Here, Proserpio et al. show that the methyltransferase SMYD3 targets myostatin and c-Met genes—two central regulators of myogenesis and muscle atrophy—by recruitment of bromodomain protein BRD4 and the transcription elongation factor p-TEFb. Small molecule inhibition of BRD4 protects cultured myotubes from glucocorticoid-induced atrophy, and SMYD3 inhibition prevents muscle loss in a mouse model of muscle atrophy. This study reveals a mechanistic link between SMYD3/BRD4-dependent transcriptional regulation and skeletal muscle atrophy.
Keywords: SMYD3, BRD4, muscle atrophy, myostatin
Abstract
Elucidating the epigenetic mechanisms underlying muscle mass determination and skeletal muscle wasting holds the potential of identifying molecular pathways that constitute possible drug targets. Here, we report that the methyltransferase SMYD3 modulates myostatin and c-Met transcription in primary skeletal muscle cells and C2C12 myogenic cells. SMYD3 targets the myostatin and c-Met genes and participates in the recruitment of the bromodomain protein BRD4 to their regulatory regions through protein–protein interaction. By recruiting BRD4, SMYD3 favors chromatin engagement of the pause–release factor p-TEFb (positive transcription elongation factor) and elongation of Ser2-phosphorylated RNA polymerase II (PolIISer2P). Reducing SMYD3 decreases myostatin and c-Met transcription, thus protecting from glucocorticoid-induced myotube atrophy. Supporting functional relevance of the SMYD3/BRD4 interaction, BRD4 pharmacological blockade by the small molecule JQ1 prevents dexamethasone-induced myostatin and atrogene up-regulation and spares myotube atrophy. Importantly, in a mouse model of dexamethasone-induced skeletal muscle atrophy, SMYD3 depletion prevents muscle loss and fiber size decrease. These findings reveal a mechanistic link between SMYD3/BRD4-dependent transcriptional regulation, muscle mass determination, and skeletal muscle atrophy and further encourage testing of small molecules targeting specific epigenetic regulators in animal models of muscle wasting.
Establishment and maintenance of skeletal muscle mass is determined by the dynamic balance between anabolic and catabolic pathways that promote increased synthesis of muscle proteins or their degradation, respectively (Glass 2010a). In physiological conditions, extracellular cues mediated by insulin growth factors, myostatin, and cytokines continuously fine-tune the rate of protein synthesis and degradation. Skeletal muscle atrophy is the unfavorable outcome of an imbalance in the two opposing signaling networks and occurs in a wide variety of settings, such as disuse, denervation, cancer cachexia, sarcopenia, and glucocorticoid administration. Signaling molecules such as myostatin, catabolic steroids, and proinflammatory cytokines have been identified as factors contributing to the atrophic process in skeletal muscle (Glass 2010b). However, the molecular mechanisms that control the expression of these key regulators of muscle mass are poorly understood. Cellular pathways activated by these molecules converge on transcription factors that are modified following catabolic/anabolic responses and translate extracellular signals into transcriptional programs. It is now recognized that transcription factors cooperate with chromatin modifiers to establish and maintain transcription networks throughout development, tissue homeostasis, and disease (Portela and Esteller 2010; Braun and Gautel 2011; Greer and Shi 2012). While the contribution of epigenetic modifiers and chromatin remodeling factors to skeletal muscle commitment and differentiation has been well documented (Perdiguero et al. 2009; Albini and Puri 2010; Sartorelli and Juan 2011), their role in skeletal muscle atrophy has only recently begun to be investigated (Moresi et al. 2010) and remains largely unknown. Identifying epigenetic mechanisms underlying skeletal muscle atrophy is of interest because the reversibility and plasticity of epigenetic modifications can be exploited to design novel strategies for therapeutic intervention. Relevant for therapeutic purposes are recent reports on the beneficial effects of blocking the myostatin/activin signaling pathways in a mouse model of cancer cachexia (Benny Klimek et al. 2010; Zhou et al. 2010) and to counter muscle wasting in muscular dystrophies (Bartoli et al. 2007; Qiao et al. 2008; Wagner et al. 2008; Krivickas et al. 2009; Qiao et al. 2009).
Members of the SMYD family of histone methyltransferases are potential regulators of skeletal muscle mass, as they control gene expression in cardiac and skeletal muscle owing to their restricted or more abundant expression in myogenic tissues (Gottlieb et al. 2002; Brown et al. 2006; Thompson and Travers 2008). To date, five members of the SMYD family have been identified: SMYD1–5 (Brown et al. 2006). They share a distinctive architecture of their SET domain, which is split into two parts by a MYND (myeloid translocation protein 8, nervy, DEAF-1) domain followed by a cysteine-rich post-SET domain. The MYND domain is a cysteine-rich structure that mediates protein–protein interactions and DNA binding (Xu et al. 2011) and is present in proteins implicated in embryonic development and cancer progression. SMYD3 has been reported to promote H3K4me3 and, more recently, H4K5me1 (Hamamoto et al. 2004; Van Aller et al. 2012). However, the relevance of SMYD3-dependent epigenetic modifications to gene expression regulation has not been addressed.
Here, we report that SMYD3 regulates expression of myostatin, a member of the TGF-β family and a negative regulator of skeletal muscle growth during embryonic development (Amthor et al. 2006; Manceau et al. 2008) and muscle homeostasis (Trendelenburg et al. 2009). In primary mouse myoblasts and C2C12 myoblasts, numerous studies have provided evidence that myostatin signals via the canonical Smad2/3 pathway and inhibits myogenesis progression through down-regulation of differentiation-related genes such as MyoD and myogenin as well as their downstream targets (Langley et al. 2002; Ríos et al. 2002; Joulia et al. 2003; Yang et al. 2006; Huang et al. 2007). In addition, myostatin plays a distinct role in adult myofibers, where it regulates muscle mass and promotes the activation of the FoxO1/3 pathway, leading to atrogene transcription and muscle mass loss in conditions related to muscle wasting (Glass 2010b; Braun and Gautel 2011). SMYD3 also controls transcription of c-Met, the hepatocyte growth factor receptor (Zou et al. 2009), involved in migration of muscle satellite cells and muscle atrophy (Anastasi et al. 1997; Gal Levi et al. 1998; Crepaldi et al. 2007).
Our findings reveal that SMYD3 favors chromatin recruitment of the bromodomain protein BRD4 and the positive transcription elongation factor complex (p-TEFb), thus facilitating early steps of transcriptional elongation. Consistently, the BRD4 inhibitor JQ1 negatively regulates expression of myostatin and atrophy-related genes and counteracts dexamethasone (Dex)-induced atrophy in cultured myotubes. Importantly, SMYD3 reduction blunted Dex-induced muscle atrophy in the animal. Thus, we identified SMYD3 as an effector of skeletal muscle atrophy and have begun to clarify the molecular mechanisms involved in its proatrophic function.
Results
SMYD3 determines myotube size through regulation of myostatin expression
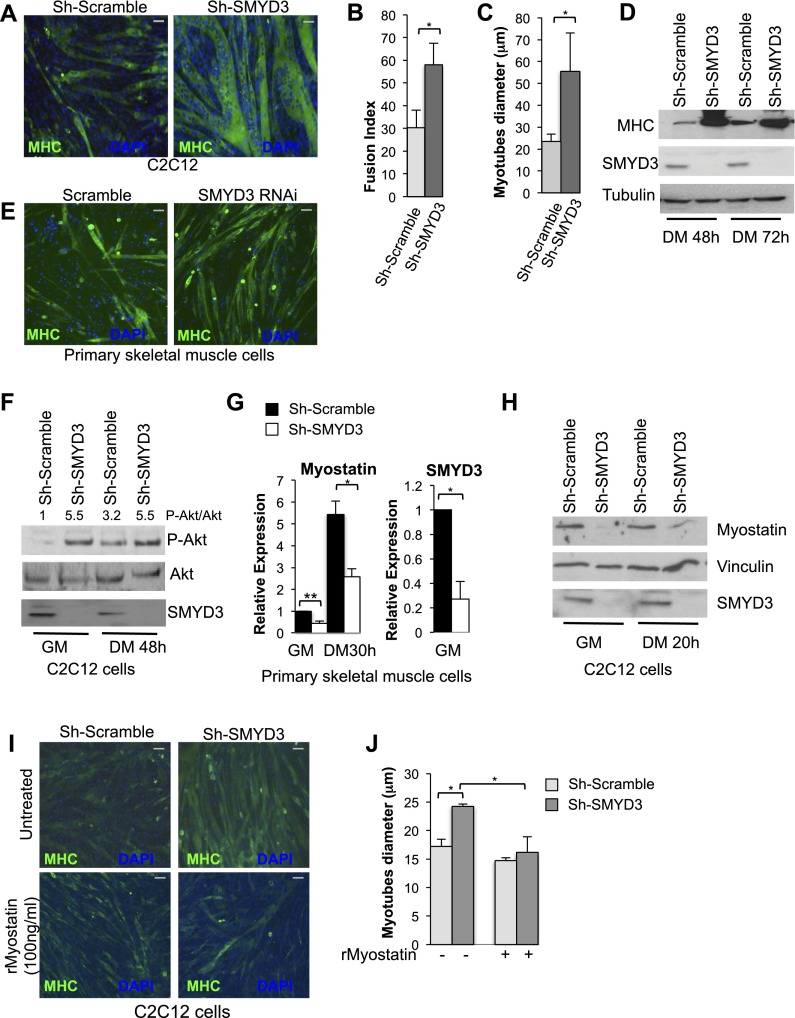
Given its function in regulating zebrafish skeletal myogenesis (Fujii et al. 2011), we investigated the role of SMYD3 in a cell culture model of mammalian myogenesis. SMYD3 expression was reduced in C2C12 cells by transducing them with a specific shRNA. SMYD3-depleted C2C12 myotubes showed a higher percentage of myosin heavy chain (MHC)-positive multinucleated myotubes (Fig. 1A,B,D; Supplemental Fig. S1A–C) and simultaneously revealed an increase in average myotube diameter (P-value 0.043) (Fig. 1C). Analogously, mouse primary skeletal muscle cells transfected with a chemically modified siRNA targeting SMYD3 showed increased ability to form MHC-positive multinucleated myotubes (Fig. 1E). Formation of C2C12 myotubes with increased MHC reactivity and diameter was prevented by retroviral expression of human SMYD3 (hSMYD3), which is refractory to murine shRNA interference (Supplemental Fig. S1D–F), excluding off-target effects of the Sh-SMYD3. Increased fusion index and myotube diameter upon SMYD3 reduction may be the result of myotube hyperplasia, hypertrophy, or a combination of the two processes.
