Abstract
The body temperature is considered a universal cue by which the master clock synchronizes the peripheral clocks in mammals, but the mechanism is not fully understood. Here we identified two cold-induced RNA-binding proteins (RBPs), Cirbp and Rbm3, as important regulators for the temperature entrained circadian gene expression. The depletion of Cirbp or Rbm3 significantly reduced the amplitudes of core circadian genes. PAR-CLIP analyses showed that the 3′UTR binding sites of Cirbp and Rbm3 were significantly enriched near the polyadenylation sites (PASs). Furthermore, the depletion of Cirbp or Rbm3 shortened 3′UTR, whereas low temperature (upregulating Cirbp and Rbm3) lengthened 3′UTR. Remarkably, we found that they repressed the usage of proximal PASs by binding to the common 3′UTR, and many cases of proximal/distal PAS selection regulated by them showed strong circadian oscillations. Our results suggested that Cirbp and Rbm3 regulated the circadian gene expression by controlling alternative polyadenylation (APA).
The circadian clock is a daily time-keeping mechanism that regulates a wide range of biochemical, physiological and behavioral processes in living organisms1. In mammals, most cells in the body contain cell-autonomous circadian clocks2,3. The circadian clocks of peripheral tissues were synchronized by the master clock residing in the suprachiasmatic nucleus (SCN)4. In this synchronization process, the circadian oscillation of body temperature controlled by the SCN, can serve as a global entrainment cue5,6. The oscillation of body temperature consists of the temperature-rising phase and the temperature-declining phase, which can be considered a heat shock process and a cold shock process, respectively. The heat shock pathway was proposed to play a critical role in synchronizing the peripheral clocks to thermal stimuli5,7. However, the modulators in the temperature-declining phase remain to be identified. Here we proposed that cold shock proteins, such as cold-induced RBPs, may regulate the circadian clock in this process.
RBPs have long been known to play important roles in the cellular response to low temperature. In E. coli, the CspA can destabilize the RNA secondary structures during hypothermia and thereby facilitate translation8. The Y-box binding proteins in Xenopus oocytes that contain a cold-shock domain can control translation during hypothermia9. In mammals, two cold-induced RBPs, Cirbp and Rbm3, were shown to function as RNA chaperones and influence both transcription and translation during hypothermia10,11,12. Cirbp and Rbm3 belong to the same family of cold-induced RBPs and are highly homologous to each other10. Although it was initially discovered in response to the UV irradiation13, Cirbp was later found to be induced by other stressors as well, such as hypothermia and hypoxia14,15. Rbm3 is one of the most significantly upregulated genes in multiple tissues during animal hibernation16.
Increasing studies indicated the importance of posttranscriptional regulation by RBPs in the mammalian circadian clock, but the regulatory mechanisms have been limited in the mRNA translation and stability control17. Rbm4 (mLark) can promote the translation of Per1 by binding to the 3′UTR18. The RBPs Ptb and Hnrnpd were shown to promote the degradation of Per2 and Cry1 mRNAs by binding to their 3′UTRs, respectively19,20. In addition, most recently, Cirbp was shown to modulate circadian gene expression posttranscriptionally, but the detailed mechanisms were not fully elucidated21. In this study, we proposed a novel posttranscriptional regulatory mechanism in circadian clock: APA regulation.
APA is one of the most important mechanisms in the posttranscriptional regulation. More than half of the mammalian genes contain tandem UTRs which are generated by multiple PASs in the terminal exon22. Without changing the mRNA coding potential, tandem UTRs can alter the length of 3′UTR, thus influence the mRNA stability, translation and localization by changing the accessibility of RBPs and miRNAs to 3′UTR23. Previous studies have shown that 3′UTRs tend to be shortened during T lymphocytes activation24 and in cancer25, but lengthened during embryonic development26. However, the regulators of APA during specific biological processes remain to be identified.
In this study, we demonstrated a new mechanism in the temperature entrainment of circadian clock. We showed that two cold-induced RBPs (Cirbp and Rbm3) can control the APA of their target genes and influence the amplitude of temperature entrained circadian clock. Cirbp and Rbm3 were required for high-amplitude oscillation of core circadian genes in the temperature synchronized cells. The 3′UTR binding sites of Cirbp and Rbm3 were enriched within 100 nucleotides upstream of the PASs, and these two RBPs repressed the usage of proximal PASs by binding to the common 3′UTR. Furthermore, we found that the usage of proximal or distal PASs which was regulated by Cirbp or Rbm3 showed strong circadian oscillations in mouse liver. These results suggest that the regulation of APA by Cirbp and Rbm3 is an important process in regulating the amplitude of temperature entrained circadian clock.
Results
Cirbp and Rbm3 are essential for the temperature entrained circadian gene expression
To identify the candidate genes which are crucial in the temperature entrainment of circadian clock, we searched for the genes that showed circadian oscillating expression in multiple tissues, and their oscillating expression should be dependent on body temperature and independent of the local molecular clock. In a meta-analysis of circadian microarray datasets covering 14 mouse tissues, 9,887 known genes exhibited circadian oscillations in at least one tissue, and 151 of them showed circadian oscillations in at least six tissues27. We divided the 151 genes into six groups based on their circadian peak times (Fig. 1a) and performed gene ontology (GO) enrichment analysis for each group using DAVID28 (Table 1). The well-known core circadian genes were significantly enriched in the CT10 and CT22 phases. Besides, the protein-folding related genes encoding heat shock proteins (HSPs) were significantly enriched in the CT18 phase (i.e. the temperature rising phase). The genes of RBPs were significantly enriched in the CT6 phase (i.e. the temperature declining phase). This result was consistent with the pre-existing knowledge of HSPs and RBPs functioning in response to high and low temperature, respectively7,14,29.
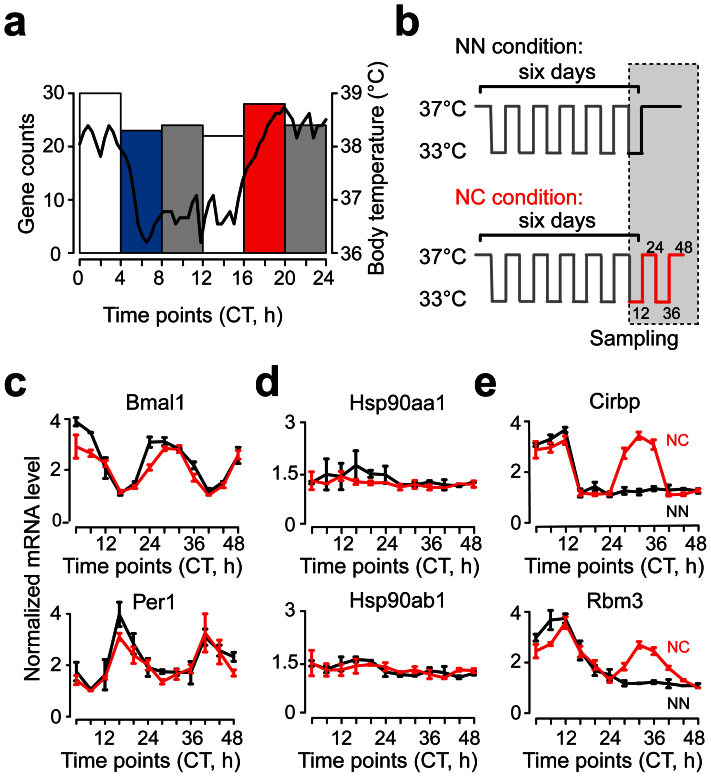
Figure 1. RBPs are potential regulators in the temperature entrainment of circadian clock.
