Summary
Nck links phosphotyrosine-based signaling to Arp2/3-dependent actin polymerization during many different cellular processes as well as actin-based motility of enteropathogenic Escherichia coli (EPEC) [1, 2], vaccinia [3, 4], and other vertebrate poxviruses [5] by interacting with N-WASP/WASP [6, 7]. Nck also binds WASP-interacting protein (WIP) [8], which inhibits the ability of N-WASP to activate the Arp2/3 complex until it receives an appropriate signaling input [9, 10]. Using mouse embryonic fibroblasts (MEFs) lacking Nck, WIP, or N-WASP [3, 11, 12], we have investigated whether an interaction of Nck with both WIP and N-WASP is required for their recruitment to vaccinia during Arp2/3-dependent actin assembly. We find that WIP or its homolog WIRE is required for N-WASP recruitment and actin-based motility of the virus. WIP contains two Nck-binding sites and is recruited to the virus, bound to N-WASP, by interacting with the second SH3 domain of Nck. N-WASP also contains two Nck-binding sites, but its recruitment is dependent on its interaction with WIP rather than Nck. The first and third SH3 domains of Nck are not required to recruit the WIP:N-WASP complex but are essential to stimulate actin assembly. We have established that WIP acts as an essential link between Nck and N-WASP. Our observations provide important insights into the hierarchy and connections in one of the major cellular signaling networks stimulating Arp2/3 complex-dependent actin polymerization.
Highlights
-
•
Vaccinia-induced actin polymerization is dependent on WIP or WIRE
-
•
WIP and N-WASP both contain two Nck-binding sites
-
•
The second SH3 domain of Nck recruits a WIP:N-WASP complex via WIP
-
•
Recruitment and activation of WIP:N-WASP is mediated by different Nck SH3 domains
Results and Discussion
WIP or WIRE Is Essential for Actin-Based Motility of Vaccinia
Fusion of newly assembled vaccinia virus particles with the plasma membrane results in Src and Abl family kinase-mediated phosphorylation of tyrosine 112 and 132 of the viral membrane protein A36 (Figure 1A) [13, 14, 15, 16]. Phosphorylation of A36 leads to the recruitment of a signaling network consisting of Grb2, Nck, WIP, and N-WASP that stimulates Arp2/3 complex-dependent actin polymerization beneath extracellular viruses attached to the plasma membrane (Figure 1A) [3, 4, 13, 17, 18, 19, 20]. The induction of actin polymerization beneath the virus ultimately enhances the spread of infection by propelling the virus onto neighboring cells [21, 22, 23, 24].
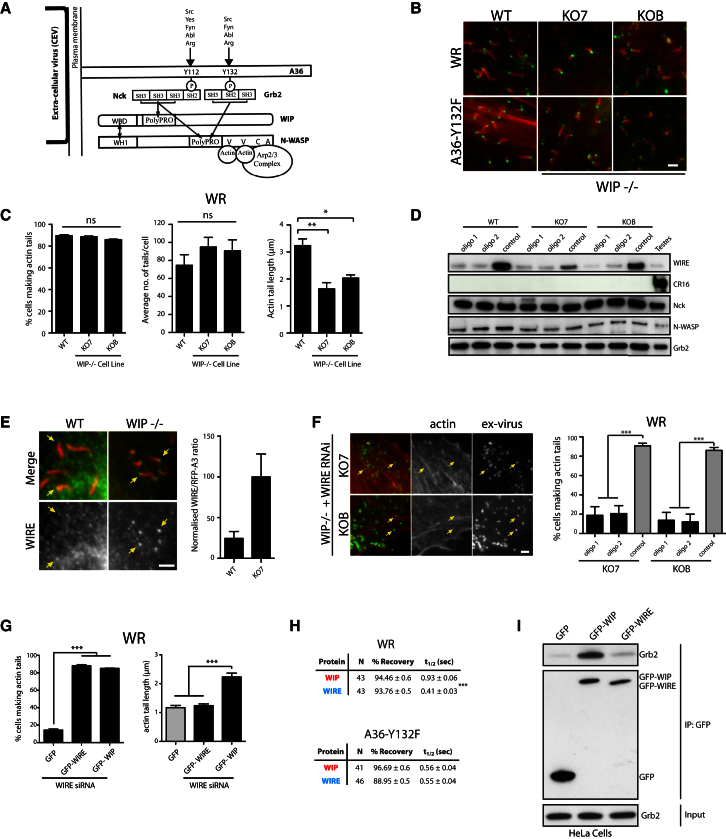
Figure 1.
WIP or WIRE Is Required for Actin Tail Formation
(A) Schematic representation of the interactions between the proteins in the vaccinia virus actin-polymerization complex. Src and Abl family kinases phosphorylating tyrosine 112 or 132 of A36 are indicated, together with motifs and domains.
(B) Immunofluorescence images showing actin tails (red) induced by Western Reserve (WR) and A36-Y132F viruses (ex-virus) in wild-type (WT) or WIP−/− (KO7 and KOB) MEFs.
(C) Quantification of the percentage of cells with actin tails, the average number actin tails, and their length in WT or WIP−/− MEFs infected with WR.