Figure 1.
SMYD3 depletion promotes myotube hypertrophy and affects myostatin expression levels. (A) MHC immunostaining (green) of C2C12 cells transduced with control or Sh-SMYD3 retroviruses and induced to differentiate for 48 h. Nuclei were visualized with DAPI (blue). (B) The fusion index was determined by counting nuclei within myotubes of two or more nuclei and dividing by the number of total nuclei and is expressed as percentage. Error bars represent standard deviation (SD) (n = 3). (*) P-value < 0.05. Bar, 50 μm. (C) Mean myotube diameter of Sh-SMYD3 and Sh-Scramble myotubes was quantified. Error bars represent SD (n = 3). (*) P-value < 0.05. (D) Immunoblot of SMYD3, MHC, and tubulin in C2C12 transduced with control or Sh-SMYD3 retroviruses at different stages of differentiation. (E) Primary skeletal muscle cells were isolated from 18-d-old mice and transfected with a double-strand RNAi oligonucleotide carrying a sequence targeting SMYD3 or its scramble sequence, and, after 24 h, cells were induced to differentiate for 36 h before immunostaining with MHC antibody (green). Nuclei were visualized by DAPI staining (blue). Bar, 100 μm. (F) Protein levels of SMYD3, phosphorylated Akt-Ser473, and total Akt were quantified by Western blot using whole-cell C2C12 extracts transduced with Sh-SMYD3 and Sh-Scramble retroviruses. The ratio of phospho-Akt/total Akt is provided after quantification of band intensity. (G) Primary skeletal muscle cells were isolated from 18-d-old mice and transfected with SMYD3-RNAi or scramble oligonucleotides. RNA was extracted from cells cultured in growth condition and induced to differentiate for 30 h, and expression levels of myostatin and SMYD3 were measured by quantitative RT–PCR (qRT–PCR). Relative expression was normalized against GAPDH. Error bars represent SD (n = 5). (**) P-value < 0.001; (*) P-value < 0.05. (H) Immunoblot of myostatin and SMYD3 in whole-cell extract of C2C12 cells transduced with Sh-SMYD3 and control retroviruses, cultured in growth medium, and induced to differentiate for 20 h. Vinculin served as a loading control. (I) C2C12 cells were transduced with Sh-SMYD3 or Sh-Scramble retroviruses and allowed to differentiate for 48 h in the presence of recombinant myostatin (100 ng/mL). Cells were fixed with 4% paraformaldehyde and immunostained with antibodies against MHC (green). Nuclei were visualized by DAPI staining (blue). Bar 50 μm. (J) Mean myotube diameter of Sh-SMYD3 and Sh-Scramble myotubes treated with recombinant myostatin or vehicle, as in I. Error bars represent SD (n = 3). (*) P-value < 0.05
With the aim of defining the molecular pathways leading to increased myotube size in Sh-SMYD3 cells, we chose to investigate the impact of SMYD3 depletion on Akt/protein kinase B (Akt). This choice was motivated by observations indicating that Akt activation represents a critical event during muscle hypertrophy (Bodine et al. 2001) and that constitutive Akt activation in adult skeletal muscle is sufficient to induce muscle hypertrophy (Lai et al. 2004). While total Akt levels were not appreciably affected, Ser473-phosphorylated (activated) Akt was increased in both Sh-SMYD3 C2C12 myoblasts and myotubes (Fig. 1F). Several lines of evidence have linked the Akt pathway to myostatin, a crucial negative regulator of muscle mass in vertebrates. Hypertrophic skeletal muscles of myostatin knockout mice display higher activation of Akt signaling (Morissette et al. 2009; Lipina et al. 2010), and in vitro studies have documented negative modulation of the Akt signaling pathway upon myostatin delivery (McFarlane et al. 2006; Morissette et al. 2009; Trendelenburg et al. 2009). Based on Akt activation and the observation that the Sh-SMYD3 myotube phenotype is reminiscent of that caused by myostatin knockdown (Morissette et al. 2009; Trendelenburg et al. 2009), we evaluated whether SMYD3 depletion may affect myostatin transcription. As shown in Figure 1G, myostatin transcripts were significantly reduced in mouse primary skeletal muscle cells depleted for SMYD3 in both proliferating and differentiating conditions (Fig. 1G). Similarly, myostatin protein and mRNA levels were substantially diminished in Sh-SMYD3 C2C12 myoblasts and myotubes (Fig. 1H; Supplemental Fig. S1I). Culturing Sh-SMYD3 C2C12 cells in the presence of recombinant myostatin (at final concentration of 100 ng/mL) reduced MHC expression and diameter, thus rendering them comparable in size with the control (Fig. 1I,J). Taken together, these data suggest that SMYD3 regulates myostatin mRNA and protein levels in both C2C12 and primary skeletal muscle cells and controls the size of C2C12 cells in a myostatin-dependent fashion by influencing either hypertrophy or hyperplasia.
SMYD3 is recruited to regulatory regions of the myostatin and c-Met genes and favors engagement of Ser2-phosphorylated RNA polymerase II (PolII)
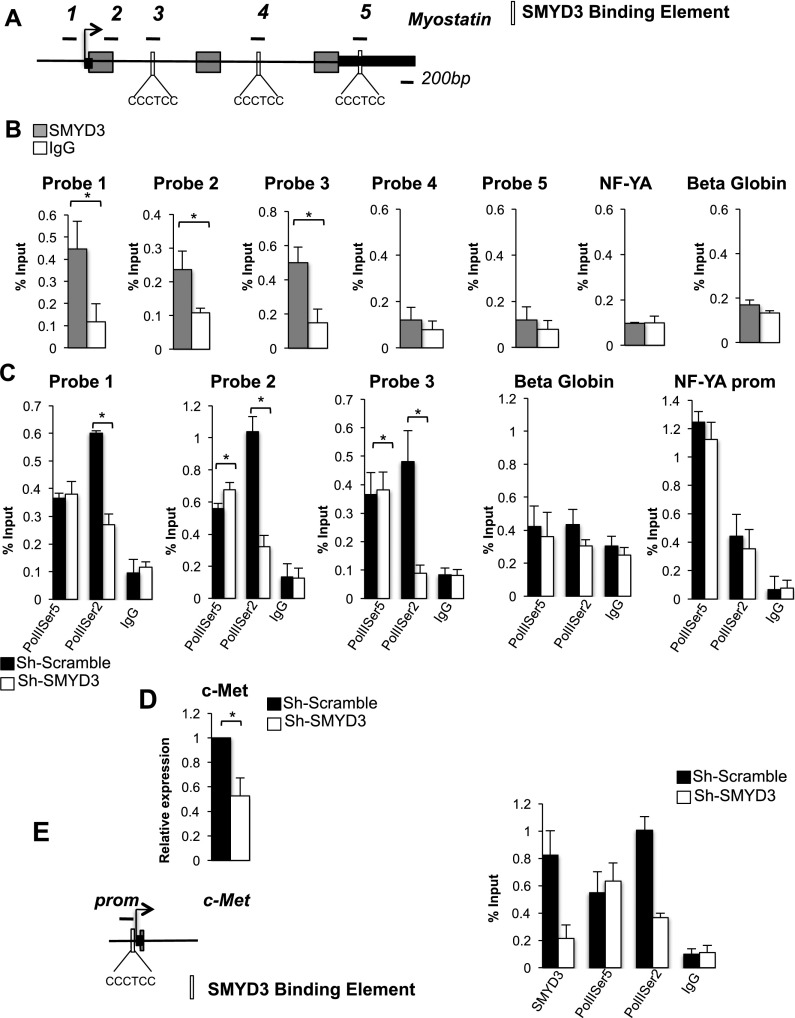
Inspection of the myostatin gene sequence revealed the presence of two putative SMYD3-binding motifs within the first and second intron and of a third motif positioned within the 3′ untranslated region (UTR) (Fig. 2A). Employing chromatin immunoprecipitation (ChIP) assays, we could not detect significant recruitment of SMYD3 to the consensus sites situated within either the second intron (probe 4) or the 3′ UTR of the myostatin gene (probe 5) (Fig. 2B). However, SMYD3 enrichment was observed at a region encompassing an evolutionarily conserved SMYD3 consensus site in the first intron (probe 3). In addition, SMYD3 binding was detected at the myostatin promoter (probe 1), which, in contrast, does not contain canonical SMYD3 consensus motifs. A chromatin region located halfway between the myostatin promoter and the first intron was also enriched for SMYD3 binding (probe 2). These findings are consistent with a direct and indirect modality of SMYD3 chromatin recruitment (Kim et al. 2009). To further characterize the SMYD3-bound myostatin regions, we employed p300 and histone H3K4me1 antibodies in ChIP to identify potential enhancer regions (Heintzman et al. 2007). The myostatin first intron (probe 3) was enriched for both p300 and H3K4me1, whereas the 3′ UTR (probe 5) was not significantly enriched for either mark (Supplemental Fig. S2A,B). As expected, the SMYD3-bound promoter region (probe 1) was occupied by p300 but was not significantly enriched for H3K4me1 (Supplemental Fig. S2A,B). These results indicate that the first intron of myostatin may host an active enhancer (Creyghton. et al. 2010; Rada-Iglesias et al. 2011). The engagement of SMYD3 at regulatory regions and the reduced myostatin transcription upon Sh-SMYD3 interference prompted us to evaluate potential effects of SMYD3 on RNA PolII recruitment. PolII phosphorylation on the Ser5 C-terminal domain (CTD) is a highlight of transcriptional initiation, while PolII phosphorylation of Ser2 is a signature of transcription elongation (Brookes and Pombo 2009). By ChIP, Ser5-phosphorylated RNA PolII (PolIISer5P) recruitment was moderately increased at the myostatin promoter and regions encompassing probes 2 and 3 in SMYD3-depleted cells compared with control cells. In contrast, PolIISer2P engagement was decreased to background levels when SMYD3 was knocked down. PolIISer2 and PolIISer5 were unchanged at control active (NF-YA) and silent (β-globin) promoters (Fig. 2C). To extend our findings to other SMYD3 targets, we chose to analyze c-Met, whose transcription is regulated by SMYD3 (Zou et al. 2009). c-Met plays a role in embryonic myogenesis and skeletal muscle regeneration (Anastasi et al. 1997; Gal Levi et al. 1998), and similar to myostatin, conditional activation of c-Met in skeletal muscle induces atrophy (Crepaldi et al. 2007). In both C2C12 myoblasts (Fig. 2D) and primary skeletal muscle cells (Supplemental Fig. S2C), c-Met transcripts decreased when SMYD3 was reduced. Previous studies have implicated the c-Met promoter in SMYD3-mediated regulation of the c-Met gene (Zou et al. 2009). Inspection of the c-Met promoter sequence revealed the presence of a SMYD3-binding site. Consistently, ChIP experiments documented SMYD3 enrichment at this region (Fig. 2E). Chromatin recruitment of both SMYD3 and PolIISer2P were reduced following SMYD3 depletion, while PolIISer5P was unaltered (Fig. 2E). Collectively, these data suggest that SMYD3 does not affect assembly of the RNA PolII preinitiation complex but is rather involved in the chromatin recruitment of elongating PolIISer2P at both the myostatin and c-Met genes.