(a) Histogram of the circadian peak times of 151 circadian oscillating genes. Black curve represented the mouse daily body temperature52. The groups of the genes with significantly enriched GO categories were highlighted. CT, circadian time in hour (h). (b) The schematic diagram of the square wave temperature synchronization model. Sampling time was indicated. (c–e) The expression patterns of the core circadian genes and the genes of HSPs and RBPs in the temperature synchronization model. The lowest expression value was normalized to 1 (black, NN condition; red, NC condition). Data are represented as mean +/− SEM (n = 3).
Table 1. Significantly enriched GO terms in each group of genes in Figure 1A.
| Phase | GO terms | Genes | Fisher's exact test P |
|---|---|---|---|
| CT6 | RNA binding | Cirbp, Fus, Hnrpdl, Rbm3, Rbms1, Srsf5, Srsf6 | 1.3E-04 |
| CT10 | rhythmic process | Dec1, Dbp, Per1, Per3, Tef, Nr1d2 | 2.7E-04 |
| CT18 | protein folding | Hspa5, Hspa8, Hspa1b, Hsph1, Hspb1, Hsp90aa1, Hsp90ab1 | 1.9E-06 |
| CT22 | rhythmic process | Bmal1, Clock, Nfil3, Bcl2 | 3.8E-04 |
We then analyzed a published circadian microarray dataset of mouse liver (ArrayExpress, E-MEXP-842), in which the liver molecular clock was either active or arrested30. This analysis identified 401 genes that exhibited a highly similar circadian oscillating expression between the liver clock active and the liver clock arrested mice (Supplementary Fig. S1a; COSOPT31, P < 0.01; Pearson correlation > 0.5). For these genes that showed independent oscillating expression of local molecular clock, they must be driven by systemic signals, such as temperature. Almost all of the RBPs and HSPs in Table 1 belonged to this gene set, which showed persistent oscillating expression in the liver clock arrested mice. (Figs. S1b and S1c). This result implied that the RBPs and HSPs are potential candidates to play important roles in the temperature entrainment of peripheral clocks. Previous studies have identified HSPs as important modulators in resetting the peripheral clocks to thermal stimuli5, but the function of RBPs in this process needs further investigation.
To identify the regulator in the temperature entrainment of circadian clock, we subjected mouse embryonic fibroblasts (MEFs) to a square wave temperature cycle between 37°C and 33°C, i.e. 12 h at a normal temperature (N) of 37°C followed by 12 h at a low temperature (C) of 33°C, to model the temperature entrainment naturally occurs in mice32 (Fig. 1b). After six days of temperature cycling, we either changed the condition to constant temperature of 37°C (NN condition) or continued the temperature cycling (NC condition). The expression of the core circadian genes and the genes of HSPs and RBPs were measured by quantitative real-time PCR (qPCR) every four hours for two days. The expression patterns of the candidate genes can be divided into three groups. The core circadian genes such as Bmal1 and Per1 oscillated in both NN and NC conditions (Fig. 1c). Once entrained by temperature cycling, their expression oscillated no matter the temperature cycling still exists or not. The genes encoding HSPs did not oscillate in either NN or NC condition (Fig. 1d). This result supported our earlier claim that the HSPs responded to high temperature but not low temperature. The two genes encoding cold-induced RBPs, Cirbp and Rbm3, oscillated only in the NC condition but not in the NN condition (Fig. 1e). Their expression levels increased following the decrease in temperature and stabilized as soon as the temperature became constant. Because the expression of Cirbp and Rbm3 were highly correlated with low temperature, oscillating in multiple tissues and independent of the local molecular clock, we proposed that the two RBPs played an important role in the temperature entrainment of circadian clock.
Our previous microarray analysis reported that the mRNA levels of Cirbp and Rbm3 oscillate in multiple mouse tissues27. We examined their mRNA levels in mouse liver and cortex (See Experimental Procedures), and found that their expression showed strong circadian oscillations in both tissues (Supplementary Fig. S2a). Moreover, the protein levels of Cirbp and Rbm3 in the liver showed strong oscillation with peaks around the CT6 and CT10, when the mouse body temperature was low (Supplementary Figs. S2b and S2c).
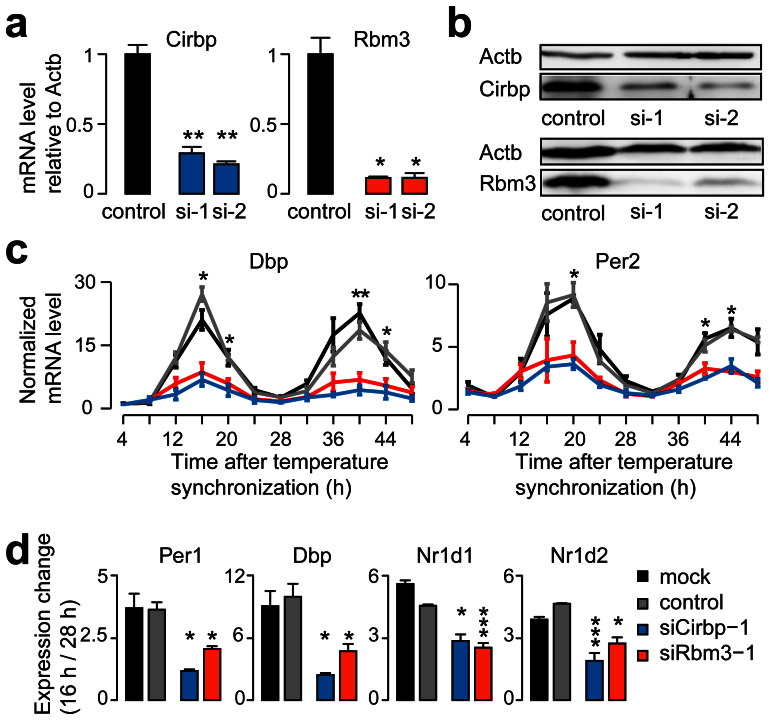
To examine whether Cirbp and Rbm3 are required for the temperature entrained circadian gene expression, we employed two independent siRNAs to deplete Cirbp or Rbm3 in MEFs (Figs. 2a and 2b). We entrained mock, control, Cirbp- and Rbm3-depleted cells with the NN condition of temperature synchronization model. We monitored the expression of Dbp and Per2 at a series of circadian time points. The oscillating amplitudes in the mRNA levels for both genes were significantly reduced in both Cirbp- and Rbm3-depleted cells comparing with the mock and control cells (Fig. 2c and Supplementary Fig. S2d). To determine the regulatory extent of Cirbp and Rbm3 on circadian gene expression, we examined the expression of core circadian genes at two time points (16 h and 28 h after temperature synchronization) in the temperature synchronized cells. Four out of 12 core circadian genes, including Per1, Dbp, Nr1d1 and Nr1d2, showed significantly reduced expression changes between the two time points in both Cirbp- and Rbm3-depleted cells comparing with the mock and control cells (Fig. 2d and Supplementary Fig. S2e). These results indicated the importance of Cirbp and Rbm3 for regulating the amplitude of the temperature entrained circadian gene expression.
Figure 2. The depletion of Cirbp or Rbm3 reduced the oscillating amplitude of core circadian genes in the temperature synchronized cells.
(a and b) The mRNA and protein levels for the control and the two independent siRNA knockdowns of Cirbp and Rbm3. Actb was used as control. (c) The oscillating mRNA levels of Dbp and Per2 in the temperature synchronized mock (black), control (gray), siCirbp-1 (blue) and siRbm3-1 (red) cells. The lowest expression value was normalized to 1. See also Fig. S2C. (d) The expression changes between 16 h and 28 h after temperature synchronization of the core circadian genes (Per1, Dbp, Nr1d1 and Nr1d2) in the temperature synchronized mock, control, siCirbp-1 and siRbm3-1 cells. The statistical significances were calculated between the control and Cirbp- or Rbm3-depleted cells. See also Fig. S2D. Data are represented as mean +/− SEM (n = 3); Student's t-test * P < 0.05, ** P < 0.01, *** P < 0.001.
