Summary
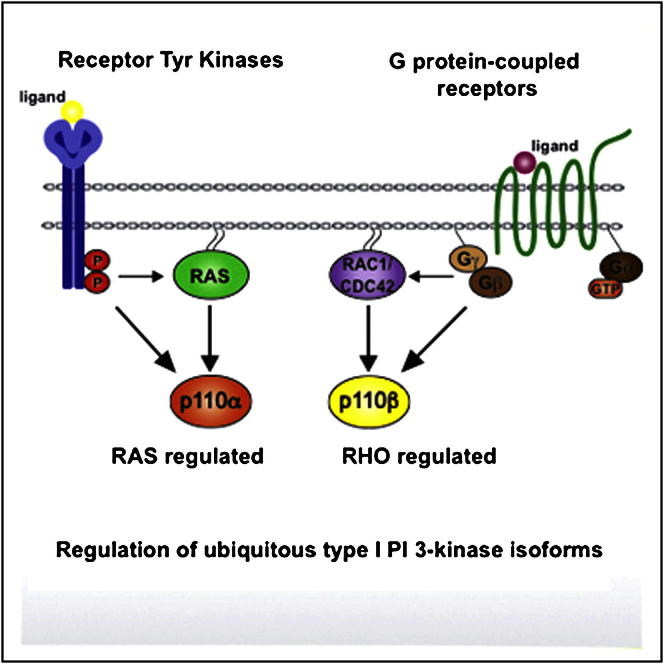
RAS proteins are important direct activators of p110α, p110γ, and p110δ type I phosphoinositide 3-kinases (PI3Ks), interacting via an amino-terminal RAS-binding domain (RBD). Here, we investigate the regulation of the ubiquitous p110β isoform of PI3K, implicated in G-protein-coupled receptor (GPCR) signaling, PTEN-loss-driven cancers, and thrombocyte function. Unexpectedly, RAS is unable to interact with p110β, but instead RAC1 and CDC42 from the RHO subfamily of small GTPases bind and activate p110β via its RBD. In fibroblasts, GPCRs couple to PI3K through Dock180/Elmo1-mediated RAC activation and subsequent interaction with p110β. Cells from mice carrying mutations in the p110β RBD show reduced PI3K activity and defective chemotaxis, and these mice are resistant to experimental lung fibrosis. These findings revise our understanding of the regulation of type I PI3K by showing that both RAS and RHO family GTPases directly regulate distinct ubiquitous PI3K isoforms and that RAC activates p110β downstream of GPCRs.
Graphical Abstract
Highlights
-
•
Unlike p110α, p110γ, and p110δ PI3K isoforms, RAS is unable to interact with p110β
-
•
The RHO family GTPases RAC1 and CDC42 directly bind and activate p110β via its RBD
-
•
GPCRs couple to PI3K via Dock180/Elmo1-mediated RAC activation and binding to p110β
-
•
Mice with RBD mutant p110β are resistant to experimental lung fibrosis
RAS proteins bind and activate multiple PI3K isoforms. The p110β isoform of PI3K, however, is instead activated by the binding of RHO family members RAC and CDC42. Blocking this interaction blunts GPCR-mediated cellular chemotaxis and confers resistance to lung fibrosis.
Introduction
The type I phosphoinositide 3-kinases (PI3Ks) are critical signaling proteins involved in the regulation of cell growth, survival, motility, and metabolism. In mammals, there exist four isoforms of the type I PI3K catalytic p110 subunits: α, β, γ, and δ. Of these, α and β are ubiquitously expressed, whereas γ and δ have more limited distribution, most notably in hematopoietic cells (Vanhaesebroeck et al., 2010). The lipid kinase activity of p110α is regulated downstream of receptor tyrosine kinases by the binding of tyrosine-phosphorylated proteins to its regulatory p85 subunit, resulting in attenuation of its autoinhibitory activity. In addition, activated RAS proteins bind directly to an N-terminal RAS-binding domain (RBD) on p110α, acting synergistically with the input from tyrosine-phosphorylated proteins to optimally activate lipid kinase activity (Rodriguez-Viciana et al., 1994, 1996). Proof of the pathophysiological importance of the direct interaction of RAS with p110α came from the generation of mice bearing germline mutations in the RBD of p110α, which were found to be highly resistant to mutant-RAS-induced lung and skin cancer formation (Gupta et al., 2007).