(D) The immunoblot shows the level of WIRE, CR16, Nck, N-WASP, and Grb2 in WT or WIP−/− MEFs after WIRE knockdown with the indicated siRNA oligos.
(E) Images and intensity quantification showing the recruitment of endogenous WIRE to WR (yellow arrows) increases in the absence of WIP.
(F) Images of WIP−/− MEFs treated with WIRE siRNA and infected with WR. The graphs show the quantification of the percentage of WR-infected cells with at least one actin tail in WIRE siRNA-treated (black bars) or control-treated (gray bar) WIP−/− cell lines.
(G) Quantification of the percentage of WR-infected cells expressing the indicated GFP-tagged protein with at least one actin tail and their average length in WIP−/− cells treated with control (gray bars) or WIRE (black bars) siRNA.
(H) Comparison of the recovery kinetics of GFP-tagged WIP or WIRE on WR or A36-Y132F after photobleaching in WIRE siRNA-treated WIP−/− cells (KO7).
(I) Immunoblot analysis shows that endogenous Grb2 coimmunoprecipitates with GFP-WIP, but not GFP-WIRE. The Grb2 input and immunoprecipitated GFP-tagged proteins are shown. All error bars in the graphs represent SEM from three independent experiments. ns, not significant; **p < 0.05; **p < 0.01; ***p < 0.001. Scale bars represent 2 μm. See also Figure S1.
Nck and N-WASP are essential for vaccinia-induced actin polymerization [3, 4]. In contrast, recent observations suggest that WIP is not required for actin-based motility of vaccinia virus [25]. We also found that the Western Reserve (WR) or its A36-Y132F mutant, which is deficient in Grb2 recruitment [4, 18], are able to induce actin tails in two independently derived mouse embryonic fibroblast (MEF) cell lines lacking WIP (Figures 1A–1C; see also Figures S1A and S1B available online). Loss of WIP did, however, reduce the length of WR-induced, but not A36-Y132F-induced, actin tails (Figures 1C and S1B). We wondered whether the function of WIP is replaced by the WIP-related protein WIRE/WICH [26, 27], which, in contrast to CR16, is expressed in both MEF cell lines lacking WIP (Figure 1D). Consistent with this notion, there is a dramatic increase in WIRE recruitment to the tips of WR-induced actin tails in the absence of WIP (Figure 1E). Knockdown of WIRE in wild-type MEFs expressing WIP had no impact on WR or A36-Y132F actin tail formation (Figure S1C). In contrast, we found that RNAi-mediated ablation of WIRE in WIP-deficient MEFs results in a dramatic reduction in the number of WR- or A36-Y132F-infected cells with actin tails (Figures 1D, 1F, and S1D). In addition, where actin tails did form, their average number per cell was decreased by over 90% (Figure S1E). Our results are contrary to those of Garber et al. [25], which were obtained using a single WIP−/− cell line. We believe the most likely explanation for this difference relates to the efficiency of WIRE knockdown, as in our experience vaccinia is very efficient at recruiting residual protein. This would also explain why we never achieve 100% inhibition of actin tail formation in infected WIP-deficient MEFs treated with WIRE RNAi (Figures 1F, S1D, and S1E).
Expression of GFP-tagged human WIP or WIRE in MEFs lacking endogenous WIP and WIRE rescues the ability of both viruses to induce actin tails (Figures 1G, S1F, and S1G). Consistent with our earlier observations, the WR-induced actin tails rescued by GFP-WIRE were shorter than those formed by GFP-WIP (Figure 1G). In contrast, A36-Y132F actin tails were equally short (Figure S1G). The most straightforward explanation for this difference in actin tail length is that Grb2, which is downstream of phosphorylated tyrosine 132 of A36 (Figure 1A), can interact with WIP, but not WIRE. Consistent with this notion, fluorescence recovery after photobleaching (FRAP) analysis reveals that the exchange rate of GFP-WIRE on WR-induced actin tails is ∼2.2 times faster than that of WIP (Figures 1H and S1H). In contrast, the turnover of both proteins on the A36-Y132F virus is similar (Figures 1H and S1H). Pull-down assays from HeLa cell extracts confirmed that endogenous Grb2 readily copurifies with GFP-tagged WIP, but not WIRE (Figure 1I). Our observations demonstrate that WIP or WIRE is required for vaccinia actin-based motility, although the presence of Grb2 only stabilizes WIP to promote the formation of longer actin tails. Our data also suggest that the absence of a phenotype in WIP−/− cells during N-WASP-dependent cellular processes should be treated with caution, as WIP and WIRE are clearly interchangeable in some circumstances. For example, the presence of WIRE may explain why N-WASP-dependent Mycobacterium marinum actin tail formation still occurs in WIP−/− MEFs [28].
WIP Links Nck to N-WASP during Actin Tail Formation
Nck can interact with the polyproline-rich regions of both WIP and N-WASP [7, 8]. However, it remains to be established whether the interaction of Nck with both of these proteins is essential for their recruitment to the virus. To investigate the importance of the interaction of Nck with WIP, we set out to define the Nck-binding sites in WIP. By probing a peptide array of WIP with His-Nck1, we identified two Nck-binding peptides containing PXXPXR class II SH3-binding motifs [29] (Figures 2A and S2A). The second of these motifs conforms to the consensus-binding site for the second SH3 domain of Nck [30]. It is also striking that both of the WIP peptides share a common PXXPXRXL motif.
