Abstract
Clinical neurologists and scientists who study multiple sclerosis face open questions regarding the integration of epidemiological data with genome-wide association studies and clinical management of patients. It is becoming evident that the interplay of environmental influences and individual genetic susceptibility modulates disease presentation and therapeutic responsiveness. The molecular mechanisms through which environmental signals are translated into changes in gene expression include DNA methylation, post-translational modification of nucleosomal histones, and non-coding RNAs. These mechanisms are regulated by families of specialised enzymes that are tissue selective and cell-type specific. A model of multiple sclerosis pathogenesis should integrate underlying risk related to genetic susceptibility with cell-type specific epigenetic changes occurring in the immune system and in the brain in response to ageing and environmental stimuli.
Introduction
This Review proposes a view of multiple sclerosis pathogenesis that involves environmental modulation of cellular mechanisms regulating gene expression. This view is based on the integration of epidemiological data1 and genome-wide association studies2 with reports of neuroimaging abnormalities in clinically unaffected individuals.3–5 Epigenetics may explain why a proportion of genetically susceptible individuals remain healthy whereas others manifest the disease,6 and even provide an explanation for patients identified as having radioimmunological syndrome, who are characterised by radiological changes without clinical symptoms.1 Here, we describe the epigenetic mechanisms of disease pathogenesis in molecular terms, and discuss multiple sclerosis as the endpoint of environmental interactions with genetic risk.
The term epigenetics has evolved to define mechanisms underlying phenotype plasticity due to environmental influences, parent-of-origin effects, gene-dosage control, imprinting, and X-chromosome inactivation (panel). At the molecular level, epigenetics comprises modification of DNA base pairs, post-translational modification of histones, and the effects of non-coding RNAs (figure 1). These mechanisms are highly conserved from plants to humans, thereby supporting a crucial function that survived phylogenetic pressure. In plants, for example, seasonal flowering is regulated by histone methylation,8,9 and stomatal development is regulated by DNA methylation.10 In healthy individuals, epigenetics mediates the response to many environmental influences, such as dietary folate intake,11 smoking,12 and ageing.13,14 In patients with multiple sclerosis, modified histones have been detected in non-lesioned white matter.15,16 The role of the epigenome in multiple sclerosis has been inferred from epidemiological studies of the effect of geographic location, month of birth, nutritional status (eg, diet and vitamin D intake), and smoking,1 whereas the importance of imprinting and X-chromosome inactivation is suggested by the maternal parent-of-origin effect17 and the longitudinal increase in female incidence—ie, more women than men develop multiple sclerosis now than in the 1950s.18–20,21 Although evidence of a direct effect of environmental signals on the epigenome in multiple sclerosis is still lacking, in this Review, we present a view of pathogenesis that integrates environmentally induced epigenetic changes with inherent genetic risk, and discuss potential treatment options.
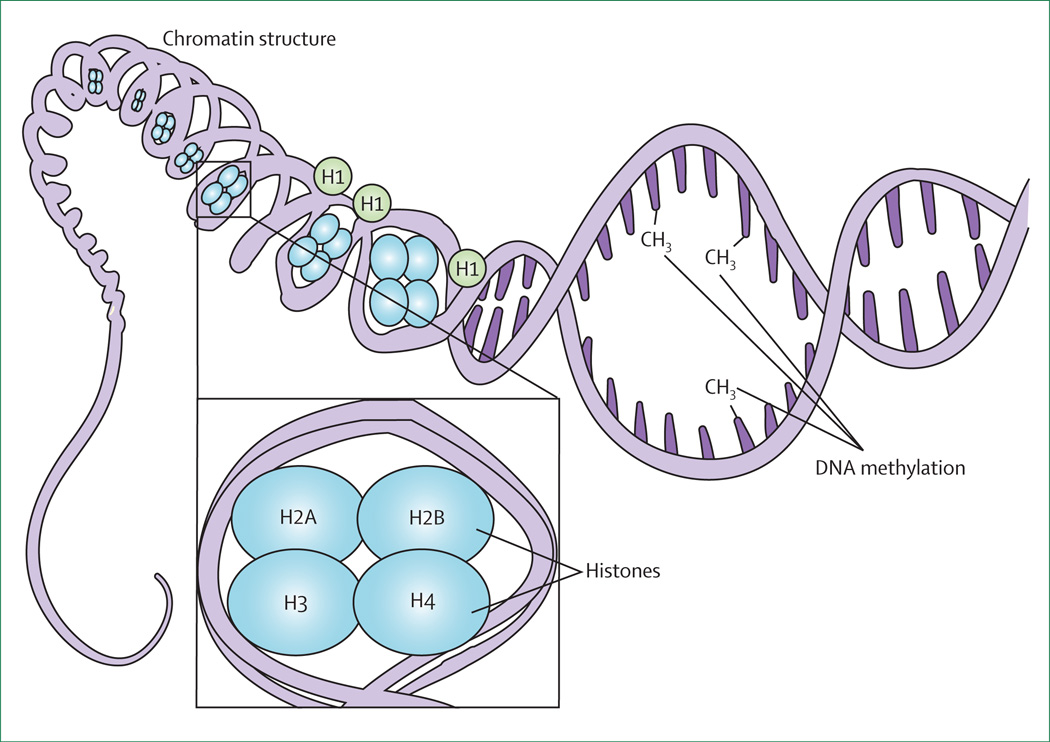
Figure 1. Epigenetic mechanisms regulating transcription.
The human genome is tightly packed into the nucleus via wrapping around histones and chromatin compaction. The on and off state of gene expression is governed by DNA accessibility and epigenetic marks. The balance between these two states is modulated by DNA methylation, post-translational modification, and microRNAs. Theoctameric structure of nucleosomes, composed of dimers of H2A, H2B, H3, and H4 subunits, is shown Histone HI, by contrast, is not part of the nucleosome but serves as a linker.
Epigenetic mechanisms regulating gene expression
A clear example of epigenetic regulation of gene expression is shown by the generation of the different cell types in the developing brain.22,23 Although the cells in all organs share the same DNA sequence, each tissue is characterised by cellular and functional specificity. Activation of cell-specific transcription is associated with the presence of activation marks on lysine and arginine residues of histone tails (figure 2, table 1). Transcriptional repression is achieved by repressive marks on aminoacid residues of histone tails (figure 2, table 2), and is possibly associated with DNA hypermethylation. Fine-tuning of transcript levels is further achieved by the presence of target-specific microRNAs (miRNAs).
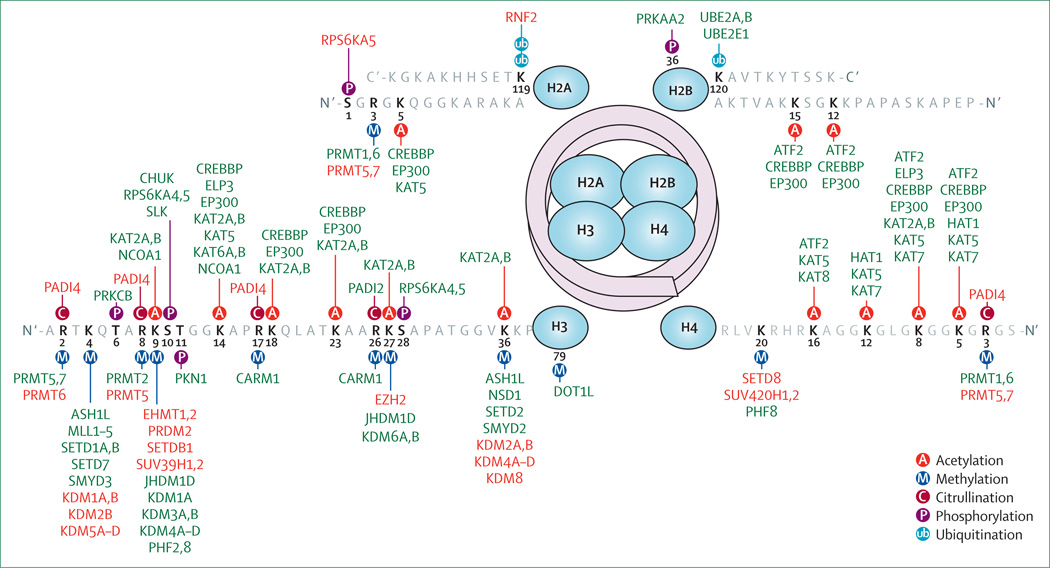
Figure 2. Major post-translational modifications on histonetails.
Post-translational modifications of lysine (K), arginine (R), serine (S), and threonine (T) residues are shown with their respective modifying enzymes. Arginine residues can be methylated symmetrically or asymmetrically; lysine residues can be monomethylated, dimethylated,ortrimethylated. Enzymes shown in green are associated with transcriptional activation (table 1), and enzymes shown in red are associated with transcriptional repression (table 2).
Table 1.