The direct binding of RAS to p110γ has also been studied in detail. The 3D structure of RAS bound to p110γ has been determined, and RAS has been shown to activate the lipid kinase activity of p110γ cooperatively with input from Gβγ subunits via the regulatory p101 subunit (Pacold et al., 2000). Mice with mutations in the RBD of p110γ show neutrophil defects in the regulation of PI3K activity by some G-protein-coupled receptors (GPCRs) (Suire et al., 2006). RAS also has been reported to bind and activate p110δ in vitro (Vanhaesebroeck et al., 1997). In addition, RBD mutations have been used to demonstrate that input of RAS binding to the single Drosophila type I PI3K is critical in insulin-pathway-controlled developmental growth (Orme et al., 2006) and that RAS binding is required for PI3K activation by chemoattractants in Dictyostelium (Funamoto et al., 2002).
p110β has been much less thoroughly studied than p110α. It appears to be relatively insensitive to activation by growth factor receptor tyrosine kinase signaling but important downstream of certain GPCRs, including those for lysophosphatidic acid (LPA) and sphingosine 1-phosphate (S1P), making p110β the only GPCR-regulated type I PI3K isoform outside the hematopoietic system (Ciraolo et al., 2008; Guillermet-Guibert et al., 2008; Jia et al., 2008). p110β may also play an important role in cancer because mouse models of breast and prostate cancer, as well as a number of human cancer cell lines, depend on p110β, particularly in the setting of PTEN loss (Ciraolo et al., 2008; Jia et al., 2008). In platelets, p110β is essential for integrin-dependent adhesion and clot formation (Jackson et al., 2005; Martin et al., 2010), leading to the intense effort to develop isoform-specific p110β inhibitors, some of which are now in clinical trials as antiplatelet and anticancer agents (NCT01458067, NCT00688714).
The molecular basis of how p110β can exert these distinct functions is poorly understood. p110β is overall structurally similar to other p110 catalytic subunits and engages the very same p85 type regulatory subunits as p110α, albeit in a somewhat different way (Zhang et al., 2011). Early reports have found p110β to associate with Gβγ subunits from heterotrimeric G proteins, which can directly stimulate its lipid kinase activity in vitro (Kurosu et al., 1997; Maier et al., 1999). It has, however, remained entirely unclear whether the p110β RBD contributes to p110β activation and function, and despite the apparently similar level of relatedness between the RBDs across the four isoforms, a systematic analysis of RAS effector proteins failed to detect any activation of p110β by RAS in cotransfected cells (Rodriguez-Viciana et al., 2004).
In this report, we explore the role of p110β regulation through its RBD for PI3K signaling and function. We present extensive in vitro work to show that p110β is the only type I PI3K isoform not regulated by RAS and to identify the RHO family GTPases RAC and CDC42 as direct isoform-specific RBD interactors and activators of p110β. We go on to show that GPCRs couple to PI3K via Dock180/Elmo1-mediated RAC activation and subsequent interaction with p110β. Mouse embryonic fibroblasts (MEFs) from p110β RBD mutant mice show reduced PI3K activity and mice are resistant to bleomycin-induced lung fibrosis, a pathology that has been linked with LPA signaling. These findings explain longstanding inconsistencies and revise our understanding of type I PI3K regulation by small GTP-binding proteins, providing molecular insight into the regulation and function of the ubiquitous p110β isoform.