Identification and analyses of Cirbp- and Rbm3-binding sites using PAR-CLIP
To study the functions of Cirbp and Rbm3, we used PAR-CLIP (photoactivatable ribonucleoside enhanced crosslinking and immunoprecipitation)33,34,35 in combination with next-generation sequencing to identify their direct binding targets. In PAR-CLIP, the photoactivatable nucleotide (4-thiouridine, 4SU) is incorporated into nascent RNA and thereby enhances the crosslinking of protein to RNA. Moreover, the crosslinking of 4SU-labeled RNA to proteins leads to specific T-to-C changes in cDNA sequences, marking the protein binding sites on the target RNA33, which facilitates the discrimination of the signal-to-noise ratio. The Cirbp- or Rbm3-bound RNA fragments were successfully recovered (Supplementary Fig. S3a), purified and converted to cDNAs after the ligation of adapters and then sequenced on Illumina GAII platform.
We observed diagnostic T-to-C transitions in PAR-CLIP cDNA libraries of Cirbp and Rbm3 (Supplementary Figs. S3b and S3c). These T-to-C changes indicated the existence of the bona fide RBP binding sites on the target RNA33. All of the sequencing data were analyzed with a recently developed computational pipeline34. Two independent PAR-CLIP experiments yielded 23.6 million and 4.98 million reads for Cirbp, 13.1 million and 12.6 million reads for Rbm3, which were aligned to the mouse mm9 genome by allowing only unique genomic hits and one nucleotide mismatch (Supplementary Table S1). Using the aligned reads, we generated 12,070 and 16,893 Cirbp- and Rbm3-binding clusters with T-to-C transitions and 20 or more overlapping reads, respectively. The mean lengths of the Cirbp- and Rbm3-binding clusters were 25.9 and 23.1 nucleotides, respectively, which indicated the high resolution of PAR-CLIP. 8,597 Cirbp- and 9,669 Rbm3-binding sites can be assigned to annotated gene regions, including 5′UTR, coding sequence (CDS), 3′UTR and intron, based on the UCSC mouse gene annotation36 (Figs. 3a and 3b, Supplementary Table S2). The majority of the Cirbp- or Rbm3-binding sites were located in the exonic regions, especially the CDS and 3′UTR. Furthermore, we found that the 3′UTR binding sites of Cirbp and Rbm3 were significantly enriched within 100 nucleotides upstream of PASs (Cirbp, 939 out of 3,520; Proportion test, P < 1.0E-15; Rbm3, 549 out of 2,081; P < 1.0E-15; Fig. 3c), which implied the potential function of the two RBPs in the polyadenylation.
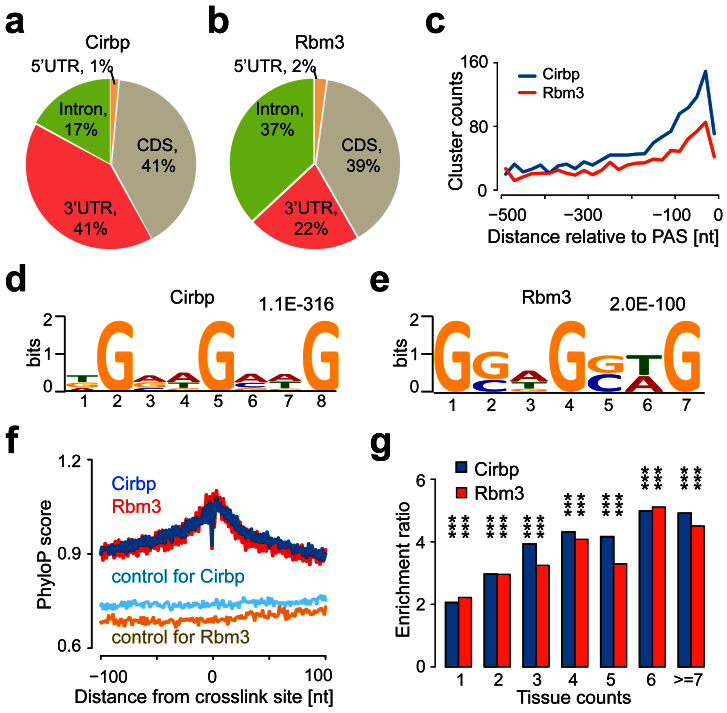
Figure 3. Identification and analyses of Cirbp- and Rbm3-binding sites using PAR-CLIP.
(a and b) The distribution of Cirbp- and Rbm3-binding sites in gene region revealed predominant binding within CDS and 3′UTR. (c) The distribution of Cirbp- and Rbm3-binding sites relative to the PASs revealed the enrichment within 100 nucleotides upstream of the PASs. (d and e) The enriched sequence motifs of Cirbp- and Rbm3-binding clusters which are located in the exonic regions were identified by the MEME software. For each motif we indicated the E-value. (f) The conservation profiles of Cirbp- and Rbm3-binding sites that are located in the exonic regions. Average PhyloP score at a given distance from the crosslinking sites was shown. Random selected regions from the same transcript were drawn to serve as control. (g) Enrichment ratios of Cirbp- or Rbm3-target circadian oscillating genes compared to the expected circadian oscillating genes by chance in the certain tissue counts. Chi-square test ** P < 0.01, *** P < 0.001.
We employed the MEME software37 to identify the sequence motifs in the Cirbp- or Rbm3-binding clusters which were located in the exonic regions. In the 7,139 Cirbp-binding clusters, MEME identified a strong motif that was present in 3,353 (47.0%) clusters (Fig. 3d). In the 6,106 Rbm3-binding clusters, a strong motif was present in 3,533 (57.9%) clusters (Fig. 3e). These results indicated that the binding motifs of Cirbp and Rbm3 are not identical, but very similar to each other. Previous studies have indicated that functional RNA binding elements tend to be more conserved than their flanking genomic regions34,38. We therefore estimated the evolutionary conservation of Cirbp- or Rbm3-binding sites across 20 placental mammalian species using the PhyloP nucleotide conservation scores39. In each binding cluster, the position with the highest frequency of T-to-C transitions was considered the preferred crosslinking site. We extracted the PhyloP scores at a given distance around the crosslinking site and averaged these over all binding sites (Fig. 3f), and found that the sequences around the crosslinking site showed highly elevated conservation, which suggests the existence of functional elements.
In total, we identified 4,376 and 5,300 target genes of Cirbp and Rbm3, respectively. Despite their different binding motifs, their target genes had significant overlap (2,539 genes; Fisher's exact test, P < 1.0E-15). GO enrichment analyses showed that their target genes were mostly involved in regulating the transcription and mRNA metabolic processes (Supplementary Table S3). In addition, we independently performed Cirbp or Rbm3 RNA-IP in MEFs and validated five Cirbp- and Rbm3-target genes out of five tested through qPCR, including the core circadian genes Nr1d2 and Tef (Supplementary Fig. S3d).