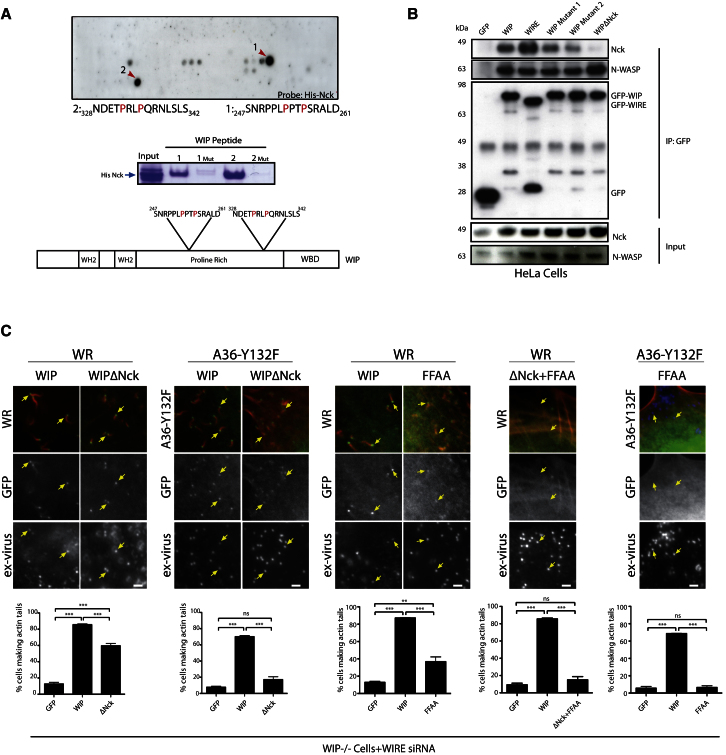
Figure 2.
WIP Is Essential to Link Nck to N-WASP
(A) Far western analysis of a peptide array of WIP probed with His-Nck1 identifies two Nck-interacting peptides (red arrows). In vitro peptide-binding assays demonstrate that the two WIP peptides 1 and 2 identified in the far western analysis retain His-Nck from an E. coli-soluble fraction. Mutation of the two prolines indicated in red to alanine (1 Mut, 2 Mut) leads to loss of Nck binding.
(B) Immunoblot analysis demonstrates that endogenous N-WASP, but not Nck, coimmunoprecipitates with GFP-WIPΔNck. The N-WASP and Nck inputs and the immunoprecipitated GFP-tagged proteins are indicated.
(C) Images showing the recruitment of GFP-tagged WIP, WIPΔNck, WIP-FFAA, or WIPΔNck+FFAA to WR and A36-Y132F viruses as well as actin tail formation in WIP−/− MEFs treated with WIRE siRNA. The graphs show the quantification of the percentage of infected WIRE siRNA-treated WIP−/− cells expressing the indicated protein with at least one actin tail. All error bars represent SEM from three independent experiments. **p < 0.01; ***p < 0.001; ns, not significant. Scale bars represent 2 μm. See also Figure S2.
In vitro binding assays demonstrate both peptides can interact with Nck and that alanine substitution of the prolines in the PXXPXR motif disrupts binding (Figure 2A). Pull-down assays on HeLa cell extracts containing the GFP-tagged WIP mutants demonstrated that both sites are functional, as loss of Nck binding was only achieved when they were both mutated (WIPΔNck) (Figure 2B). Disrupting the binding of WIP to Nck did not impact its interaction with N-WASP. It did, however, result in the loss of Nck binding to N-WASP complexed to WIPΔNck (Figure 2B). Nevertheless, GFP-WIPΔNck is still recruited to WR but is not as effective as WIP in rescuing actin tail formation (Figure 2C). Those actin tails that did form were also significantly shorter (Figure S2B). Loss of the ability of WIP to bind Nck also resulted in a 1.65-fold increase in its rate of exchange (half-life of recovery = 0.55 ± 0.05 s compared to 0.91 ± 0.07 s) (Figure S2C). In the absence of Grb2, GFP-WIPΔNck recruitment was considerably weaker, and the A36-Y132F virus induced very few short actin tails in WIP−/− MEFs treated with siRNA against WIRE (Figures 2C and S2B).
These observations with the A36-Y132F virus suggest that WIP plays an important role in connecting Nck to N-WASP. To test this hypothesis, we investigated the consequences of disrupting the interaction between WIP and N-WASP on the ability of vaccinia to induce actin tails. We found that GFP-WIP-FFAA, which cannot bind the WH1 domain of N-WASP (Figure S2D) [19, 31], is poorly recruited to WR, has a faster rate of exchange (half-life of recovery = 0.27 ± 0.04 s), and induces even lower numbers of short actin tails than WIPΔNck (Figures 2C and S2E). Moreover, when the FFAA mutation was combined with ΔNck, WIP was largely incapable of rescuing WR-induced actin tails, consistent with its lack of recruitment on the majority of virus particles (Figure 2C). In agreement with a role for WIP in linking Nck to N-WASP, we found that N-WASP was also weakly recruited to very few virus particles in cells lacking WIP and WIRE but expressing GFP-WIPΔNck+FFAA (Figure S2F). The residual recruitment of N-WASP and formation of small numbers of actin tails is likely due to incomplete knockdown of WIRE (Figure 1D).
