Abstract
Many of the mutations contributing to leukemogenesis in acute myeloid leukemia have been identified. A common activating mutation is an internal tandem duplication (ITD) mutation in the FLT3 gene that is found in approximately 25% of patients and confers a poor prognosis. FLT3 inhibitors have been developed and have some efficacy, but patients often relapse. Levels of FLT3 ligand (FL) are significantly elevated in patients during chemotherapy and may be an important component contributing to relapse. We used a mouse model to investigate the possible effect of FL expression on leukemogenesis involving FLT3-ITD mutations in an in vivo system. FLT3ITD/ITD FL−/− (knockout) mice had a statistically significant increase in survival compared with FLT3ITD/ITD FL+/+ (wildtype) mice, most of which developed a fatal myeloproliferative neoplasm. These findings suggest that FL levels may have prognostic significance in human patients. We also studied the effect of FL expression on survival in a FLT3-ITD NUP98–HOX13 (NHD13) fusion mouse model. These mice develop an aggressive leukemia with short latency. We asked whether FL expression played a similar role in this context. The NUP98-HOX13 FLT3ITD/wt FL−/− mice did not have a survival advantage, compared with NUP98-HOX13 FLT3ITD/wt FL+/+ mice (normal FL levels). The loss of the survival advantage of the FL knockout group in the NUP98–HOX13 model suggests that adding a second mutation changes the effect of FL expression in the context of more aggressive disease.
Abbreviations: AML, acute myeloid leukemia; FL, FLT3 ligand; FLT3, FMS-like tyrosine kinase 3; ITD, internal tandem duplication; MPN, myeloproliferative neoplasm
FMS-like tyrosine kinase 3 (FLT3) is normally activated by binding of its ligand (FL) to 2 FLT3 molecules, causing them to dimerize, autophosphorylate, and activate downstream targets.20,26,31 Although FL expression is relatively ubiquitous, the FLT3 receptor is found predominantly on hematopoietic cells and has an important role in hematopoiesis.6,13,24 Several mutations in the FLT3 gene can lead to constitutive activation that occurs independent of ligand binding and leads to activation of downstream targets; these mutations typically are found in patients with acute myeloid leukemia (AML). The most common mutation described in AML is an internal tandem duplication (ITD) that occurs in the juxtamembrane domain of FLT3. The ITD mutations vary in length,17,25 but these forms all constitutively activate FLT3 kinase activity to result in autophosphorylation and phosphorylation of its downstream targets.4,14,28,32 The ITD mutation is seen in approximately 25% of adult AML cases and is associated with a poor prognosis.18,19,23
Despite the fact that FTL3-ITD is constitutively activated, some evidence indicates that FL may continue to play a role in FLT3 signaling and affect AML prognosis.35 Elevated plasma levels of FL have been reported in patients that have undergone chemotherapy.2,30 In addition, elevated levels of FL have been shown to increase the amount of FLT3 inhibitor needed to reduce the levels of phosphorylated FLT3-ITD in a cell line (Molm14) model.8,21,34 When a lentivirus was used to introduce a FLT3-ITD mutation into mouse embryonic fibroblast cells from FL-knockout mice, the addition of FL to the culture media resulted in an increase in the level of phosphorylated FLT3, further supporting the idea that FL may play a role in FLT3-ITD–associated AML.33 These previous models have all used cell lines, cultured cells, and plasma from patient samples to address the potential importance of FL expression in cases where an ITD mutation is present.
Here we use primary hematopoietic cells from a combination of genetically engineered mouse models to investigate the role of FLT3 and FL in the pathogenesis of AML. The first model is a FLT3-ITD knockin mouse model with an 18-bp insertion in the juxtamembrane domain of FLT3 that was generated and characterized by our lab. This mouse model consistently and predictably develops myeloproliferative neoplasia (MPN) with moderately elevated WBC counts, splenomegaly, and myeloid expansion in the bone marrow, as evidenced by histopathologic changes and increased granulocytic/ monocytic fractions by flow cytometry.11 A small percentage (7%; 9 of 129) of the FLT3-ITD homozygous (FLT3ITD/ITD) mice spontaneously developed fully transformed leukemia.10 The second mouse model uses transgenic expression of a Nup98–Hox13 fusion (NHD13) that is expressed primarily in hematopoietic tissues. Mice that carry this mutation typically develop a myelodysplastic syndrome that often progresses to acute leukemia after a long lag time.12 When these mice were bred to our FLT3-ITD mice, the resulting double-mutant Nup98–Hox13 (NHD13) FLT3wt/ITD mice predominantly developed an AML with minimal differentiation and demonstrated a markedly shorter latency to disease. Interestingly, a subset of mice display loss of heterozygosity of the wildtype Flt3 allele in the bone marrow7 as occurs in a fraction of human FLT3-ITD AML patients.22,29 The third model is a FL-knockout mouse model that was developed at Immunex (Seattle, WA) and is currently commercially available. These mice have the majority of the FL extracellular domain coding region disrupted by insertion of a PKG–Neo cassette. These mice demonstrated reduced cellularity in the bone marrow and an overall reduction in hematopoietic precursors, especially of the myeloid and lymphoid lineages.16
To examine the effect of FL expression on disease conferred by a FLT3-ITD mutation, we used 2 genetically engineered mouse models: the first is the model of MPN generated by the FLT3ITD/ITD mutation alone. The second was a leukemia model that is generated by the combination of a FLT3wt/ITD together with a NHD13 mutation. Into both of these models, we bred mice that were either wildtype for FL or that had FL knocked out. We then characterized survival and disease phenotype data from each cohort to ascertain the effect of FL expression on MPN and AML generated by FLT3-ITD expression.
Materials and Methods
Mouse (Mus musculus) models used for experiments.