To evaluate the function of Cirbp and Rbm3 in the circadian clock, we compared their target genes with the circadian gene database27 and found that the circadian oscillating genes are significantly enriched. 2,458 (56.4%; Chi-square test, P < 1.0E-15) Cirbp-target genes and 2,762 (52.1%; Chi-square test, P < 1.0E-15) Rbm3-target genes exhibited oscillating expression in at least one tissue. When we compared oscillating Cirbp- or Rbm3-target gene counts to the expected oscillating gene counts by chance, we found the enrichment ratios were 2.1 and 2.2 for the genes oscillating in one tissue, and the enrichment ratios increased with the number of tissues in which the genes oscillated (Fig. 3g). We also found that the circadian peak times of the Cirbp- or Rbm3-target genes were significantly enriched in the CT6 phase (i.e. the temperature declining phase; Supplementary Figs. S3e and S3f). Furthermore, many core circadian genes, including Bmal1, Clock, Per1, Cry1, Nr1d2 and Tef, were shown to be bound by Cirbp or Rbm3. The Cirbp- or Rbm3-target genes that showed oscillating expression in at least eight tissues were shown in Supplementary Table S4. These results suggested that the two RBPs are important regulators in the circadian clock.
Cirbp and Rbm3 repress the usage of proximal PASs by binding to the common 3′UTR
To assess the functions of Cirbp and Rbm3 at transcriptome level, we applied RNA-seq to mock cells, Cirbp-depleted cells, Rbm3-depleted cells and mock cells that were treated with cold shock (32°C) (Supplementary Table S5). The analyses of the RNA-seq datasets showed that the results between the biological replicates were highly reproducible (Supplementary Figs. S4a–d). Using stringent criteria (FPKM > 1 in every sample, and log2 [fold change] > 0.8), we identified 876 upregulated and 454 downregulated genes upon Cirbp depletion, and 237 upregulated and 293 downregulated genes upon Rbm3 depletion (Supplementary Table S6). The GO enrichment analyses showed that the genes which were upregulated upon Cirbp or Rbm3 depletion were significantly enriched in cell cycle (Supplementary Table S7). This result was consistent with the previous studies that Cirbp and Rbm3 regulated cellular proliferation40,41,42,43. To assess the influence of Cirbp or Rbm3 on the stability of their target transcripts, we compared the mRNA levels from knockdown and mock cells. The expression levels of transcripts with Cirbp-binding sites in the intron significantly decreased upon Cirbp depletion, but the levels of transcripts with Cirbp-binding sites in the CDS and 3′UTR were not significantly changed upon Cirbp depletion (Supplementary Fig. S4e). The expression levels of transcripts with Rbm3 binding sites in CDS, 3′UTR and intron significantly decreased upon Rbm3 depletion (Supplementary Fig. S4f). These results suggested that Cirbp stabilized its target pre-mRNAs, and Rbm3 stabilized its target pre-mRNAs and mRNAs.
Because the PAR-CLIP results implied the potential functions of Cirbp and Rbm3 in the polyadenylation, we systematically screened our RNA-seq data to identify the role of Cirbp and Rbm3 in the APA regulation. After combining the PolyA_DB2 database44 with the latest polyA-seq datasets22, we annotated 12,250 genes with reliable tandem UTRs from the UCSC known genes in mouse. We then compared the relative expression of the common 3′UTR (between stop codon and proximal PAS) and of the extended 3′UTR (between proximal PAS and distal PAS) upon Cirbp or Rbm3 depletion, and identified the switching events in the usage of proximal or distal PAS (PAS-usage switching events) which were regulated by Cirbp or Rbm3. We identified 247 and 140 genes that showed significantly changed ratios of extended to common 3′UTR (ext/com UTR-ratio) upon Cirbp or Rbm3 depletion under the criteria (Fisher's exact test, P < 0.01; the ext/com UTR-ratio change > 0.6). Moreover, 166 (67.2%) and 119 (85.0%) of the genes showed downregulated ext/com UTR-ratio upon Cirbp or Rbm3 depletion, respectively (Supplementary Figs. S4a and S4b, Supplementary Table S8). These results indicated that 3′UTRs tended to be shortened upon Cirbp or Rbm3 depletion.
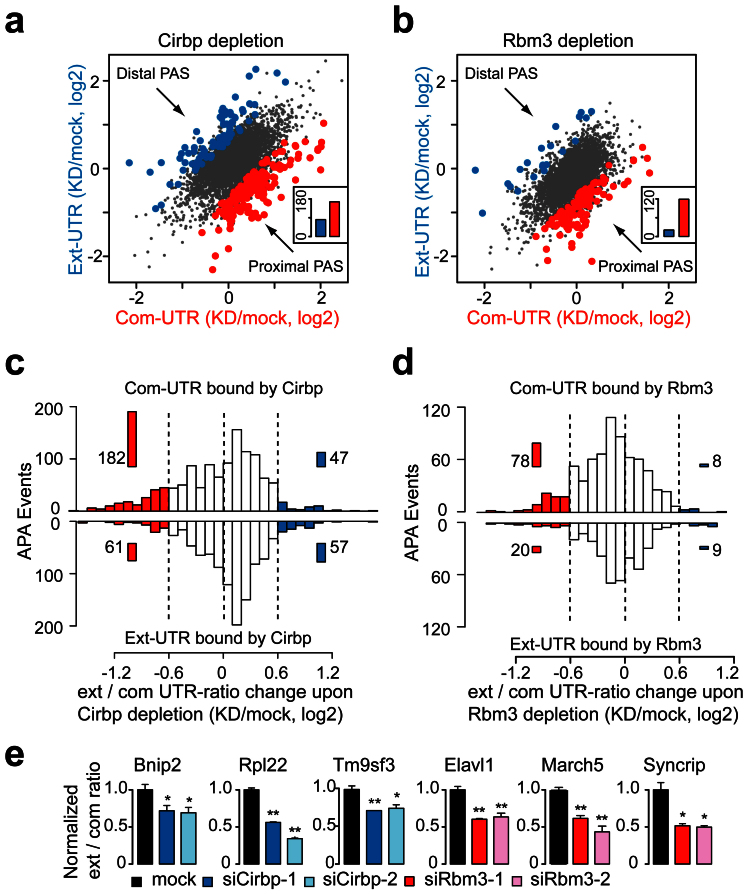
To examine the relationship of Cirbp- or Rbm3-binding sites and the PAS-usage switching, we compared the Cirbp- or Rbm3-binding sites on the tandem UTRs with the ext/com UTR-ratio changes upon Cirbp or Rbm3 depletion (Figs. 4c and 4d). Among the APA events that contain Cirbp-binding sites in the common 3′UTRs (Fig. 4c), 182 events showed downregulated ext/com UTR-ratio (Proportion test, P < 1.0E-15), and only 47 events showed upregulated ext/com UTR-ratio upon Cirbp depletion under the criteria (Fisher's exact test, P < 0.01; the ext/com UTR-ratio change > 0.6). If Cirbp bound to the extended 3′UTRs, there were few PAS-usage switching events upon Cirbp depletion (61 downregulated and 57 upregulated). Similarly, the APA events containing Rbm3 binding sites in the common 3′UTRs were more likely to show downregulated (78 events; Proportion test, P < 1.0E-15) than upregulated ext/com UTR-ratio (8 events) upon Rbm3 depletion (Fig. 4d), whereas with Rbm3-binding sites in the extended 3′UTRs, there were rare PAS-usage switching events upon Rbm3 depletion (20 downregulated and 9 upregulated). These results suggested that Cirbp and Rbm3 repressed the usage of proximal PASs by binding to the common 3′UTRs. The selected genes which showed PAS-usage switching upon Cirbp or Rbm3 depletion were validated by qPCR (Fig. 4e and Supplementary Fig. S4g).
Figure 4. RNA-seq and PAR-CLIP analyses indicated that Cirbp and Rbm3 repressed the usage of proximal PASs by binding to the common 3′UTRs.