To investigate the contribution of Grb2 in the system, we examined whether the A36-Y132F virus can induce actin tails in WIP−/− cells lacking WIRE (RNAi treated) and expressing GFP-WIP-FFAA. Consistent with the role of Grb2 in stabilizing the vaccinia-signaling complex [4], we found that GFP-WIP-FFAA and endogenous N-WASP were not recruited to the A36-Y132F virus and no actin tails were formed (Figures 2C and S2G). Our observations demonstrate that, in the absence of Grb2, the simultaneous interaction of WIP with both Nck and N-WASP is critical for vaccinia to induce actin polymerization.
How Important Is the Interaction of Nck with N-WASP?
Using a far western approach followed by in vitro peptide-binding assays, we identified two Nck-binding peptides with PXXPXR class II SH3-binding motifs in N-WASP (Figures 3A and S3A). Alanine substitution of both prolines in the PXXPXR motifs weakened but did not fully abrogate Nck binding, presumably due to the presence of additional PXXP motifs in the peptides (Figure 3A). Pull-down assays on HeLa cell lysates reveal that Nck still associates with the N-WASP mutants, albeit more weakly than with the wild-type protein (Figure 3B). This residual Nck binding is likely to be largely mediated by WIP, as it still binds N-WASPΔNck (Figure 3B). Consistent with this, Nck binding to N-WASPΔNck is severely reduced in the absence of WIP and WIRE (Figure 3C). GFP-N-WASPΔNck is still recruited to WR and weakly to the A36-Y132F virus (no Grb2) in N-WASP−/− MEFs (Figure 3D). In contrast to the situation with WIP, the loss of Nck binding to N-WASP did not impact the ability of either virus to induce actin polymerization (Figures 3D and 3E). The actin tails induced by both viruses were, however, reduced in length (Figure 3E). There was also a small but significant increase in the rate of N-WASP exchange on WR in the absence of Nck binding (Figure 3F). An interaction with Nck clearly helps stabilize N-WASP, but it does not appear to be essential for its recruitment to the virus or actin tail formation even in the absence of Grb2.
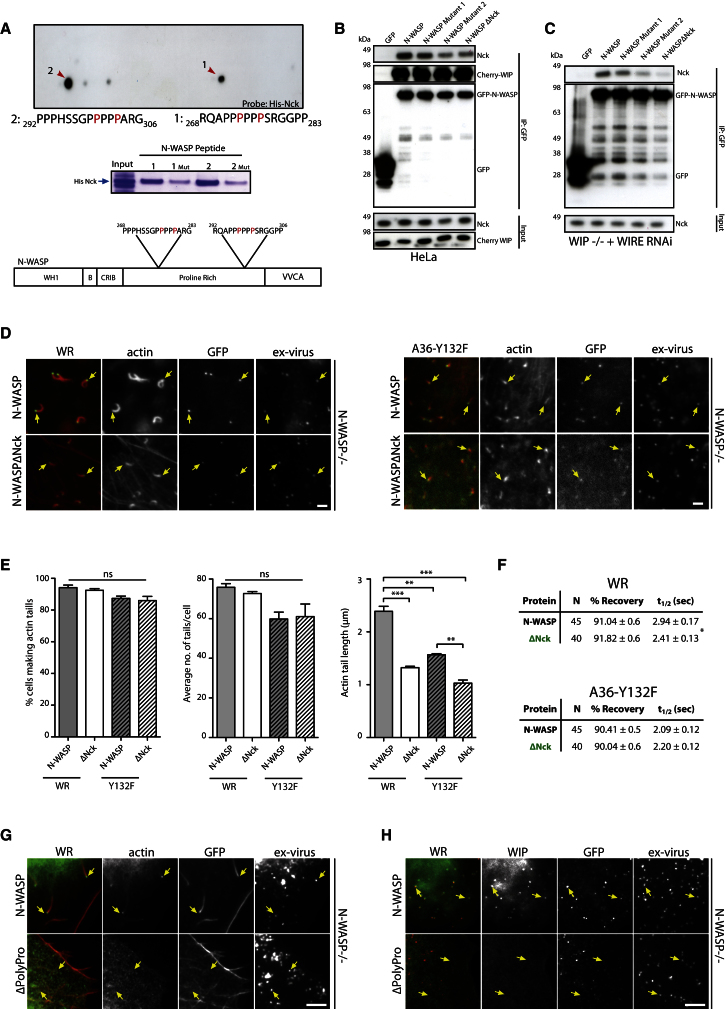
Figure 3.
The Interaction of Nck and N-WASP Is Not Required for Actin Tail Formation
(A) Far western analysis of an N-WASP peptide array with His-Nck1 identifies two Nck-interacting peptides (red arrows). In vitro peptide-binding assays demonstrate that the two peptides identified in the far western analysis bind His-Nck. Substitution of the two prolines indicated in red to alanine (1 Mut, 2 Mut) reduce but do not abrogate Nck binding.
(B) Immunoblot analysis demonstrates that Nck coimmunoprecipitates with GFP-tagged N-WASP and N-WASPΔNck from HeLa cell extracts in the presence of WIP.
