block in fusion after increases in the numbers of autophagosomes, and therefore levels of LC3-II, already occurred. Static levels of LC3-II therefore correlate with autophagosome number, not function. True autophagic function can be measured by autophagic flow or the change in LC3-II levels in the presence and absence of general lysosomal inhibitors, which measures the rate of LC3-II degradation.22 Although considerable information exists about the process of autophagosome formation in immortalized fibroblasts and transformed cells, it is important to note that whether these mechanisms are all operative in hepatic and pancreatic cells remains to be determined.
Once formed, autophagosomes traffic along microtubules by a dynein-dependent mechanism to reach perinuclear lysosomes located near the microtubule-organizing center. Another method to monitor the induction of macroautophagy is therefore to detect perinuclear, LC3-positive aggregates by immunofluorescence.23 Before they fuse with lysosomes, autophagosomes can fuse with early and late endosomes to form an amphisome. This process allows for a point of convergence between the pathways of autophagy and endocytosis. Autophagosomes dock and fuse to form an autophagolysosome or autolysosome by a process that has not been well defined in mammalian cells. The term “autophagic vacuole” has been used for autophagosomes, amphistomes, and autolysosomes, which can be indistinguishable even by electron microscopy. In yeast, fusion is mediated by soluble N-ethylmaleimide–sensitive factor attachment protein receptors.24 The soluble N-ethylmaleimide–sensitive factor attachment protein receptor protein vti1b mediates autophagosome-endosome fusion in mouse hepatocytes.25 Other factors that regulate the endocytic pathway, such as the guanosine triphosphate binding protein Rab7, have also been shown to function in the fusion process.26 Autophagosome-lysosome fusion allows mixing of their contents and degradation of the cargo of the autophagosome by lysosomal acid hydrolases, which include proteases, nucleases, lipases, glycosidases, and phosphatases. The degraded products are then transported back to the cytosol for reuse. Little is known about this process in mammalian cells, although putative efflux proteins have been identified in yeast.27
Macroautophagy is induced by a number of physiologic stimuli in addition to nutrient deprivation; considerable focus has been placed on identifying cellular events associated with increases in macroautophagy that might serve as markers of autophagic function. However, constitutive levels of autophagy are required for cell survival and function in most organs, including the liver and pancreas.5,28–30 Macroautophagy therefore has vital functions, even when not activated.
CMA and Microautophagy
The 2 other forms of autophagy are CMA and microautophagy (Figure 1). In CMA, soluble proteins with a specific pentapeptide motif are recognized by the chaperone Hsc70 for translocation to the lysosome, where binding to the lysosome-associated membrane protein type 2A (LAMP-2A) receptor leads to protein internalization and degradation.7 Similar to macroautophagy, CMA is constitutively active and increases with cellular stresses. CMA function in the liver and pancreas has not been well studied, although CMA has been shown to mediate hepatocyte resistance to oxidant stress.31,32 Although macroautophagy and CMA are distinct pathways, they interact and a reduction in one pathway can lead to activation of the other.33,34 This interaction can complicate interpretation of the effects of inhibiting either one.35 Microautophagy is a noninducible, lysosomal internalization of cellular constituents that occurs by invagination of the lysosomal membrane. In the absence of significant information on the function of CMA or microautophagy in the liver or pancreas, the remainder of this review focuses on macroautophagy (hereafter also referred to as autophagy).
Autophagy Removes Cytosolic Organelles and Proteins
Mitophagy Is a Selective Form of Macroautophagy
Autophagy performs a quality control function in the cell by removing aged or damaged cytosolic organelles and proteins. The engulfment of cytosol by the autophagosome has been considered a nonselective process, but there is evidence for selective autophagy of several types of organelles. Prominent among the structures targeted for autophagic degradation are damaged mitochondria, which can compromise cellular energy homeostasis or trigger apoptosis. The trafficking of mitochondria to lysosomal structures has long been known to occur in liver,36 and the livers of mice with a conditional knockout of atg7 develop massive hepatomegaly marked by the accumulation of deformed mitochondria.5 The process of lysosomal removal of mitochondria by autophagy was first characterized in hepatocyte models by Lemasters et al and termed “mitophagy.”3,37 Using fluorescence confocal microscopy, they showed that induction of autophagy by starvation or glucagon promotes mitochondrial engulfment by autophagosomes.3,38 Before removal, mitochondria undergo the mitochondrial permeability transition (MPT), in which inner mitochondrial membrane pores open, leading to adenosine triphosphate (ATP) depletion from the uncoupling of oxidative phosphorylation and outer membrane rupture with release of pro-apoptotic factors. The MPT induces mitophagy, because the MPT inhibitor cyclosporine blocks this process.38 After localized, laser-induced photo damage, LC3-positive structures appeared in hepatocytes in the area of injury.3 These findings indicate that a mechanism exists for the selective targeting of MPT-damaged mitochondria for autophagic degradation. Recent studies in nonhepatic cells have implicated Atg32,39,40 the Bcl-2 family member Nix,41,42 and the ubiquitin ligase Parkin43 in this process, but further studies are required to determine whether these proteins mediate selective mitophagy in hepatocytes. Mitochondrial damage that induces the MPT leads to death of hepatocytes, and these findings provide a mechanism by which insufficient or impaired autophagy can promote hepatocyte cell death. Selective mitophagy might be a mechanism to protect against hepatocyte death, because removal of mitochondria that have undergone the MPT could prevent mitochondrial oxidative stress, ATP depletion, or the release of pro-apoptotic factors. The contribution of mitophagy to the response of the hepatocyte to various death stimuli needs to be more carefully examined.
Macroautophagy Removes Protein Aggregates in a1-Antitrypsin Deficiency
Removing abnormal protein aggregates and damaged organelles by autophagy is particularly important in nondividing cells such as neurons, whose long life span might lead to an excessive and harmful accumulation of cellular components in the absence of autophagy. Neurodegenerative disease research has focused on the role of autophagy in the accumulation of abnormal protein aggregates in neurons of patients with Alzheimer’s and Parkinson’s disease.45 In the absence of a regenerative stimulus, hepatocytes are essentially nondividing cells. Atg7-null mice, which do not undergo autophagy, accumulate polyubiquitinated proteins that contribute to their massive hepatomegaly.5 One of these proteins is p62/sequestosome-1/SQSTM1 (p62),46 and a knockout of p62 in Atg7-null mice decreased the liver injury that accompanied the loss of autophagy.47 p62 activates the transcription factor nuclear factor erythroid 2-related factor (Nrf2); up-regulation of this pathway can lead to overproduction of Nrf2 target proteins that form aggregates in hepatocytes.48
Mutations in α1-antitrypsin (AT) can cause its accumulation in hepatocytes and the liver disease α1-antitrypsin deficiency.49 AT is synthesized by hepatocytes and secreted into the blood, where it functions as an important inhibitor of neutrophil proteases.50 A common mutant form of the protein, ATZ, undergoes abnormal polymerization and aggregation that blocks its secretion.51 The aggregated ATZ is retained in the ER, where it has toxic effects on hepatocytes that lead to the injury and inflammation associated with AT deficiency. Although initial studies reported that AT was degraded in the proteasome,52 subsequent studies found that numbers of autophagosomes increased in fibroblasts and mouse liver cells that expressed the mutant form of AT and in the livers of patients with this disease.53 Mutant AT was identified in autophagosomes, and its degradation was found to be blocked by pharmacologic inhibitors of autophagy.54 Furthermore, ATZ expressed in Atg5-null fibroblasts has a decreased rate of degradation compared with wild-type cells.55 Autophagy therefore functions in the removal of mutant forms of AT.