Figure 2.
SMYD3 is recruited at regulatory regions of the myostatin and c-Met genes, and its depletion impacts chromatin engagement of RNA PolIISer2P. (A) Schematic representation of myostatin gene illustrating exons (gray rectangles), introns (black lines between exons), 5′ and 3′ UTRs (black rectangles), and putative SMYD3-binding sequences (white rectangles). Probes used in the ChIP assays are indicated above the gene scheme. (B) ChIP assay was performed with chromatin from C2C12 myoblasts using normal rabbit IgG and antibodies recognizing SMYD3. n = 4; (*) P-value < 0.05. (C) ChIP assay was performed with normal rabbit IgG and antibodies recognizing PolIISer5P and PolIISer2P on chromatin derived from control and Sh-SMYD3 proliferating C2C12 myoblasts. NF-YA and β-globin promoter regions were amplified as control genes not affected by SMYD3 depletion. n = 3; (*) P-value < 0.05. (D) c-Met expression levels were measured by qRT–PCR in Sh-Scramble and Sh-SMYD3 myoblast cells. Relative expression was normalized against GAPDH. Error bars represent SD (n = 3). (*) P-value < 0.05. (E) Schematic representation of the c-Met promoter gene. A putative SMYD3-binding site is depicted (white rectangle). ChIP DNA as in C was used to amplify regions within the c-Met promoter.
SMYD3 regulates myostatin transcription by favoring engagement of the bromodomain protein BRD4 and the p-TEFb-CDK9 subunit
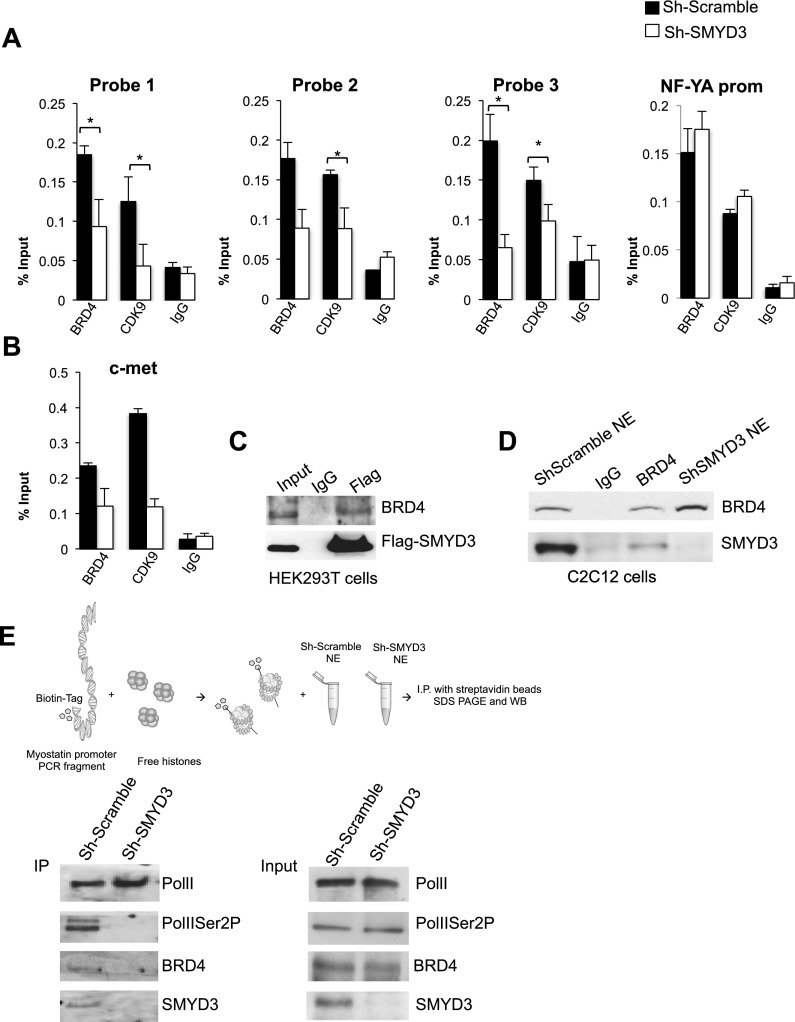
The p-TEFb complex (CycT1/CDK9) mediates PolIISer2 phosphorylation during the early elongation steps and can be recruited to promoter regions and the gene body by the bromodomain protein BRD4 (Brès et al. 2008). We therefore asked whether recruitment of BRD4 and p-TEFb was itself affected by SMYD3. Antibodies against the p-TEFb subunit CDK9 and BRD4 were employed in ChIP, which revealed that recruitment of both CDK9 and BRD4 was impaired upon SMYD3 depletion at the promoter and first intron of the myostatin gene (Fig. 3A) as well as the promoter of the c-Met gene (Fig. 3B). CDK9 and BRD4 proteins were not affected by SMYD3 reduction (Supplemental Fig. S3A). To evaluate whether SMYD3 may associate with BRD4, extracts from HEK293T cells transiently transfected with Flag-SMYD3 plasmid were immunoprecipitated with anti-Flag-agarose beads, and the precipitated material was immunoblotted with an antibody recognizing BRD4. The outcome of this experiment indicated that endogenous BRD4 interacted with ectopically expressed SMYD3 (Fig. 3C). As a control, we showed that the unrelated Flag-tagged Ezh2 protein was not able to associate with BRD4 (Supplemental Fig. S3B). In addition, endogenous SMYD3 associated with BRD4 in C2C12 whole-cell extracts (Fig. 3D). In immunoprecipitation experiments performed with HEK293T extracts overexpressing Flag-SMYD3, we also uncovered an interaction between SMYD3 and both PolIISer5P and PolIISer2P (Supplemental Fig. S3C,D). These results not only confirmed a previous report describing an interaction between SMYD3 and RNA PolII (Hamamoto et al. 2004), but also revealed an association between SMYD3 and PolII isoforms that participate in the early steps of transcriptional elongation.
Figure 3.
SMYD3 interacts with BRD4 and recruits BRD4 and pTEFb to the myostatin regulatory regions. (A) ChIP assay was performed with normal rabbit IgG and antibodies recognizing BRD4 and CDK9 on chromatin derived from control and Sh-SMYD3 proliferating C2C12 myoblasts. The location of the myostatin probe sets is indicated in Figure 2A. The NF-YA promoter region was amplified as a control gene. n = 3; (*) P < 0.05. (B) ChIP, performed as in A, was analyzed for the c-Met promoter. (C) Protein complexes were immunoprecipitated from HEK293T cells overexpressing Flag-SMYD3 with an antibody recognizing either IgG or Flag. The immunoblot was detected with antibodies recognizing BRD4 and the Flag epitope. (D) Immunoprecipitation assay of nuclear extracts (NEs) obtained from Sh-Scramble cells. Protein complexes were immunoprecipitated using either IgG or an antibody against BRD4. Sh-SMYD3 NEs were loaded after immunoprecipitated samples. The Western blot was detected with antibodies against BRD4 and SMYD3. (E) In vitro reconstituted nucleosomes assembled at the myostatin promoter were incubated with nuclear extracts derived from C2C12 infected with either Sh-Scramble or Sh-SMYD3 retroviral supernatants. Biotinylated nucleosomes were captured with streptavidin, and interacting proteins were analyzed by immunoblot with the indicated antibodies.