(C) Immunoblot analysis shows that GFP-tagged N-WASP, but not N-WASPΔNck, can interact with Nck in the absence of WIP and WIRE.
(D) Images showing actin tail formation and the recruitment of GFP-tagged N-WASP and N-WASPΔNck to WR or the A36-Y132F virus in N-WASP−/− cells.
(E) Quantification of the percentage of WR and A36-Y132F-infected cells with actin tails, the average number of tails per cell, and their length in N-WASP−/− cells expressing N-WASP and N-WASPΔNck.
(F) The recovery kinetics of GFP-N-WASP or GFP-N-WASPΔNck on WR and the A36-Y132F virus after photobleaching in N-WASP−/− cells is shown.
(G) Images showing that GFP-N-WASPΔpolyPro is not recruited to WR (ex-virus) in N-WASP−/− cells.
(H) Endogenous WIP is not recruited to WR (ex-virus) in N-WASP−/− cells expressing GFP-N-WASPΔpolyPro. All error bars are the SEM from three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant. Scale bars represent 2 μm. See also Figure S3.
Given this unexpected result, we examined whether N-WASP lacking its proline-rich region would rescue WR actin tail formation in N-WASP−/− MEFs. We found that GFP-N-WASPΔpolyPro is not recruited to WR, nor is it able to rescue actin tail formation in N-WASP−/− MEFs (Figure 3G). Endogenous WIP is also not recruited to virus particles in N-WASP−/− MEFs expressing GFP-N-WASPΔpolyPro (Figure 3H). The most straightforward explanation for the difference between N-WASPΔpolyPro and N-WASPΔNck is that an additional unknown protein binds the latter to contribute to N-WASP recruitment. We do not believe this unknown component is Grb2, as N-WASPΔNck is still recruited to the A36-Y132F virus and also has the same exchange rate as N-WASP in the absence of Grb2 (Figure 3F).
The Second SH3 Domain of Nck Is Essential to Recruit WIP
Our previous observations have shown that Nck, but not WIP, is still recruited to WR in N-WASP−/− MEFs [4]. Similarly, we found that Nck is still recruited to WR and A36-Y132F in the absence of WIP and WIRE (Figure S4A). This demonstrates that Nck is recruited independently from the WIP:N-WASP complex. Given that Nck is upstream of WIP:N-WASP, we performed in vitro peptide pull-down assays to investigate whether the three different Nck SH3 domains have a preference for the two different Nck-binding sites in WIP and N-WASP (Figures 4A and 4B). Consistent with the presence of a common PXXPXRXL motif, we found that mutation of the second SH3 domain eliminated Nck binding to both WIP peptides (Figure 4B). In contrast, the N-WASP peptides showed the greatest preference for the third SH3 domain of Nck (Figure 4B). To investigate whether these pull-down assays reflect the situation in cells, we examined the ability of WR and the A36-Y132F virus to induce actin tails in Nck1/2−/− cells expressing GFP-tagged Nck SH3 mutants. Mutation of individual SH3 domains had no impact on the ability of WR to induce actin tails (Figures 4C and 4D). In contrast, in the absence of Grb2 recruitment, the A36-Y132F virus was unable to induce actin tails in Nck1/2−/− cells expressing the NckΔ2 mutant (Figures 4C and 4D). A similarly dramatic loss of WR-induced actin tails is only observed when any pair of SH3 domains is disrupted (Figure 4D). The loss of A36-Y132F virus-induced actin tails in Nck and WIP null cells expressing NckΔ2 and WIPΔNck, respectively, but not N-WASP−/− cells expressing N-WASPΔNck, confirms that an interaction of the second Nck SH3 domain with WIP is required to recruit a complex of WIP:N-WASP to the virus. Consistent with this, pull-downs demonstrate that NckΔ2 is deficient in its ability to interact with N-WASP (Figure 4E).
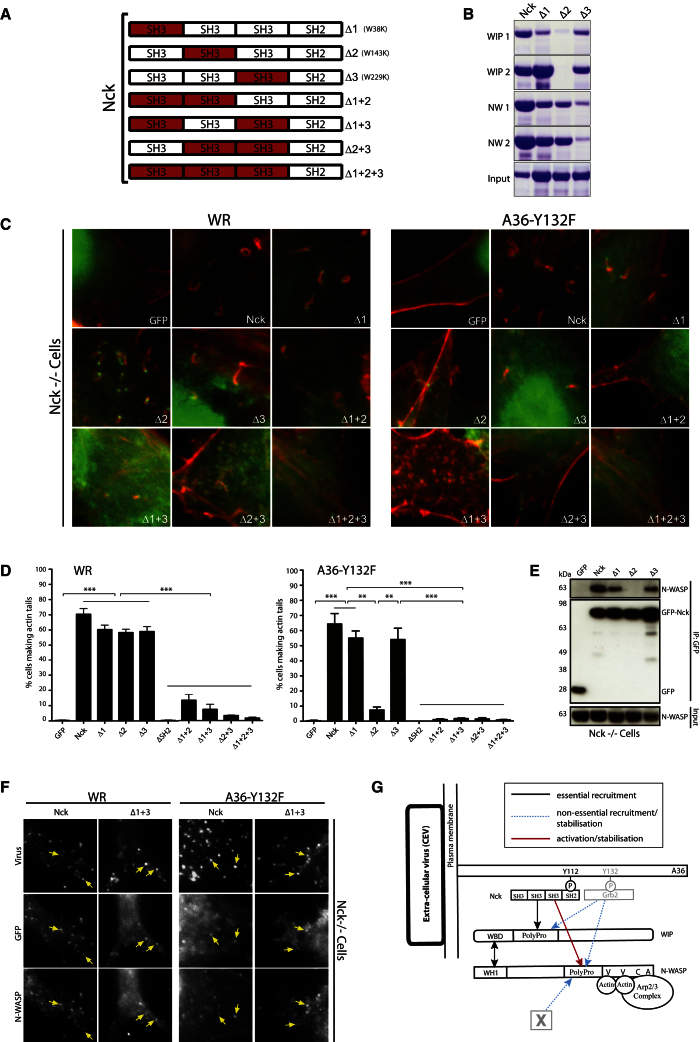
Figure 4.