FLT3 ligand knockout (FL−/−) mice (C57BL/6-flt3ltm1Imx) are currently commercially available from Taconic (Hudson, NY). These mice were generated by deleting most of the extracellular domain of FL and replacing it with a neomycin-resistance cassette under control of the PGK promoter.16 FLT3-ITD mice (B6.129S6-Flt3tm1Dosm ) were generated and characterized by our laboratory previously. These mice have an extra 18-bp ITD insertion in the juxtamembrane domain of the Flt3 gene.11 The FLT3-ITD mice have been backcrossed to a C57BL/6 background for more than 7 generations and bred to obtain FLT3ITD/ITD FL+/+ and FLT3ITD/ITDFL−/− mice.
Mice that carry a NUP98–HOX13 (NHD13) transgene (C57BL/6-Tg[Vav1-NUP98/HOXD13]G2Apla/N ) were obtained from Dr Peter Aplan (NIH, Bethesda, MD). These mice are transgenic for a fusion of the N-terminal of NUP98 gene and the DNA binding homeodomain of HoxD13.12 They were backcrossed to a C57BL/6 background, and bred to both FLT3wt/ITD mice and FL−/− mice to obtain FLT3wt/ITD NHD13 FL+/+ and FLT3wt/ITD NHD13 FL−/− mice for the follow-up study.
All mice were bred and housed in a barrier facility in filter-top microisolation cages (Thoren Caging, Hazelton, PA) with Tek-fresh bedding and were fed Teklad Global 2018SX diet (Harlan, Madison, WI). Water was delivered via an automated hyperchlorinated reverse-osmosis-treated watering system. Rooms were on a 12:12-h light:dark cycle. Mouse disease status was monitored by use of rotating sentinels (1 cage of 2 per rack). Mice were found to be free of a wide range of pathogens including Sendai virus, mouse hepatitis virus, mouse parvovirus 1 and 2, Theiler mouse encephalomyelitis virus, reovirus, epizootic diarrhea of infant, mice, lymphocytic choriomeningitis virus, ectromelia virus, murine adenovirus, and murine cytomegalovirus. The facility is considered to be Helicobacter-positive. Procedures and protocols were approved by the IACUC at Johns Hopkins in accordance with the Animal Welfare Act.1
The mice were ear-tagged and genotyped when weaned and followed over time to generate a survival curve. On showing overt signs of clinical illness including hunching, lethargy, and increased respiratory rate, mice were euthanized and necropsied. Spleen weight and gross lesions were noted. Tissue samples were collected for further analysis. All mice were genotyped based on the possible outcomes of the cross by using the primers described following. C57BL/6 mice that did not have any of the described mutations were used as wildtype controls. Littermates were used whenever possible.
Primers used for genotyping DNA from tail snips were: mITD-F, 5′ TGC AGA TGA TCC AGG TGA CT 3′,; mITD-R, 5′ CTC TCG GGA ACT CCC ACT TA 3′; Nup98-F, 5′ TGG AGG GCC TCT TGG TAC AGG 3′; HoxD13-R, 5′ GGC TTC TAA GCT GTC TGT GGC C 3′; FL-F: 5′GCC CAA ATG TGC GTA TAC CT 3′; FL-R, 5′ CCC AGC ACA GTA TGG GAA CT 3′; NeoFL-F, 5′ ACA CTT CGA AGC TGG AAA GC 3′; and NeoFL-R, 5′ GGG GAA CTT CCT GAC TAG GG 3′. All genotyping was done by PCR (Bio-Rad, Hercules, CA) according to a program that had been optimized for each primer set and that used a hot-start master mix (Denville Scientific, Metuchen, NJ). All products were run on agarose gels (3.5% for ITD genotyping; 1% for the other primer sets) with ethidium bromide for band visualization.
Characterization of disease.
CBC of peripheral blood from moribund mice (those showing overt signs of severe clinical illness such as hunching, lethargy, or dyspnea) were performed by using an automated system (Hemavet950, Drew Scientific, Dallas, TX). Tissues collected at necropsy were stored in 10% buffered formalin. Peripheral blood smears and bone marrow cytospins were stained by using a modified Wright–Giemsa stain. Images were obtained by using a BX46 microscope (Olympus, Irving, TX) with a 100× oil objective and a mounted digital camera (DP72, Olympus). FACS analysis was performed on bone marrow and spleen cells from each of the euthanized mice as previously described by using 4-color FACS (FACS Calibur, Becton Dickinson, San Jose, CA).11 Bone marrow and spleen cells were suspended in 2.5% fetal bovine serum in 1× PBS and stained with the antibody sets outlined in Table 1. Antibody clones were: lineage mixture, MLM20; CD135, A2F10.1; Sca1, D7; c-Kit, 2B8; CD41, MWReg30; Ter119, TER-119; Mac1, M1/70; Gr1, RB6-8C5; CD24, M1/69; CD43, S7; B220, RA3-6B2; IgM, II/41; CD4, L3T4; CD3, 17A2; CD8a, Ly2; CD90, 53-2.1; CD86, GL1; and CD11c, HL3. All antibodies were obtained from BD Biosciences (San Jose, CA) except for the lineage mixture (Invitrogen, Carlsbad, CA) and Sca-1 (eBiosciences, San Diego, CA). FACS data were analyzed by using FlowJo (version 9.3.3, Tree Star, Ashland, OR).
Table 1.