Collectively, our data suggest that SMYD3 recruitment to the myostatin and c-Met regulatory regions is crucial for BRD4 engagement and subsequent association of the pTEFb complex. To further support these results, we investigated SMYD3 and BRD4 recruitment to an in vitro reconstituted nucleosome template encompassing the myostatin promoter. We first in silico analyzed the most probable nucleosome positioning at the myostatin promoter (Xi et al. 2010) and experimentally confirmed that the selected DNA fragment containing the myostatin promoter was able to assemble a nucleosome template by salt dialysis using recombinant histones (see the Materials and Methods) (Caretti et al. 1999; data not shown). We next tested the ability of the reconstituted biotinylated nucleosomes to recruit SMYD3, BRD4, and PolII using nuclear extracts derived from C2C12 myoblasts depleted of SMYD3 and control cells (Fig. 3E). After incubation with nuclear extracts, nucleosomes were immobilized on streptavidin beads, extensively washed, and analyzed by immunoblot. Notably, BRD4 and PolIISer2P were recruited to the nucleosomes only when SMYD3 was present in the extracts. Conversely, total PolII was recruited at levels comparable with nucleosomes incubated with protein extracts derived from either Sh-SMYD3 or Sh-Scramble C2C12 cells (Fig. 3E). Therefore, we were able to recapitulate the interplay between SMYD3 and BRD4 at the myostatin promoter in an in vitro assembled nucleosome system. Taken together, these results indicate that SMYD3 association with the myostatin regulatory regions is a critical step for BRD4 recruitment, implying a role of SMYD3 in the transition from transcription initiation to early elongation.
The N-terminal regions of BRD4 and SMYD3 mediate protein interaction
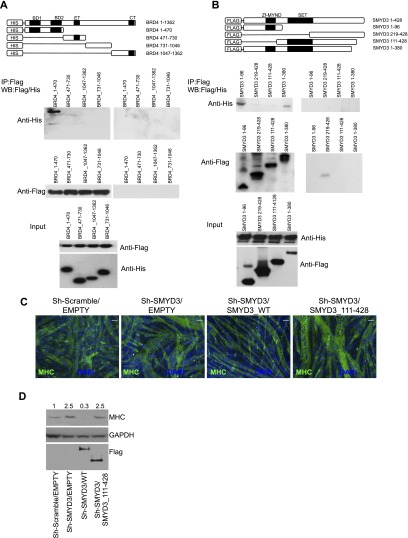
To map the surfaces mediating SMYD3 and BRD4 interaction, we tested the ability of four truncated forms of BRD4 to interact with full-length SMYD3 in coimmunoprecipitation assays. We found that a truncated version of His-tagged BRD4 solely encompassing the two tandem N-terminal bromodomains BD1 and BD2 (BRD4_1–470) maintained the ability to pull down Flag-SMYD3, while the remaining BRD4 regions failed to do so (Fig. 4A, anti-His, middle panel). We also generated four Flag-tagged SMYD3 deletion constructs and tested their capability to associate with His-BRD4 (Fig. 4B). The region encompassing the first 96 amino acids of SMYD3 mediated the interaction with BRD4. Conversely, the N-terminal SMYD3 mutants (SMYD3_111–428 and SMYD3_219–428) did not associate with BRD4 (Fig. 4B, anti-His, middle panel). Interestingly, the N-terminal region of SMYD3 encompasses the MYND domain, which is a protein–protein interaction module (Sirinupong et al. 2010).
Figure 4.
The N-terminal regions of BRD4 and SMYD3 mediate protein interaction. (A, top panel) Schematic diagram of BRD4 constructs. Protein complexes were immunoprecipitated with M2-Flag beads or IgG from HEK293T cells cotransfected with wild-type Flag-SMYD3 and His-tagged BRD4 deletion mutants. Inputs are shown in the bottom panel. Immunoblotting was performed with HRP-conjugated antibodies recognizing the His and the Flag epitope for BRD4 and SMYD3, respectively. (B, top panel) Schematic diagram of SMYD3 constructs. Total extracts of HEK293 overexpressing the full-length His-BRD4 and SMYD3 truncated mutants were immunoprecipitated with M2-Flag beads or IgG, and the immunoblots were detected using anti-His and anti-Flag antibodies for BRD4 and SMYD3, respectively. Inputs are shown in the bottom panel. (C) MHC immunostaining (green) of Sh-Scramble and Sh-SMYD3 C2C12 cells transduced with empty vector or SMYD3 and SMYD3_111-428 retroviruses induced to differentiate for 48 h. Nuclei were visualized with DAPI (blue). Bar, 50 μm. (D) Immunoblot of MHC, Flag, and GAPDH of Sh-Scramble and Sh-SMYD3 C2C12 cells transduced with empty vector or SMYD3 and SMYD3_111-428 retroviruses induced to differentiate for 48 h. Numbers represent quantification of MHC band intensity.
To investigate whether the SMYD3 N-terminal region is relevant for SMYD3 function, we performed rescue experiments in Sh-SMYD3 and control cells (Supplemental Fig. S4A) with a retrovirus carrying either full-length or a SMYD3 mutant incapable of interacting with BRD4 (SMYD3_111–428). While SMYD3 wild type could rescue the phenotype of Sh-SMYD3 cells, the SMYD3_111–428 mutant (containing the methylase SET domain) did not revert Sh-SMYD3 cell hypertrophy (Fig. 4C,D; Supplemental Fig. S4B–D). These results suggest that the sole SET methyltransferase domain of SMYD3 is not sufficient to induce the atrophic response. We conclude that the N-terminal region of SMYD3 mediates BRD4 interaction and is crucial in regulating myotube size.
SMYD3 expression is increased in Dex-induced atrophy of C2C12 myotubes
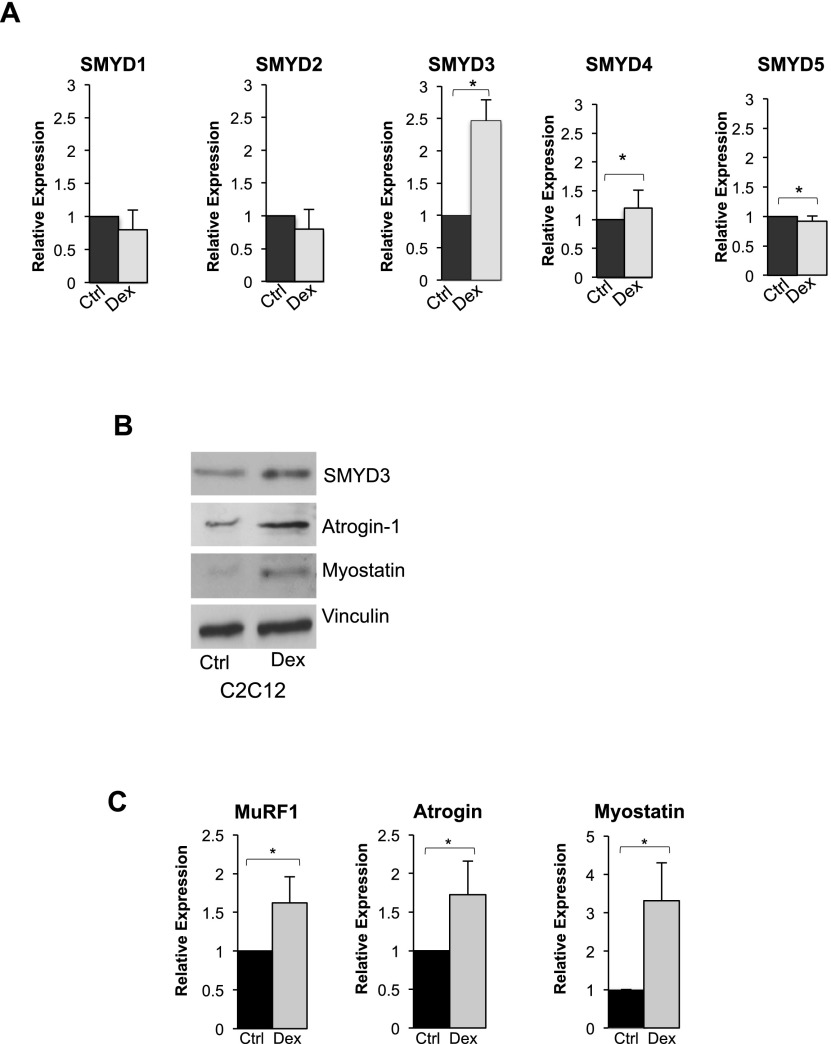
The experiments presented above indicate a role of SMYD3 in regulating skeletal muscle cell differentiation and myotube size. To directly test whether SMYD3 may be involved in skeletal muscle atrophy, we employed a cell culture model where differentiated C2C12 myotubes are treated with the synthetic glucocorticoid Dex, leading to phenotypic reduction of myotube size (Clarke et al. 2007). C2C12 cells were allowed to form myotubes and were then treated with Dex (100 μM) for 24 h. Transcript levels of SMYD3 increased following Dex treatment, while mRNA levels of other SMYD family members were unaffected except for a modest SMYD4 increase (Fig. 5A). SMYD3 protein levels were also increased by Dex (Fig. 5B). As expected, expression of myostatin and the atrogenes atrogin-1 (Fig. 5B,C) and MuRF1 was induced by Dex treatment (Fig. 5C).
Figure 5.