(a) Scatterplot of the common and extended 3′UTR expression of tandem UTR genes upon Cirbp depletion. Significantly increased expression of isoforms resulting from usage of distal (blue) and proximal PAS (red) are highlighted. Inset: total number of the different changes (Binomial test, P = 3.4E-08). (b) Scatterplot of the common and extended 3′UTRs expression of tandem UTR genes upon Rbm3 depletion as in (a) (Binomial test, P < 1.0E-15). (c) Histogram showed the ext/com UTR-ratio changes of the genes with Cirbp-binding sites in the tandem UTRs upon Cirbp depletion (Top, common 3′UTRs bound by Cirbp; Bottom, extended 3′UTRs bound by Cirbp). The PAS-usage switching events with up (blue) or downregulated (red) ext/com UTR-ratio were highlighted. Inset: the total number of cases in each region. (d) Histogram showed the ext/com UTR-ratio changes of the genes with Rbm3-binding sites in the tandem UTRs upon Rbm3 depletion as in (c). (e) QPCR analyses of the selected genes that showed downregulated ext/com UTR-ratio upon Cirbp or Rbm3 depletion. The statistical significance was calculated against the mock cells. See also Fig. S4G. Data are represented as mean +/- SEM (n = 3); Student's t-test * P < 0.05, ** P < 0.01.
Cirbp and Rbm3 are important in the APA regulation upon cold shock and in the circadian clock
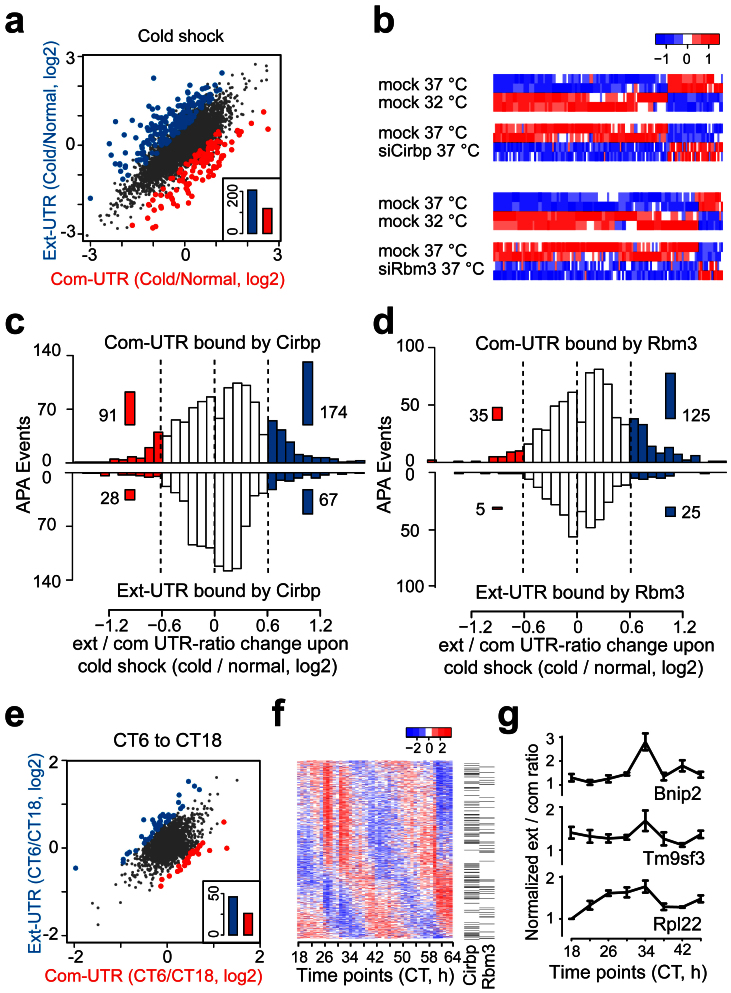
Considering that the expression of Cirbp and Rbm3 were significantly upregulated upon cold shock (Supplementary Fig. S5a), we examined the PAS-usage switching events in response to cold shock. Under the statistical criteria (Fisher's exact test, P < 0.01; the ext/com UTR-ratio change > 0.6), 325 genes showed significant PAS-usage switching upon cold shock, and 63.4% (206) of them showed upregulated ext/com UTR-ratio, indicating that 3′UTRs tended to be lengthened upon cold shock (Fig. 5a). Considering that Cirbp and Rbm3 were upregulated upon cold shock, we supposed that the genes regulated by Cirbp or Rbm3 should show opposite direction of PAS-usage switching upon Cirbp or Rbm3 depletion comparing to cold shock. We identified the genes which showed opposite direction of PAS-usage switching between upon cold shock and RBP depletion (Cirbp: 129 genes; Rbm3: 123 genes; the ext/com UTR-ratio change > 0.4). Among these genes, 98 genes for Cirbp and 110 genes for Rbm3 showed 3′UTR lengthening upon cold shock and shortening upon RBP depletion (Fig. 5b). The selected genes that exhibited opposite PAS-usage switching were validated by qPCR (Supplementary Fig. S5b).
Figure 5. Cirbp and Rbm3 are important in the APA regulation upon cold shock and in the circadian clock.
(a) Scatterplot of the common and extended 3′UTRs expression of tandem UTR genes between the mock at 32°C (Cold) and at 37°C (Normal). Significantly increased expression of isoforms resulting from usage of distal (blue) and proximal PAS (red) are highlighted. Inset: total number of the different changes (Binomial test, P = 8.0E-07). (b) Heatmap of the genes that showed opposite direction of PAS-usage switching between upon cold shock and RBP depletion. (c) Histogram showed the ext/com UTR-ratio changes of the genes with Cirbp binding sites in the tandem UTRs upon cold shock. The PAS-usage switching events with up (blue) or downregulated (red) ext/com UTR-ratio were highlighted. Inset: the total number of cases in each region. (d) Histogram showed the ext/com UTR-ratio changes of the genes with Rbm3 binding sites in the tandem UTRs upon cold shock as in (c). (e) Scatterplot of the common and extended 3′UTR expression of tandem UTR genes between CT6 and CT18 in circadian mouse liver as in (a) (Binomial test, P = 0.01). (f) Heatmap of the circadian oscillating ext/com UTR-ratio of 1153 genes in mouse liver. The genes that contained Cirbp- or Rbm3-binding sites in the tandem UTRs were indicated. See also Fig. S6A. (g) QPCR analyses of the ext/com UTR-ratio of the selected genes in circadian mouse liver. Data are represented as mean +/− SEM (n = 3).
To examine the relationship of Cirbp- and Rbm3-binding sites and the PAS-usage switching upon cold shock, we compared the Cirbp- or Rbm3-binding sites on the tandem UTRs with the ext/com UTR-ratio changes upon cold shock (Figs. 5c and 5d). Among the APA events that contain Cirbp-binding sites in the common 3′UTRs (Fig. 5c), 174 events showed upregulated ext/com UTR-ratio (Proportion test, P < 1.0E-15), and only 91 events showed downregulated ext/com UTR-ratio upon cold shock under the criteria (Fisher's exact test, P < 0.01; the ext/com UTR-ratio change > 0.6). In comparison, if Cirbp bound to the extended 3′UTRs, there were few PAS-usage switching events upon cold shock (67 upregulated and 28 downregulated). Similarly, the APA events containing Rbm3-binding sites in the common 3′UTRs were more likely to show upregulated ext/com UTR-ratio (125 events; Proportion test, P < 1.0E-15) than downregulated ext/com UTR-ratio (35 events) upon cold shock, whereas with Rbm3-binding sites in the extended 3′UTRs, there were rare PAS-usage switching events upon cold shock (25 upregulated and 5 downregulated, Fig. 5d). These results suggested that 3′UTRs tend to be lengthened upon cold shock and upregulated Cirbp and Rbm3 play an important role in this process by binding to the common 3′UTRs and repressing the usage of proximal PASs. Several examples of depth-of-coverage profiles in RNA-seq were shown (Supplementary Figs. S5c–S5e).