The Second SH3 Domain of Nck Is Essential for Actin Tail Formation
(A) Schematic representation of the SH3-disrupting point mutations (highlighted in red) introduced into Nck.
(B) Pull-down of recombinant His-tagged wild-type or mutant Nck with the indicated peptides (left) identified in the WIP and N-WASP peptide arrays.
(C) Immunofluorescence images of actin tails induced by WR or A36-Y132F in Nck−/− cells expressing the indicated GFP-tagged Nck mutant.
(D) Quantification of the percentage of WR or A36-Y132F-infected Nck−/− cells expressing the indicated GFP-Nck mutant inducing actin tails. Error bars represent the SEM from three independent experiments. **p < 0.01; ***p < 0.001.
(E) Immunoblot analysis demonstrates that endogenous N-WASP does not coimmunoprecipitate with GFP-NckΔ2. The N-WASP input and the immunoprecipitated GFP-tagged proteins are indicated.
(F) Immunofluorescence images demonstrating that endogenous N-WASP is recruited to WR or A36-Y132F viruses in Nck−/− cells expressing the indicated GFP-tagged Nck or NckΔ1+3.
(G) Schematic representation of the interactions and their importance in the Nck:WIP:N-WASP-signaling network. Light gray (Grb2 and X) help but are not essential for Nck:WIP:N-WASP-dependent actin tail formation. Scale bars represent 2 μm. See also Figure S4.
Our conclusion is consistent with recent observations showing that WIP is required to recruit N-WASP to Nck SH3 aggregates [32]. The same study also suggested that WIP not only links Nck to N-WASP but also allows the latter to bind and be activated by a second Nck molecule [32]. This suggestion was based on the observation that the second Nck SH3 domain alone, while sufficient to recruit N-WASP, presumably via WIP, was unable to induce actin polymerization unless all three Nck SH3 domains were present [32]. Our observations, however, suggest that the reason the second Nck SH3 domain alone cannot induce robust actin polymerization is because it recruits but does not activate the WIP:N-WASP complex. In agreement with this notion, we found that endogenous N-WASP is recruited to the virus even in the absence of Grb2 (A36-Y132F virus) in Nck1/2−/− cells expressing GFP-NckΔ1+3, despite the absence of actin tail formation (Figures 4D and 4F). We suggest that the second Nck SH3 domain interacts with WIP to recruit the WIP:N-WASP complex, and the subsequent binding of the first, or more likely the third, SH3 domain to N-WASP is required to activate the Arp2/3 complex (Figure 4G). Such a model explains why mutation of the second Nck SH3 domain leads to a loss of A36-Y132F actin tails and why WR is unable to induce actin polymerization when any pair of Nck SH3 domains is mutated, even though Grb2 is present. The interaction of Nck with WIP is clearly essential for recruitment of the WIP/N-WASP complex. However, our observations also demonstrate that, in the absence of Grb2, an interaction between N-WASP and WIP is critical for complex recruitment and actin tail formation (Figures 2C and S2G). Given this, it is striking that the majority of mutations resulting in Wiskott-Aldrich syndrome are located in the WIP-binding WH1 domain of WASP [33]. In summary, our analysis, which has important implications for a variety of Nck:N-WASP/WASP-dependent cellular processes, has established that WIP is essential to link Nck-dependent signaling to the Arp2/3 complex via N-WASP. The task ahead is now to understand the molecular events involved in activating the WIP:N-WASP complex once it has been recruited to the virus by Nck.
Acknowledgments
We wish to thank Tony Pawson (Samuel Lunenfeld Research Institute, Toronto), Raif Geha (Harvard Medical School, Boston), and Scott Snapper (Department of Medicine and Immunology, Massachusetts General Hospital, Boston) for Nck-, WIP-, and N-WASP-null MEFs, respectively. We also thank Shiro Suetsugu (University of Tokyo, Tokyo) for the CR16 polyclonal antibody and Jasmine Abella (of the Way laboratory) for the pLVX-GFP-puro lentivirus vector. We also wish to thank all members of the Way lab for constructive comments on the manuscript and Nicola O’Reilly (LRI Peptide Synthesis Laboratory) for the peptide arrays. This work was supported by Cancer Research UK and LRI Ph.D. studentships to S.K.D., I.W., and M.Z.