Post-translational histone modifications associated with transcriptional activation
| Histone modification | Residues modified | Enzymes | |
|---|---|---|---|
| Wang et al (2008);24 Kouzarides (2007)25 | Lysine acetylation | H2AK5, H2AK9, H2BK5, H2BK12, H2BK15, H2BK20, H2BK120, H3K4, H3K9, H3K14, H3K18, H3K23, H3K27, H3K36, H3K56, H4K5, H4K8, H4K12, H4K16, H4K91 | ATF2, CREBBP, ELP3, EP300, HAT1, KAT2A, KAT2B, KAT5, KAT6A, KAT6B, KAT7, KAT8, NCOA1 |
| Greer and Shi (2012)26 | Lysine methylation | H3K4, H3K36, H3K79 | ASH1L, DOT1L, MLL, MLL2, MLL3, MLL5, NSD1, SETD1A, SETD1B, SETD2, SETD7, SMYD2, SMYD3 |
| Kooistra and Helin (2012)27 | Lysi ne demethylation | H3K9, K3K27,H4K20 | JHDM1D, KDM1A, KDM3A, KDM3B, KDM4A, KDM4B, KDM4C, KDM4D, KDM6A, KDM6B,PHF2,PHF8 |
| Di Lorenzo and Bedford (2011)28 | Arginine methylation | H2AR3, H3R2, H3R8, H3R17, H3R26, H4R3 | PRMT1, PRMT2, CARM1, PRMT5, PRMT6, PRMT7 |
| Zhang etal(2012)29 | Arginine citrullination | H3R26 | PADI2 |
| Banerjee and Chakravarti (2011)30 | Serine phosphorylation | H2BS36, H3S10, H3S28 | CHUK, PRKAA2, RPS6KA4, RPS6KA5, SLK |
| Banerjee and Chakravarti (2011)30 | Threonine phosphorylation | H3T6, H3T11 | PKN1, PRKCB |
| Liu et al(2012)31 | Lysine sumoylation | Specific residues notyetdetermined | SUMO1 |
| Zhu et al (2005);32 Kim et al (2009)33 | Lysine ubiquitylation | H2BK120 | UBE2A,UBE2B,UBE2E1 |
Table 2.
Post-translational histone modifications associated with transcriptional repression
| Histone modification | Residues modified | Enzymes | |
|---|---|---|---|
| Wang et al (2008);24 Kouzarides (2007)25 | Lysine deacetylation | H2AK5, H2AK9, H2BK5, H2BK12, H2BK15, H2BK20, H2BK120, H3K4, H3K9, H3K14, H3K18, H3K23, H3K27, H3K36, H3K56, H4K5, H4K8, H4K12, H4K16, H4K91 | HDAC1, HDAC2, HDAC3, HDAC4, HDAC5, HDAC7, HDAC8, HDAC9, HDAC11, SIRT1, SIRT2, SIRT6, SIRT7 |
| Greer and Shi (2012)26 | Lysine methylation | H3K9, K3K27, H4K20 | EHMT1, EHMT2, EZH2, PRDM2, SETD8, SETDB1, SUV39H1, SUV39H2, SUV420H1, SUV420H2 |
| Kooistra and Helin (2012)27 | Lysine demethylation | H3K4, H3K36 | KDM1A, KDM1B, KDM2A, KDM2B, KDM4A, KDM4B, KDM4C, KDM4D, KDM5A, KDM5B, KDM5C, KDM5D, KDM8 |
| Di Lorenzo and Bedford (2011)28 | Arginine methylation | H2AR3, H3R2, H3R8, H4R3 | PRMT5, PRMT6, PRMT7 |
| Thompson and Fast (2006)34 | Arginine citrullination | H3R2, H3R8, H3R17, H4R3 | PADI4 |
| Banerjee and Chakravarti (2011)30 | Serine phosphorylation | H2AS1 | RPS6KA5 |
| Shiio and Eisenman (2003)35 | Lysine sumoylation | Specific residues not yet determined | UBE2I |
| Wang et al (2004)36 | Lysine ubiquitylation | H2AK119 | RNF2 |
Another example of epigenetic regulation of gene expression is the differentiation of progenitors into myelin-forming cells. In this case, the changes in transcription that lead to myelination are characterised by the presence of activating epigenetic marks at myelin genes,37 marks of repression at the transcriptional inhibitors of myelin genes,38,39 and fine-tuning by specific miRNAs.40–42
DNA modifications
One of the best-characterised examples of an epigenetic modification, traditionally associated with transcriptional repression, is DNA methylation. This process refers to the addition of methyl groups to cytosines by DNA methyltransferases (DNMTs),43 enzymes that are prevalent in the brain.44,45 Methylated cytosines near the transcriptional start site interfere with sequence recognition by transcription factors, resulting in stable transcriptional repression.46,47 Recent molecular and biological advances, however, have challenged the functional role and the concept of stability of DNA methylation. Genome-wide sequencing has shown that DNA methylation is not only localised in regions functionally related to repression of expression, but is also distributed throughout the genome and possibly contributes to chromatin remodelling,48 gene splicing,45 and allele-specific transcriptional elongation.50 Additionally, the stability of DNA methylation has been questioned by evidence of DNA demethylation during somatic cell reprogramming,51 in fertilised eggs,52,53 and in primordial germ cells,54,55 eliciting a search for potential DNA demethylase enzymes. The first candidates were the mammalian homologues of plant glycosylases (ie, thymine DNA glycosylase and methyl-CpG-binding domain protein 4 [MBD4]), which remove methyl groups from plant DNA.56 However, in mammals these enzymes demethylate 5-hydroxymethyluracil or repair G–T mismatches through the base excision-repair mechanism.57 Demethylation is achieved by the ten-eleven translocation (TET) enzymes, which catalyse hydroxy-methylation, formylation, and carboxylation of cytosine residues,58,59 or by the DNA excision-repair system after 5-methylcytosine or 5-hydroxymethylcytosine is converted into thymine or 5-hydroxymethyluracil, catalysed by the AID/APOBEC family of enzymes.60–62 These studies reveal a more complex role for DNA methylation than originally thought, and warrant caution regarding the interpretation of study results related to DNA methylation in disease.
Histone modifications
DNA does not exist as a double strand that is randomly distributed within the cell nucleus; roughly 2 m of DNA have to fit within a nuclear sphere with an average diameter of 5–10 microns. This packaging is achieved by tightly wrapping the DNA within chromatin (figure 1). The basic unit of chromatin is the nucleosome, composed of 147 base pairs of DNA wrapped around dimers of the histone proteins H2A, H2B, H3, and H4,63 and inter-nucleosomal DNA bound to linker histones.64 The tails of nucleosomal histones are rich in lysine and arginine residues that can be modified in response to extracellular signals, thereby allowing modulation of gene expression simply by modifying the interaction between DNA and the other chromatin components.
At least three types of modification of the nucleosomal organisation have been described: changes in the composition of the nucleosomal octamers (ie, histone variants); rapid sliding movements that occur locally using ATP hydrolysis to expose specific DNA regions (ie, ATP-dependent chromatin remodelling complexes); and post-translational modifications of aminoacids on histone tails (eg, histone acetylation, methylation, citrullination, sumoylation, and ubiquitylation; table 1 and table 2).
Histone variants
The nucleosome is the basic chromatin unit, composed of octamers of histone proteins. Variants have been identified for histone H2A (eg, H2A·Z, macroH2A, H2A·Bbd, H2A·X) and H3 (eg, H3·3, CenH3).65 Although it seems that these variants do not physically disrupt the basic nucleosomal structure, it is increasingly clear they might modify the surrounding chromatin structure.66 H2A·Z is detected in proliferative cells and is mutually exclusive with DNA methylation.67 MacroH2A is enriched on the X chromosome, and has been shown to block tumour progression and act as a barrier to nuclear reprogramming.68,69 Finally, H2A·Bbd, like its murine homologue H2A·Lap, probably has important roles in CNS development or pathology, due to its enrichment in the brain and testes.70,71 Although nucleosomal structural changes are envisioned as permanent ways to regulate developmental states, future studies are needed to elucidate their role in neurology, particularly for variants that are highly enriched in the CNS.
ATP-dependent chromatin remodelling complexes
ATP-dependent chromatin remodelling complexes can be best understood in molecular terms as engines that allow transcription to occur in a localised way. These large complexes bind nucleosomes in an energy-independent manner and then use ATP hydrolysis to allow nucleosome repositioning and sliding. This in turn allows transcription factors and large transcriptional complexes to access the DNA, which is tightly wrapped in chromatin fibres, and favours transcription.72 In human cells, the best-characterised complexes include the DNA-dependent ATPase–helicase subunits BRG1 (also known as SMARCA4) and BRM (also known as SMARCA2), which are homologues to yeast SWI (also known as SNF), and associated proteins called BAFs.73 The functional output of these remodelling complexes might be transcriptional activation or repression, depending on the recruitment of coactivator or corepressor complexes. Clapier and Cairns have published a detailed description of nucleosome remodelling complexes.72
Post-translational modification of nucleosomal histones
Epigenetic regulation of gene expression on aminoacid residues in nucleosomal histones includes the processes of acetylation, methylation, citrullination, sumoylation, and ubiquitination. Transcriptionally competent chromatin has been associated with high levels of lysine acetylation, and trimethylation of lysine residues at aminoacid positions 4 (K4), 36 (K36), and 79 (K79) on histone H3 (figure 2, table 1). Transcriptionally incompetent chromatin has been associated with lysine deacetylation and repressive trimethylation of lysine residues at positions 9 (K9) and 27 (K27) on histone H3, as well as monomethylation of lysine 20 (K20) on histone H4 (figure 2, table 2). The final transcriptional outcome is the result of a careful balance of opposing enzymatic activities that are directly regulated by environmental stimuli, and that modulate several aminoacid residues on histone tails.