FACS surface antibodies and fluorophore combinations used to analyze bone marrow and spleen cell suspensions to characterize the disease outcome of sick mice
| Classification guideline | FITC | PE | PerCP | APC |
| Hematopoietic stem cell | Lineage | CD135 (FLT3) | Sca1 | c-Kit |
| Myeloid | CD41 | Ter119 | Mac1 | Gr1 |
| B cell | CD24 | CD43 | B220 | IgM |
| T cell | CD4 | CD3 | CD8a | CD90 |
| Dendritic cell | Mac1 | CD86 | B220 | CD11c |
| Leukemic cell (simplified) | Gr1 | Mac1 | B220 | c-Kit |
APC, allophycocyanin; PerCP, peridinin chlorophyll protein complex.
This list of cell surface markers is not exhaustive but represents generally accepted markers for the characterization of hematopoietic disorders in mice.
Statistical analysis.
Prism 5 (Graph Pad Software, San Diego, CA) was used to generate many of the graphs and to perform statistical analysis. The survival curves were compared and a P value generated by using both a log-rank (Mantel–Cox) test and a Gehan–Breslow–Wilcoxon test. The P values from the bone marrow and spleen data was calculated by using an unpaired 2-tailed T test.
Results
Lack of FL confers a survival advantage in a FLT3 ITD/ITD mouse model of MPN.
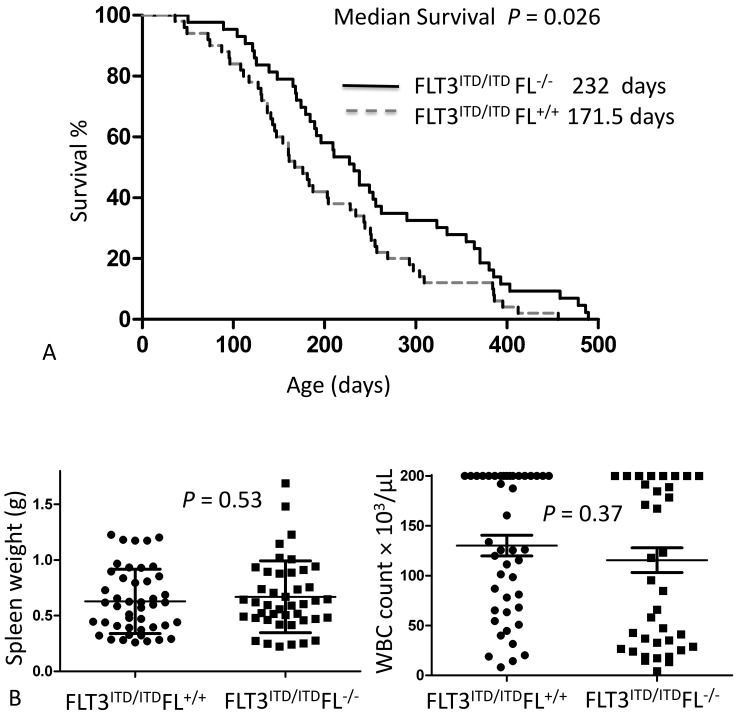
We first investigated the role that FL plays in FLT3-ITD–mediated disease by breeding FLT3/ITD knockin mice with FL knockout mice to obtain FLT3ITD/ITDFL+/+ and FLT3ITD/ITDFL−/− mice. When both cohorts were followed for survival, the mice that did not express FL demonstrated a small but statistically significant (P = 0.025, log-rank [Mantel–Cox] test, or P = 0.026, Gehan–Breslow–Wilcoxon test) survival advantage compared with that of mice with normal FL levels (Figure 1 A).33
Figure 1.
(A) Completed survival curve demonstrating the difference between homozygous FLT3ITD/ITD knockin mice with (FL+/+; median survival, 171.5 d; n = 50) and without (FL−/−; median survival, 232 d; n = 43) FL expression. Some of these data were reported previously in a nearly completed survival curve.33 (B) Spleen weights and WBC counts are not statistically significant between the 2 groups. Note: the automated analyzer reports all values greater than 200 × 103/µL as ‘high;’ for the purpose of data analysis, ‘high’ values were recorded as 200 × 103/µL.
At the time of necropsy, a CBC analysis of the peripheral blood was performed. WBC counts ranged from 4.2 × 103/µL to greater than 200 × 103/µL in sick mice. The spleen weights recorded at the time of death ranged from 0.22 to 1.69 g. The differences in WBC counts and spleen weights were not statistically significant between the 2 cohorts (Figure 1 B).