Hidvegi et al56 provided definitive evidence for the role of autophagy in AT deficiency by using carbamazepine to increase autophagy in a mouse model of the disease. This agent increased the formation of autophagosomes and reduced the soluble and insoluble forms of ATZ in the liver. The effect on protein aggregates was sufficient to significantly reduce hepatic fibrosis. This study indicated for the first time that pharmacologic modifiers of autophagic function might be used to successfully treat patients with hepatic and pancreatic diseases such as AT deficiency. Other chemical modifiers of autophagy are likely to be developed for their potential therapeutic value.57 Carbamazepine is already in widespread use in humans and has very low toxicity, although the mice received higher than normal doses, and clinical trials of this agent in AT deficiency could begin without extensive preclinical toxicity studies. The effects of an increase in autophagy on the clinical manifestations of AT deficiency in mice suggest that individual variation in levels of autophagy might determine the severity of this disorder in humans. A minority of individuals with a genetic defect in AT develop chronic liver disease, so other genetic and/or environmental factors determine disease development. Individual variation in hepatic autophagic function and therefore effectiveness in removal of mutant AT might determine the severity of disease. It might be possible to therapeutically regulate autophagy in specific individuals with the disease who have impaired autophagy.
Autophagy as a Metabolic Pathway: Hepatocyte Lipids Are Degraded by Lipophagy
Lysosomal lipases can degrade lipoproteins that are internalized by endocytosis. However, cellular lipids stored as triglycerides (TGs) were not identified as lysosomal substrates until studies showed that autophagy mediates the breakdown of these lipid stores in hepatocytes by a process termed “lipophagy.”58 Rat hepatocytes were presented with increased lipids in the form of supplemental fatty acids or culture in methionine- and choline-deficient medium. In response to these lipid stimuli, the cells accumulated significantly greater amounts of TG and cholesterol following genetic or pharmacologic inhibition of autophagy. Imaging studies showed that the excess lipid was in lipid droplets that increased in size and number when macroautophagy was inhibited. Neutral lipid colocalized with markers of autophagic vacuoles and was visualized in these vacuoles by electron microscopy, confirming the trafficking of TGs through the autophagic pathway. A decreased rate of lipolysis in the absence of autophagy led to the increased cellular TG content. Although autophagy was once believed to be nonselective, the removal of hepatocellular lipid stores by autophagy was specifically altered by the nutrient supply. Serum deprivation to stimulate lipolysis increased the number of autophagosomes that contained lipid, and with prolonged serum deprivation, the percentage of autophagosomes with other cargo, such as mitochondria, decreased. These findings indicate that hepatocytes specifically target lipids for autophagic degradation in response to nutritional stress.
In vivo studies confirmed the association of lipid and lipid droplet proteins with autophagic vacuoles and of the autophagosomal protein LC3 with lipid droplets in mouse liver.58 Mice with a hepatocyte-specific knockout of Atg7 had marked increases in hepatic lipids, based on biochemical and histologic analyses. Autophagy therefore limits hepatocyte accumulation of lipid by mediating the breakdown of TGs stored in lipid droplets (Figure 3). This alternative pathway for lipolysis might account for the ability of hepatocytes to rapidly mobilize large amounts of lipid, despite their low levels of cytosolic lipases compared with fat-storing adipocytes.59 It is not clear whether this pathway is specific for hepatocytes or exists in all cell types, including those in the pancreas. Similar findings were reported in mouse embryonic fibroblasts,58 but not in adipocytes, although an inhibition of autophagy in adipocytes altered their differentiation, which secondarily affected lipid storage.60,61 Defects in this alternative pathway of lipid metabolism might contribute to hepatic steatosis.62 The incidence of the metabolic syndrome, and accompanying nonalcoholic fatty liver disease, increases with age,63 in parallel with a decrease in autophagic function.64 Variants in genes that regulate autophagic function or an overall decrease in autophagy, which occurs with aging, might in part underlie the pathogenesis of steatosis.
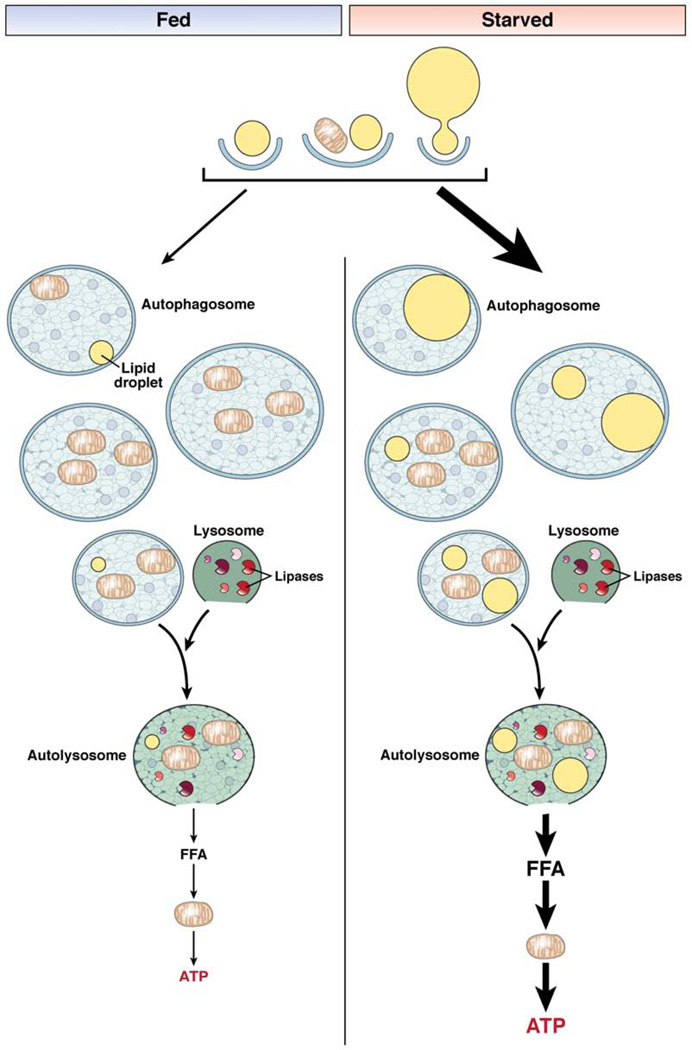
Figure 3.
Autophagic degradation of lipids. In lipophagy, lipid droplets are sequestered in autophagosomes for delivery to lysosomes for breakdown. Small lipid droplets can be taken up whole by an autophagosome, or alternatively portions of large lipid droplets can be degraded. The autophagosome can contain lipid, as the only substrate, or together with other cellular constituents such as mitochondria. The autophagosome eventually fuses with a hydrolase-containing lysosome whose lipases degrade the lipid. This process results in the release of free fatty acids (FFA) that can be oxidized in mitochondria to supply the cell with ATP. In the fed state, lipid breakdown by autophagy is not needed for nutrient supply and autophagy is suppressed. As a result, movement of lipids into the autophagic pathway is minimal. Most of the autophagosomes contain cargo other than lipid or occasionally lipid mixed with other cellular components. The amount of ATP generated by this process in the fed state is minor. In the starved state, autophagy is induced and selectively targets lipid droplets to utilize lipid stores to supply the cell with energy in the absence of other nutrients. As a result, more autophagosomes contain lipid, either alone or in combination with other cargo, enabling the cell to generate ATP from FFA breakdown to maintain energy homeostasis.