Histone acetylation of lysine residues, is catalysed by histone acetyltransferases (HATs), and is associated with transcriptional competence, whereas deacetylation is catalysed by histone deacetylases (HDACs) and is associated with repression.74 The histone acetyltransferase family includes five main groups: KAT2A (formerly known as GCN5), KAT2B (formerly known as PCAF), KAT6–8 (including the MYST family members), CREBBP (formerly known as CBP), and EP300 (formerly known as P300). The histone deacetylase family includes 11 members. Class I (HDAC1, 2, 3, 8) is predominantly nuclear, classes II and IV are nuclear or cytoplasmic, and class III comprises NAD+-dependent enzymes, called sirtuins, that can be nuclear (SIRT1, 2, 6, 7), mitochondrial (SIRT3, 4, 5), or cytoplasmic (SIRT1, 2).75 The sirtuins, particularly SIRT1, have gained attention because they are regulated by caloric restriction and have shown anti-ageing effects related to the regulation of genes involved in antioxidant, anti-inflammatory, and antiapoptotic responses.76 The functional outcome of histone acetylation is remarkably cell specific, and the use of pharmacological compounds that increase acetylation must take into account the effects of acetylation on many cell types. In oligodendrocytes, for example, histone acetylation favours the expression of transcriptional inhibitors of myelin gene expression, thereby interfering with myelin formation.39,77 In neurons, by contrast, histone acetylation is important for memory,78 and increased acetylation protects against age-related cognitive decline.75
Histone methylation is a complex event that occurs on arginine and lysine residues, with varying functional output depending on the number of methyl groups added and the position of specific aminoacids. For lysine residues, specific methyltransferases and demethylases are responsible for monomethylation, dimethylation, trimethylation, or demethylation at particular locations (figure 2). Trimethylation of histone 3 lysine 4 (H3K4me3) is associated with transcriptional activation and is regulated by enzymes in the MLL family of methyltransferases, whereas demethylation is catalysed by LSD1 (also known as KDM1A; figure 2, table 1). Similarly, trimethylation of histone 3 lysine 9 (H3K9me3) or lysine 27 (H3K27me3) has been associated with transcriptional repression and is modulated by enzymes specific for each residue (figure 2, table 2). The presence of both H3K4me3 and H3K27me3 on a particular gene is regarded as the signature for poised expression (ie, genes that are currently not expressed, but are not silenced and therefore could be activated by specific signals), and is characteristically found in stem cells that are uncommitted and pluripotent.80 As cells become committed to a specific lineage, only silenced genes retain the repressive H3K27me3 mark, and only expressed genes retain the H3K4me3 mark.80
Arginine residues can also be monomethylated, asymmetrically dimethylated, and symmetrically dimethylated by protein arginine methyltransferases (PRMTs), with type I (PRMT1, 2, 3, 4, 6, 8) enzymes responsible for asymmetric dimethylation and type II (PRMT5, 7) enzymes responsible for symmetric dimethylation. The transcriptional role of these changes depends on the targeted residue (eg, asymmetrically dimethylated H3R2 is a repressive mark, whereas asymmetrically dimethylated H4R3 is an activating mark).28
By contrast with lysine methylation, there are few accounts of direct demethylation of methylarginine. Instead, a second histone mark, citrullination, directly competes with methylation of arginine residues. Citrullination refers to the deimination of arginine into citrulline, which is mediated by the family of peptidylarginine deiminase enzymes (PADI family; table 1, table 2 and figure 2).34,81 The transcriptional consequences of citrullination depend on the specific arginine residue involved, the transcriptional outcome of the original methylation mark, and the presence of additional histone marks of repression82 or activation.29 Conversion into citrulline seems to be very stable; there is currently no known enzyme that converts it back into arginine.
In addition to acetylation and methylation, lysine residues can be modified by histone sumoylation, a process by which 11-kDa ubiquitin-related modifier peptides (SUMO)83 are added in an ATP-dependent manner, catalysed by specific conjugating enzymes (UBE21). A clear understanding of the functional role of sumoylation is currently lacking, although enzymes with the ability to proteolytically remove this small peptide have been identified in humans (SUMO-specific and sentrin-specific proteases).84 The consequences of this modification have been traditionally associated with repression of transcription,85 although recent reports have also identified sumoylated lysine residues in the histone tails of transcriptionally active genes, characterised by the presence of H3K4me3.31
Histone ubiquitination is a process that requires energy for the conjugation of a 9 kDa ubiquitin peptide to lysine residues. In chromatin, this process has been mainly found on histone H2B and is thought to participate in transcriptional elongation. Ubiquitination might also favour the creation of other activating marks on the tails of histone H3.86
Non-coding RNAs
Recent studies by the Encyclopedia of DNA Elements (ENCODE) consortium to characterise the human genome have revealed that a large portion of the genome encodes the broad family of RNA molecules,87 which include non-coding RNAs. These non-protein-coding RNAs have been implicated in regulatory functions related to brain development88 and neurological disorders.89 They are transcribed from almost every element of the genome, including intergenic regions, centromeric and telomeric repeats, enhancer domains, and even within the splice site of coding genes.88 Non-coding RNAs have a broad capacity to modulate gene expression, which is exerted in a spatiotemporal manner and with the cooperation of chromatin-modifying complexes. In the brain,90 the RNA encoded by HOTAIR recruits histone-modifying complexes containing PRC2 and CoREST,91 which are responsible for coordinating repressive methylation on H3K27 with demethylation on H3K4.92,93 This is an example of the high tissue-specificity of long non-coding RNAs, which are envisioned as important players in human diseases.89
The best-characterised members of the non-coding RNA family are miRNAs. These are encoded by genes, located either as an independent unit or within the coding region of other genes, and are processed by RNA polymerase II into an initial primary miRNA (pri-miRNA).94 This roughly 100-nucleotide pri-miRNA folds into a hairpin structure that is cleaved by the enzyme DROSHA into a 70-nucleotide precursor miRNA (pre-miRNA), which is transported by Exportin-5 into the cytoplasm.95,96 After additional processing by DICER, a 20–24 nucleotide, double-stranded miRNA is formed; one strand binds Argonaute proteins to form an RNA-induced silencing complex,97 and the other strand is typically degraded. This complex regulates gene expression mainly through translational repression or the degradation of mRNA targets with sequences that are complementary with the seed region of the miRNA. With the short length of the seed region on miRNAs, these molecules have the unique capacity to target hundreds of mRNA transcripts that share sequence similarities, and thereby fine-tune the expression of entire gene networks and possibly many cell types.98
Epigenetics in multiple sclerosis
In the past decade, genome-wide associatin studies in large cohorts of patients with multiple sclerosis2 have led to the identification of specific single-nucleotide polymorphisms in genes encoding for at least three MHCs and several non-MHC susceptibility alleles, which include cytokines and their receptors or downstream effectors.99 These loci define the genetic risk of developing multiple sclerosis, and together with epidemiological data,100 suggest a model of disease incidence resulting from the balance between genetic susceptibility and interaction with the environment. The importance of epigenetics in the manifestation of multiple sclerosis is also suggested by studies of the parent-of-origin effect,17 and the increase in female-to-male ratio in diagnosis reported in longitudinal studies.18–20 These studies underscore the importance of epigenetic modifications on the X chromosome in modulating clinical manifestations of the disease. Because no susceptibility genes have been identified on the X chromosome, alternative mechanisms are probably accountable, including X-chromosome inactivation and imprinting.
The role of genes and environment in pathogenesis
A large series of studies on multiple sclerosis incidence and geographic location, month of birth, and dietary intake in adolescence supports the role of environment and gene regulation in multiple sclerosis.101 Past studies have shown that multiple sclerosis is more prevalent in some geographic locations than in others,102–104 and that migrant children younger than 15 years can acquire the incidence of the geographic location where they migrate to.105,106 These data led to the hypothesis that geographic distribution of infectious agents, dietary habits, and sun exposure could contribute to disease manifestation.103,107–109 Detailed epidemiological studies in France supported the importance of sun exposure,110 since the prevalence of disease was significantly higher in areas with lower UV irradiation. A potential explanation for the association between incidence of multiple sclerosis and geographic location is the effect of sun exposure on synthesis of vitamin D.103 Low vitamin D levels are associated with high risk of developing multiple sclerosis,111 and vitamin D supplementation reduces the risk.112 Epidemiological evidence on the effect of month of birth further supports the concept that maternal deficiency of vitamin D could increase disease risk, since greater incidence of the disease has been reported for May births compared with November births in the Northern hemisphere.113,114 The precise mechanism of action for vitamin D in modulating disease onset and course remains to be determined. However, recent studies using chromatin immunoprecipitation followed by deep-sequencing analysis showed that the vitamin D receptor has an important role in modulating the transcription of several genes involved in autommunity.115 In addition to vitamin D levels, maternal dietary intake affects the levels of folate and DNA methylation, with direct consequences on fetal CNS development11,116 and overall metabolism.117,118 Although the association between diet, metabolism, and multiple sclerosis has not been adequately addressed, several studies have reported an association between body-mass index in adolescence and incidence of multiple sclerosis.119 These studies, together with previous reports of higher mortality rates among patients with multiple sclerosis on a diet high in animal protein and fat, compared with those on a low-fat diet,107,120 suggest that food intake might have a role in disease manifestation. New studies have also proposed an important role for the microbiome in animal models of multiple sclerosis.121–123 The proposed model suggests that differences in the composition of the microbiota in individuals might affect the immune system by modulating the equilibrium between subclasses of lymphocytes.124 Future studies will be needed to carefully address whether the microbiome might modulate the risk of developing multiple sclerosis, as well as the disease course, through epigenetic changes.