Dex induces SMYD3 up-regulation in C2C12 myotubes. (A) SMYD family member expression was analyzed by qRT–PCR in C2C12 myotubes treated with 100 μM Dex for 24 h. Data are normalized against GAPDH (n = 4). Error bars represent SD. (*) P-value < 0.05. (B) SMYD3, atrogin-1, and myostatin protein levels were analyzed by Western blot in C2C12 myotubes treated as in A. Vinculin served as a loading control. (C) MuRF1, atrogin, and myostatin transcript levels were quantified by qRT–PCR, and relative expression was normalized against GAPDH. Error bars represent SD (n = 3). (*) P < 0.05.
SMYD3 inhibition or BRD4 pharmacological blockade prevents myostatin and atrogene induction and counteracts Dex-induced atrophy in cultured myotubes
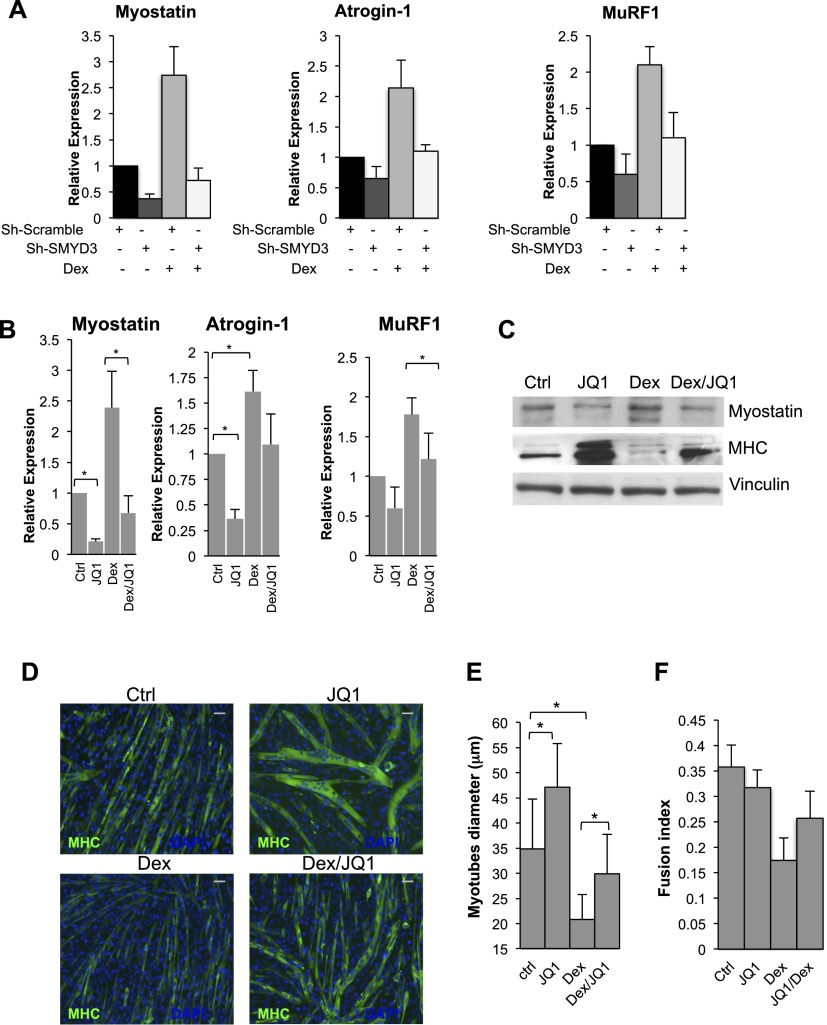
The critical function exerted by myostatin in skeletal muscle atrophy (Ma et al. 2003; Glass 2010a) further prompted us to evaluate SMYD3's role in a setting of skeletal muscle atrophy. As previously described (Ma et al. 2001; McFarlane et al. 2006) and as reported in Figure 5, myostatin levels were induced by Dex treatment, and this phenomenon was accompanied by up-regulation of the atrophy-related genes atrogin-1 and MuRF1. However, the transcript levels of myostatin, atrogin-1, and MuRF1 failed to increase in C2C12 cells depleted of SMYD3 following Dex treatment (Fig. 6A). These data suggested that SMYD3 is involved in the transcriptional up-regulation of myostatin and atrophy-related genes in Dex-induced skeletal muscle atrophy.
Figure 6.
SMYD3 reduction or the bromodomain inhibitor JQ1 tempers Dex-induced atrophy in C2C12 myotubes. (A) C2C12 myotubes from Sh-Scramble and Sh-SMYD3 C2C12 cells were treated with Dex (100 μM) for 24 h, and mRNA levels of myostatin, atrogin-1, and MuRF1 were measured by qRT–PCR. Relative expression was normalized against GAPDH. Error bars represent SD (n = 3). (B) C2C12 myoblasts were exposed to the bromodomain inhibitor JQ1 (50 nM) or DMSO for 12 h, allowed to differentiate for 24 h in the presence of JQ1, and then treated with Dex (100 μM) or DMSO for 24 h before qRT–PCR analysis of transcript levels of myostatin, atrogin-1, and muRF1. Relative expression was normalized against GAPDH. Error bars represent SD (n = 3). (*) P-value < 0.05. (C) Protein levels of myostatin, MHC, and vinculin were analyzed in C2C12 myotube extracts, cultured, and treated as in B. (D) C2C12 myoblasts treated as in B were immunostained with MHC antibody (green). Nuclei were visualized by DAPI staining (blue). Bar, 50 μm. (E) Mean myotube diameter of Sh-SMYD3 and Sh-Scramble myotubes was quantified. Error bars represent SD (n = 3). (*)P-value < 0.05. (F) Fusion index of Sh-SMYD3 and Sh-Scramble myotubes. Error bars represent SD (n = 3).
To evaluate whether the role of BRD4 in regulating myostatin transcription described in Figure 3 is functionally relevant also for myostatin expression during muscle atrophy, we employed the recently developed small molecule JQ1. JQ1 is a selective inhibitor of bromodomain and extraterminal (BET) family members; it occupies the acetyl-binding pockets of bromodomains and determines BRD4 dissociation from chromatin and loss of downstream signaling events to PolII (Delmore et al. 2011). We reasoned that JQ1 could interfere with myostatin transcription and hence may counteract skeletal muscle atrophy induced by Dex. Indeed, C2C12 myotubes treated with Dex and exposed to JQ1 (50 nM) showed decreased levels of myostatin, MuRF1, and atrogin-1 and increased MHC expression (Fig. 6B,C). Myoblasts cultured in the presence of JQ1 and prompted to differentiate gave rise to myotubes with larger diameter and increased levels of MHC when compared with control cells (Fig. 6C–F) and were resistant to the Dex-induced reduction of cell diameter (Fig. 6D,E). The BRD4 target c-myc has been previously shown to be down-regulated by JQ1 administration in several cellular models and served as an internal control (Supplemental Fig. S5A). These in vitro results indicate that pharmacological BRD4 inhibition prevents transcription of myostatin and atrophy-related genes, thus effectively counteracting Dex-induced myotube atrophy.