Published: May 23, 2013
Footnotes
Supplemental Information includes four figures and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.cub.2013.04.051.
Supplemental Information
References
- 1.Gruenheid S., DeVinney R., Bladt F., Goosney D., Gelkop S., Gish G.D., Pawson T., Finlay B.B. Enteropathogenic E. coli Tir binds Nck to initiate actin pedestal formation in host cells. Nat. Cell Biol. 2001;3:856–859. doi: 10.1038/ncb0901-856. [DOI] [PubMed] [Google Scholar]
- 2.Lommel S., Benesch S., Rottner K., Franz T., Wehland J., Kühn R. Actin pedestal formation by enteropathogenic Escherichia coli and intracellular motility of Shigella flexneri are abolished in N-WASP-defective cells. EMBO Rep. 2001;2:850–857. doi: 10.1093/embo-reports/kve197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snapper S.B., Takeshima F., Antón I., Liu C.H., Thomas S.M., Nguyen D., Dudley D., Fraser H., Purich D., Lopez-Ilasaca M. N-WASP deficiency reveals distinct pathways for cell surface projections and microbial actin-based motility. Nat. Cell Biol. 2001;3:897–904. doi: 10.1038/ncb1001-897. [DOI] [PubMed] [Google Scholar]
- 4.Weisswange I., Newsome T.P., Schleich S., Way M. The rate of N-WASP exchange limits the extent of ARP2/3-complex-dependent actin-based motility. Nature. 2009;458:87–91. doi: 10.1038/nature07773. [DOI] [PubMed] [Google Scholar]
- 5.Dodding M.P., Way M. Nck- and N-WASP-dependent actin-based motility is conserved in divergent vertebrate poxviruses. Cell Host Microbe. 2009;6:536–550. doi: 10.1016/j.chom.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 6.Rivero-Lezcano O.M., Marcilla A., Sameshima J.H., Robbins K.C. Wiskott-Aldrich syndrome protein physically associates with Nck through Src homology 3 domains. Mol. Cell. Biol. 1995;15:5725–5731. doi: 10.1128/mcb.15.10.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rohatgi R., Nollau P., Ho H.Y., Kirschner M.W., Mayer B.J. Nck and phosphatidylinositol 4,5-bisphosphate synergistically activate actin polymerization through the N-WASP-Arp2/3 pathway. J. Biol. Chem. 2001;276:26448–26452. doi: 10.1074/jbc.M103856200. [DOI] [PubMed] [Google Scholar]
- 8.Antón I.M., Lu W., Mayer B.J., Ramesh N., Geha R.S. The Wiskott-Aldrich syndrome protein-interacting protein (WIP) binds to the adaptor protein Nck. J. Biol. Chem. 1998;273:20992–20995. doi: 10.1074/jbc.273.33.20992. [DOI] [PubMed] [Google Scholar]
- 9.Martinez-Quiles N., Rohatgi R., Antón I.M., Medina M., Saville S.P., Miki H., Yamaguchi H., Takenawa T., Hartwig J.H., Geha R.S., Ramesh N. WIP regulates N-WASP-mediated actin polymerization and filopodium formation. Nat. Cell Biol. 2001;3:484–491. doi: 10.1038/35074551. [DOI] [PubMed] [Google Scholar]
- 10.Ho H.Y., Rohatgi R., Lebensohn A.M., Le Ma, Li J., Gygi S.P., Kirschner M.W. Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP-WIP complex. Cell. 2004;118:203–216. doi: 10.1016/j.cell.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 11.Bladt F., Aippersbach E., Gelkop S., Strasser G.A., Nash P., Tafuri A., Gertler F.B., Pawson T. The murine Nck SH2/SH3 adaptors are important for the development of mesoderm-derived embryonic structures and for regulating the cellular actin network. Mol. Cell. Biol. 2003;23:4586–4597. doi: 10.1128/MCB.23.13.4586-4597.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antón I.M., Saville S.P., Byrne M.J., Curcio C., Ramesh N., Hartwig J.H., Geha R.S. WIP participates in actin reorganization and ruffle formation induced by PDGF. J. Cell Sci. 2003;116:2443–2451. doi: 10.1242/jcs.00433. [DOI] [PubMed] [Google Scholar]
- 13.Frischknecht F., Moreau V., Röttger S., Gonfloni S., Reckmann I., Superti-Furga G., Way M. Actin-based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature. 1999;401:926–929. doi: 10.1038/44860. [DOI] [PubMed] [Google Scholar]
- 14.Newsome T.P., Scaplehorn N., Way M. SRC mediates a switch from microtubule- to actin-based motility of vaccinia virus. Science. 2004;306:124–129. doi: 10.1126/science.1101509. [DOI] [PubMed] [Google Scholar]
- 15.Reeves P.M., Bommarius B., Lebeis S., McNulty S., Christensen J., Swimm A., Chahroudi A., Chavan R., Feinberg M.B., Veach D. Disabling poxvirus pathogenesis by inhibition of Abl-family tyrosine kinases. Nat. Med. 2005;11:731–739. doi: 10.1038/nm1265. [DOI] [PubMed] [Google Scholar]
- 16.Newsome T.P., Weisswange I., Frischknecht F., Way M. Abl collaborates with Src family kinases to stimulate actin-based motility of vaccinia virus. Cell. Microbiol. 2006;8:233–241. doi: 10.1111/j.1462-5822.2005.00613.x. [DOI] [PubMed] [Google Scholar]