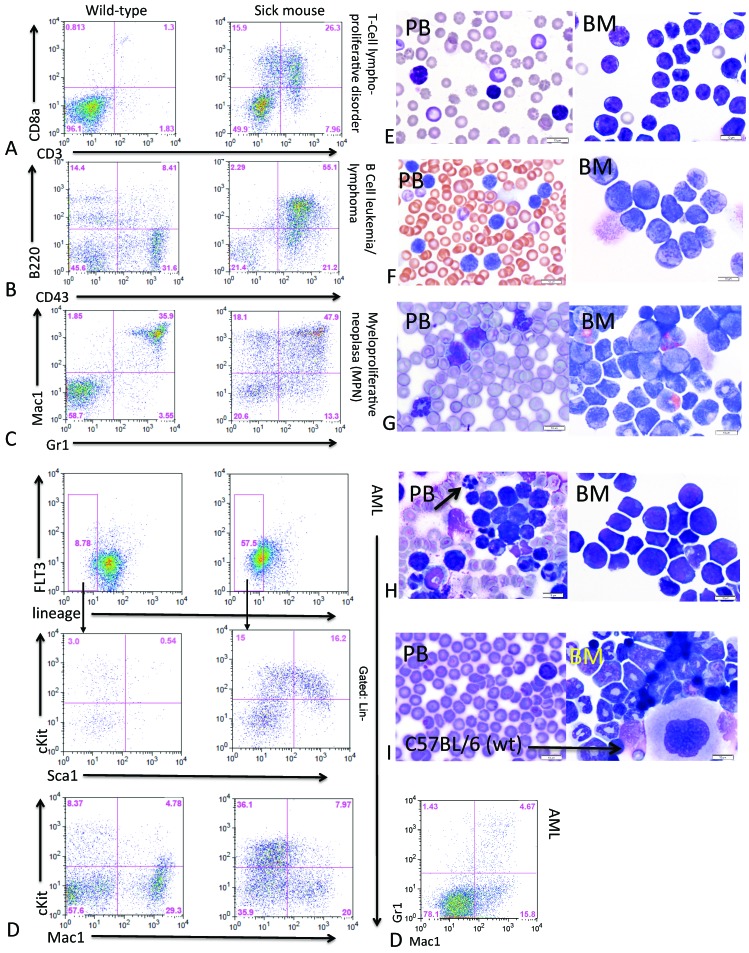
The FACS staining profile (Table 1) of the bone marrow and spleen cells revealed that the FLT3ITD/ITDFL+/+ and FLT3ITD/ITDFL−/− mice developed hematopoietic neoplasms that could be classified into 1 of 4 categories. Specifically, 9.6% of the mice developed a T-cell lymphoproliferative disorder that typically showed increased CD8a+ and CD3+ fractions (Figure 2 A). Another 8.6% of the mice developed a B-cell leukemia–lymphoma and often had enlarged lymph nodes; Flow cytometric analysis of these mice showed increased B220+ and CD43+ fractions (Figure 2 B). Many of the mice with B and T cell disorders appeared to demonstrate more than one hematopoietic abnormalitie, with expansion of the Mac1+ and Gr1+ compartments and some myeloid expansion in the bone marrow, suggesting a concomitant MPN. The predominant disease these mice developed was a classic MPN (75.2% of mice) as demonstrated by increased in levels of lineage-negative cells (not shown) and increased fractions of the myeloid–granulocytic markers Mac1+ and Gr1+ (Figure 2 C). The remaining 6.5% of mice developed AML, which could be divided into 2 subtypes. One subtype showed an expansion of the lineage–Sca1+cKit+ population with reduced Mac1+Gr1+ fractions; this subtype is most consistent with AML with minimal differentiation in humans (M0 in the French–American–British classification system).27 The other subtype of AML was characterized by expansion of the lineage–Sca1–cKit+ population and increased Mac1+Gr1+ fractions, thus closely approximating cases of human AML with some maturation. An example of a minimally differentiated AML with a lineage–Sca1+c-Kit+ expansion, c-Kit greater than 20%, and decreased Mac1+/Gr1+ fractions is shown in Figure 2 D. Figure 2 (A through D) shows a wildtype control mouse alongside a sick mouse for each disease outcome.
Figure 2.
Immunophenotyping summary of a typical diagnostic FACS profile used to classify mice into 4 disease categories. A wildtype mouse (C57BL/6 littermate lacking mutations) is shown (left side) for comparison with the moribund mouse (right side) in each panel (A through D). Representative peripheral blood (PB) and bone marrow (BM) cytospin photomicrographs are included also (panels E through I); bar, 10 μM. (A and E) T-cell lymphoproliferative disorder. (B and F) B-cell leukemia–lymphoma. (C and G) Myeloproliferative neoplasia (MPN). (D and H) AML with minimal differentiation; the low levels of Mac1+Gr1+ cells in the additional FACS plot (D, right) support the diagnosis of minimal differentiation. (H) Arrow indicates an apoptotic cell. (I) Peripheral blood and bone marrow cytospin from a wildtype mouse.
Images of the peripheral blood and bone marrow cytospins from a representative mouse for each classification are shown in Figure 2 (E through H). The peripheral blood from wildtype controls showed predominantly RBC, with readily identified platelets, scattered lymphocytes, and occasional neutrophils (Figure 2 I). The bone marrow of a wildtype mouse demonstrated trilineage hematopoiesis, with progressive maturation of the myeloid and erythroid lineages (Figure 2 I). The peripheral blood and bone marrow of mice that developed T- and B-cell disorders (Figures 2 E and F, respectively) showed increased atypical lymphocytes, and FACS analysis using surface markers readily identifies the abnormal lymphoid populations. In bone marrow cytospins from mice with B- and T-cell disorders, both abnormal lymphoid cells and myeloid precursors were increased, consistent with concomitant lymphoproliferative disorder and MPN. FACS and morphologic analysis of peripheral blood and bone marrow from mice with MPN (Figure 2 G) showed expansion of myeloid cells in various stages of maturity in the peripheral blood and a marked myeloid predominance in the bone marrow. In mice that developed AML (Figure 2 H), numerous blasts were present in the peripheral blood, and the bone marrow was largely replaced by immature myeloid cells, consistent with a diagnosis of AML with minimal differentiation.