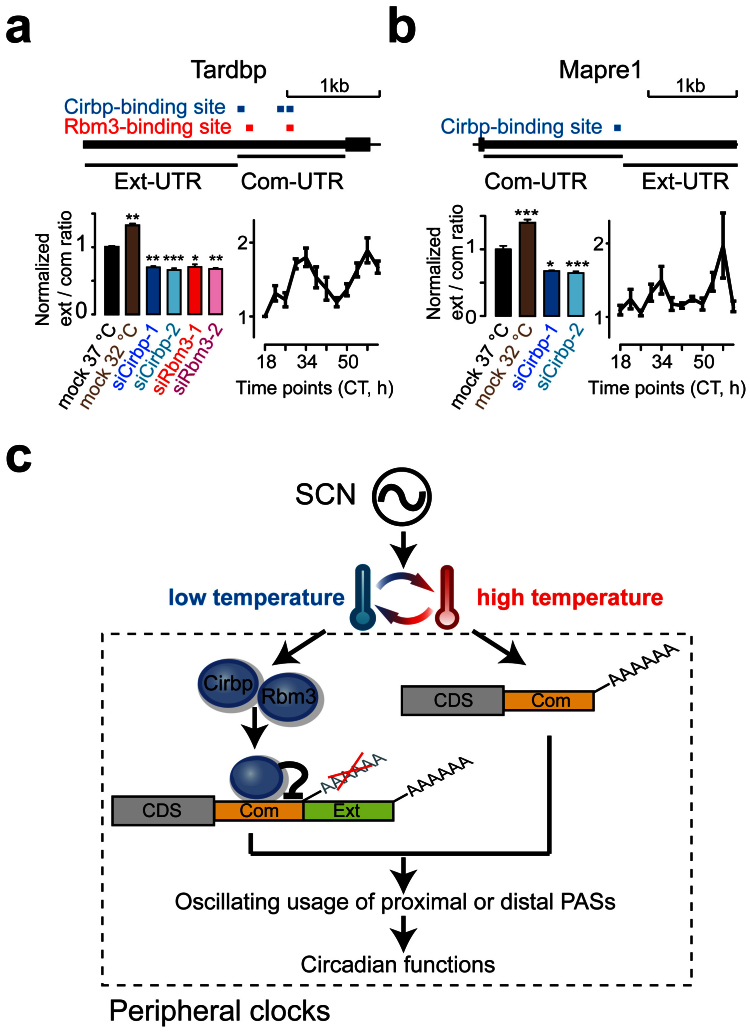
Because the mouse body temperature oscillates daily with a trough at the CT6 phase and a peak at the CT18 phase, we quantified the expression changes in the common and extended 3′UTRs between these two time points based on the circadian microarray data of mouse liver (Gene Expression Omnibus, GSE11923)45. Out of the 72 genes that showed significant PAS-usage switching (Fisher's exact test, P < 0.01; the ext/com UTR-ratio change > 0.6), 63.9% of them showed upregulated ext/com UTR-ratio (indicating 3′UTR lengthening) at the CT6 phase (Fig. 5e). Furthermore, to estimate the PAS-usage in a circadian manner, we calculated the ext/com UTR-ratio for each gene in the microarray every one hour for two days45, and identified 1153 genes that exhibited circadian oscillations of the ext/com UTR-ratio (COSOPT, P < 0.01, Fig. 5f), including many core circadian genes, such as Clock, Rorc and Tef (Supplementary Table S9). Moreover, we observed that the circadian peak times of these genes were significantly enriched in the CT6 phase (236, 40.1%; Proportion test, P < 1.0E-15; Supplementary Fig. S6a). Interestingly, we found that a significant proportion of these genes containing Cirbp- or Rbm3-binding sites in the tandem UTRs (164 and 104; Fisher's exact test, P = 5.0E-4 and 1.4E-5, respectively). These results indicated that the usage of PASs was under strong circadian regulation, and longer 3′UTRs were preferred at low body temperature. Cirbp and Rbm3 are potential regulators in this process. In addition, we estimated the PAS-usage of the genes which were regulated by Cirbp or Rbm3 (Fisher's exact test, P < 0.01; the ext/com UTR-ratio change > 0.6) in circadian mouse liver, and found that 35 and 23 genes regulated by Cirbp and Rbm3, respectively, showed strong circadian oscillations of the ext/com UTR-ratio (COSOPT, P < 0.01, Supplementary Figs. S6b and S6c). The selected genes that exhibited circadian oscillations of the ext/com UTR-ratio in mouse liver were validated by qPCR (Fig. 5g). We verified that two genes, Tardbp (Tdp43) and Mapre1, which contained Cirbp- or Rbm3-binding sites in the common 3′UTRs, showed 3′UTR lengthening upon cold shock and 3′UTR shortening upon Cirbp or Rbm3 depletion, and exhibited strong circadian oscillations of the ext/com UTR-ratio in mouse liver (Figs. 6a and 6b). Collectively, these results suggest a novel posttranscriptional mechanism that Cirbp and Rbm3 regulate the amplitude of temperature entrained circadian gene expression by controlling APA.
Figure 6. Two examples of the PAS-usage switching events regulated by Cirbp or Rbm3 and a model of the body temperature entraining the peripheral clocks.
(a and b) Tandem UTRs for Tardbp and Mapre1 are shown together with Cirbp- (blue) or Rbm3- (red) binding sites (Top). QPCR analyses of the ext/com UTR-ratio of Tardbp and Mapre1 upon cold shock and Cirbp or Rbm3 depletion (Bottom left). The statistical significance was calculated against the mock cells. QPCR analyses of the ext/com UTR-ratio of Tardbp and Mapre1 in circadian mouse liver (Bottom right). Data are represented as mean +/− SEM (n = 3 or 4); Student's t-test * P < 0.05, ** P < 0.01, *** P < 0.001. (c) Model representing the role of Cirbp and Rbm3 in the body temperature entraining the peripheral clocks. For simplicity, additional factors are not depicted.
Discussion
Temperature has long been suggested as an important entrainment cue in the circadian clock through posttranscriptional regulation46. For example, in Neurospora, temperature changes induce a thermosensitive alternative splicing of a core circadian gene FRQ (FREQUENCY)47. In Drosophila, a thermosensitive splicing event in the 3′UTR of per (period) plays an important role in temperature compensation and seasonal adaptation48. However, in mammals, the posttranscriptional mechanisms in the temperature entrainment of circadian clock remain unclear.
In this study, we identified two cold-induced RBPs, Cirbp and Rbm3, as important regulators in the temperature entrained circadian gene expression, and proposed a novel posttranscriptional mechanism in this process. The depletion of Cirbp or Rbm3 significantly reduced the oscillating amplitudes of core circadian genes in the temperature synchronized cells. With a combination of PAR-CLIP and RNA-seq analyses, we observed that the 3′UTR binding sites of Cirbp and Rbm3 were enriched within 100 nucleotides upstream of the polyadenylation sites, and the two RBPs repressed the usage of proximal PASs by binding to the common 3′UTRs. Furthermore, in mouse liver, we estimated the PAS-usage of the genes which were regulated by Cirbp or Rbm3, and found that many of them indeed showed strong circadian oscillations. In summary, our results indicate that Cirbp and Rbm3 modulate the amplitude of temperature entrained circadian gene expression by controlling APA.