SMYD3 depletion prevents Dex-induced in vivo skeletal muscle atrophy
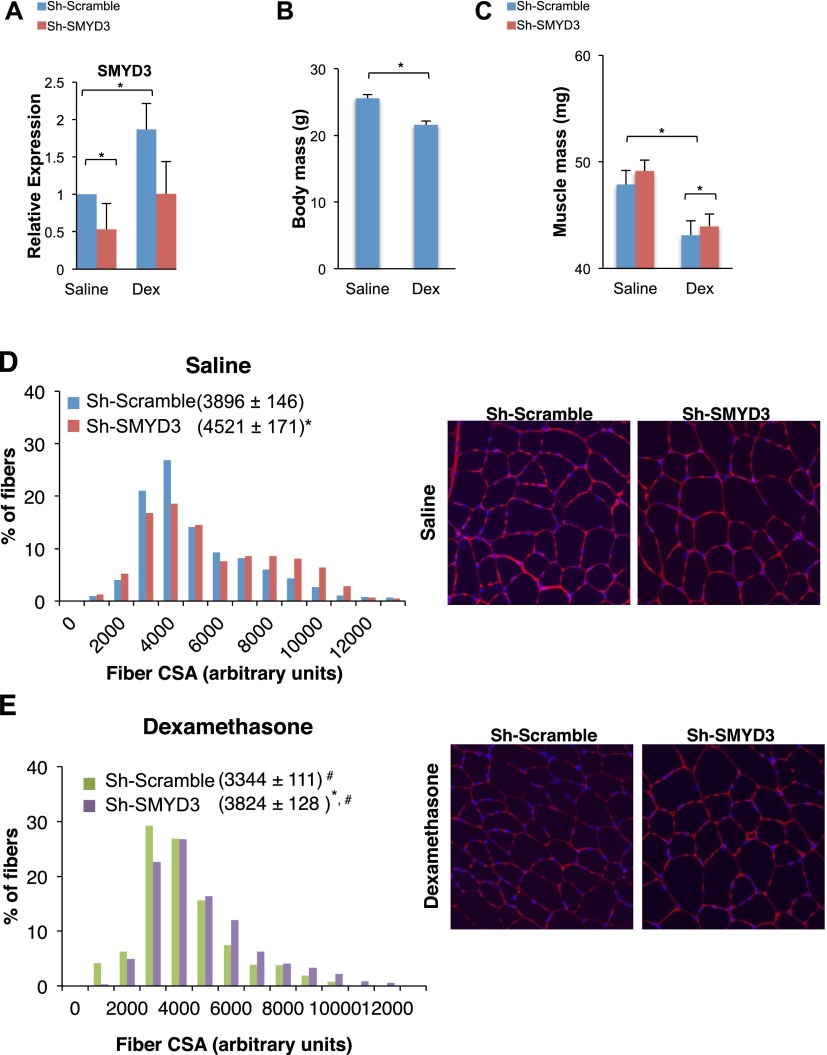
To determine whether SMYD3 is implicated in muscle atrophy in vivo, we induced skeletal muscle atrophy by Dex administration in C57BL/6 mice (n = 8 saline, n = 9 Dex) and reduced SMYD3 levels by electroporation with a plasmid carrying a shRNA targeting SMYD3 in the tibialis anterior (TA). A scrambled sequence was injected in the contralateral TA muscle to serve as a control. In agreement with SMYD3's expression profile in C2C12 myotubes treated with Dex (Fig. 5A), endogenous SMYD3 mRNA levels were also increased by Dex exposure in TAs (Fig. 7A), while SMYD1, SMYD2, SMYD4, and SMYD5 transcripts were unaltered or negatively affected by Dex administration (Supplemental Fig. S6). As expected, SMYD3 mRNA levels were reduced in muscle electroporated with Sh-SMYD3 (Fig. 7A), and Dex delivery reduced mouse body mass by 14.8% (Fig. 7B). Notably, we found that SMYD3 knockdown increased the weight of TA muscles following either saline or Dex administration, and muscle mass sparing was significant only in Dex-treated mice (P = 0.047) (Fig. 7C). These results suggest that SMYD3 plays a role in muscle mass control in the animal. Next, we stained cross-sections of TA muscles with an antibody recognizing laminin and measured cross-sectional areas in the four animal groups (∼200 fibers per muscle). As shown in Figure 7, D and E, median fiber size was higher in fibers electroporated with the Sh-SMYD3 plasmid than in fibers expressing the control scramble shRNA in both control (4521 ± 171 vs. 3896 ± 146 arbitrary units) and Dex-treated (3824 ± 128 vs. 3344 ± 111 arbitrary units) mice. Furthermore, fibers of bigger size were prevalent in TA muscles depleted of SMYD3 in either saline- or Dex-treated animals. The myofiber size of TA muscles depleted of SMYD3 and treated with Dex (3824 ± 128) was comparable with that of control myofibers treated with saline solution (3896 ± 146), thus suggesting that SMYD3 depletion prevents myofiber size reduction triggered by Dex treatment (Fig. 7D,E). These results are consistent with a pivotal role played by SMYD3 in regulating myofiber size in the adult skeletal muscle in vivo and indicate that SMYD3 knockdown prevents myofiber size reduction induced by the atrophic agent Dex.
Figure 7.
SMYD3 depletion prevents Dex-induced muscle atrophy in vivo. (A) Total RNA was extracted from fixed TA muscles, and SMYD3 transcript levels were measured by qRT–PCR. Relative expression was normalized against GAPDH. Error bars represent SD (n = 4). (*) P-value < 0.05. (B) Body mass of saline- and Dex-treated mice (n Sal = 8, n Dex = 9). (C) Muscle mass of control and Sh-SMYD3 TA electroporated muscles from either saline- or Dex-treated mice. (D) TA muscle fiber cross-sectional area (CSA) from control and Sh-SMYD3 electroporated muscles from saline-treated mice (values are median ± 95% CI). (E) TA muscle fiber CSA from control and Sh-SMYD3 electroporated muscles from Dex-treated mice (values are median ± 95% CI). (*) P < 0.05 versus relative control group; (#) P < 0.05 versus saline group.
Discussion
Our findings reveal a previously unappreciated role of SMYD3 in regulating skeletal muscle mass maintenance and glucocorticoid-induced skeletal muscle atrophy. SMYD3 function in cell culture and the animal is explained at least in part by its transcriptional regulation of myostatin, a critical negative regulator of cell differentiation and muscle mass (Lee 2004).
Myostatin plays a prominent role in muscle mass maintenance and several conditions related to muscle wasting, such as glucocorticoid-induced skeletal muscle atrophy. Glucocorticoids have broad-spectrum immunosuppressive and anti-inflammatory effects and are thought to be important regulators of muscle mass in catabolic diseases (Hanaoka et al. 2012). Nevertheless, extended oversecretion or exogenous administration of glucocorticoids has catabolic effects on skeletal muscle, which are ascribed to myostatin up-regulation (Ma et al. 2003; Hanaoka et al. 2012). In addition to its role in atrophy, myostatin up-regulation is a hallmark of different forms of muscle wasting, such as sarcopenia (McKay et al. 2012), cancer cachexia (Fearon et al. 2012; Lokireddy et al. 2012), and muscle disuse (Reardon et al. 2001). Strategies aiming at muscle size increase and strength through myostatin blockade hold promise for application in pathological conditions associated with progressive and irreversible muscle loss, such as cancer and muscular dystrophies (Kemaladewi et al. 2011; Pistilli et al. 2011). In addition to its role in regulating muscle mass, myostatin is expressed in developing somites and modulates muscle size during embryonic development by shifting precursor skeletal muscle cell fate toward myogenic differentiation instead of cell proliferation (Manceau et al. 2008). Furthermore, myostatin is also expressed in quiescent satellite cells (McCroskery et al. 2003).
Our experiments provide evidence for a role of SMYD3 in regulating processes involved in transcriptional elongation of the myostatin and c-Met genes in skeletal muscle cells. By recruiting a complex containing the acetyl-lysine recognition domain bromodomain BRD4 protein and pause–release factor pTEFb, SMYD3 promotes PolIISer2P chromatin engagement. Control of transcriptional elongation is emerging as a major means of gene regulation in development and tissue homeostasis (Zhou et al. 2012). Whereas a plethora of transcription factors promotes the assembly of a preinitiation complex (Juven-Gershon and Kadonaga 2010), growing evidence indicates that a subset of transcriptional activators, such as c-Myc (Rahl et al. 2010) and NF-κB (Barboric et al. 2001), promotes early elongation steps (Levine 2011). The availability of selective small molecules targeting proteins implicated in regulating transcriptional initiation and elongation makes this process amenable to pharmacological intervention (Delmore et al. 2011). We successfully employed the bromodomain inhibitor JQ1 in proof-of-concept experiments to reduce glucocorticoid-induced myostatin and atrophy-related gene expression and prevent myotube atrophy. We cannot exclude that other BET proteins might be affected, even though, at the molarity employed here (50 nM), JQ1 is expected to preferentially inhibit BRD3 and BRD4 (Filippakopoulos et al. 2010). These results lend further support to, and expand the potential application of, muscle atrophy and wasting of bromodomain inhibitors as therapeutic agents (Delmore et al. 2011; Zuber et al. 2011; Matzuk et al. 2012). SMYD3's abnormal overexpression in certain tumors (Hamamoto et al. 2004; Van Aller et al. 2012) has been linked to uncontrolled expression of genes that promote cancer progression and invasion. Interestingly, the recent reports that BRD4 plays a pivotal role in acute myeloid leukemia maintenance and multiple myeloma (Delmore et al. 2011; Zuber et al. 2011) raise the possibility that SMYD3 may collaborate with BRD4 in regulating growth-promoting genes by dynamics similar to the ones we describe here. BRD4 is a bromodomain protein that regulates a broad range of cellular processes and is ubiquitously expressed (Belkina and Denis 2012). Conversely, SMYD3 appears to be more abundantly expressed in skeletal muscle compared with other tissues (Hamamoto et al. 2004). The preferential tissue distribution of SMYD3 renders it an attractive target for the development of epigenetic drugs with increased specificity for the treatment of muscle diseases.
Materials and methods
Plasmids construction
Plasmids were constructed as detailed in the Supplemental Material.
Cell cultures and satellite cell isolation
C2C12 cells and HEK293T cells were obtained from American Type Culture Collection. Mouse C2C12 myoblasts were cultured in DMEM without Na-Pyruvate (Invitrogen) supplemented with 20% fetal bovine serum (FBS), 2 mM L-glutamine, 100 U of penicillin, and 0.1 mg/mL streptomycin at 37°C and 5% CO2 and induced to differentiate by switching the medium to DM medium, which is DMEM (Invitrogen) supplemented with 2% horse serum (Invitrogen), 1× insulin, transferrin, and selenium (Invitrogen).
For myostatin treatment, C2C12 was induced to differentiate in DM supplemented with recombinant myostatin (R&D Systems) at a final concentration of 100 ng/mL. C2C12 was allowed to differentiate in DM for 36 h, and then forming myotubes were treated with 100 μM Dex (Sigma-Aldrich) for 24 h before RNA extraction. C2C12 was treated with 50 nM JQ1 (Cayman Chemical) for 12 h and allowed to differentiate, maintaining JQ1 in differentiation medium. After 24 h in differentiation medium, forming myotubes were treated with 100 μM Dex or vehicle for an additional 24 h.
Satellite cells were isolated from muscles of 18-d-old mice as describe in Joe et al. (2010) with minor modifications. Isolated cells were resuspended in growth medium composed of F-10 HAM (Sigma), 20% FBS, 1% chicken embryo extract (SeraLab), 2.5 ng/mL human fibroblast growth factor (Cell Signaling), 2 mM L-glutamine, 100 U of penicillin, and 0.1 mg/mL streptomycin. Cells were preplated overnight to eliminate contaminating fibroblasts, and the nonadherent cells were plated in growth medium on Matrigel-coated plates (ECM gel, Sigma). After 2–3 d of culture, cells were used for experiments.
siRNA transfection and retroviral transduction
Retroviral transduction and primary skeletal muscle stem cell transfections were performed as described (see the Supplemental Material; Caretti et al. 2004).