Epigenetic changes in blood
DNA methylation
Baranzini and colleagues125 tested the hypothesis that discordance of multiple sclerosis in monozygotic twins can be attributed to methylation changes in susceptibility genes. Reduced representation bisulfite sequencing of DNA isolated from CD4+ lymphocytes of three monozygotic twin pairs was compared with results from their transcriptome to define potential transcriptional correlates.125 Of 2 million CpG sites assayed, only two (in the genes TMEM1 and PEX14) showed a complete shift from hypomethylation to hypermethylation in one pair of siblings, and from hypermethylation to hypomethylation in the other. Thus, changes in DNA methylation in the lymphocytes of twins with multiple sclerosis do not seem to be as substantial as those induced by age13 or cancer.126 The potential contribution of smaller changes in DNA methylation could not be assessed because of the small sample size of the study and the differences in sex and ethnicity among the three twin pairs.125 Other researchers have investigated DNA methylation as a potentially useful biomarker of disease activity, by analysing the methylation of 56 genes previously shown to be differentially methylated in patients with cancer from DNA in cell-free plasma obtained from healthy individuals and from patients with multiple sclerosis. 15 of 56 genes showed significant differences in methylation patterns between control individuals and patients with multiple sclerosis. For five of these 15 genes, the methylation status of the promoter was able to discriminate between patients in remission and those in exacerbation.127 Together, these results highlight the need for additional studies of DNA methylation in multiple sclerosis.
miRNAs
miRNAs are essential for normal cellular function and development, and their dysregulation can disrupt homoeostasis and even contribute to autoimmunity.128 Most of the studies to identify miRNAs with a potential role in the pathogenesis of multiple sclerosis have been done in blood-derived cells (table 3). We did a global evaluation of the results and found a lack of consensus regarding miRNAs that are common to all the studies. There was little overlap among studies done in the same tissue, and in some cases, even opposing results. A potential explanation for the heterogeneity of results is the use of different platforms between studies and the complexity of the tissues being investigated. However, the results of several studies agree that miRNA dysregulation favours a proinflammatory state and promotes disease progression.129,131,135,136 Additionally, specific miRNAs (so-called NeurimmiRs) modulate neuronal and immune processes and are possibly responsible for the crosstalk between these two systems.143 These include miR-155 and miR-326, which are dysregulated in peripheral blood mononuclear cells135 and CD4+ T cells,129 respectively, in patients with multiple sclerosis. Both miR-155 and miR-326 have an important role in the immune system by regulating T-cell development, and have been shown to decrease the severity of experimental autoimmune encephalomyelitis when they are silenced.129,144 Thus, upregulation of miR-155 and miR-326 in multiple sclerosis might modulate distinct targets in the brain and blood, possibly coordinating gene expression profiles in the two compartments to drive disease progression. In addition to understanding how miRNAs contribute to the pathogenesis and progression of multiple sclerosis, studies have investigated miRNA profiles as a potential diagnostic and prognostic biomarker for multiple sclerosis. For example, a study of whole-blood samples from patients with relapsing-remitting multiple sclerosis found that 165 miRNAs were differentially expressed compared with healthy controls.139 miR-145 was identified statistically as the best candidate to discriminate individuals with multiple sclerosis from controls, with a high level of specificity, sensitivity, and accuracy.139 Other studies that have focused on differences between patients with relapsing-remitting multiple sclerosis and controls have noted upregulation of miR-18b, miR-493, and miR-599 during the relapse phase.138 One study introduced the concept that current therapies for multiple sclerosis might modify miRNA levels. Sievers and colleagues142 identified 49 miRNAs that were downregulated in B cells of untreated patients with relapsing-remittig multiple sclerosis compared with healthy controls and 10 miRNAs that were upregulated in B cells from natalizumab-treated patients compared with untreated individuals with relapsing-remitting multiple sclerosis. Although these studies hint at the possibility of using miRNA profiles to identify individuals with multiple sclerosis or to direct a treatment course, they provide only a glimpse of the disease process, and longitudinal studies of large populations must be undertaken to identify the stability of these changes and their validity as biomarkers.
Table 3.
Dysregulated miRNAs in multiple sclerosis
| Sample | Tissue | Dysregulated miRNAs | |
|---|---|---|---|
| Disease pathogenesis | |||
| Du et al (2009)129 | 43 RR MS, 42 healthy individuals | PBL,CD4+T cells | miR-326 |
| Junker et al (2009)130 | 20RRMS, SPMS, PPMS,or Marburg variant; nine healthy individuals | Multiple sclerosis lesions (active and inactive) | Active lesions: miR-650, miR-155, miR-326, miR-142-3p, miR-146a, miR-146b, miR-34a, miR-21, miR-23a, miR-199a, miR-27a, miR-142-5p, miR-193a, miR-15a, miR-200c, miR-130a, miR-223, miR-22, miR-320, miR-214, miR-656, miR-184, miR-139, miR-23b, miR-328, miR-487b, miR-l8lc, miR-340 |
| Lindberg et al (2010)131 | Eight RR MS, ten healthy individuals | CD4+T cells, CD8+T cells, B cells | CD4+T cells: miR-485-3p, miR-376a, miR-1, miR-497, miR-193a, miR-200b, miR-126, miR-486, miR-17-5p, miR-34a |
| De Santis et al (2010)132 | 12 RR MS, 14 healthy individuals | CD4+CD25+ T cells | miR-29c, miR-107, miR-210, let-7i, miR-15a, miR-19a, miR-19b, miR-138-2*, miR-324-3p, miR-301a, miR-338-5p, miR-22, miR-512-3p, miR-564, miR-886-3p, miR-106b, miR-29a, miR-93, miR-489, miR-148a, miR-590-5p, miR-223, miR-221 |
| Cox et al (2010)133 | 59 RR MS, SPMS, or PP MS; 37 healthy individuals | Whole blood | miR-768-3p, HS_265.1, let-7d, let-7f, let-7g, let-7i, miR-106a, miR-126, miR-126*, miR-140-5p, miR-15a, miR-15b, miR-16, miR-17, miR-20a, miR-20b, miR-211, miR-27a, miR-27b, miR-374a, miR-454, miR-510, miR-579, miR-623, miR-624*, miR-93, miR-98 |
| Fenoglio etal (2011)134 | 29 RR MS, SPMS, or PP MS; 19 healthy individuals | PBMCs | miR-21, miR-146a, miR-146b |
| Paraboschi et al (2011)135 | Ten RR MS, six healthy individuals | PBMCs | miR-155, miR-92a, let-7f, miR-19a |
| Guerau-de-Arellano et al (2011)136 | 22 RR MS, SPMS, or PP MS; 16 healthy individuals | Naive CD4+ T cells | miR-660, miR-5l8d-3p, miR-586, miR-128, miR-564, miR-708, miR-378, miR-346, miR-645, miR-566 |
| Martinelli-Boneschi et al (2012)137 | 19 RR MS, SPMS, or PP MS; 14 healthy individuals | PBMCs | miR-363, miR-31*, miR-524-3p, miR-876-3p, let-7g, miR-223*, miR-550*, miR-l8lc, miR-374a*, miR-150 |
|
Disease course | |||
| Otaegui et al (2009)138 | 13 RR MS, eight healthy individuals | PBMCs | miR-l8b,miR-493, miR-599 |
| Keller etal(2009)139 | 20 RR MS, 19 healthy individuals | Whole blood | miR-145, miR-186, miR-664, miR-20b, miR-422a, miR-142-3p, miR-584, miR-223, miR-1275, miR-491-5p |
| Siegel et al (2012)140 | Four multiple sclerosis, four healthy individuals | Plasma | miR-614, miR-572, miR-1979, miR-648, miR-422a, miR-1826, miR-22 |
| Haghikia et al (2012)141 | 53 RR MS, SPMS, or PP MS, 39 patients with other neurologica (non-multiple sclerosis) disease | CSF | miR-922, miR-l8lc, miR-633 |
|
Treatment (natalizumab) | |||
| Sieversetal(2012)142 | 20 RR MS, ten healthy individuals | B cells | Untreated multiple sclerosis: miR-515, miR-411*, miR-25, miR-16, miR-297a, miR-329, miR-299-5p, miR-520g, miR-486-5p, miR-363 |
For each study, only a subset of miRNAs are listed. Refer to the reference for the full list. In miRNA nomenclature, an asterisk refers to microRNAs corresponding to complementary strands of DNA. As such, the two microRNAs have different sequences and probably have distinct targets, despite similar genomic coordinates. miRNAs=microRNAs. RR MS=relapsing-remitting multiple sclerosis. PBL=peripheral blood leucocytes. SP MS=secondary-progressive multiple sclerosis. PP MS=primary-progressive multiple sclerosis. PBMCs=peripheral blood mononuclear cells.