Results
PI3K p110β Is Not a RAS Target Protein
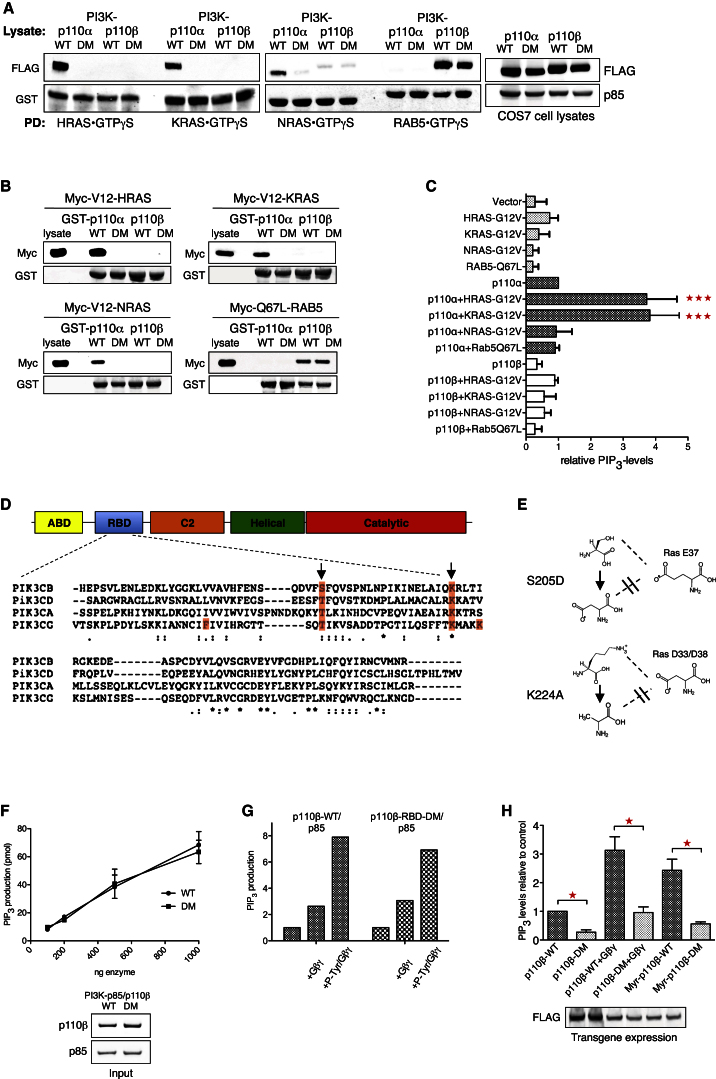
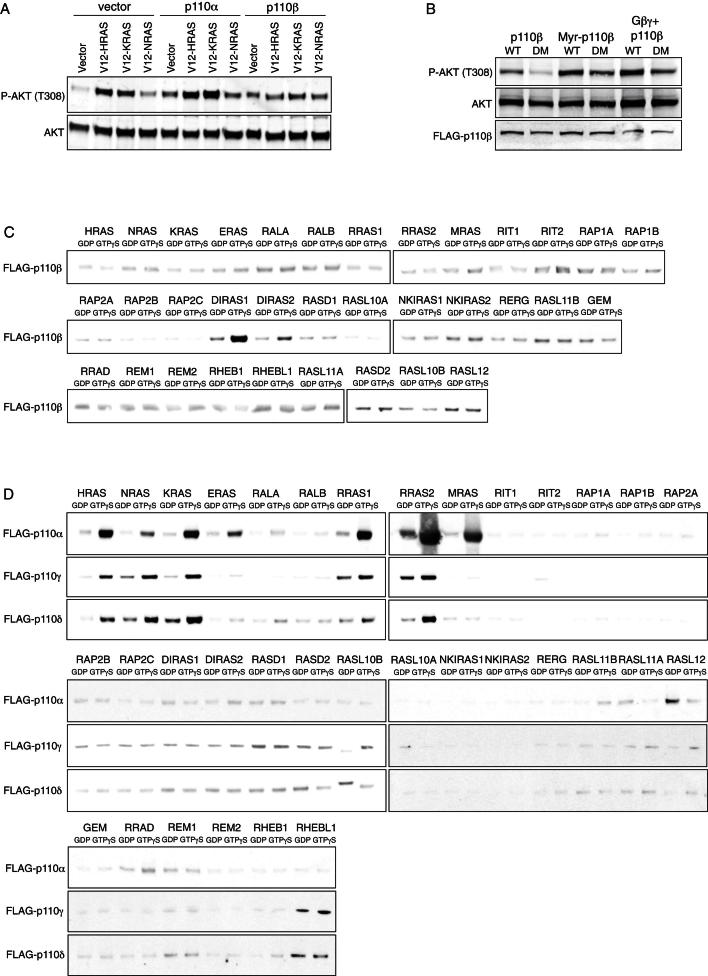
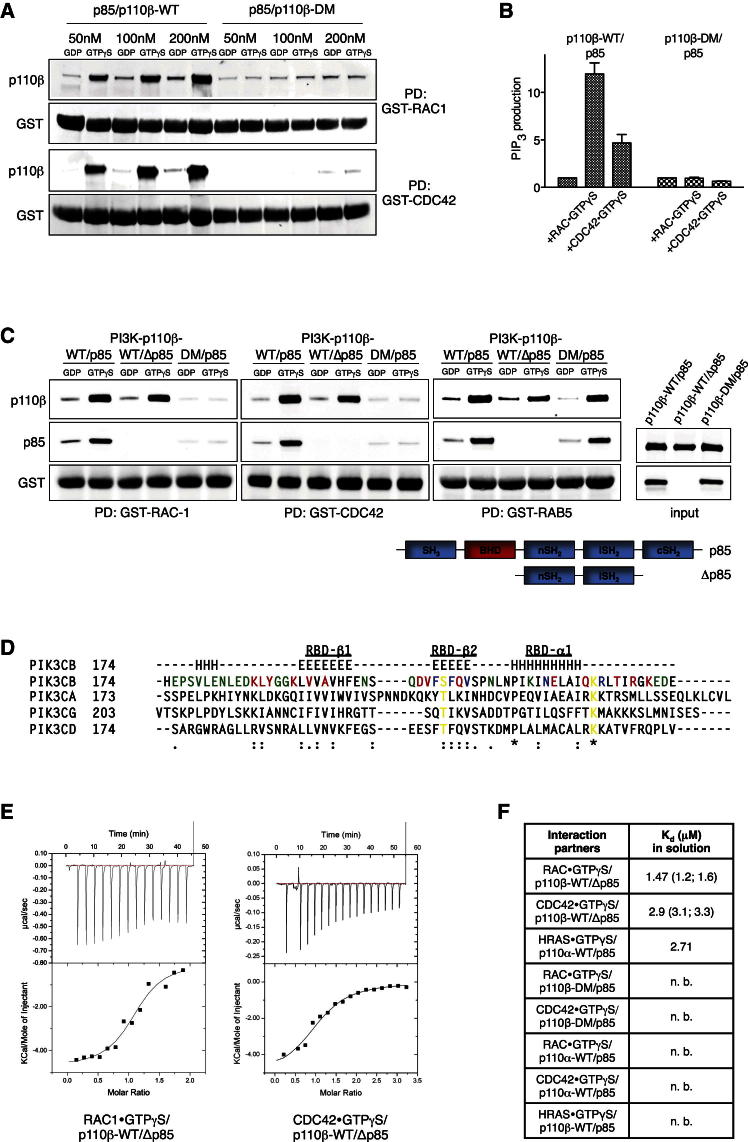
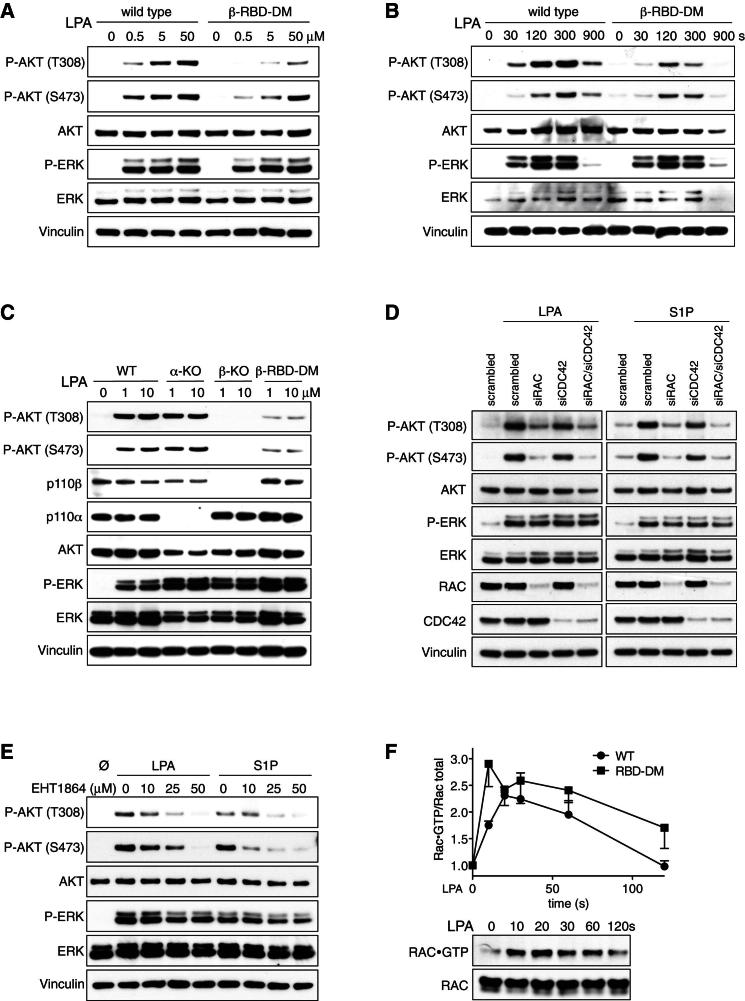
To investigate the role of RAS in regulating p110β, we set out to characterize the biochemical interaction between the two in vitro. In glutathione S-transferase (GST) pull-down studies using recombinant, GTPγS-loaded HRAS, KRAS, and NRAS as baits, we found strong and specific interaction between all three RAS proteins and p110α (Figure 1A). In contrast, p110β bound to none of the RAS proteins, but did bind to RAB5, a previously identified GTPase interactor of p110β (Christoforidis et al., 1999). Mutating key residues within the RBD of p110α (T208D/K227A) abrogated RAS binding, whereas introduction of analogous mutations (see below for details) into p110β did not affect RAB5 binding. Similar results were obtained when we used recombinant full-length GST-p110/p85 complexes to pull down active RAS or RAB5 proteins (Figure 1B). Moreover, when we expressed constitutively active RAS or RAB5, along with p110α/p85 or p110β/p85 in COS7 cells, and measured PIP3-levels (Figure 1C) or steady-state phospho-AKT (Figure S1A available online) as indicators of PI3K activity, HRAS and KRAS strongly enhanced p110α activity, whereas p110β was not stimulated by either RAS proteins or RAB5.
Figure 1.
PI 3-Kinase p110β Is Unable to Interact with RAS
(A) p110β does not bind to GTP-loaded RAS proteins. Purified recombinant, GTPγS-loaded HRAS, KRAS, NRAS, and RAB5 were incubated with lysates from COS7 cells expressing FLAG-p110α/p85 or FLAG-p110β/p85, wild-type (WT), or RBD double mutant (DM).
(B) Active RAS proteins do not bind to immobilized p110β/p85. Purified recombinant GST-p110α/p85 and GST-p110β/p85 were incubated with lysates from COS7 cells expressing Myc-tagged, constitutively active HRAS, KRAS, NRAS, and RAB5.
(C) Active RAS proteins do not stimulate p110β in cells. Lipids were extracted and PIP3 levels measured from COS7 cells expressing constitutively active mutants of HRAS, KRAS, NRAS, and RAB5, alone or in combination with p110α/p85 or p110β/p85 (n = 3; mean with SD; one-way ANOVA).
(D) Type I PI3K RBDs show moderate sequence similarity. Alignment of amino acid sequences of all four type I PI3K RBDs (PIK3CB = p110β, PIK3CD = p110δ, PIK3CA = p110α, PIK3CG = p110γ). Orange represents RAS-binding residues in p110γ, and arrows represent conserved “RAS-binding” residues.