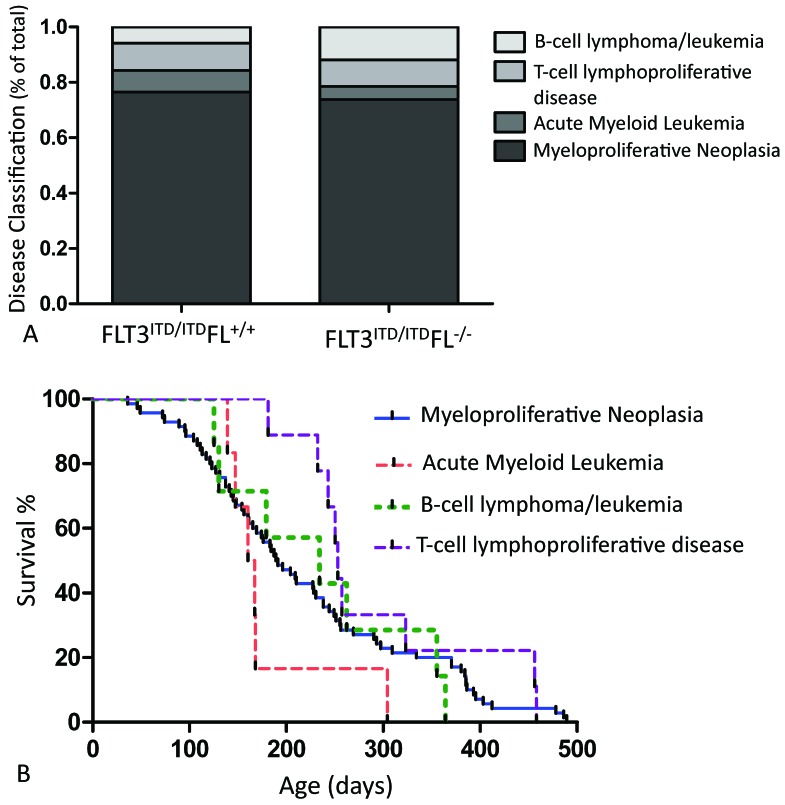
The distribution of disease types was similar between the FLT3ITD/ITDFL+/+ and FLT3ITD/ITDFL−/− cohorts of mice and is shown in Figure 3 A. We also examined survival according to disease subtype by combining data from both genotypes. Mice with MPN succumbed to disease beginning at day 36, and continued to demonstrate signs of significant clinical illness throughout the entire 500-d study at a steady rate. The first mice with AML did not show overt clinical signs until day 139. Most of the AML cases occurred in a relatively short period of time (between day 139 and 168), long after several MPN mice had already succumbed to disease. Compared with those with AML, mice with the B- and T-cell disorders showed longer survival (Figure 3 B).
Figure 3.
(A) Disease classification of FLT3ITD/ITD FL+/+ (n = 55) and FLT3ITD/ITD FL−/− (n = 44) mice expressed as a percentage of total. (B) Pooled outcome of FLT3ITD/ITD mice. Disease outcome is plotted in relationship to time to disease. There is a steady incidence of MPN; after a brief lag, cases of AML tended to occur early compared with those of B- and T-cell disease.
In conclusion, these data demonstrate that FL may play a role in survival even when FLT3 is constitutively activated as in our ITD mouse model. Our survival data suggest that the absence of FL confers a survival advantage in this context. FL levels do not, however, appear to have noticeable effects on disease phenotype or manifestations.
FL plays no role in survival of the myeloid leukemia that develops in FLT3wt/ITDNHD13 mice.
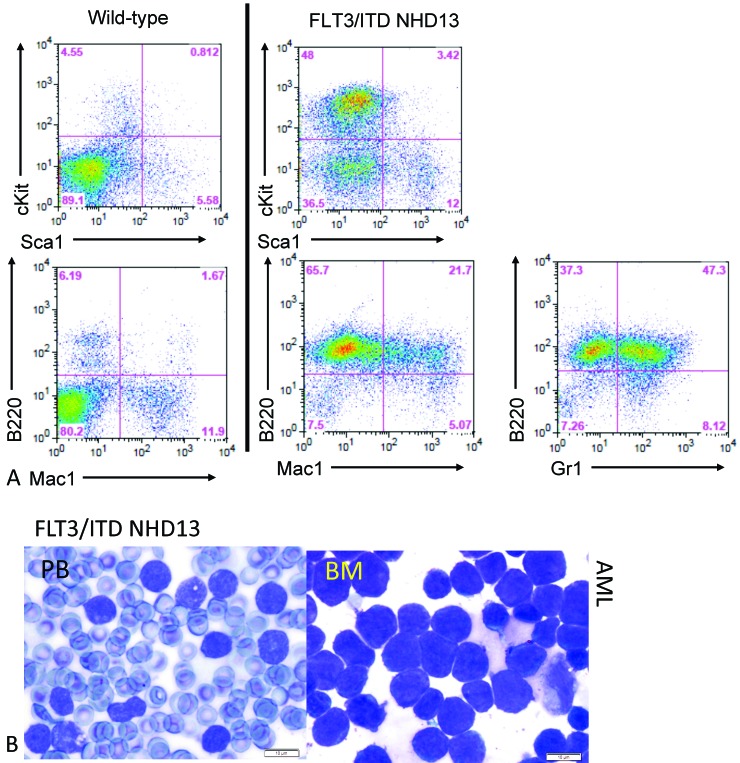
FLT3-ITD mutations alone are insufficient to cause full-blown leukemia, and cooperative events are required.7,11 Our lab previously demonstrated that double-mutant mice that carry both the NHD13 fusion gene and FLT3-ITD (FLT3wt/ITD NHD13) developed full-blown leukemia after a short latency and with 100% penetrance, supporting the idea that additional events can cooperate with FLT3-ITD to cause leukemia. These FLT3wt/ITDNHD13 mice developed aggressive transplantable myeloid leukemias that express increased fractions of Gr1+ and B220+ but not the mature B-cell markers CD19 and IgM.7 An example of a typical FACS profile is shown in Figure 4 A. In this example, c-Kit is elevated (>50%), consistent with the elevated fractions of blastic cells that are typically seen in leukemia. In addition, there is increased B220+ expression in the FLT3wt/ITDNHD13 that includes B220+Mac1+ and B220+Gr1+ cells. Photomicrographs of the peripheral blood from a representative sick FLT3wt/ITDNHD13 mouse shows numerous circulating blasts and an expansion of blasts in the bone marrow (Figure 4 B).
Figure 4.
(A) Representative FACS profile of a FLT3wt/ITD NDH13 mouse with leukemia. (B) Samples from a sick FLT3wt/ITD NHD13 mouse show blasts in the bone marrow (BM) and peripheral blood (PB).