TA electroporation and Dex treatment
All experiments were performed in accordance with the Animal Care and Use regulations of the National Institutes of Health. Seventeen male C57BL6 mice (8–10 wk of age) were anaesthetized with 1.5%––2.5% isoflurane. Two hours prior to electroporation, hyaluronidase (HA; 30 μL of 0.5 U/μL) was administered via intramuscular (i.m.) injection into the TA muscle of the left and right hindlimbs. Immediately prior to electroporation, a small (<5-mm) incision was made, and the TA muscle was surgically exposed. The left TA muscle was injected with vector alone (40 μL of 2 μg/μL i.m.), while the right TA received the Smyd3 shRNA plasmid (40 μL of 2 μg/μL i.m.). Electroporation was conducted as previously described (Schertzer and Lynch 2008). Briefly, electric pulses were applied by two stainless steel pin electrodes (each with an area of 27 mm2) placed on either side of the muscle. Square-wave electric pulses (three pulses of 20-msec duration) were delivered by an ECM 830 electroporation unit (BTX; Harvard) at a field strength of 70 V/cm. The polarity was then reversed, and a further three pulses were delivered to the muscle. Following electroporation, the wound was closed with surgical clips.
To induce muscle wasting, mice received 14 d of either continuous Dex infusion (2 mg/kg/d, Dex, n = 9) or saline (saline, n = 8) via a subcutaneously implanted miniosmotic pump (model 2002, Alzet). Dex (or saline) infusion began on the same day as electroporation. At the completion of 14 d, mice were sacrificed via CO2 followed by cervical dislocation. Mice were weighed, and the TA muscles were surgically excised, weighed, mounted, and frozen in thawing isopentane for immunofluorescence.
RNA extraction and quantitative RT–PCR (qRT–PCR)
Total RNA from either whole muscle or cells was reverse-transcribed using a cDNA synthesis kit (Applied Biosystems) and subjected to qPCR analysis. The experimental procedure is detailed in the Supplemental Material.
ChIP
ChIP assays were performed as previously described (Caretti et al. 2004), with minor modifications (see the Supplemental Material).
Immunoprecipitation, immunoblot, and immunostaining
Cells were lysed, and their extracts were resolved by SDS-PAGE and transferred onto nitrocellulose filters (see the Supplemental Material). Specific antibodies used to perform immunoprecipitations, immunoblots, and immunostaining are described in Supplemental Table S3.
Nucleosome reconstitution assay
Calf thymus histones (Roche) were used to in vitro reconstitute nucleosomes on a biotinylated PCR-amplified fragment encompassing the myostatin promoter. Nucleosome reconstitution was performed as in Caretti et al. (1999). The reconstituted nucleosome core particles were analyzed by EMSA on a 4.5% nondenaturing acrylamide–bisacrylamide gel as in Caretti et al. (1999). Seven-hundred-fifty micrograms of nuclear extracts from Sh-scramble or Sh-SMYD3 C2C12 cells was incubated for 2 h at 4°C with nucleosome core particles that were previously bound to steptavidin magnetic beads in the presence of 0.1 mM acetyl-CoA and 0.1 μM ATP. Unbound proteins were eliminated by three washes with the lysis and immunoprecipitation buffer. Interacting proteins were eluted by boiling the samples and were analyzed by immunoblot.
Statistical methods
Statistical significance was determined by Student's t-test ([*] P-value < 0.05; [**] P-value < 0.001).
Acknowledgments
We thank A. Derfoul for help with satellite cell isolation protocol. The support of Associazione Italiana per la Ricerca sul Cancro (AIRC) MFAG 5386 and Marie Curie PIRGES-224833 to G.C. is kindly acknowledged. This work was supported in part by the Intramural Research Program of the National Institute of Arthritis, Musculoskeletal, and Skin Diseases of the National Institutes of Health.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.217240.113.
References
- Albini S, Puri PL 2010. SWI/SNF complexes, chromatin remodeling and skeletal myogenesis: It's time to exchange! Exp Cell Res 316: 3073–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amthor H, Otto A, Macharia R, McKinnell I, Patel K 2006. Myostatin imposes reversible quiescence on embryonic muscle precursors. Dev Dyn 235: 672–680 [DOI] [PubMed] [Google Scholar]
- Anastasi S, Giordano S, Sthandier O, Gambarotta G, Maione R, Comoglio P, Amati P 1997. A natural hepatocyte growth factor/scatter factor autocrine loop in myoblast cells and the effect of the constitutive Met kinase activation on myogenic differentiation. J Cell Biol 137: 1057–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM 2001. NF-κB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell 8: 327–337 [DOI] [PubMed] [Google Scholar]
- Bartoli M, Poupiot J, Vulin A, Fougerousse F, Arandel L, Daniele N, Roudaut C, Noulet F, Garcia L, Danos O, et al. 2007. AAV-mediated delivery of a mutated myostatin propeptide ameliorates calpain 3 but not α-sarcoglycan deficiency. Gene Ther 14: 733–740 [DOI] [PubMed] [Google Scholar]
- Belkina AC, Denis GV 2012. BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer 12: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benny Klimek ME, Aydogdu T, Link MJ, Pons M, Koniaris LG, Zimmers TA 2010. Acute inhibition of myostatin-family proteins preserves skeletal muscle in mouse models of cancer cachexia. Biochem Biophys Res Commun 391: 1548–1554 [DOI] [PubMed] [Google Scholar]
- Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, et al. 2001. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol 3: 1014–1019 [DOI] [PubMed] [Google Scholar]
- Braun T, Gautel M 2011. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol Cell Biol 12: 349–361 [DOI] [PubMed] [Google Scholar]
- Brès V, Yoh SM, Jones KA 2008. The multi-tasking P-TEFb complex. Curr Opin Cell Biol 20: 334–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes E, Pombo A 2009. Modifications of RNA polymerase II are pivotal in regulating gene expression states. EMBO Rep 10: 1213–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MA, Sims RJ 3rd, Gottlieb PD, Tucker PW 2006. Identification and characterization of Smyd2: A split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol Cancer 5: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caretti G, Motta MC, Mantovani R 1999. NF-Y associates with H3-H4 tetramers and octamers by multiple mechanisms. Mol Cell Biol 19: 8591–8603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V 2004. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev 18: 2627–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke BA, Drujan D, Willis MS, Murphy LO, Corpina RA, Burova E, Rakhilin SV, Stitt TN, Patterson C, Latres E, et al. 2007. The E3 ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab 6: 376–385 [DOI] [PubMed] [Google Scholar]
- Crepaldi T, Bersani F, Scuoppo C, Accornero P, Prunotto C, Taulli R, Forni PE, Leo C, Chiarle R, Griffiths J, et al. 2007. Conditional activation of MET in differentiated skeletal muscle induces atrophy. J Biol Chem 282: 6812–6822 [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. 2011. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146: 904–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon KC, Glass DJ, Guttridge DC 2012. Cancer cachexia: Mediators, signaling, and metabolic pathways. Cell Metab 16: 153–166 [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. 2010. Selective inhibition of BET bromodomains. Nature 468: 1067–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T, Tsunesumi S, Yamaguchi K, Watanabe S, Furukawa Y 2011. Smyd3 is required for the development of cardiac and skeletal muscle in zebrafish. PLoS ONE 6: e23491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Levi R, Leshem Y, Aoki S, Nakamura T, Halevy O 1998. Hepatocyte growth factor plays a dual role in regulating skeletal muscle satellite cell proliferation and differentiation. Biochim Biophys Acta 1402: 39–51 [DOI] [PubMed] [Google Scholar]
- Glass DJ 2010a. PI3 kinase regulation of skeletal muscle hypertrophy and atrophy. Curr Top Microbiol Immunol 346: 267–278 [DOI] [PubMed] [Google Scholar]
- Glass DJ 2010b. Signaling pathways perturbing muscle mass. Curr Opin Clin Nutr Metab Care 13: 225–229 [DOI] [PubMed] [Google Scholar]
- Gottlieb PD, Pierce SA, Sims RJ, Yamagishi H, Weihe EK, Harriss JV, Maika SD, Kuziel WA, King HL, Olson EN, et al. 2002. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat Genet 31: 25–32 [DOI] [PubMed] [Google Scholar]
- Greer EL, Shi Y 2012. Histone methylation: A dynamic mark in health, disease and inheritance. Nat Rev Genet 13: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, Yagyu R, Nakamura Y 2004. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol 6: 731–740 [DOI] [PubMed] [Google Scholar]
- Hanaoka BY, Peterson CA, Horbinski C, Crofford LJ 2012. Implications of glucocorticoid therapy in idiopathic inflammatory myopathies. Nat Rev Rheumatol 8: 448–457 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Huang Z, Chen D, Zhang K, Yu B, Chen X, Meng J 2007. Regulation of myostatin signaling by c-Jun N-terminal kinase in C2C12 cells. Cell Signal 19: 2286–2295 [DOI] [PubMed] [Google Scholar]