Epigenetic changes in brain tissue
The role of miRNAs in the brain of patients with multiple sclerosis has also been investigated. Junker and colleagues130 identified 28 miRNAs in active lesions and 35 miRNAs in inactive lesions that were dysregulated compared with those in white matter specimens from healthy controls (table 3). miR-34a, miR-155, and miR-326 were upregulated in active multiple sclerosis lesions. Although the regulatory profile of these miRNAs in the brain is still unknown, a putative target is CD47, a molecule that inhibits macrophage activity directed at resident brain cells. CD47 transcripts are decreased in active lesions.130
Post-translational modifications of nucleosomal histones
When considering epigenetic changes in the brain in patients with multiple sclerosis, it is important to reiterate the cell specificity of these modifications and the fact that the same modification (eg, post-translational modification of histone H3) could have distinct roles in different cell types. An example is histone acetylation. Studies have shown that high levels of histone acetylation in hippocampal neurons are associated with increased transcriptional activity during learning,78 whereas decreased histone acetylation is associated with cognitive decline.79 In the oligodendrocyte lineage, by contrast, high levels of histone acetylation were characteristic of undifferentiated progenitor cells77,145 and were associated with high levels of transcriptional repressors of myelin gene expression.38,39 Myelination during development required decreased expression of transcriptional inhibitors of myelin genes,77 and mice deficient in Hdacl and Hdac2 completely lacked myelin in the CNS and peripheral nervous system.146,147 Deacetylation is also important for repair after demyelination and its efficiency decreases with age. Deacyetylation is a naturally occurring event that is associated with defective repair of myelin.39 Higher levels of histone acetylation and transcriptional inhibitors were detected in the brains of older compared with younger mice,39 and in normal-appearing white matter of patients with multiple sclerosis compared with patients without multiple sclerosis.16 Histone citrullination was increased in animal models of demyelination and in patients with multiple sclerosis compared with patients without multiple sclerosis,15 and was associated with increased concentrations of PADI4, although the precise function of this protein remains to be investigated. Aberrant citrullination has been detected in myelin proteins (ie, MBP),148 and is proposed to contribute to myelin sheath instability and increased proteolysis with release of immunogenic peptides,149,150 eventually leading to oligodendrocyte apoptosis.151
Histone deacetylase inhibitors
The concept of cell specificity is important in evaluating epigenetic modulators as therapeutic strategies for demyelinating disorders. The use of broad spectrum histone deacetylase inhibitors, for example, was originally proposed as a treatment option for multiple sclerosis because of the reduction of inflammatory infiltrates in animal models.152 Use of histone deacetylase inhibitors has also been advocated to counteract the cognitive decline associated with Alzheimer’s disease153,154 or traumatic brain injury,155,156 and to protect from axonal damage in conditions characterised by impaired axonal transport.157 However, systemic use of histone deacetylase inhibitors also negatively affects the generation of new myelin. Histone deacetylation is important for developmental myelination77,145 and myelin repair in adults.39 The use of histone deacetylase inhibitors has a detrimental effect on these processes by preventing myelination of white matter tracts when given during development,77 and by decreasing the efficiency of endogenous myelin repair if administered to adult mice after demyelination.35
Together, these data caution against the use of broad inhibitors of histone deacetylases in demyelinating disorders, because the potential deleterious effects on myelin might counteract the beneficial effects on neurons, and suggest the need to develop more targeted approaches.
Conclusions
We have discussed molecular mechanisms of the regulation of gene expression that occur independently of changes in the DNA sequence. These epigenetic mechanisms include post-translational modifications of nucleosomal histones, DNA methylation, and regulation by non-coding miRNAs, and can be affected by the environment and lifestyle (eg, diet, smoking habits, exercise, and drug addiction). We propose that environmental factors might regulate disease manifestation by modulating the epigenome, and that multiple sclerosis might result from cumulative changes imposed by environmental factors on the immune system and in the brain (figure 3). Distinct cell populations might differentially respond to the same stimulus, by changing the organisation of the nuclear structure in a cell-specific manner. In immune cells, epigenetic regulation might result in increased expression of genes associated with disease manifestation, whereas in the brain, the same stimuli might impair the ability of progenitors to form new myelin. For example, social interaction was recently shown to profoundly affect myelin content in the prefrontal cortex of adult mice, and to modulate post-translational modifications of nucleosomal histones.158 The combination of epigenetic changes in immune cells and oligodendrocytes might modulate disease onset (by modulating expression of susceptibility genes) and disease course (by modulating myelin formation). However, caution is warranted regarding use of epigenetic marks as disease biomarkers for neurological disorders, since epigenetic changes identified in a specific tissue population (eg, peripheral blood monocytes) cannot be easily generalised to a different organ (eg, brain) or cell type (eg, CD4+ lymphocytes). Nevertheless, within a single tissue, epigenetic mechanisms need to be studied longitudinally to determine the effect of environmental stimuli on disease course or therapeutic responsiveness. The presence of repressive marks in regions containing sequences specific for vitamin D receptor binding, for example, might explain the lack of patient responsiveness to vitamin D supplementation. It can be envisioned that maps of responsive genes will be created and used for screening patients in the future. This will be possible only if precise signatures of epigenetic maps are accurately defined for each cell type. The integration of the wealth of information from genome-wide association studies with the longitudinal analysis of epigenetic marks on selected patient populations will possibly lead to effective personalised therapeutic approaches.
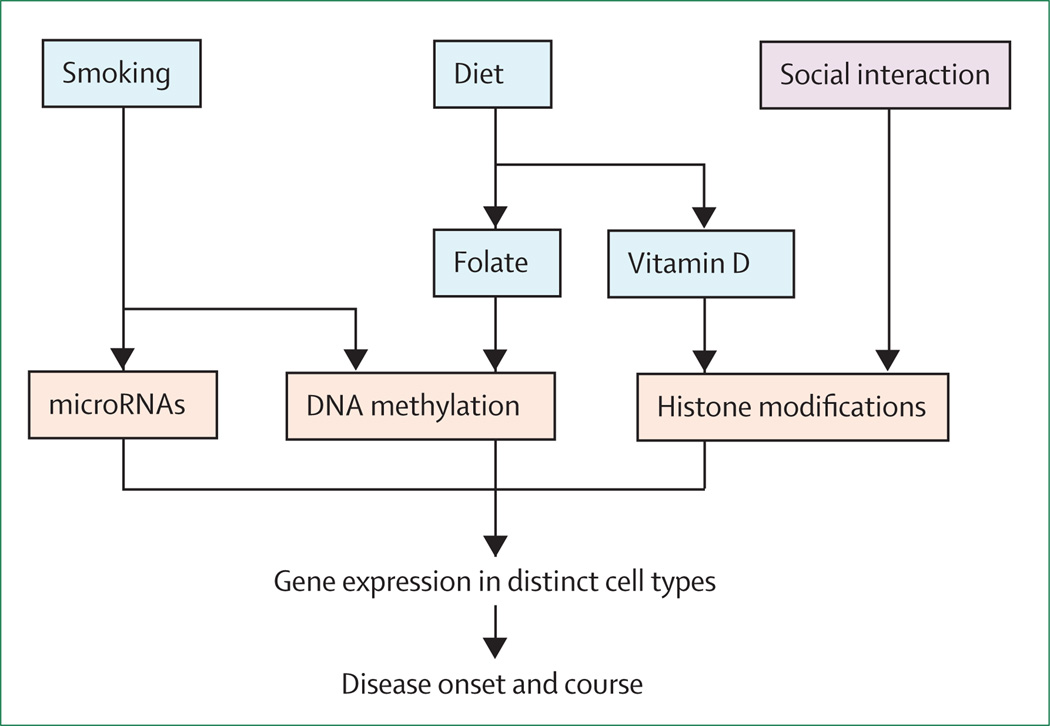
Figure 3. Environmental stimuli influence gene expression through epigenetic mechanisms.
Putative environmental insults associated with multiple sclerosis alter the epigenetic landscape, which can ultimately affect gene expression in a cell-specific manner. These changes can affect disease onset or progression, depending on the timing of the accumulating stimuli. Environmental signals that affect human disease are shown in blue, evidence of an environmental effect from animal studies in purple, and the molecular mechanisms responsible for outcomes are in red.
Panel: Glossary of common terms used in epigenetics.
Parent-of-origin effects
Although the contribution of autosomal chromosomes from each parent to their offspring is equivalent, some diseases are inherited only throughthe maternal or paternal lineage. For example, mitochondria are always inherited from the mother; therefore, mitochondrial disorders always show maternal, but never paternal, inheritance. Many other parent-of-origin effects are caused by imprinted genes, a special class of genes that function differentially depending on their parental origin. Diseases caused by imprinted genes (see below) will therefore have distinct disease manifestations depending on whether they are inherited from the motherorfrom the father.
Imprinting
An epigenetic mechanism by which the activity of a gene depends on the parent of origin (see above). It is estimated that 1–2% of genes in humans are imprinted, and identification of these genes is an active field of investigation. Well-known diseases associated with imprinting are characterised by mental retardation associated with delayed sexual development and obesity (Prader-Willi syndrome) or with ataxia and seizures (Angel man syndrome).
Gene-dosage control
For some genes, producing the correct amount of protein product is very important to maintain a healthy state—having too muchortoo little protein can result in disease, so regulation of transcript levels istightly controlled. In some cases, both alleles are needed to guarantee biological function (eg, the presence of two functioning alleles encoding for tumour-suppressor genes, such as P27KIP1 or TPS3, is an important mechanism against aberrant proliferation). In other cases, only one allele needs to be active and the activity of both alleles results in pathology (eg, the amount of proteolipid protein, a myelin constituent expressed by the PLP gene on the X chromosome, is tightly regulated by mechanisms responsible for chromosome inactivation. When these mechanisms are defective, the contribution of both alleles results in a disease called Pelizaeus-Merzbacher, characterised by dysmyelination).