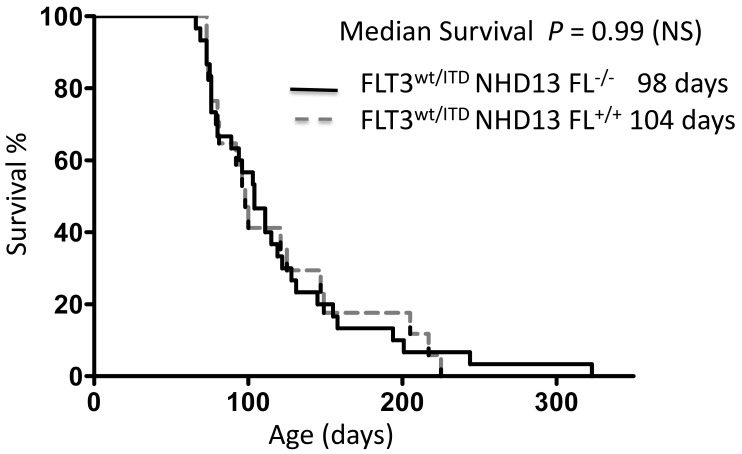
To investigate the possible effect of FL expression in the process of leukemogenesis and in the maintenance of leukemic cells carrying FLT3-ITD mutations, we bred the FLT3wt/ITDNHD13 mice onto the FL knockout background to obtain both FLT3wt/ITDNHD13FL+/+ and FLT3wt/ITDNHD13FL−/− mice. These mice were followed for assessment of survival and were euthanized when they showed signs of significant clinical illness. Both cohorts demonstrated substantially shorter latency to disease than that of the FLT3ITD/ITD group and developed aggressive leukemia with 100% penetrance. The presence or absence of FL did not affect overall survival, with FLT3wt/ITDNHD13FL+/+ mice surviving a median of 104 d compared with FLT3wt/ITDNHD13FL−/− which survived a median of 98 d (P = 0.99, log-rank [Mantel–Cox] test) or P = 0.94 [Gehan–Breslow–Wilcoxon test]; Figure 5). The lack of any survival advantage for the leukemic mice in the absence of FL suggests that the addition of a ‘second hit’ may negate the effect of FL stimulation in more aggressive hematopoietic neoplasms.
Figure 5.
Survival data from FLT3wt/ITD Nup98–Hox13 FL+/+ (median survival, 104 d; n = 17) and FLT3wt/ITD Nup98–Hox13 Fl−/− (median survival, 98 d; n = 30). Survival did not differ (P = 0.99) between the 2 cohorts.
Discussion
The increased availability of knockin, knockout, and transgenic mouse models offers a powerful tool that facilitates the investigation of the complex interactions of different genes in a system of nearly identical subjects.3,5,15 In the current study, we combined different mouse models to investigate the effects of FL expression on disease states rendered by FLT3-ITD signaling, a constitutively active form of FLT3. We were particularly interested in discovering whether FL affects outcomes of in vivo model systems, given that observational studies of patients treated with FLT3 inhibitors and studies using cell lines suggest that FL alters leukemogenesis and response to FLT3 inhibitors.9,21 The mechanism by which FL further stimulates constitutively activated FLT3 mutants has not been elucidated, but the phenomenon has been observed frequently.33 FL often is coexpressed with FLT3 in leukemic cells, and the increased levels of FL elevate phospho-FLT3 levels, rendering the inhibition of FLT3 signaling by FLT3 TKI more difficult in clinical trials.21
In the FLT3ITD/ITD mouse model, FL levels do affect survival and disease progression even when driven by constitutively activated FLT3 signaling. In the current study, we examined in depth the spectrum of disease phenotypes that developed from cohorts of mice with both the FLT3ITD/ITDFL+/+ and FLT3ITD/ITDFL−/− genotypes that had been followed predominantly for survival previously.33 We also highlighted strategies that can be used for disease characterization. We found that the presence or absence of FL in this setting did not affect the proportions of the various diseases developing in the FLT3ITD/ITD mice in both groups.
Although all of the FLT3ITD/ITD mice displayed a MPN phenotype as early as 8 to 12 wk, they also developed other disease phenotypes. MPN likely was present in these mice, but other genetic events probably emerged to affect the disease outcome. When mice developed AML, it developed relatively early. Perhaps the hematopoietic stem or progenitor cells in cases of AML acquired a second mutation during the lag time and thus were fully transformed to leukemic cells. Mice with B- and T-cell disorders generally developed disease later than did mice with either a MPN or an AML. We speculate that other genetic events emerged that cooperate with the FLT3-ITD mutation to transform the normal hematopoietic stem or progenitor cells to lead to a different disease phenotype (B- or T-cell disorders).
We had not yet looked at the effect of FL expression in the FLT3wt/ITDNHD13 model that results in AML with 100% penetrance and wanted to learn whether FL levels have an effect in the face of a more aggressive disease.7 In fact, in the leukemic FLT3wt/ITD NHD13 model, the survival advantage previously observed by eliminating FL in the MPN model disappeared, suggesting that the addition of a ‘second hit’ decreases the effect of FL.
One possible explanation for this decrease is that the effects of FL on the activation of the FLT3-ITD mutations are relatively weak compared with the constitutive activation caused by the mutation itself.33 The relatively small differences in survival between the FLT3ITD/ITDFL+/+ and FLT3ITD/ITDFL−/− mice are observable because MPN takes a long time to develop in both genotypes; a small change in the degree of FLT3 activation may result in this difference. However, when FLT3-ITD is combined with NHD13, disease latency is so short that a small change in FLT3 activation does not result in an observable survival difference. In this regard, the cooperation between the 2 mutations (FLT3-ITD and NHD13) is dominant and may undermine the relatively weak effect caused by further FL stimulation of FLT3-ITD signaling. Nonetheless, methods to inhibit the FL stimulation of patients with FLT3-mutant AML (for example, through antiFL antibodies) might still have an effect on outcome of therapy.