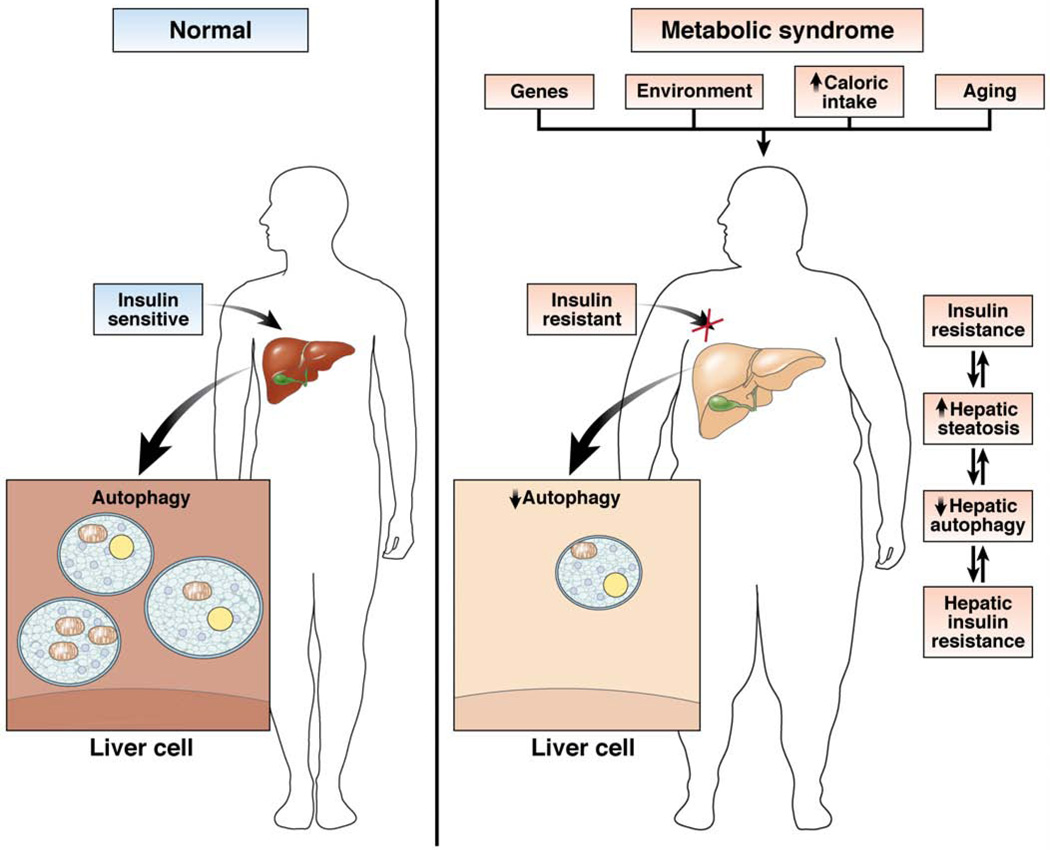
Hepatocyte lipid accumulation also decreases autophagy, indicating that steatosis might be a mechanism of defective hepatic autophagy.58 A decrease in fusion efficiency between autophagosomes and lysosomes, due to membrane lipid alterations in response to a high-fat diet, might underlie the inhibitory effect of diet-induced steatosis on macroautophagy.65 Hyperinsulinemia could compound the defect in autophagy induced by lipid accumulation. The ability of insulin to suppress autophagy is retained despite the presence of insulin resistance in hepatoma cells or mouse liver, indicating that hyperinsulinemia might contribute to impaired autophagy in the obese state.66 Conversely, a decrease in autophagy might impair hepatic insulin signaling. Restoration of autophagy that was decreased in the livers of obese mice increased insulin sensitivity in this organ.67 These findings indicate that a self-perpetuating cycle might exist in which hepatic lipid accumulation promotes insulin resistance, and both impair autophagy, which then worsens steatosis and insulin insensitivity (Figure 4). Hepatic levels of autophagy might mediate in part physiologic changes that lead to the clinical manifestations of the metabolic syndrome. With the development of better agents to increase autophagy, this possibility should be tested by studies of the effects of an increase in autophagy on various organ-specific manifestations of the metabolic syndrome in animal models and patients.
Figure 4.
Relationship of altered hepatic autophagy to other manifestations of the metabolic syndrome. Numerous factors can contribute to a decrease in hepatic autophagic function, including genetic and environmental factors and the effects of aging. In individuals with the metabolic syndrome, obesity and resultant peripheral insulin resistance can decrease levels of hepatic autophagy. Obesity, insulin resistance, and defective hepatic autophagy can all act to promote the development of hepatic steatosis and hepatic insulin resistance. Increased lipid content in the liver can then further impair hepatic autophagy, leading to increased insulin resistance and its manifestations in other organs.
Impaired Autophagy Mediates the Development of Pancreatitis
Acute pancreatitis is an inflammatory disorder believed to be initiated by self-digestion of the pancreatic acinar cell following inappropriate activation of pancreatic enzymes, particularly trypsin.68 However, little is known about the cellular location and mechanism of activation of trypsin, which is normally safely stored in granules as the inactive zymogen trypsinogen. Early pathologic studies of human and rodent acute pancreatitis reported accumulation of cytoplasmic vacuoles in acinar cells69,70 , that were subsequently shown to contain activated trypsin.71 The first indication that autophagy was involved in this process was the finding that these vacuoles contain LC3 and are therefore of autophagosomal origin.72 The function of autophagy was then examined in mice with acute cerulein-induced pancreatitis, which was attenuated in Atg5-null mice. Cultured Atg5-null acinar cells had decreased activation of trypsinogen, indicating that increased autophagy promoted enzyme activation and disease development.72 Studies in a rat alcohol-induced model of pancreatitis revealed that a subtoxic dose of alcohol or lipopolysaccharide (LPS) induced formation of autophagosomes and autolysosomes in acinar cells.73 However, the combination of alcohol and LPS, which induced pancreatitis, increased numbers of autophagosomes but not autolysosomes.73 These results indicated that the combination of alcohol and LPS blocked autophagosome-lysosome fusion, and findings of decreased levels of LAMP-2, a lysosomal membrane protein that promotes fusion,73,74 provided a possible mechanism for this effect. However, LPS alone caused a similar decrease in LAMP-2 yet increased the numbers of autolysosomes, and the LAMP-2 effect was not specific because the lysosomal protein granule membrane protein 92 (Gramp-92) was also decreased. Studies of human pancreatitis confirmed a decrease in LAMP-2 but also in LAMP-1. In contrast to the conclusions from studies of mice with cerulein-induced pancreatitis, the findings in the alcohol model indicated that autophagic function in pancreatitis is blocked by an unclear mechanism and that this impairment promotes disease.