X-chromosome inactivation
A mechanism of dosage compensation. Whereas females carry two copies of the X chromosome, which is large and contains roughly 1000 genes, males carry a single X and a Y chromosome, which is small and gene poor. X-chromosome inactivation effectively silences one of the two X chromosomes in each cell in females, switching off most genes on the chromosome and thereby restoring the balance of gene dosage between the sexes.
Search strategy and selection criteria.
References for this Reviewwere identified through searchesof PubMed with the terms “histone” AND “acetylation”, “methylation”, “citrullination”, “phosphorylation”, “sumoylation”, “ubiquitination”, “DNA methylation”, “TET”, “non-coding RNA”, “microRNA”, “environment” AND “epigenetics”, and “multiple sclerosis” from January, 1960, until October, 2012. Articles were also identified through searchesof the authors′own files. Only papers published in English were reviewed.The final reference I ist was generated on the basis of originality and relevance to the broad scope of this Review.
Acknowledgments
This report was funded by NIH-NINDS grants (2R37NS042925-10, R01NS52738, and R01NS69835) to PC, and by a fellowship from NIH-NINDS (1F31NS077504-01) and a grant from the Foundation of the Consortium of Multiple Sclerosis Centers’ MS Workforce of the Future programme to JLH. The content is solely the responsibility of the authors and does not necessarily represent official views of the National Institutes of Health. We thank Andrew Sharp for reviewing and editing the definitions in the panel.
Footnotes
Contributors
Both authors did the literature search. PC had the idea for the Review, provided the figures, and wrote the manuscript. JLH provided the tables, revised the figures, edited the manuscript, and wrote the section on microRNAs.
Conflicts of interest
We declare that we have no conflicts of interest.
Contributor Information
Jimmy L Huynh, Department of Neuroscience, Mount Sinai School of Medicine, New York, NY, USA; Department of Genetics and Genomic Sciences, Mount Sinai School of Medicine, New York, NY, USA.
Patrizia Casaccia, Department of Neuroscience, Mount Sinai School of Medicine, New York, NY, USA; Department of Genetics and Genomic Sciences, Mount Sinai School of Medicine, New York, NY, USA; Department of Neurology, Mount Sinai School of Medicine, New York, NY, USA.
References
- 1.Ramagopalan SV, Dobson R, Meier UC, Giovannoni G. Multiple sclerosis: risk factors, prodromes, and potential causal pathways. Lancet Neurol. 2010;9:727–739. doi: 10.1016/S1474-4422(10)70094-6. [DOI] [PubMed] [Google Scholar]
- 2.Gourraud PA, Harbo HF, Hauser SL, Baranzini SE. The genetics of multiple sclerosis: an up-to-date review. Immunol Rev. 2012;248:87–103. doi: 10.1111/j.1600-065X.2012.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siger-Zajdel M, Filippi M, Selmaj K. MTR discloses subtle changes in the normal-appearing tissue from relatives of patients with MS. Neurology. 2002;58:317–320. doi: 10.1212/wnl.58.2.317. [DOI] [PubMed] [Google Scholar]
- 4.De Stefano N, Cocco E, Lai M, et al. Imaging brain damage in first-degree relatives of sporadic and familial multiple sclerosis. Ann Neurol. 2006;59:634–639. doi: 10.1002/ana.20767. [DOI] [PubMed] [Google Scholar]
- 5.Okuda DT, Mowry EM, Beheshtian A, et al. Incidental MRI anomalies suggestive of multiple sclerosis: the radiologically isolated syndrome. Neurology. 2009;72:800–805. doi: 10.1212/01.wnl.0000335764.14513.1a. [DOI] [PubMed] [Google Scholar]
- 6.Poser CM. Multiple sclerosis trait: the premorbid stage of multiple sclerosis A hypothesis. Acta Neurol Scand. 2004;109:239–843. doi: 10.1111/j.1600-0404.2004.00277.x. [DOI] [PubMed] [Google Scholar]
- 7.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 8.Sung S, Amasino RM. Vernalization and epigenetics: how plants remember winter. Curr Opin Plant Biol. 2004;7:4–10. doi: 10.1016/j.pbi.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 9.Kim DH, Sung S. Environmentally coordinated epigenetic silencing of FLC by protein and long noncoding RNA components. Curr Opin Plant Biol. 2012;15:51–56. doi: 10.1016/j.pbi.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Tricker PJ, Gibbings JG, Rodriguez Lopez CM, Hadley P, Wilkinson MJ. Low relative humidity triggers RNA-directed de novo DNA methylation and suppression of genes controlling stomatal development. J Exp Bot. 2012;63:3799–3813. doi: 10.1093/jxb/ers076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang H, Zhang T, Zhang Z, et al. Tissue-specific distribution of aberrant DNA methylation associated with maternal low-folate status in human neural tube defects. J Nutr Biochem. 2011;22:1172–1177. doi: 10.1016/j.jnutbio.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am J Hum Genet. 2011;88:450–457. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraga MF, Ballestar E, Paz MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heyn H, Li N, Ferreira HJ, et al. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci USA. 2012;109:10522–10527. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mastronardi FG, Wood DD, Mei J, et al. Increased citrullination of histone H3 in multiple sclerosis brain and animal models of demyelination: a role for tumor necrosis factor-induced peptidylarginine deiminase 4 translocation. J Neurosci. 2006;26:11387–11396. doi: 10.1523/JNEUROSCI.3349-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pedre X, Mastronardi F, Bruck W, Lopez-Rodas G, Kuhlmann T, Casaccia P. Changed histone acetylation patterns in normal-appearing white matter and early multiple sclerosis lesions. J Neurosci. 2011;31:3435–3435. doi: 10.1523/JNEUROSCI.4507-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ebers GC, Sadovnick AD, Dyment DA, Yee IM, Wilier CJ, Risch N. Parent-of-origin effect in multiple sclerosis: observations in half-siblings. Lancet. 2004;363:1773–1774. doi: 10.1016/S0140-6736(04)16304-6. [DOI] [PubMed] [Google Scholar]
- 18.Wallin MT, Page WF, Kurtzke JF. Multiple sclerosis in US veterans of the Vietnam era and later military service: race, sex, and geography. Ann Neurol. 2004;55:65–71. doi: 10.1002/ana.10788. [DOI] [PubMed] [Google Scholar]
- 19.Orton SM, Herrera BM, Yee IM, et al. Sex ratio of multiple sclerosis in Canada: a longitudinal study. Lancet Neurol. 2006;5:932–936. doi: 10.1016/S1474-4422(06)70581-6. [DOI] [PubMed] [Google Scholar]
- 20.Hirst C, Ingram G, Pickersgill T, Swingler R, Compston DA, Robertson NP. Increasing prevalence and incidence of multiple sclerosis in South East Wales. J Neurol Neurosurg Psychiatry. 2009;80:386–391. doi: 10.1136/jnnp.2008.144667. [DOI] [PubMed] [Google Scholar]
- 21.Kaliszewska A, De Jager PL. Exploring the role of the epigenome in multiple sclerosis: a window onto cell-specific transcriptional potential. J Neuroimmunol. 2012;248:2–9. doi: 10.1016/j.jneuroim.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 22.Hirabayashi Y, Gotoh Y. Epigenetic control of neural precursor cell fate during development. Nat Rev Neurosci. 2010;11:377–388. doi: 10.1038/nrn2810. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Casaccia P. Epigenetic regulation of oligodendrocyte identity. Trends Neurosci. 2010;33:193–201. doi: 10.1016/j.tins.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Zang C, Rosenfeld JA, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 28.Di Lorenzo A, Bedford MT. Histone arginine methylation. FEBS Lett. 2011;585:2024–2031. doi: 10.1016/j.febslet.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Bolt M, Guertin MJ, et al. Peptidylarginine deiminase 2-catalyzed histone H3 arginine 26 citrullination facilitates estrogen receptor alpha target gene activation. Proc Natl Acad Sci USA. 2012;109:13331–13336. doi: 10.1073/pnas.1203280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banerjee T, Chakravarti D. A peek into the complex realm of histone phosphorylation. Mol Cell Biol. 2011;31:4858–4873. doi: 10.1128/MCB.05631-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu HW, Zhang J, Heine GF, et al. Chromatin modification by SUMO-1 stimulates the promoters of translation machinery genes. Nucleic Acids Res. 2012;40:10172–10186. doi: 10.1093/nar/gks819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu B, Zheng Y, Pham AD, et al. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 33.Kim J, Guermah M, McGinty RK, et al. RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson PR, Fast W. Histone citrullination by protein arginine deiminase: is arginine methylation a green light or a roadblock? ACS Chem Biol. 2006;1:433–441. doi: 10.1021/cb6002306. [DOI] [PubMed] [Google Scholar]
- 35.Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci USA. 2003;100:13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Wang L, Erdjument-Bromage H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 37.Liu J, Sandoval J, Doh ST, Cai L, Lopez-Rodas G, Casaccia P. Epigenetic modifiers are necessary but not sufficient for reprogramming non-myelinating cells into myelin gene-expressing cells. PLoS One. 2010;5:13023. doi: 10.1371/journal.pone.0013023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He Y, Dupree J, Wang J, et al. The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron. 2007;55:217–230. doi: 10.1016/j.neuron.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen S, Sandoval J, Swiss VA, et al. Age-dependent epigenetic control of differentiation inhibitors is critical for remyelination efficiency. Nat Neurosci. 2008;11:1024–1034. doi: 10.1038/nn.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lau P, Verrier JD, Nielsen JA, Johnson KR, Notterpek L, Hudson LD. Identification of dynamically regulated micro RNA and mRNA networks in developing oligodendrocytes. J Neurosci. 2008;28:11720–11730. doi: 10.1523/JNEUROSCI.1932-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dugas JC, Cuellar TL, Scholze A, et al. Dicerl and miR-219 are required for normal oligodendrocyte differentiation and myelination. Neuron. 2010;65:597–611. doi: 10.1016/j.neuron.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao X, He X, Han X, et al. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron. 2010;65:612–626. doi: 10.1016/j.neuron.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 44.Goto K, Numata M, Komura JI, Ono T, Bestor TH, Kondo H. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation. 1994;56:39–44. doi: 10.1046/j.1432-0436.1994.56120039.x. [DOI] [PubMed] [Google Scholar]