In conclusion, lack of FL expression confers a survival advantage in FLT3-ITD mice with MPN but not in FLT3-ITD NHD13 mice that have particularly aggressive AML. These data suggest that FL levels play a less important role when additional mutations are present. Follow-up studies may help further determine the importance of the greatly elevated FL levels that have been observed clinically when AML patients undergo chemotherapy.21 A transgenic model of a mouse overexpressing FL has not yet been generated but would be useful to model the overexpression that occurs in AML patients. We currently are investigating how the FLT3-ITD mutation interacts with other oncogenic mutations found in leukemias, using other available mouse models and bone marrow transduction and subsequent transplantation techniques. Comparing the efficacy of FLT3 inhibitors and antiFL antibodies in FLT3-ITD leukemia models with FL overexpression compared with FL-knockout mice might be a useful way of further dissecting the importance of FL in leukemia.
Acknowledgments
Thanks to Rui Zheng for the early cell line data that led to the mouse portion of this project. Thanks to Kelly Pate for comments and suggestions after reading the manuscript.
This work was supported by the National Cancer Institute (CA90668, CA70970), Leukemia and Lymphoma Society, the Giant Food Pediatric Cancer Research Fund, and the Intramural Research Program of the NIH National Cancer Institute.
References
- 1.Animal Welfare Act as Amended. 2008. 7 USC §2131–2159.
- 2.Bojko P, Pawloski D, Stellberg W, Schroder JK, Seeber S. 2002. Flt3 ligand and thrombopoietin serum levels during peripheral blood stem cell mobilization with chemotherapy and recombinant human glycosylated granulocyte colony-stimulating factor (rhu-G-CSF, lenograstim) and after high-dose chemotherapy. Ann Hematol 81:522–528 [DOI] [PubMed] [Google Scholar]
- 3.Cheon DJ, Orsulic S. 2011. Mouse models of cancer. Annu Rev Pathol 6:95–119 [DOI] [PubMed] [Google Scholar]
- 4.Choudhary C, Muller-Tidow C, Berdel WE, Serve H. 2005. Signal transduction of oncogenic Flt3. Int J Hematol 82:93–99 [DOI] [PubMed] [Google Scholar]
- 5.Frese KK, Tuveson DA. 2007. Maximizing mouse cancer models. Nat Rev Cancer 7:645–658 [DOI] [PubMed] [Google Scholar]
- 6.Gilliland DG, Griffin JD. 2002. The roles of FLT3 in hematopoiesis and leukemia. Blood 100:1532–1542 [DOI] [PubMed] [Google Scholar]
- 7.Greenblatt S, Li L, Slape C, Nguyen B, Novak R, Duffield A, Huso D, Desiderio S, Borowitz MJ, Aplan P, Small D. 2012. Knock-in of a FLT3–ITD mutation cooperates with a NUP98–HOXD13 fusion to generate acute myeloid leukemia in a mouse model. Blood 119:2883–2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levis M. 2011. FLT3–ITD AML and the law of unintended consequences. Blood 117:6987–6990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levis M, Ravandi F, Wang ES, Baer MR, Perl A, Coutre S, Erba H, Stuart RK, Baccarani M, Cripe LD, Tallman MS, Meloni G, Godley LA, Langston AA, Amadori S, Lewis ID, Nagler A, Stone R, Yee K, Advani A, Douer D, Wiktor-Jedrzejczak W, Juliusson G, Litzow MR, Petersdorf S, Sanz M, Kantarjian HM, Sato T, Tremmel L, Bensen-Kennedy DM, Small D, Smith BD. 2011. Results from a randomized trial of salvage chemotherapy followed by lestaurtinib for patients with FLT3 mutant AML in first relapse. Blood 117:3294–3301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L, Bailey E, Greenblatt S, Huso D, Small D. 2011. Loss of the wild-type allele contributes to myeloid expansion and disease aggressiveness in FLT3-ITD knockin mice. Blood 118:4935–4945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li L, Piloto O, Nguyen HB, Greenberg K, Takamiya K, Racke F, Huso D, Small D. 2008. Knock-in of an internal tandem duplication mutation into murine FLT3 confers myeloproliferative disease in a mouse model. Blood 111:3849–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin YW, Slape C, Zhang Z, Aplan PD. 2005. NUP98–HOXD13 transgenic mice develop a highly penetrant, severe myelodysplastic syndrome that progresses to acute leukemia. Blood 106:287–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyman SD, Jacobsen SE. 1998. c-Kit ligand and Flt3 ligand: stem–progenitor cell factors with overlapping yet distinct activities. Blood 91:1101–1134 [PubMed] [Google Scholar]
- 14.Marchetto S, Fournier E, Beslu N, Aurran-Schleinitz T, Dubreuil P, Borg JP, Birnbaum D, Rosnet O. 1999. SHC and SHIP phosphorylation and interaction in response to activation of the FLT3 receptor. Leukemia 13:1374–1382 [DOI] [PubMed] [Google Scholar]
- 15.McCormack E, Bruserud O, Gjertsen BT. 2008. Review: genetic models of acute myeloid leukaemia. Oncogene 27:3765–3779 [DOI] [PubMed] [Google Scholar]
- 16.McKenna HJ, Stocking KL, Miller RE, Brasel K, De Smedt T, Maraskovsky E, Maliszewski CR, Lynch DH, Smith J, Pulendran B, Roux ER, Teepe M, Lyman SD, Peschon JJ. 2000. Mice lacking Flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood 95:3489–3497 [PubMed] [Google Scholar]