Based on these results, we proposed a model for the posttranscriptional mechanism in the temperature entrainment of circadian clock (Fig. 6c). In mammals, the master clock in SCN sets the daily body temperature cycle that affects the peripheral clocks. In the low-temperature phase, the cold-induced RBPs (Cirbp and Rbm3) are upregulated. They repress the usage of the proximal PASs of their target genes through binding to the common 3′UTRs, thus lengthen the 3′UTR and increase the posttranscriptional cis elements28. In the high-temperature phase, the downregulation of cold-induced RBPs leads to the preferred use of the proximal PASs and the shortening of 3′UTR, thus increases mRNA stability through reducing microRNA-mediated repression23. The APA regulation in the circadian clock could control the mRNA stability, translation and localization of the circadian oscillating genes, and influence their circadian functions.
RBPs were shown to cross-regulate each other49. To assess the RBPs which are the targets of Cirbp and Rbm3, we obtained 415 mouse RBP genes from the RBPDB database50, and found that 199 of them (Fisher's exact test, P < 1.0E-15) were bound by either Cirbp or Rbm3. This result is consistent with the GO category enrichment of RNA binding in the Cirbp- or Rbm3-target genes. Moreover, RNA-seq analyses showed that 56 Cirbp- or Rbm3-target RBP genes (Fisher's exact test, P < 1.0E-15) exhibited expression changes at either transcript (log2 [fold change] > 0.8) or APA level (Fisher's exact test, P < 0.01; the ext/com UTR-ratio change > 0.6) upon Cirbp or Rbm3 depletion (Supplementary Fig. S6d). Intriguingly, several Cirbp- or Rbm3-target RBP genes were responsible for polyadenylation and 3′UTR processing, including Cpsf6, Cpsf7, Hnrnph1, Pabpn1 and Ptbp1. In particular, Hnrnph1 was shown to bind to Cirbp and Rbm3 mRNA, and stabilized their mRNA level49. We found Hnrnph1 was bound by Cirbp and upregulated upon Cirbp depletion, indicating that Hnrnph1 and Cirbp/Rbm3 formed a regulatory feedback loop. Moreover, we found that Rbm3 was bound by Cirbp, and the expression of Rbm3 was upregulated upon Cirbp depletion. Analysis of the mRNA sequences of Cirbp and Rbm3 suggests that while both of them are highly conserved across a wide range of species, Rbm3 is only present in mammals and is likely a result of gene duplication of Cirbp in mammals (data not shown). Future work should reveal the regulatory and evolutionary relationship of these two RBPs. In addition, another Cirbp-target gene, Sfpq (Psf), was shown to play a critical role in regulating circadian clock through the recruitment of Sin3a and rhythmically delivering histone deacetylases to the Per1 promoter to repress its transcription51. Therefore, these findings suggest that Cirbp and Rbm3 reside in an interconnected RBP network that regulates the RNA metabolism and the circadian clock.
For the Cirbp- or Rbm3-binding sites, in addition to falling in the tandem UTRs, we also identified more than 3000 binding sites located in the CDS region and non-tandem UTRs. Besides, our RNA-seq analyses revealed that Cirbp and Rbm3 stabilized their target transcripts. These results implied that Cirbp and Rbm3 may have other mechanisms in regulating the circadian clock besides the APA regulation, such as modulating the mRNA localization and stability of their associated circadian genes. Morf et al. recently reported that Cirbp is required for high-amplitude circadian gene expression. Using a biotin-streptavidin–based cross-linking and immunoprecipitation (CLIP) procedure, they found that Cirbp-binding sites are significantly enriched near the PASs. They found that Cirbp bound several transcripts encoding core circadian oscillator proteins, including CLOCK. They surmised that Cirbp influenced the circadian clock through increasing the cytoplasmic accumulation of mRNAs encoding Clock and other circadian regulators21. Collectively, these results suggested that Cirbp and Rbm3 regulated circadaian clock in diverse posttranscriptional mechanisms.
For the core circadian genes whose ext/com UTR-ratios showed strong circadian oscillations in mouse liver, their ext/com UTR-ratios seem not influenced by the depletion of Cirbp or Rbm3 in the cells. This is perhaps due to the differences between tissue and cell line, or there are still other RBPs which may have potential functions in the APA regulation of circadian clock. As shown in Table 1, several other RBPs such as Fus and Hnrnpdl, also showed oscillating expression patterns in more than six different mouse tissues, and independent expresion of the local molecular clock. Further studies on the function and interaction of these remaining RBPs and the detailed mechanisms of APA regulation on the circadian genes will provide valuable information on the posttranscriptional regulation in the mammalian circadian clock.
Methods
Animals
Male BALB/c mice were housed in 12 h light/12 h dark (LD) cycles for two weeks, then subjected to complete darkness (DD) as a continuation of the dark phase of the last LD cycle. Mice were sacrificed from CT18 every four hours during two days in continuous dark phase. Tissues were immediately frozen in liquid nitrogen and stored at −80°C until the extraction of total RNA. The age of the animals was between 3 and 4 months. The use and care of animals and the experiments complied with the guidelines of the Animal Advisory Committee at the Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences.
Cell culture
Immortalized MEFs and 293 T cells were cultivated at 37°C with 5% CO2 in high glucose Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) (Hyclone) and antibiotic (100 U/ml penicillin and 100 μg/ml streptomycin). Cell lines stably expressing Flag-tagged proteins were generated by retroviruses co-transfection. For the treatment of cold shock, the cells were cultivated at 32°C for 12 h. Prior to harvest, cells were rinsed with ice-cold PBS twice.
Square wave temperature synchronization model
5 × 105 MEFs were plated into six cm dishes four days before the treatment was applied. Once the MEFs were fully confluent (90–95%), the temperature cycles that are 12 h at 37°C and 12 h at 33°C were used to entrain the circadian clock of the cells. After six days of entrainment, we either kept the temperature at constant 37°C, or continued the temperature cycles. The cells were harvested every four hours from the beginning of the sixth 33°C during two days. At indicated time, the cells were washed twice with ice-cold PBS and harvested in 1 ml of TRIzol reagent (Invitrogen). These samples were frozen and stored at −80°C until the extraction of total RNA.
Accession number
The Gene Expression Omnibus accession number for the PAR-CLIP and RNA-seq raw data reported in this paper is GSE40468.
Retrovirus plasmids and infection, Western blot and antibodies, RNA extraction and qPCR, Tandem UTRs database construction, RNA-seq, RIP and qPCR, Microarray analyses, PAR-CLIP library construction, sequencing and analyses are available in the Supplementary information.
Author Contributions
Y.L. and J.Y. conceived, designed and interpreted the experiments. Y.L. and Y.M. performed experiments, including PAR-CLIP. Y.L. and W.H. performed all bioinformatics analyses, including mapping of the PAR-CLIP tags to the genome and statistical analyses. J.Y., G.W. and M.L. provided the expertise and instrumentation required for the experiments. W.H. constructed the tandem UTRs database. J.Y. supervised the project. Y.L. and J.Y. wrote the manuscript. All authors reviewed the manuscript.
Supplementary Material
Supplementary information
Supplementary Dataset
Acknowledgments
We would like to express our gratitude to Dr. Nikolaus Rajewsky (MDC, Berlin) and members of his lab for sharing the PAR-CLIP computational analysis pipeline for this study as well as Claudia Langnick and Mirjam Feldkamp from the Dr. Wei Chen lab (MDC, Berlin) for sequencing. We thank Dr. Peter W. Vanderklish (The Scripps Research Institute, La Jolla) for providing us the Rbm3 specific antibody and Dr. Wenqin Hu (Institute of Neuroscience, Shanghai) for comments and suggestions. This research was supported by both Chinese Academy of Sciences and German Max-Planck Society.