- 45.Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- 46.Watt F, Molloy PL. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988;2:1136–1143. doi: 10.1101/gad.2.9.1136. [DOI] [PubMed] [Google Scholar]
- 47.Takizawa T, Nakashima K, Namihira M, et al. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell. 2001;1:749–758. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- 48.Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol. 2004;11:1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- 49.Shukla S, Kavak E, Gregory M, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas BJ, Rubio ED, Krumm N, et al. Allele-specific transcriptional elongation regulates monoallelic expression of the IGF2BP1 gene. Epigenetics Chromatin. 2011;4:14. doi: 10.1186/1756-8935-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simonsson S, Gurdon J. DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nat Cell Biol. 2004;6:984–990. doi: 10.1038/ncb1176. [DOI] [PubMed] [Google Scholar]
- 52.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 53.Oswald J, Engemann S, Lane N, et al. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 54.Hajkova P, Erhardt S, Lane N, et al. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 55.Lee J, Inoue K, Ono R, et al. Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development. 2002;129:1807–1817. doi: 10.1242/dev.129.8.1807. [DOI] [PubMed] [Google Scholar]
- 56.Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 57.Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2011;13:7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 59.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 61.Popp C, Dean W, Feng S, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saitou M, Kagiwada S, Kurimoto K. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development. 2012;139:15–31. doi: 10.1242/dev.050849. [DOI] [PubMed] [Google Scholar]
- 63.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 64.Tremethick DJ. Higher-order structures of chromatin: the elusive 30 nm fiber. Cell. 2007;128:651–654. doi: 10.1016/j.cell.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 65.Henikoff S, Furuyama T, Ahmad K. Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet. 2004;20:320–406. doi: 10.1016/j.tig.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 66.Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol. 2012;13:436–447. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kapoor A, Goldberg MS, Cumberland LK, et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature. 2010;468:1105–1109. doi: 10.1038/nature09590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pasque V, Gillich A, Garrett N, Gurdon JB. Histone variant macroH2A confers resistance to nuclear reprogramming. EMBOJ. 2011;30:2373–2387. doi: 10.1038/emboj.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ishibashi T, Li A, Eirin-Lopez JM, et al. H2A.Bbd: an X-chro mo some-encoded histone involved in mammalian spermiogenesis. Nucleic Acids Res. 2010;38:1780–1789. doi: 10.1093/nar/gkp1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Soboleva TA, Nekrasov M, Pahwa A, Williams R, Huttley GA, Tremethick DJ. A unique H2A histone variant occupies the transcriptional start site of active genes. Nat Struct Mol Biol. 2011;19:25–30. doi: 10.1038/nsmb.2161. [DOI] [PubMed] [Google Scholar]
- 72.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 73.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hassig CA, Schreiber SL. Nuclear histone acetylases and deacetylases and transcriptional regulation: HATs off to HDACs. Curr Opin Chem Biol. 1997;1:300–308. doi: 10.1016/s1367-5931(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 75.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bonda DJ, Lee HG, Camins A, et al. The sirtuin pathway in ageing and Alzheimer disease: mechanistic and therapeutic considerations. Lancet Neurol. 2011;10:275–279. doi: 10.1016/S1474-4422(11)70013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shen S, Li J, Casaccia-Bonnefil P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J Cell Biol. 2005;169:577–589. doi: 10.1083/jcb.200412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Levenson JM, O’Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- 79.Peleg S, Sananbenesi F, Zovoilis A, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 80.Mikkelsen TS, Ku M, Jaffe DB, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang Y, Wysocka J, Sayegh J, et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 82.Denis H, Deplus R, Putmans P, Yamada M, Metivier R, Fuks F. Functional connection between deimination and deacetylation of histones. Mol Cell Biol. 2009;29:4982–4993. doi: 10.1128/MCB.00285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Melchior F. SUMO—nonclassical ubiquitin. Annu Rev Cell Dev Biol. 2000;16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- 84.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 85.Nathan D, Sterner DE, Berger SL. Histone modifications: now summoning sumoylation. Proc Natl Acad Sci USA. 2003;100:13118–13120. doi: 10.1073/pnas.2436173100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 87.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci. 2012;13:528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 90.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rossbach M. Non-coding RNAs in neural networks, REST-assured. Front Genet. 2011;2:8. doi: 10.3389/fgene.2011.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee Y, Kim M, Han J, et al. Micro RNA genes are transcribed by RNA polymerase II. EMBOJ. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates micro RNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 96.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 98.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 99.Baranzini SE. Revealing the genetic basis of multiple sclerosis: are we there yet? Curr Opin Genet Dev. 2011;21:317–324. doi: 10.1016/j.gde.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ebers GC. Environmental factors and multiple sclerosis. Lancet Neurol. 2008;7:268–277. doi: 10.1016/S1474-4422(08)70042-5. [DOI] [PubMed] [Google Scholar]
- 101.Handel AE, Giovannoni G, Ebers GC, Ramagopalan SV. Environmental factors and their timing in adult-onset multiple sclerosis. Nat Rev Neurol. 2010;6:156–166. doi: 10.1038/nrneurol.2010.1. [DOI] [PubMed] [Google Scholar]
- 102.Davenport CB. Multiple sclerosis: from the standpoint of geographic distribution and race. Arch Neur Psych. 1922;8:51–58. [Google Scholar]
- 103.Acheson ED, Bachrach CA, Wright FM. Some comments on the relationship of the distribution of multiple sclerosis to latitude, solar radiation, and other variables. Acta Psychiatr Scand Suppl. 1960;35:132–147. doi: 10.1111/j.1600-0447.1960.tb08674.x. [DOI] [PubMed] [Google Scholar]
- 104.Bulman DE, Ebers GC. The geography of multiple sclerosis reflects genetic susceptibility. J Trop Geogr Neurol. 1992;2:66–72. [Google Scholar]
- 105.Alter M, Leibowitz U, Speer J. Risk of multiple sclerosis related to age at immigration to Israel. Arch Neurol. 1966;15:234–237. doi: 10.1001/archneur.1966.00470150012002. [DOI] [PubMed] [Google Scholar]
- 106.Dean G, Elian M. Age at immigration to England of Asian and Caribbean immigrants and the risk of developing multiple sclerosis. J Neurol Neurosurg Psychiatry. 1997;63:565–568. doi: 10.1136/jnnp.63.5.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Swank RL, Dugan BB. Effect of low saturated fat diet in early and late cases of multiple sclerosis. Lancet. 1990;336:37–39. doi: 10.1016/0140-6736(90)91533-g. [DOI] [PubMed] [Google Scholar]
- 108.Lauer K. Ecologic studies of multiple sclerosis. Neurology. 1997;49(suppl 2):18–26. doi: 10.1212/wnl.49.2_suppl_2.s18. [DOI] [PubMed] [Google Scholar]
- 109.Guggenmos J, Schubart AS, Ogg S, et al. Antibody cross-reactivity between myelin oligodendrocyte glycoprotein and the milk protein butyrophilin in multiple sclerosis. J Immunol. 2004;172:661–668. doi: 10.4049/jimmunol.172.1.661. [DOI] [PubMed] [Google Scholar]
- 110.Vukusic S, Van Bockstael V, Gosselin S, Confavreux C. Regional variations in the prevalence of multiple sclerosis in French farmers. J Neurol Neurosurg Psychiatry. 2007;78:707–709. doi: 10.1136/jnnp.2006.101196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Munger KL, Levin LI, Hollis BW, Howard NS, Ascherio A. Serum 25-hydroxyvitamin D levels and risk of multiple sclerosis. JAMA. 2006;296:2832–2838. doi: 10.1001/jama.296.23.2832. [DOI] [PubMed] [Google Scholar]
- 112.Munger KL, Zhang SM, O’Reilly E, et al. Vitamin D intake and incidence of multiple sclerosis. Neurology. 2004;62:60–65. doi: 10.1212/01.wnl.0000101723.79681.38. [DOI] [PubMed] [Google Scholar]