(E) Mutation of RBD key residues in p110β. The two point mutations are shown together with hypothetical interactor residues modeled on the RAS-p110γ interaction.
(F) Unaltered lipid kinase activity of recombinant p110β-RBD-DM protein. Lipid kinase assay assessing basal activities of purified recombinant p110β/p85 complexes (n = 3; mean with SEM).
(G) p110β-RBD-DM protein remains sensitive to Gβγ and phosphotyrosine. A representative lipid kinase assay assessing effect of recombinant Gβγ and a PDGFR-derived phosphotyrosine peptide (pY740) on the activity of purified recombinant p85/p110β-WT and p85/p110β-RBD-DM is shown.
(H) Activity of p110β-RBD-DM in living cells is reduced. Lipids were extracted and PIP3 levels measured from COS7 cells expressing wild-type or RBD mutant p110β/p85. Gβγ, coexpression of Gβ2 and Gγ1; Myr, myristoylated p110β (n = 3; mean with SEM; paired t test).
See also Figure S1.
Figure S1.
PI 3-Kinase p110β Is Unable to Interact with RAS, Related to Figure 1
(A) Active RAS proteins fail to stimulate p110β in cells. Constitutively active mutants of HRAS, KRAS and NRAS were expressed in COS7 cells along with empty vector, p110α/p85 or p110β/p85. Cells were serum-starved and protein lysates were made for western blot analysis.
(B) p110β-RBD-DM is functionally compromised in intact cells. Wild-type p110β or p110β-RBD-DM were expressed at low levels in COS7 cells, along with p85. Cells were serum-starved and protein lysates were made for western blot analysis. Gβγ, coexpression of Gβ2 and Gγ1; Myr, myristoylated p110β.
(C) DIRAS1 and DIRAS2 bind p110β in a GTP-dependent manner. cDNAs encoding all 34 murine members of the RAS subfamily of small GTPases (RFGs) were cloned into pGEX-2T and verified by sequencing. GST-tagged RFGs were expressed in E. coli, purified on glutathione agarose, loaded with GDP/GTPγS in vitro and incubated with lysates from transfected COS7 cells, expressing FLAG-p110β/p85.
(D) Non-β isoforms bind to RAS proteins and a number of closely related RFGs. GST-tagged RFGs were purified from E. coli lysates and loaded with GDP/GTPγS in vitro. FLAG-tagged p110α, p110γ and p110δ were expressed in COS7 cells along with their respective regulatory subunits p85 or p101. COS7 cell lysates were incubated with GTPases for 1 hr and bound p110 was detected by western blot for FLAG.
An Intact RBD Is Essential for p110β Activity in Cells
The modest RBD sequence similarity among the four paralogs of type I PI3K is shown in Figure 1D. Even though the overall structural organization of the p110β RBD is conserved (Zhang et al., 2011), we speculated that because we cannot detect any interaction with RAS, it might have lost or altered its function. We therefore mutated two highly conserved key residues within the p110β RBD to generate a p110β-S205D/K224A double mutant (p110β-RBD-DM; Figure 1E). Analogous mutations in p110α and p110γ disrupt RAS binding (Gupta et al., 2007; Pacold et al., 2000). In vitro, the basal lipid kinase activity of purified recombinant p110β-RBD-DM protein was indistinguishable from its wild-type counterpart prepared in parallel (Figure 1F). Moreover, p110β-RBD-DM was still stimulated by the addition of purified recombinant Gβγ subunits, alone or in combination with a platelet-derived growth factor receptor (PDGFR)-derived phosphotyrosine peptide (pY740), indicating that the RBD mutations had no intrinsic effect on p110β lipid kinase activity or RBD-independent stimulatory input (Figure 1G). However, RBD mutant p110β was much less active than wild-type when expressed in COS7 cells (Figures 1H and S1B) even when Gβγ subunits were coexpressed or a myristoylation signal was added, pointing to a critical role of the RBD for p110β activity in living cells.