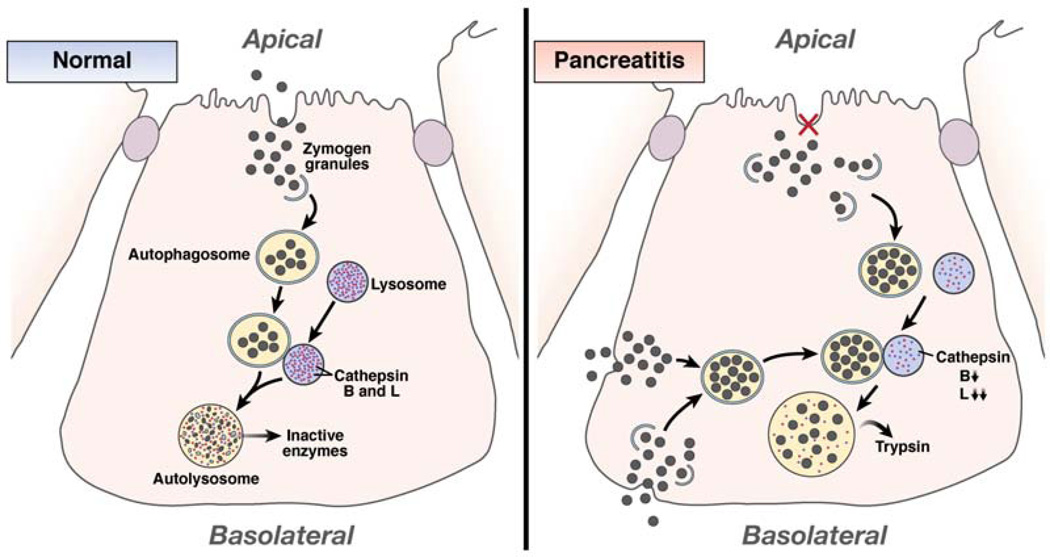
These discrepant findings were resolved in studies by Mareninova et al.75 Using cultured cells, multiple rodent models of pancreatitis, and human tissue, they found that autophagic function was impaired in pancreatitis, because acinar cells accumulated abnormally large autophagic vacuoles that contained undigested cargo. Furthermore, an in vitro model of pancreatitis showed reduced autophagy-dependent degradation of long-lived proteins and the zymogen-rich subcellular fraction from animals with pancreatitis had increased levels of lysosomal proteins. The existence of very large autophagic vacuoles with undigested material that contained lysosomal proteins and LC3 indicated that the defect in autophagy did not occur during fusion. Processing, and therefore activity, of the lysosomal enzymes cathepsin B and L were reduced in models of pancreatitis, and inhibitors of these enzymes increased the numbers of autophagic vacuoles in acinar cells. These results indicate that decreased cathepsin activity impairs protein degradation, blocking autophagy at the autolysosome stage. This possibility is supported by findings of a defect in autolysosomal degradation in cathepsin L null mice.76 Consistent with prior studies,72 inhibition of autophagy, either in vitro or in vivo, decreased trypsin activation.75 In contrast, pancreatic trypsin was not activated in rats or mice that were fasted to induce autophagy, presumably because these animals lacked the defect in cathepsin activity.75 These findings are consistent with impaired autophagy promoting trypsin activation. Trypsinogen activation peptide, a surrogate marker for activated trypsin, colocalized with autophagosomal and lysosomal markers, thus identifying autolysosomes as the site of trypsin activation. The lysosome therefore provides the acidic pH needed for zymogen activation. The defect in cathepsin L, an inactivator of trypsin, was found to be greater than that of the trypsinogen activator cathepsin B, indicating that an imbalance between the inactivating and activating enzymes resulted in inappropriate activation of trypsin. Therefore, the findings of Mareninova et al75 indicate that defects in lysosomal enzyme function disrupt the autophagic pathway at the level of protein degradation in autolysosomes. This defect results in accumulation of abnormally large autophagic vacuoles where trypsinogen is activated due to a defect in cathepsin L activity and then released into the cell (Figure 5).
Figure 5.
Autophagy mediates trypsin activation in pancreatitis. In a normal acinar cell, some trypsinogen-containing zymogen granules that are not secreted from the apical membrane can be taken up by autophagosomes for removal. These autophagosomes fuse with lysosomes to form autolysosomes in which the contents of the granules, including pancreatic enzymes, are degraded by lysosomal enzymes, preventing their release. In pancreatitis, a block in enzyme secretion leads to an accumulation of intracellular zymogen granules, which increases uptake of these structures by autophagosomes. Alternatively or additively, granules improperly released from the basolateral surface of the acinar cell are taken back up by the cell and sequestered by autophagosomes. Following autophagosome-lysosome fusion, content degradation fails to occur because of decreased levels of cathepsin B and L. The more pronounced defect in cathepsin L, which inactivates trypsin, than in the activating enzyme cathepsin B leads to trypsin generation within the autolysosome. Active enzymes are released from these enlarged autolysosomes into the cytoplasm to initiate pancreatic cellular injury.
It is not clear how cathepsin processing becomes defective or how this relates to reported decreases in other lysosomal proteins. The findings indicate that etiology-specific or multifactorial lysosomal dysfunction is central in the pathogenesis of acute pancreatitis, and further studies are required to examine this possibility. It is not clear how trypsinogen granules, normally a substrate for the secretory pathway, enter the autophagic pathway. Mareninova et al75 proposed that zymogen granules are normally degraded by the autophagic pathway, and with defective cathepsin function this process goes awry. Alternatively, pancreatitis is associated with a decrease in the capacity of the acinar cell for zymogen secretion. Simple overload of the cell with zymogen granules that contain a potentially toxic protease might initiate or potentiate their engulfment by autophagosomes as a protective mechanism to sequester and degrade the enzyme. In support of this model, experimental and human pancreatitis are associated with reduced secretion of pancreatic enzymes, although it is not clear whether this is a cause or consequence of the disease.77,78 Alternatively, in pancreatitis, disruption of cellular apical junctional barriers and release of zymogen granules through the basolateral membrane could lead to the presence of enzymes in the interstitial space that enter the autophagic pathway following endocytosis (Figure 5).68 Finally, individual variation in genes that regulate autophagy might influence the tendency to develop pancreatitis. Mutations in serine protease inhibitor Kazal type 1 (SPINK1), which inhibits trypsin activity, are associated with familial pancreatitis.79 Mice with a knockout of the homologous gene spink3 form increased numbers of acinar cell autophagosomes, similar to patients with acute pancreatitis, but in the absence of an increase in activated trypsin.80 Spink3 might therefore somehow directly regulate autophagy, in addition to its effect on trypsin. If additional studies confirm that inhibition of autophagy impairs the development of acute pancreatitis, or more importantly improves its course once the process has started, then agents to decrease autophagy might be developed as a treatment for acute pancreatitis. Current therapy might also be altered because the practice of resting the pancreas by withholding oral feedings in the absence of other nutritional support might actually worsen pancreatitis by further inducing autophagy.