- 17.Moreau V., Frischknecht F., Reckmann I., Vincentelli R., Rabut G., Stewart D., Way M. A complex of N-WASP and WIP integrates signalling cascades that lead to actin polymerization. Nat. Cell Biol. 2000;2:441–448. doi: 10.1038/35017080. [DOI] [PubMed] [Google Scholar]
- 18.Scaplehorn N., Holmström A., Moreau V., Frischknecht F., Reckmann I., Way M. Grb2 and Nck act cooperatively to promote actin-based motility of vaccinia virus. Curr. Biol. 2002;12:740–745. doi: 10.1016/s0960-9822(02)00812-6. [DOI] [PubMed] [Google Scholar]
- 19.Zettl M., Way M. The WH1 and EVH1 domains of WASP and Ena/VASP family members bind distinct sequence motifs. Curr. Biol. 2002;12:1617–1622. doi: 10.1016/s0960-9822(02)01112-0. [DOI] [PubMed] [Google Scholar]
- 20.Humphries A.C., Dodding M.P., Barry D.J., Collinson L.M., Durkin C.H., Way M. Clathrin potentiates vaccinia-induced actin polymerization to facilitate viral spread. Cell Host Microbe. 2012;12:346–359. doi: 10.1016/j.chom.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Cudmore S., Cossart P., Griffiths G., Way M. Actin-based motility of vaccinia virus. Nature. 1995;378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 22.Cudmore S., Reckmann I., Griffiths G., Way M. Vaccinia virus: a model system for actin-membrane interactions. J. Cell Sci. 1996;109:1739–1747. doi: 10.1242/jcs.109.7.1739. [DOI] [PubMed] [Google Scholar]
- 23.Ward B.M., Moss B. Vaccinia virus intracellular movement is associated with microtubules and independent of actin tails. J. Virol. 2001;75:11651–11663. doi: 10.1128/JVI.75.23.11651-11663.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doceul V., Hollinshead M., van der Linden L., Smith G.L. Repulsion of superinfecting virions: a mechanism for rapid virus spread. Science. 2010;327:873–876. doi: 10.1126/science.1183173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garber J.J., Takeshima F., Antón I.M., Oyoshi M.K., Lyubimova A., Kapoor A., Shibata T., Chen F., Alt F.W., Geha R.S. Enteropathogenic Escherichia coli and vaccinia virus do not require the family of WASP-interacting proteins for pathogen-induced actin assembly. Infect. Immun. 2012;80:4071–4077. doi: 10.1128/IAI.06148-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aspenström P. The WASP-binding protein WIRE has a role in the regulation of the actin filament system downstream of the platelet-derived growth factor receptor. Exp. Cell Res. 2002;279:21–33. doi: 10.1006/excr.2002.5576. [DOI] [PubMed] [Google Scholar]
- 27.Kato M., Miki H., Kurita S., Endo T., Nakagawa H., Miyamoto S., Takenawa T. WICH, a novel verprolin homology domain-containing protein that functions cooperatively with N-WASP in actin-microspike formation. Biochem. Biophys. Res. Commun. 2002;291:41–47. doi: 10.1006/bbrc.2002.6406. [DOI] [PubMed] [Google Scholar]
- 28.Stamm L.M., Pak M.A., Morisaki J.H., Snapper S.B., Rottner K., Lommel S., Brown E.J. Role of the WASP family proteins for Mycobacterium marinum actin tail formation. Proc. Natl. Acad. Sci. USA. 2005;102:14837–14842. doi: 10.1073/pnas.0504663102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S.S. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem. J. 2005;390:641–653. doi: 10.1042/BJ20050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Z.S., Manser E., Lim L. Interaction between PAK and nck: a template for Nck targets and role of PAK autophosphorylation. Mol. Cell. Biol. 2000;20:3906–3917. doi: 10.1128/mcb.20.11.3906-3917.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peterson F.C., Deng Q., Zettl M., Prehoda K.E., Lim W.A., Way M., Volkman B.F. Multiple WASP-interacting protein recognition motifs are required for a functional interaction with N-WASP. J. Biol. Chem. 2007;282:8446–8453. doi: 10.1074/jbc.M609902200. [DOI] [PubMed] [Google Scholar]
- 32.Ditlev J.A., Michalski P.J., Huber G., Rivera G.M., Mohler W.A., Loew L.M., Mayer B.J. Stoichiometry of Nck-dependent actin polymerization in living cells. J. Cell Biol. 2012;197:643–658. doi: 10.1083/jcb.201111113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin Y., Mazza C., Christie J.R., Giliani S., Fiorini M., Mella P., Gandellini F., Stewart D.M., Zhu Q., Nelson D.L. Mutations of the Wiskott-Aldrich Syndrome Protein (WASP): hotspots, effect on transcription, and translation and phenotype/genotype correlation. Blood. 2004;104:4010–4019. doi: 10.1182/blood-2003-05-1592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.