- 17.Nakao M, Yokota S, Iwai T, Kaneko H, Horiike S, Kashima K, Sonoda Y, Fujimoto T, Misawa S. 1996. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia 10:1911–1918 [PubMed] [Google Scholar]
- 18.Ravandi F, Kantarjian H, Faderl S, Garcia-Manero G, O'Brien S, Koller C, Pierce S, Brandt M, Kennedy D, Cortes J, Beran M. 2010. Outcome of patients with FLT3-mutated acute myeloid leukemia in first relapse. Leuk Res 34:752–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rombouts WJ, Blokland I, Lowenberg B, Ploemacher RE. 2000. Biological characteristics and prognosis of adult acute myeloid leukemia with internal tandem duplications in the FLT3 gene. Leukemia 14:675–683 [DOI] [PubMed] [Google Scholar]
- 20.Rosnet O, Buhring HJ, deLapeyriere O, Beslu N, Lavagna C, Marchetto S, Rappold I, Drexler HG, Birg F, Rottapel R, Hannum C, Dubreuil P, Birnbaum D. 1996. Expression and signal transduction of the FLT3 tyrosine kinase receptor. Acta Haematol 95:218–223 [DOI] [PubMed] [Google Scholar]
- 21.Sato T, Yang X, Knapper S, White P, Smith BD, Galkin S, Small D, Burnett A, Levis M. 2011. FLT3 ligand impedes the efficacy of FLT3 inhibitors in vitro and in vivo. Blood 117:3286–3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shih LY, Huang CF, Wu JH, Lin TL, Dunn P, Wang PN, Kuo MC, Lai CL, Hsu HC. 2002. Internal tandem duplication of FLT3 in relapsed acute myeloid leukemia: a comparative analysis of bone marrow samples from 108 adult patients at diagnosis and relapse. Blood 100:2387–2392 [DOI] [PubMed] [Google Scholar]
- 23.Small D. 2006. FLT3 mutations: biology and treatment. Hematology Am Soc Hematol Educ Program 2006:178–184 [DOI] [PubMed] [Google Scholar]
- 24.Small D, Levenstein M, Kim E, Carow C, Amin S, Rockwell P, Witte L, Burrow C, Ratajczak MZ, Gewirtz AM, Civin CI. 1994. STK1, the human homolog of Flk2/Flt3, is selectively expressed in CD34+ human bone marrow cells and is involved in the proliferation of early progenitor–stem cells. Proc Natl Acad Sci USA 91:459–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stirewalt DL, Kopecky KJ, Meshinchi S, Engel JH, Pogosova-Agadjanyan EL, Linsley J, Slovak ML, Willman CL, Radich JP. 2006. Size of FLT3 internal tandem duplication has prognostic significance in patients with acute myeloid leukemia. Blood 107:3724–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stirewalt DL, Radich JP. 2003. The role of FLT3 in haematopoietic malignancies. Nat Rev Cancer 3:650–665 [DOI] [PubMed] [Google Scholar]
- 27.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. 2008. WHO classification of tumours of haematopoietic and lymphoid tissues, 4th ed. Lyon (France): International Agency for Research on Cancer (IARC) [Google Scholar]
- 28.Thiede C, Steudel C, Mohr B, Schaich M, Schakel U, Platzbecker U, Wermke M, Bornhauser M, Ritter M, Neubauer A, Ehninger G, Illmer T. 2002. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood 99:4326–4335 [DOI] [PubMed] [Google Scholar]
- 29.Whitman SP, Archer KJ, Feng L, Baldus C, Becknell B, Carlson BD, Carroll AJ, Mrozek K, Vardiman JW, George SL, Kolitz JE, Larson RA, Bloomfield CD, Caligiuri MA. 2001. Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: a Cancer and Leukemia Group B study. Cancer Res 61:7233–7239 [PubMed] [Google Scholar]
- 30.Wodnar-Filipowicz A, Lyman SD, Gratwohl A, Tichelli A, Speck B, Nissen C. 1996. Flt3 ligand level reflects hematopoietic progenitor cell function in aplastic anemia and chemotherapy-induced bone marrow aplasia. Blood 88:4493–4499 [PubMed] [Google Scholar]
- 31.Zhang S, Fukuda S, Lee Y, Hangoc G, Cooper S, Spolski R, Leonard WJ, Broxmeyer HE. 2000. Essential role of signal transducer and activator of transcription (Stat)5a but not Stat5b for Flt3-dependent signaling. J Exp Med 192:719–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang S, Mantel C, Broxmeyer HE. 1999. Flt3 signaling involves tyrosyl phosphorylation of SHP2 and SHIP and their association with Grb2 and Shc in Baf3–Flt3 cells. J Leukoc Biol 65:372–380 [DOI] [PubMed] [Google Scholar]
- 33.Zheng R, Bailey E, Nguyen B, Yang X, Piloto O, Levis M, Small D. 2011. Further activation of FLT3 mutants by FLT3 ligand. Oncogene 30:4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou J, Bi C, Janakakumara JV, Liu SC, Chng WJ, Tay KG, Poon LF, Xie Z, Palaniyandi S, Yu H, Glaser KB, Albert DH, Davidsen SK, Chen CS. 2009. Enhanced activation of STAT pathways and overexpression of survivin confer resistance to FLT3 inhibitors and could be therapeutic targets in AML. Blood 113:4052–4062 [DOI] [PubMed] [Google Scholar]
- 35.Zwierzina H, Anderson JE, Rollinger-Holzinger I, Torok-Storb B, Nuessler V, Lyman SD. 1999. Endogenous FLT3 ligand serum levels are associated with disease stage in patients with myelodysplastic syndromes. Leukemia 13:553–557 [DOI] [PubMed] [Google Scholar]