Autophagy Regulates Hepatic and Pancreatic Cell Death
Although autophagy functions in cell death pathways, there is controversy over whether this lysosomal degradative process promotes or prevents cell death. Autophagy has been characterized as a type of cell death, along with apoptosis and necrosis.81 However, a number of studies have identified pro-survival effects of autophagy after injurious stresses that induce cell death.31,82–84 It is logical that some functions of autophagy would prevent cell death, including the removal of damaged organelles or proteins and the supply of substrates to maintain cellular levels of ATP. To complicate the relationship between the autophagic and cell death pathways, components of the autophagic pathway also participate directly in apoptosis.85,86 Overall, evidence indicates that autophagy is a cell survival pathway that can also mediate cell death under certain conditions, such as when autophagy is overactivated. Studies have begun to delineate the roles of autophagy in hepatic and pancreatic resistance to injury and cell death.
Basal Autophagy Maintains Hepatocyte and Pancreatic Beta Cell Structure and Function
Macroautophagy is an inducible pathway initially believed to function only in response to stimuli such as starvation or oxidant stress. The development of mice with conditional Atg knockouts overcame the problem of postnatal lethality from an embryonic Atg disruption and permitted studies of the effects of an organ-specific loss of autophagy.5,87 As expected, the normal decrease in liver protein failed to occur in hepatocyte-specific Atg7 knockout mice after the induction of autophagy by starvation. However, the loss of basal levels of autophagy also had major effects. Within 90 days after the induction of a hepatocyte-specific, Cre-mediated deletion of Atg7, massive hepatomegaly developed with a 4-fold increase in liver mass.5 Histologic studies showed that the livers had lobular disorganization, with swollen hepatocytes full of abnormal vacuoles that contained cellular organelles such as mitochondria and lipid droplets. The hepatocytes had increased numbers of abnormal mitochondria, peroxisomes, and polyubiquitinated protein aggregates. These abnormalities resulted in hepatocyte death, demonstrated by increased levels of serum transaminases. These findings indicate that not only is autophagy an important response to stress, but also basal levels of autophagy are required to maintain hepatocyte homeostasis. This is not surprising because hepatocytes have low rates of proliferation and long life spans, so they accumulate organelles and proteins that must be degraded by autophagy. Factors that inhibit autophagy in hepatocytes might therefore amplify any form of liver injury through effects on basic cellular homeostasis. Hepatic levels of autophagy decrease with aging,64 and this reduction may be sufficient to induce liver abnormalities such as the buildup of oxidized proteins.32
Basal levels of autophagy are also required for maintenance of beta cells in the pancreas.29,30 A beta cell–specific knockout of atg7 resulted in increased apoptosis and reduced proliferation of these cells as mice aged, resulting in a 40% decrease in beta cell mass. The cells had mitochondrial abnormalities and large aggregates of polyubiquitinated proteins, which included p62. Although aggregates of undegraded proteins could be toxic to beta cells, this finding could simply be an epiphenomenon, and neither study defined the mechanism of increased beta cell destruction. Autophagy might also be required to maintain energy homeostasis or to directly counteract apoptotic pathways. Pancreatic islets from mice with beta cell–specific disruption of Atg7 had decreased ATP production,30 although this might have been secondary to mitochondrial damage. Islet cell destruction had functional consequences, because the remaining cells were depleted of insulin and the mice developed hyperglycemia that was worsened by a high-fat diet.29,30 Similar to findings in hepatocytes, autophagy is required to maintain beta cell function and viability. It will be important to determine whether reduced autophagy contributes to loss of beta cells in patients with diabetes.
Autophagy Is Induced by Alcohol and Prevents Liver Injury
The ability of autophagy to regulate hepatocyte steatosis and cell death, particularly death from oxidant stress and mitochondrial damage,31,37 indicates that this pathway might modulate the development of the fatty liver diseases of alcoholic and nonalcoholic steatohepatitis.62 Recent studies by Ding et al88 showed that autophagy protects against liver injury from alcohol. In an acute mouse model of binge alcohol consumption, increased autophagosome formation was shown by LC3 imaging, electron microscopy, and measurement of autophagic flux. Primary hepatocytes treated with ethanol had increased numbers of autophagosomes compared with control hepatocytes, possibly as a result of oxidant-mediated suppression of mTOR. This increase in autophagosome number did not alter the degradation of long-lived proteins, although levels of p62 decreased. These data indicate a selective increase in organelle removal but not protein degradation by autophagy. This conclusion was supported by findings of increased numbers of autophagosomes that contained mitochondria or lipid droplets. In response to alcohol, the liver might increase autophagy to selectively eliminate damaged mitochondria and limit lipid accumulation. Autophagy protects cells against injury from alcohol, because chemical and genetic inhibition of autophagy increased the levels of injury in cultured hepatocytes and mouse liver. In vivo, increasing autophagy with rapamycin reduced alcohol-induced lipid accumulation and autophagic inhibition worsened steatosis, confirming that autophagy regulates hepatic lipid content. Autophagy could limit alcohol-induced liver damage by simply reducing levels of cellular lipids that promote injury, but it is more likely that autophagy has direct inhibitory effects on cell death pathways. Animal models of acute alcohol-induced injury might not accurately reflect the pathophysiology of chronic alcoholic liver disease, and the function of autophagy needs to be examined in a chronic alcohol injury model and in models of nonalcoholic steatohepatitis. Individual variation in the development of human alcoholic liver disease might result from differences in genes that control autophagic function, but this requires further study, along with the possible therapeutic use of inducers of autophagy for this disease.
Regulation of Infectious Diseases by Autophagy
Autophagy disposes of unwanted cellular constituents, which includes the removal of intracellular microbes as part of the innate immune response.89 However, some microorganisms have developed the ability to circumvent autophagy to ensure their own survival. Viruses can even exploit this pathway for their own benefit, and the hepatitis C and B viruses (HCV and HBV) regulate levels of autophagy to promote their own replication.
HCV Induces Autophagy to Initiate Replication
HCV is an enveloped, positive-strand RNA virus with a genome that contains a single open reading frame that encodes multiple proteins flanked by 2 noncoding regions required for translation and replication.90 HCV replication occurs in association with intracellular structures composed of vesicles and a membranous matrix, termed the membranous web, that seems to be associated with the ER.91 HCV replication begins with translation of the incoming viral RNA to produce proteins that mediate viral RNA replication.90 Autophagy has an important role in the replication process, and HCV infection alters autophagic function. Immortalized human hepatocytes infected with the HH7 strain of HCV and Huh7 hepatoma cells transfected with RNA or infected with the JFH1 viral strain have increased autophagosome number, based on imaging and LC3 immunoblot analyses.92–94 However, studies of JFH1-infected cells showed no increase in protein degradation and an apparent decrease in autolysosome number, consistent with an induction of autophagosome formation but a block in autophagosome maturation and lysosomal fusion.94 This activation of the autophagic response could result from viral induction of ER stress, but the mechanism that led to a block in pathway progression was not delineated.94 Studies of JFH1-transfected or infected Huh7 cells in which macroautophagy was inhibited by knockdown of autophagy genes revealed that a loss of autophagy inhibited the production of viral messenger RNA and infective particles.93,94 An elegant dissection of the effects of a loss of autophagy on the viral life cycle showed that cellular uptake and secretion of the virus were unaffected, but viral replication was blocked, secondary to a failure in translation of the incoming viral RNA.93 Subsequent replication was unaffected, indicating that autophagy was specifically required for initiation of the replicative process but not further replication. A second study using the same JFH1-Huh7 in vitro model showed ER stress-mediated induction of autophagy by HCV.95 However, autophagic flow and electron microscopy analyses indicated that autophagy proceeded to autolysosome formation, although protein degradation was not assessed.95 Thus, controversy exists over the effects of HCV infection on autophagic function. These discrepant findings likely represent limitations of the in vitro HCV culture systems and indicate the need for a good animal model of HCV infection to address this question or, alternatively, studies in human samples.