- Joe AW, Yi L, Natarajan A, Le Grand F, So L, Wang J, Rudnicki MA, Rossi FM 2010. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol 12: 153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joulia D, Bernardi H, Garandel V, Rabenoelina F, Vernus B, Cabello G 2003. Mechanisms involved in the inhibition of myoblast proliferation and differentiation by myostatin. Exp Cell Res 286: 263–275 [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T, Kadonaga JT 2010. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol 339: 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemaladewi DU, Hoogaars WM, van Heiningen SH, Terlouw S, de Gorter DJ, den Dunnen JT, van Ommen GJ, Aartsma-Rus A, ten Dijke P, ‘t Hoen PA 2011. Dual exon skipping in myostatin and dystrophin for Duchenne muscular dystrophy. BMC Med Genomics 4: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Heo K, Kim JH, Kim K, Choi J, An W 2009. Requirement of histone methyltransferase SMYD3 for estrogen receptor-mediated transcription. J Biol Chem 284: 19867–19877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivickas LS, Walsh R, Amato AA 2009. Single muscle fiber contractile properties in adults with muscular dystrophy treated with MYO-029. Muscle Nerve 39: 3–9 [DOI] [PubMed] [Google Scholar]
- Lai KM, Gonzalez M, Poueymirou WT, Kline WO, Na E, Zlotchenko E, Stitt TN, Economides AN, Yancopoulos GD, Glass DJ 2004. Conditional activation of akt in adult skeletal muscle induces rapid hypertrophy. Mol Cell Biol 24: 9295–9304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley B, Thomas M, Bishop A, Sharma M, Gilmour S, Kambadur R 2002. Myostatin inhibits myoblast differentiation by down-regulating MyoD expression. J Biol Chem 277: 49831–49840 [DOI] [PubMed] [Google Scholar]
- Lee SJ 2004. Regulation of muscle mass by myostatin. Annu Rev Cell Dev Biol 20: 61–86 [DOI] [PubMed] [Google Scholar]
- Levine M 2011. Paused RNA polymerase II as a developmental checkpoint. Cell 145: 502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipina C, Kendall H, McPherron AC, Taylor PM, Hundal HS 2010. Mechanisms involved in the enhancement of mammalian target of rapamycin signalling and hypertrophy in skeletal muscle of myostatin-deficient mice. FEBS Lett 584: 2403–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokireddy S, Wijesoma IW, Bonala S, Wei M, Sze SK, McFarlane C, Kambadur R, Sharma M 2012. Myostatin is a novel tumoral factor that induces cancer cachexia. Biochem J 446: 23–36 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ma K, Mallidis C, Artaza J, Taylor W, Gonzalez-Cadavid N, Bhasin S 2001. Characterization of 5′-regulatory region of human myostatin gene: Regulation by dexamethasone in vitro. Am J Physiol Endocrinol Metab 281: E1128–E1136 [DOI] [PubMed] [Google Scholar]
- Ma K, Mallidis C, Bhasin S, Mahabadi V, Artaza J, Gonzalez-Cadavid N, Arias J, Salehian B 2003. Glucocorticoid-induced skeletal muscle atrophy is associated with upregulation of myostatin gene expression. Am J Physiol Endocrinol Metab 285: E363–E371 [DOI] [PubMed] [Google Scholar]
- Manceau M, Gros J, Savage K, Thome V, McPherron A, Paterson B, Marcelle C 2008. Myostatin promotes the terminal differentiation of embryonic muscle progenitors. Genes Dev 22: 668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzuk MM, McKeown MR, Filippakopoulos P, Li Q, Ma L, Agno JE, Lemieux ME, Picaud S, Yu RN, Qi J, et al. 2012. Small-molecule inhibition of BRDT for male contraception. Cell 150: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCroskery S, Thomas M, Maxwell L, Sharma M, Kambadur R 2003. Myostatin negatively regulates satellite cell activation and self-renewal. J Cell Biol 162: 1135–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane C, Plummer E, Thomas M, Hennebry A, Ashby M, Ling N, Smith H, Sharma M, Kambadur R 2006. Myostatin induces cachexia by activating the ubiquitin proteolytic system through an NF-κB-independent, FoxO1-dependent mechanism. J Cell Physiol 209: 501–514 [DOI] [PubMed] [Google Scholar]
- McKay BR, Ogborn DI, Bellamy LM, Tarnopolsky MA, Parise G 2012. Myostatin is associated with age-related human muscle stem cell dysfunction. FASEB J 26: 2509–2521 [DOI] [PubMed] [Google Scholar]
- Moresi V, Williams AH, Meadows E, Flynn JM, Potthoff MJ, McAnally J, Shelton JM, Backs J, Klein WH, Richardson JA, et al. 2010. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell 143: 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morissette MR, Cook SA, Buranasombati C, Rosenberg MA, Rosenzweig A 2009. Myostatin inhibits IGF-I-induced myotube hypertrophy through Akt. Am J Physiol Cell Physiol 297: C1124–C1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdiguero E, Sousa-Victor P, Ballestar E, Munoz-Canoves P 2009. Epigenetic regulation of myogenesis. Epigenetics 4: 541–550 [DOI] [PubMed] [Google Scholar]
- Pistilli EE, Bogdanovich S, Goncalves MD, Ahima RS, Lachey J, Seehra J, Khurana T 2011. Targeting the activin type IIB receptor to improve muscle mass and function in the mdx mouse model of Duchenne muscular dystrophy. Am J Pathol 178: 1287–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela A, Esteller M 2010. Epigenetic modifications and human disease. Nat Biotechnol 28: 1057–1068 [DOI] [PubMed] [Google Scholar]
- Qiao C, Li J, Jiang J, Zhu X, Wang B, Xiao X 2008. Myostatin propeptide gene delivery by adeno-associated virus serotype 8 vectors enhances muscle growth and ameliorates dystrophic phenotypes in mdx mice. Hum Gene Ther 19: 241–254 [DOI] [PubMed] [Google Scholar]
- Qiao C, Li J, Zheng H, Bogan J, Yuan Z, Zhang C, Bogan D, Kornegay J, Xiao X 2009. Hydrodynamic limb vein injection of adeno-associated virus serotype 8 vector carrying canine myostatin propeptide gene into normal dogs enhances muscle growth. Hum Gene Ther 20: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA 2010. c-Myc regulates transcriptional pause release. Cell 141: 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon KA, Davis J, Kapsa RM, Choong P, Byrne E 2001. Myostatin, insulin-like growth factor-1, and leukemia inhibitory factor mRNAs are upregulated in chronic human disuse muscle atrophy. Muscle Nerve 24: 893–899 [DOI] [PubMed] [Google Scholar]
- Ríos R, Carneiro I, Arce VM, Devesa J 2002. Myostatin is an inhibitor of myogenic differentiation. Am J Physiol Cell Physiol 282: C993–C999 [DOI] [PubMed] [Google Scholar]
- Sartorelli V, Juan AH 2011. Sculpting chromatin beyond the double helix: Epigenetic control of skeletal myogenesis. Curr Top Dev Biol 96: 57–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertzer JD, Lynch GS 2008. Plasmid-based gene transfer in mouse skeletal muscle by electroporation. Methods Mol Biol 433: 115–125 [DOI] [PubMed] [Google Scholar]
- Sirinupong N, Brunzelle J, Ye J, Pirzada A, Nico L, Yang Z 2010. Crystal structure of cardiac-specific histone methyltransferase SmyD1 reveals unusual active site architecture. J Biol Chem 285: 40635–40644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson EC, Travers AA 2008. A Drosophila Smyd4 homologue is a muscle-specific transcriptional modulator involved in development. PLoS ONE 3: e3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trendelenburg AU, Meyer A, Rohner D, Boyle J, Hatakeyama S, Glass DJ 2009. Myostatin reduces Akt/TORC1/p70S6K signaling, inhibiting myoblast differentiation and myotube size. Am J Physiol Cell Physiol 296: C1258–C1270 [DOI] [PubMed] [Google Scholar]
- Van Aller GS, Reynoird N, Barbash O, Huddleston M, Liu S, Zmoos AF, McDevitt P, Sinnamon R, Le B, Mas G, et al. 2012. Smyd3 regulates cancer cell phenotypes and catalyzes histone H4 lysine 5 methylation. Epigenetics 7: 340–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner KR, Fleckenstein JL, Amato AA, Barohn RJ, Bushby K, Escolar DM, Flanigan KM, Pestronk A, Tawil R, Wolfe GI, et al. 2008. A phase I/II trial of MYO-029 in adult subjects with muscular dystrophy. Ann Neurol 63: 561–571 [DOI] [PubMed] [Google Scholar]
- Xi L, Fondufe-Mittendorf Y, Xia L, Flatow J, Widom J, Wang JP 2010. Predicting nucleosome positioning using a duration hidden Markov model. BMC Bioinformatics 11: 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Wu J, Sun B, Zhong C, Ding J 2011. Structural and biochemical studies of human lysine methyltransferase Smyd3 reveal the important functional roles of its post-SET and TPR domains and the regulation of its activity by DNA binding. Nucleic Acids Res 39: 4438–4449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Chen Y, Zhang Y, Wang X, Yang N, Zhu D 2006. Extracellular signal-regulated kinase 1/2 mitogen-activated protein kinase pathway is involved in myostatin-regulated differentiation repression. Cancer Res 66: 1320–1326 [DOI] [PubMed] [Google Scholar]
- Zhou X, Wang JL, Lu J, Song Y, Kwak KS, Jiao Q, Rosenfeld R, Chen Q, Boone T, Simonet WS, et al. 2010. Reversal of cancer cachexia and muscle wasting by ActRIIB antagonism leads to prolonged survival. Cell 142: 531–543 [DOI] [PubMed] [Google Scholar]
- Zhou Q, Li T, Price DH 2012. RNA polymerase II elongation control. Annu Rev Biochem 81: 119–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou JN, Wang SZ, Yang JS, Luo XG, Xie JH, Xi T 2009. Knockdown of SMYD3 by RNA interference down-regulates c-Met expression and inhibits cells migration and invasion induced by HGF. Cancer Lett 280: 78–85 [DOI] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. 2011. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478: 524–528 [DOI] [PMC free article] [PubMed] [Google Scholar]