p110β Interacts with Distinct RAS Subfamily GTPases
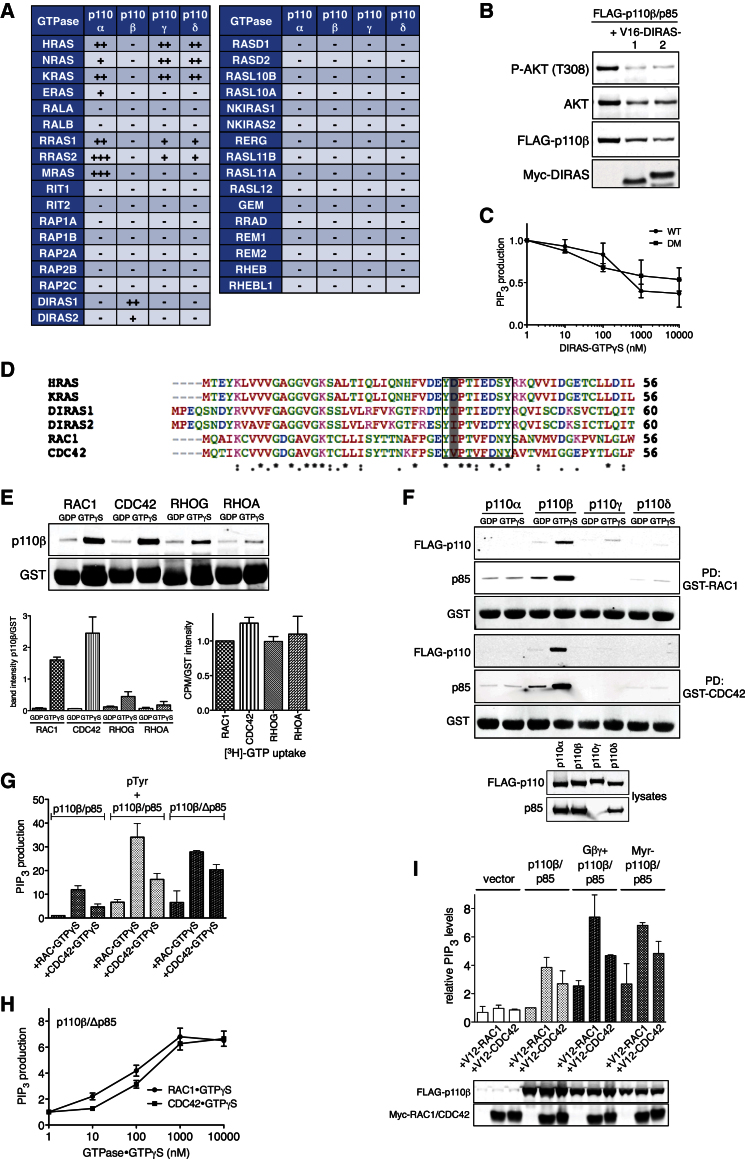
To identify RBD interactors of p110β, we probed all 34 murine members of the RAS subfamily of small GTPases (RFGs) for binding to p110β/p85 (Figure S1C). Parallel experiments were performed with p110α/p85, p110γ/p101, and p110δ/p85, respectively (Figure S1D). Strikingly, whereas all non-β isoforms interacted with the three prototypical RAS proteins and a partially overlapping subset of closely related RFGs (RRAS1, RRAS2, MRAS, and ERAS), p110β bound to none of those (Figure 2A). Instead, p110β exclusively bound to the more distantly related DIRAS1 and DIRAS2 proteins in a GTP-dependent manner (Figure S1C). DIRAS selectively bound wild-type and not RBD mutant p110β (Figure S2B), suggesting binding to the RBD. However, DIRAS failed to stimulate p110β lipid kinase activity in vitro (Figure 2C) and in cells, where constitutively active DIRAS proteins seemed to repress rather than elevate phospho-AKT when coexpressed along with p110β (Figure 2B), making DIRAS an unlikely in vivo activator of p110β.
Figure 2.