The finding that autophagy promotes HCV replication, together with the possible common origin of both the membranous web and autophagosome in the ER, suggests the possibility that HCV utilizes the autophagic machinery in the viral replication complex to form the membranous web. Although initial studies failed to find colocalization of any viral and autophagic proteins,92,93 a yeast 2-hybrid study recently identified an interaction between Atg5 and the HCV NS5B RNA-dependent RNA polymerase.96 Atg5, and possibly other autophagic proteins, might regulate formation of the membrane web or actually attract viral proteins required for replication such as NS5B. Consistent with the involvement of autophagic proteins in membranous web formation or assembly, Atg5 and NS5B both colocalize to the membranous web, but only during the early stages of infection.96
Other possible mechanisms for the regulation of HCV replication by autophagy exist, and the effects of autophagy may be multifactorial. Autophagy might promote replication through an effect on lipid droplets, which are strongly associated with chronic HCV infection. The HCV core protein attaches to lipid droplets for unknown reasons, but possibly to promote viral assembly.97 The recent evidence of a role of autophagy in lipid droplet breakdown suggests that HCV replication could be mediated through an increase in autophagy that metabolizes lipids into free fatty acids required to supply energy for the replication process. Autophagy was recently shown to perform this function for dengue virus, another positive-strand RNA virus.98 Alternatively, an ability of HCV to inhibit autophagy at the autolysosome stage might promote lipid droplet accumulation, which is required for HCV replication.99 Finally, 2 recent reports show a suppression of interferon beta signaling and therefore the innate immune response by autophagy following in vitro HCV infection that may permit replication.95,100 Although questions remain about the mechanism by which autophagy promotes HCV replication, the current investigations indicate that inhibition of autophagy might be a novel approach to treating patients with HCV infection. However, a therapeutic inhibition of autophagy might also promote detrimental hepatic lipid accumulation, rendering such an approach impractical.
Hepatitis B Also Utilizes Autophagy for Virus Replication
RNA, but not DNA, viruses commonly alter autophagic function to promote replication.101 Surprisingly, HBV, a partially double-stranded DNA virus, has also been reported to regulate autophagy to promote replication.102 In findings similar to those for HCV, Huh7 hepatoma cells transfected with HBV genomic DNA, HBV DNA–containing livers of transgenic mice, and HBV-infected human livers all had evidence of increased autophagosome number. The HBV-infected cells also had increased colocalization of LC3 and a lysosomal marker, and the mouse livers had increased numbers of autolysosomes by electron microscopy, suggesting that autophagic function increased. Still, no increase occurred in the degradation of long-lived proteins, indicating the induction of a substrate-selective increase in autophagy but also raising the possibility that viral infection had not increased autophagic function. Pharmacologic and genetic inhibition of autophagy slightly reduced the levels of pregenomic RNA and more significantly decreased subsequent DNA synthesis, so the primary effect of autophagy was not clear. The induction of autophagy was mediated by HBV X protein (HBx). Wild-type, but not mutant, HBx induced LC3 conjugation, apparently by the mechanism of association of HBx with Vps34, which increased function of this PI3K. However, HBx had been previously described to up-regulate autophagy by increasing levels of beclin-1, which also binds Vps34,103 so the mechanism of the effect of HBx is unknown but seems to involve this PI3K. It is not clear how induction of autophagy promotes HBV replication, because the process does not involve a membranous web or association with lipids. These findings do indicate that HBV and HCV each target the autophagic pathway for viral replication. Despite major structural differences, both viruses exploit autophagy to promote their own replication. These similarities indicate that the ability to circumvent autophagic pathways may be evolutionarily important to establishing chronic viral infections of the liver.
Conclusions
Autophagy is more than a cellular waste disposal pathway, and macroautophagy performs a variety of functions with widespread effects on hepatic and pancreatic physiology. Autophagic functions in the liver and pancreas are complex, with components of the autophagic machinery interacting with those from other cell processes, such as apoptosis or formation of the membranous web during HCV infection. Further studies are needed to identify the roles of autophagy in other aspects of hepatic and pancreatic cellular function and increase our understanding of disease pathogenesis in these organs. The autophagic pathway provides an exciting new therapeutic target for a number of hepatic and pancreatic diseases (Table 1).
Table 1.
Hepatic and Pancreatic Diseases Potentially Involving Autophagy.
| Disease | Autophagy | Pathophysiologic consequences |
Therapeutic intervention |
|---|---|---|---|
| Liver | |||
| α1-antitrypsin deficiency | Decreased | Increased liver injury from decreased mutant protein removal | Increase autophagy to decrease liver injury and fibrosis |
| Nonalcoholic fatty liver disease | Decreased | Increased steatosis, hepatocyte injury, and hepatic insulin resistance | Increase autophagy to reduce steatosis and hepatitis and improve insulin sensitivity |
| Acute alcoholic liver injury | Increased | Reduced steatosis and liver injury | Further increase autophagy to decrease steatosis and hepatitis in chronic disease |
| Hepatitis C | Increased autopha gosomes Deceased fusion? | Promotes viral replication | Decrease autophagy to block viral replication |
| Hepatitis B | Increased? | Promotes viral replication | Decrease autophagy to block viral replication |
| Pancreas | |||
| Pancreatitis | Increase autolysosomes Decrease cargo degradation | zymogen accumulation in autolysosomes , increased trypsin activation and cellular injury | decrease autophagy to prevent or treat pancreatitis |
Acknowledgments
Funding
Supported by National Institutes of Health grants DK044234, DK061498, and AG031782.
Abbreviations used in this paper
- AT
α1-antitrypsin
- Atg
autophagy-related gene
- CMA
chaperone-mediated autophagy
- ER
endoplasmic reticulum
- HBx
hepatitis B virus protein X
- LAMP-2
lysosome-associated membrane protein 2
- LC3
microtubule-associated protein 1 light chain 3
- LPS
lipopolysaccharide
- MPT
mitochondrial permeability transition
- mTOR
mammalian target of rapamycin
- p62
p62/sequestosome-1/SQSTM1
- PI3K
phosphatidylinositol 3-kinase
- TG
triglyceride
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
The author discloses no conflicts.