References
- Bell-Pedersen D. et al. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet 6, 544–56 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh D. K., Yoo S. H., Liu A. C., Takahashi J. S. & Kay S. A. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol 14, 2289–95 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi E. et al. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119, 693–705 (2004). [DOI] [PubMed] [Google Scholar]
- Dibner C., Schibler U. & Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol 72, 517–49 (2010). [DOI] [PubMed] [Google Scholar]
- Ethan D. B., Yoo S.-H. & Takahashi J. S. Temperature as a Universal Resetting Cue for Mammalian Circadian Oscillators. Science 330, 379–385 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham U. et al. Coupling governs entrainment range of circadian clocks. Molecular systems biology 6, 438–438 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol 11, 441–69 (1995). [DOI] [PubMed] [Google Scholar]
- Bae W., Jones P. G. & Inouye M. CspA, the major cold shock protein of Escherichia coli, negatively regulates its own gene expression. J Bacteriol 179, 7081–8 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K. & Wolffe A. P. Gene regulation by Y-box proteins: coupling control of transcription and translation. Trends Cell Biol 8, 318–23 (1998). [DOI] [PubMed] [Google Scholar]
- Lleonart M. E. A new generation of proto-oncogenes: cold-inducible RNA binding proteins. Biochim Biophys Acta 1805, 43–52 (2010). [DOI] [PubMed] [Google Scholar]
- Dresios J. et al. Cold stress-induced protein Rbm3 binds 60 S ribosomal subunits, alters microRNA levels, and enhances global protein synthesis. Proc Natl Acad Sci U S A 102, 1865–70 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart F. et al. Two isoforms of the cold-inducible mRNA-binding protein RBM3 localize to dendrites and promote translation. J Neurochem 101, 1367–79 (2007). [DOI] [PubMed] [Google Scholar]
- Sheikh M. S. Identification of Several Human Homologs of Hamster DNA Damage-inducible Transcripts. CLONING AND CHARACTERIZATION OF A NOVEL UV-INDUCIBLE cDNA THAT CODES FOR A PUTATIVE RNA-BINDING PROTEIN. Journal of Biological Chemistry 272, 26720–26726 (1997). [DOI] [PubMed] [Google Scholar]
- Fujita J. Cold shock response in mammalian cells. J Mol Microbiol Biotechnol 1, 243–55 (1999). [PubMed] [Google Scholar]
- Wellmann S. et al. Oxygen-regulated expression of the RNA-binding proteins RBM3 and CIRP by a HIF-1-independent mechanism. J Cell Sci 117, 1785–94 (2004). [DOI] [PubMed] [Google Scholar]
- Yan J., Barnes B. M., Kohl F. & Marr T. G. Modulation of gene expression in hibernating arctic ground squirrels. Physiol Genomics 32, 170–81 (2008). [DOI] [PubMed] [Google Scholar]
- Kojima S., Shingle D. L. & Green C. B. Post-transcriptional control of circadian rhythms. J Cell Sci 124, 311–20 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S. et al. LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc Natl Acad Sci U S A 104, 1859–64 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo K. C. et al. Mouse period 2 mRNA circadian oscillation is modulated by PTB-mediated rhythmic mRNA degradation. Nucleic Acids Res 37, 26–37 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo K. C. et al. Circadian amplitude of cryptochrome 1 is modulated by mRNA stability regulation via cytoplasmic hnRNP D oscillation. Mol Cell Biol 30, 197–205 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morf J. et al. Cold-Inducible RNA-Binding Protein Modulates Circadian Gene Expression Posttranscriptionally. Science (2012). [DOI] [PubMed] [Google Scholar]
- Derti A. et al. A quantitative atlas of polyadenylation in five mammals. Genome Res 22, 1173–83 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino D. C., Nishida K. & Manley J. L. Mechanisms and consequences of alternative polyadenylation. Mol Cell 43, 853–66 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R., Neilson J. R., Sarma A., Sharp P. A. & Burge C. B. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320, 1643–7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C. & Bartel D. P. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138, 673–84 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z., Lee J. Y., Pan Z., Jiang B. & Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci U S A 106, 7028–33 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Wang H., Liu Y. & Shao C. Analysis of gene regulatory networks in the mammalian circadian rhythm. PLoS Comput Biol 4, e1000193 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T. & Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- Al-Fageeh M. B., Marchant R. J., Carden M. J. & Smales C. M. The cold-shock response in cultured mammalian cells: harnessing the response for the improvement of recombinant protein production. Biotechnol Bioeng 93, 829–35 (2006). [DOI] [PubMed] [Google Scholar]
- Kornmann B., Schaad O., Bujard H., Takahashi J. S. & Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol 5, e34 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straume M. DNA microarray time series analysis: automated statistical assessment of circadian rhythms in gene expression patterning. Methods Enzymol 383, 149–66 (2004). [DOI] [PubMed] [Google Scholar]
- Brown S. A., Zumbrunn G., Fleury-Olela F., Preitner N. & Schibler U. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol 12, 1574–83 (2002). [DOI] [PubMed] [Google Scholar]
- Hafner M. et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–41 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva S. et al. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell 43, 340–52 (2011). [DOI] [PubMed] [Google Scholar]
- Mukherjee N. et al. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell 43, 327–39 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita P. A. et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res 39, D876–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L. et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37, W202–8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G. W. et al. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol 16, 130–7 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard K. S., Hubisz M. J., Rosenbloom K. R. & Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 20, 110–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Kulkarni P., Inoue T. & Getzenberg R. H. Down-regulating cold shock protein genes impairs cancer cell survival and enhances chemosensitivity. J Cell Biochem 107, 179–88 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artero-Castro A. et al. Cold-inducible RNA-binding protein bypasses replicative senescence in primary cells through extracellular signal-regulated kinase 1 and 2 activation. Mol Cell Biol 29, 1855–68 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K. et al. Moderate low temperature preserves the stemness of neural stem cells and suppresses apoptosis of the cells via activation of the cold-inducible RNA binding protein. Brain Res 1358, 20–9 (2010). [DOI] [PubMed] [Google Scholar]
- Masuda T. et al. Cold-inducible RNA-binding protein (Cirp) interacts with Dyrk1b/Mirk and promotes proliferation of immature male germ cells in mice. Proc Natl Acad Sci U S A 109, 10885–90 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Y., Yeh I., Park J. Y. & Tian B. PolyA_DB 2: mRNA polyadenylation sites in vertebrate genes. Nucleic Acids Res 35, D165–8 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M. E. et al. Harmonics of circadian gene transcription in mammals. PLoS Genet 5, e1000442 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing L. & Ruoff P. Temperature effect on entrainment, phase shifting, and amplitude of circadian clocks and its molecular bases. Chronobiol Int 19, 807–64 (2002). [DOI] [PubMed] [Google Scholar]
- Diernfellner A. C., Schafmeier T., Merrow M. W. & Brunner M. Molecular mechanism of temperature sensing by the circadian clock of Neurospora crassa. Genes Dev 19, 1968–73 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majercak J., Sidote D., Hardin P. E. & Edery I. How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron 24, 219–30 (1999). [DOI] [PubMed] [Google Scholar]
- Huelga S. C. et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep 1, 167–178 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook K. B., Kazan H., Zuberi K., Morris Q. & Hughes T. R. RBPDB: a database of RNA-binding specificities. Nucleic Acids Res 39, D301–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong H. A., Robles M. S., Knutti D. & Weitz C. J. A molecular mechanism for circadian clock negative feedback. Science 332, 1436–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousel M. R., Stroup W. W. & Nielsen M. K. Locomotor activity, core body temperature, and circadian rhythms in mice selected for high or low heat loss. Journal of Animal Science 79, 861–868 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information
Supplementary Dataset