- 113.Wilier CJ, Dyment DA, Sadovnick AD, Rothwell PM, Murray TJ, Ebers GC. Timing of birth and risk of multiple sclerosis: population based study. BMJ. 2005;330:120. doi: 10.1136/bmj.38301.686030.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bayes HK, Weir CJ, O’Leary C. Timing of birth and risk of multiple sclerosis in the Scottish population. Eur Neurol. 2010;63:36–40. doi: 10.1159/000268163. [DOI] [PubMed] [Google Scholar]
- 115.Ramagopalan SV, Heger A, Berlanga AJ, et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome Res. 2010;20:1352–1360. doi: 10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Blom HJ. Folic acid, methylation and neural tube closure in humans. Birth Defects Res A Clin Mol Teratol. 2009;85:295–302. doi: 10.1002/bdra.20581. [DOI] [PubMed] [Google Scholar]
- 117.Carone BR, Fauquier L, Habib N, et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143:1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lillycrop KA, Phillips ES, Torrens C, Hanson MA, Jackson AA, Burdge GC. Feeding pregnant rats a protein-restricted diet persistently alters the methylation of specific cytosines in the hepatic PPAR alpha promoter of the offspring. Br J Nutr. 2008;100:278–282. doi: 10.1017/S0007114507894438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Munger KL, Chitnis T, Ascherio A. Body size and risk of MS in two cohorts of US women. Neurology. 2009;73:1543–1550. doi: 10.1212/WNL.0b013e3181c0d6e0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ghadirian P, Jain M, Ducic S, Shatenstein B, Morisset R. Nutritional factors in the aetiology of multiple sclerosis: a case-control study in Montreal, Canada. Int J Epidemiol. 1998;27:845–852. doi: 10.1093/ije/27.5.845. [DOI] [PubMed] [Google Scholar]
- 121.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Nai Acad Sci USA. 2011;108(suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Berer K, Mues M, Koutrolos M, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 123.Collins SM, Surette M, Bercik P. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol. 2012;10:735–742. doi: 10.1038/nrmicro2876. [DOI] [PubMed] [Google Scholar]
- 124.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Baranzini SE, Mudge J, van Velkinburgh JC, et al. Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis. Nature. 2010;464:1351–1356. doi: 10.1038/nature08990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 127.Liggett T, Melnikov A, Tilwalli S, et al. Methylation patterns of cell-free plasma DNA in relapsing-remitting multiple sclerosis. J Neurol Sci. 2010;290:16–21. doi: 10.1016/j.jns.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pauley KM, Cha S, Chan EK. MicroRNA in autoimmunity and autoimmune diseases. J Autoimmun. 2009;32:189–194. doi: 10.1016/j.jaut.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Du C, Liu C, Kang J, et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10:1252–1259. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- 130.Junker A, Krumbholz M, Eisele S, et al. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain. 2009;132:3342–3352. doi: 10.1093/brain/awp300. [DOI] [PubMed] [Google Scholar]
- 131.Lindberg RL, Hoffmann F, Mehling M, Kuhle J, Kappos L. Altered expression of miR-17-5p in CD4+ lymphocytes of relapsing-remitting multiple sclerosis patients. Eur J Immunol. 2010;40:888–898. doi: 10.1002/eji.200940032. [DOI] [PubMed] [Google Scholar]
- 132.De Santis G, Ferracin M, Biondani A, et al. Altered miRNA expression in T regulatory cells in course of multiple sclerosis. J Neuroimmunol. 2010;226:165–171. doi: 10.1016/j.jneuroim.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 133.Cox MB, Cairns MJ, Gandhi KS, et al. MicroRNAs miR-17 and miR-20a inhibit T cell activation genes and are under-expressed in MS whole blood. PLoS One. 2010;5:12132. doi: 10.1371/journal.pone.0012132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fenoglio C, Cantoni C, De Riz M, et al. Expression and genetic analysis of miRNAs involved in CD4+ cell activation in patients with multiple sclerosis. Neurosci Lett. 2011;504:9–12. doi: 10.1016/j.neulet.2011.08.021. [DOI] [PubMed] [Google Scholar]
- 135.Paraboschi EM, Solda G, Gemmati D, et al. Genetic association and altered gene expression of mir-155 in multiple sclerosis patients. IntJ Mol Sci. 2011;12:8695–8712. doi: 10.3390/ijms12128695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Guerau-de-Arellano M, Smith KM, Godlewski J, et al. Micro-RNA dysregulation in multiple sclerosis favours pro-inflammatory T-cell-mediated autoimmunity. Brain. 2011;134:3578–3589. doi: 10.1093/brain/awr262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Martinelli-Boneschi F, Fenoglio C, Brambilla P, et al. MicroRNA and mRNA expression profile screening in multiple sclerosis patients to unravel novel pathogenic steps and identify potential biomarkers. Neurosci Lett. 2012;508:4–8. doi: 10.1016/j.neulet.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 138.Otaegui D, Baranzini SE, Armananzas R, et al. Differential micro RNA expression in PBMC from multiple sclerosis patients. PLoS One. 2009;4:6309. doi: 10.1371/journal.pone.0006309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Keller A, Leidinger P, Lange J, et al. Multiple sclerosis: micro RNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS One. 2009;4:7440. doi: 10.1371/journal.pone.0007440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Siegel SR, Mackenzie J, Chaplin G, Jablonski NG, Griffiths L. Circulating microRNAs involved in multiple sclerosis. Mol Biol Rep. 2012;39:6219–6225. doi: 10.1007/s11033-011-1441-7. [DOI] [PubMed] [Google Scholar]
- 141.Haghikia A, Hellwig K, Baraniskin A, et al. Regulated microRNAs in the CSF of patients with multiple sclerosis: a case-control study. Neurology. 2012;79:2166–2170. doi: 10.1212/WNL.0b013e3182759621. [DOI] [PubMed] [Google Scholar]
- 142.Sievers C, Meira M, Hoffmann F, Fontoura P, Kappos L, Lindberg RL. Altered microRNA expression in B lymphocytes in multiple sclerosis: towards a better understanding of treatment effects. Clin Immunol. 2012;144:70–79. doi: 10.1016/j.clim.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 143.Soreq H, Wolf Y. NeurimmiRs: microRNAs in the neuroimmune interface. Trends Mol Med. 2011;17:548–555. doi: 10.1016/j.molmed.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 144.Murugaiyan G, Beynon V, Mittal A, Joller N, Weiner HL. Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. J Immunol. 2011;187:2213–2221. doi: 10.4049/jimmunol.1003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci. 2002;22:10333–10345. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ye F, Chen Y, Hoang T, et al. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci. 2009;12:829–838. doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jacob C, Christen CN, Pereira JA, et al. HDAC1 and HDAC2 control the transcriptional program of myelination and the survival of Schwann cells. Nat Neurosci. 2011;14:429–436. doi: 10.1038/nn.2762. [DOI] [PubMed] [Google Scholar]
- 148.Moscarello MA, Wood DD, Ackerley C, Boulias C. Myelin in multiple sclerosis is develop mentally immature. J Clin Invest. 1994;94:146–154. doi: 10.1172/JCI117300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pritzker LB, Joshi S, Gowan JJ, Harauz G, Moscarello MA. Deimination of myelin basic protein. Effect of deimination of arginyl residues of myelin basic protein on its structure and susceptibility to digestion by cathepsin D. Biochemistry. 2000;39:5374–5381. doi: 10.1021/bi9925569. [DOI] [PubMed] [Google Scholar]
- 150.Harauz G, Ishiyama N, Hill CM, Bates IR, Libich DS, Fares C. Myelin basic protein-diverse conformational states of an intrinsically unstructured protein and its roles in myelin assembly and multiple sclerosis. Micron. 2004;35:503–542. doi: 10.1016/j.micron.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 151.Shanshiashvili LV, Kalandadze IV, Ramsden JJ, Mikeladze DG. Adhesive properties and inflammatory potential of citrullinated myelin basic protein peptide 45–89. Neurochem Res. 2012;37:1959–1966. doi: 10.1007/s11064-012-0816-z. [DOI] [PubMed] [Google Scholar]
- 152.Camelo S, Iglesias AH, Hwang D, et al. Transcriptional therapy with the histone deacetylase inhibitor trichostatin A ameliorates experimental autoimmune encephalomyelitis. J Neuroimmunol. 2005;164:10–21. doi: 10.1016/j.jneuroim.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 153.Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- 154.Kilgore M, Miller CA, Fass DM, et al. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer’s disease. Neuropsychopharmacology. 2010;35:870–880. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dash PK, Orsi SA, Moore AN. Histone deactylase inhibition combined with behavioral therapy enhances learning and memory following traumatic brain injury. Neuroscience. 2009;163:1–8. doi: 10.1016/j.neuroscience.2009.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Shein NA, Grigoriadis N, Alexandrovich AG, et al. Histone deacetylase inhibitor ITF2357 is neuroprotective, improves functional recovery, and induces glial apoptosis following experimental traumatic brain injury. FASEB J. 2009;23:4266–4275. doi: 10.1096/fj.09-134700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kim JY, Shen S, Dietz K, et al. HDAC1 nuclear export induced by pathological conditions is essential for the onset of axonal damage. Nat Neurosci. 2010;13:180–189. doi: 10.1038/nn.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Liu J, Dietz K, Deloyht JM, et al. Impaired adult myelination in the prefrontal cortex of socially isolated mice. Nat Neurosci. 2012;15:1621–1623. doi: 10.1038/nn.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]