References
- 1.Finn PF, Dice JF. Proteolytic and lipolytic responses to starvation. Nutrition. 2006;22:830–844. doi: 10.1016/j.nut.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Iwata J, Ezaki J, Komatsu M, et al. Excess peroxisomes are degraded by autophagic machinery in mammals. J Biol Chem. 2006;281:4035–4041. doi: 10.1074/jbc.M512283200. [DOI] [PubMed] [Google Scholar]
- 3.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cuervo AM, Knecht E, Terlecky SR, et al. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol. 1995;269:C1200–C1208. doi: 10.1152/ajpcell.1995.269.5.C1200. [DOI] [PubMed] [Google Scholar]
- 5.Komatsu M, Waguri S, Ueno T, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mehrpour M, Esclatine A, Beau I, et al. Autophagy in health and disease. 1. Regulation and significance of autophagy: an overview. Am J Physiol Cell Physiol. 2010;298:C776–C785. doi: 10.1152/ajpcell.00507.2009. [DOI] [PubMed] [Google Scholar]
- 7.Orenstein SJ, Cuervo AM. Chaperone-mediated autophagy: molecular mechanisms and physiological relevance. Semin Cell Dev Biol. 2010;21:719–726. doi: 10.1016/j.semcdb.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamasaki M, Yoshimori T. Where do they come from? Insights into autophagosome formation. FEBS Lett. 2010;584:1296–1301. doi: 10.1016/j.febslet.2010.02.061. [DOI] [PubMed] [Google Scholar]
- 10.Neufeld TP. TOR-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol. 2010;22:157–168. doi: 10.1016/j.ceb.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheong H, Yorimitsu T, Reggiori F, et al. Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki K, Kubota Y, Sekito T, et al. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 13.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 14.Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem J. 2008;410:1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- 15.Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 16.Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–606. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei Y, Pattingre S, Sinha S, et al. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizushima N, Noda T, Yoshimori T, et al. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 20.Ohsumi Y, Mizushima N. Two ubiquitin-like conjugation systems essential for autophagy. Semin Cell Dev Biol. 2004;15:231–236. doi: 10.1016/j.semcdb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Ichimura Y, Kirisako T, Takao T, et al. A ubiquitin-lie syste m mediates rotein lipidatio. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 22.Tanida I, Minematsu-Ikeguchi N, Ueno T, et al. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 23.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sudhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Atlashkin V, Kreykenbohm V, Eskelinen EL, et al. Deletion of the SNARE vti1b in mice results in the loss of a single SNARE partner, syntaxin 8. Mol Cell Biol. 2003;23:5198–5207. doi: 10.1128/MCB.23.15.5198-5207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jager s, Bucci c, Tanida I, et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 27.Yang Z, Huang J, Geng J, et al. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol Biol Cell. 2006;17:5094–5104. doi: 10.1091/mbc.E06-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 29.Jung HS, Chung KW, Won Kim J, et al. Loss of autophagy diminishes pancreatic β cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8:318–324. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 30.Ebato C, Uchida T, Arakawa M, et al. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab. 2008;8:325–332. doi: 10.1016/j.cmet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Singh R, Xiang Y, et al. Macroautophagy and chaperone-mediated autophagy are required for hepatocyte resistance to oxidant stress. Hepatology. 2010;52:266–277. doi: 10.1002/hep.23645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008;14:959–965. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaushik S, Massey AC, Mizushima N, et al. Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol Biol Cell. 2008;19:2179–2192. doi: 10.1091/mbc.E07-11-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massey AC, Kaushik S, Sovak G, et al. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci U S A. 2006;103:5805–5810. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Singh R, Massey AC, et al. Loss of macroautophagy promotes or prevents fibroblast apoptosis depending on the death stimulus. J Biol Chem. 2008;283:4766–4777. doi: 10.1074/jbc.M706666200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol. 1962;12:198–202. doi: 10.1083/jcb.12.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 38.Elmore SP, Qian T, Grissom SF, et al. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J. 2001;15:2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- 39.Kanki T, Wang K, Cao Y, et al. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 41.Schweers RL, Zhang J, Randall MS, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Novak I, Kirkin V, McEwan DG, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narendra D, Tanaka A, Suen DF, et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lemasters JJ, Qian T, He L, et al. Role of mitochondrial inner membrane permeabilization in necrotic cell death, apoptosis, and autophagy. Antioxid Redox Signal. 2002;4:769–781. doi: 10.1089/152308602760598918. [DOI] [PubMed] [Google Scholar]
- 45.Martinez-Vicente M, Cuervo AM. Autophagy and neurodegeneration: when the cleaning crew goes on strike. Lancet Neurol. 2007;6:352–361. doi: 10.1016/S1474-4422(07)70076-5. [DOI] [PubMed] [Google Scholar]
- 46.Ichimura Y, Kumanomidou T, Sou YS, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 47.Riley BE, Kaiser SE, Shaler TA, et al. Ubiquitin accumulation in autophagy-deficient mice is dependent on the Nrf2-mediated stress response pathway: a potential role for protein aggregation in autophagic substrate selection. J Cell Biol. 2010;191:537–552. doi: 10.1083/jcb.201005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Komatsu M, Kurokawa H, Waguri S, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 49.Perlmutter DH. Liver injury in α1-antitrypsin deficiency: an aggregated protein induces mitochondrial injury. J Clin Invest. 2002;110:1579–1583. doi: 10.1172/JCI16787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perlmutter DH. Pathogenesis of chronic liver injury and hepatocellular carcinoma in α1-antitrypsin deficiency. Pediatr Res. 2006;60:233–238. doi: 10.1203/01.pdr.0000228350.61496.90. [DOI] [PubMed] [Google Scholar]
- 51.Lomas DA, Evans DL, Finch JT, et al. The mechanism of Z α1-antitrypsin accumulation in the liver. Nature. 1992;357:605–607. doi: 10.1038/357605a0. [DOI] [PubMed] [Google Scholar]
- 52.Qu D, Teckman JH, Omura S, et al. Degradation of a mutant secretory protein, α1-antitrypsin Z, in the endoplasmic reticulum requires proteasome activity. J Biol Chem. 1996;271:22791–22795. doi: 10.1074/jbc.271.37.22791. [DOI] [PubMed] [Google Scholar]
- 53.Teckman JH, An JK, Loethen S, et al. Fasting in α1-antitrypsin deficient liver: constitutive activation of autophagy. Am J Physiol Gastrointest Liver Physiol. 2002;283:G1156–G1165. doi: 10.1152/ajpgi.00041.2002. [DOI] [PubMed] [Google Scholar]
- 54.Teckman JH, Perlmutter DH. Retention of mutant α1-antitrypsin Z in endoplasmic reticulum is associated with an autophagic response. Am J Physiol Gastrointest Liver Physiol. 2000;279:G961–G974. doi: 10.1152/ajpgi.2000.279.5.G961. [DOI] [PubMed] [Google Scholar]
- 55.Kamimoto T, Shoji S, Hidvegi T, et al. Intracellular inclusions containing mutant α1-antitrypsin Z are propagated in the absence of autophagic activity. J Biol Chem. 2006;281:4467–4476. doi: 10.1074/jbc.M509409200. [DOI] [PubMed] [Google Scholar]
- 56.Hidvegi T, Ewing M, Hale P, et al. An autophagy-enhancing drug promotes degradation of mutant α1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329:229–232. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- 57.Rubinsztein DC, Gestwicki JE, Murphy LO, et al. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- 58.Singh R, Kaushik S, Wang Y, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zechner R, Madeo F. Cell biology: Another way to get rid of fat. Nature. 2009;458:1118–1119. doi: 10.1038/4581118a. [DOI] [PubMed] [Google Scholar]
- 60.Baerga R, Zhang Y, Chen PH, et al. Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy. 2009;5:1118–1130. doi: 10.4161/auto.5.8.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh R, Xiang Y, Wang Y, et al. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009;119:3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Czaja MJ. Autophagy in health and disease. 2. Regulation of lipid metabolism and storage by autophagy: pathophysiological implications. Am J Physiol Cell Physiol. 2010;298:C973–C978. doi: 10.1152/ajpcell.00527.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ford ES, Giles WH, Dietz WH. Prevalence of the metabolic syndrome among US adults findings from the third National Health and Nutrition Examination Survey. JAMA. 2002;287:356–359. doi: 10.1001/jama.287.3.356. [DOI] [PubMed] [Google Scholar]
- 64.Cuervo AM, Bergamini E, Brunk UT, et al. Autophagy and aging: the importance of maintaining “clean” cells. Autophagy. 2005;1:131–140. doi: 10.4161/auto.1.3.2017. [DOI] [PubMed] [Google Scholar]
- 65.Koga H, Kaushik S, Cuervo AM. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010;24:3052–3065. doi: 10.1096/fj.09-144519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu HY, Han J, Cao SY, et al. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J Biol Chem. 2009;284:31484–31492. doi: 10.1074/jbc.M109.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang L, Li P, Fu S, et al. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gaisano HY, Gorelick FS. New insights into the mechanisms of pancreatitis. Gastroenterology. 2009;136:2040–2044. doi: 10.1053/j.gastro.2009.04.023. [DOI] [PubMed] [Google Scholar]
- 69.Adler G, Rohr G, Kern HF. Alteration of membrane fusion as a cause of acute pancreatitis in the rat. Dig Dis Sci. 1982;27:993–1002. doi: 10.1007/BF01391745. [DOI] [PubMed] [Google Scholar]
- 70.Helin H, Mero M, Markkula H, et al. Pancreatic acinar ultrastructure in human acute pancreatitis. Virchows Arch A Pathol Anat Histol. 1980;387:259–270. doi: 10.1007/BF00454829. [DOI] [PubMed] [Google Scholar]
- 71.Sherwood MW, Prior IA, Voronina SG, et al. Activation of trypsinogen in large endocytic vacuoles of pancreatic acinar cells. Proc Natl Acad Sci U S A. 2007;104:5674–5679. doi: 10.1073/pnas.0700951104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hashimoto D, Ohmuraya M, Hirota M, et al. Involvement of autophagy in trypsinogen activation within the pancreatic acinar cells. J Cell Biol. 2008;181:1065–1072. doi: 10.1083/jcb.200712156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fortunato F, Burgers H, Bergmann F, et al. Impaired autolysosome formation correlates with Lamp-2 depletion: role of apoptosis, autophagy, and necrosis in pancreatitis. Gastroenterology. 2009;137:350–360. doi: 10.1053/j.gastro.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 74.Huynh KK, Eskelinen EL, Scott CC, et al. LAMP proteins are required for fusion of lysosomes with phagosomes. EMBO J. 2007;26:313–324. doi: 10.1038/sj.emboj.7601511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mareninova OA, Hermann K, French SW, et al. Impaired autophagic flux mediates acinar cell vacuole formation and trypsinogen activation in rodent models of acute pancreatitis. J Clin Invest. 2009;119:3340–3355. doi: 10.1172/JCI38674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dennemarker J, Lohmuller T, Muller S, et al. Impaired turnover of autophagolysosomes in cathepsin L deficiency. Biol Chem. 2010;391:913–922. doi: 10.1515/BC.2010.097. [DOI] [PubMed] [Google Scholar]
- 77.Boreham B, Ammori BJ. A prospective evaluation of pancreatic exocrine function in patients with acute pancreatitis: correlation with extent of necrosis and pancreatic endocrine insufficiency. Pancreatology. 2003;3:303–308. doi: 10.1159/000071768. [DOI] [PubMed] [Google Scholar]
- 78.Niederau C, Niederau M, Luthen R, et al. Pancreatic exocrine secretion in acute experimental pancreatitis. Gastroenterology. 1990;99:1120–1127. doi: 10.1016/0016-5085(90)90633-c. [DOI] [PubMed] [Google Scholar]
- 79.Witt H, Luck W, Hennies HC, et al. Mutations in the gene encoding the serine protease inhibitor, Kazal type 1 are associated with chronic pancreatitis. Nat Genet. 2000;25:213–216. doi: 10.1038/76088. [DOI] [PubMed] [Google Scholar]
- 80.Ohmuraya M, Hirota M, Araki M, et al. Autophagic cell death of pancreatic acinar cells in serine protease inhibitor Kazal type 3-deficient mice. Gastroenterology. 2005;129:696–705. doi: 10.1016/j.gastro.2005.05.057. [DOI] [PubMed] [Google Scholar]
- 81.Gozuacik D, Kimchi A. Autophagy and cell death. Curr Top Dev Biol. 2007;78:217–245. doi: 10.1016/S0070-2153(06)78006-1. [DOI] [PubMed] [Google Scholar]
- 82.Maiuri MC, Zalckvar E, Kimchi A, et al. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 83.Hara T, Nakamura K, Matsui M, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 84.Pua HH, Dzhagalov I, Chuck M, et al. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yousefi S, Perozzo R, Schmid I, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 86.Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 87.Kuma A, Hatano M, Matsui M, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 88.Ding WX, Li M, Chen X, et al. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology. 2010;139:1740–1752. doi: 10.1053/j.gastro.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saitoh T, Akira S. Regulation of innate immune responses by autophagy-related proteins. J Cell Biol. 2010;189:925–935. doi: 10.1083/jcb.201002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lindenbach BD, Rice CM. Molecular biology of flaviviruses. Adv Virus Res. 2003;59:23–61. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- 91.Gosert R, Egger D, Lohmann V, et al. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol. 2003;77:5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ait-Goughoulte M, Kanda T, Meyer K, et al. Hepatitis C virus genotype 1a growth and induction of autophagy. J Virol. 2008;82:2241–2249. doi: 10.1128/JVI.02093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dreux M, Gastaminza P, Wieland SF, et al. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106:14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sir D, Chen WL, Choi J, et al. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48:1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ke PY, Chen SS. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest. 2011;121:37–56. doi: 10.1172/JCI41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Guevin C, Manna D, Belanger C, et al. Autophagy protein ATG5 interacts transiently with the hepatitis C virus RNA polymerase (NS5B) early during infection. Virology. 2010;405:1–7. doi: 10.1016/j.virol.2010.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McLauchlan J. Lipid droplets and hepatitis C virus infection. Biochim Biophys Acta. 2009;1791:552–559. doi: 10.1016/j.bbalip.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 98.Heaton NS, Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe. 2010;8:422–432. doi: 10.1016/j.chom.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lonardo A, Adinolfi LE, Loria P, et al. Steatosis and hepatitis C virus: mechanisms and significance for hepatic and extrahepatic disease. Gastroenterology. 2004;126:586–597. doi: 10.1053/j.gastro.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 100.Shrivastava S, Raychoudhuri A, Steele R, et al. Knockdown of autophagy enhances the innate immune response in hepatitis C virus-infected hepatocytes. Hepatology. 2011;53:406–414. doi: 10.1002/hep.24073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sir D, Ou JH. Autophagy in viral replication and pathogenesis. Mol Cells. 2010;29:1–7. doi: 10.1007/s10059-010-0014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sir D, Tian Y, Chen WL, et al. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc Natl Acad Sci U S A. 2010;107:4383–4388. doi: 10.1073/pnas.0911373107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tang H, Da L, Mao Y, et al. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of beclin 1 expression. Hepatology. 2009;49:60–71. doi: 10.1002/hep.22581. [DOI] [PubMed] [Google Scholar]