RAC and CDC42 Directly Bind and Active p110β
(A) p110β interacts with distinct RAS subfamily GTPases to non-β isoforms. Table summarizes results from GST pull-down assays (Figures S1C and S1D) probing 34 murine RAS subfamily GTPases for GTP-dependent binding of type I PI3K isoforms (−, no binding; +, specific binding; ++, strong binding; +++, very strong binding).
(B) DIRAS1/-2 do not stimulate p110β in cells. Constitutively active DIRAS1 or DIRAS2 were coexpressed with FLAG-p110β/p85 in COS7 cells.
(C) DIRAS does not stimulate p110β lipid kinase activity in vitro. Shown is a lipid kinase assay assessing the effect of purified recombinant, GTPγS-loaded DIRAS1 on p85/p110β (WT) and p85/p110β-RBD-DM (DM) activity (n = 3; mean with SEM).
(D) G2 box sequences of H-/KRAS, DIRAS1/-2 and RAC1/CDC42. Color-coded amino acid sequence alignment of the N termini of indicated small GTPases. A square frame highlights the G2 box sequence, and the variant residue (D33 in RAS) is shown in gray.
(E) RHO GTPases directly bind p110β in a GTP-dependent manner. Purified recombinant, GDP/GTPγS-loaded RHO GTPases were incubated with purified recombinant p110β/p85 (50 nM). Top: representative experiment; left: quantification (n = 2; mean with SEM); right: 3[H]-GTP uptake of GTPase preparations (n = 2; mean with SEM).
(F) Binding of p110β to RAC1 and CDC42 is isoform specific. Purified recombinant GDP/GTPγS-loaded RAC1 and CDC42 were incubated with lysates from COS7 cells expressing FLAG-tagged p110α/p85, p110β/p85, p110γ/p101, and p110δ/p85.
(G) RAC1 and CDC42 directly stimulate p110β lipid kinase activity. Purified recombinant GTPγS-loaded RAC1/CDC42 (1 μM) were added to purified recombinant p110β/p85, and lipid kinase activity was assessed in vitro. pTyr, phosphotyrosine peptide (pY740, 10 μM); Δp85, truncated p85, schematic in Figure 3C (n = 2; mean with SD).
(H) RAC1 and CDC42 dose-dependently stimulate p110β lipid kinase activity. Increasing concentrations of purified recombinant GTPγS-loaded RAC1/CDC42 were added to purified recombinant p110β/Δp85, and lipid kinase activity was assessed in vitro (n = 3; mean with SEM).
(I) RAC1 and CDC42 activate p110β in cells. Lipids were extracted and PIP3 levels measured from COS7 cells expressing constitutively active RAC1 or CDC42, alone or in combination with FLAG-p110β/p85. Gβγ, coexpression of Gβ2 and Gγ1; Myr, myristoylated p110β (n = 2; mean with SD). Western blots show expression levels of FLAG-p110β and Myc-tagged GTPases.
See also Figure S2.
Figure S2.
RAC and CDC42 Directly Bind and Active p110β, Related to Figure 2
(A) RAS-D33/DIRAS-I37 are essential residues for isoform-specific PI3K binding. Left: GST-tagged wild-type and D33I mutant HRAS were expressed in E. coli, purified on glutathione agarose beads, loaded with GDP/GTPγS in vitro, and incubated with lysates from transfected COS7 cells expressing FLAG-p110α/p85 or FLAG-p110β/p85. Right: GST-tagged wild-type and I37D mutant DIRAS1 were probed for binding FLAG-p110α/p85 or FLAG-p110β/p85. Bound p110 was identified by western blot for FLAG.
(B) DIRAS is unable to bind p110β-RBD-DM protein. GST pulldown assay assessing binding of purified recombinant p110β/p85 complexes to immobilized, GDP-/GTPγS-loaded DIRAS1. Lanes 1/2: p85/p110β wild-type; Lanes 3/4: Δp85/p110β wild-type; Lanes 5/6: p85/p110β-RBD-DM. Δp85 is depicted in Figure 3C.
(C) G2 box sequences of RHO GTPases. Color-coded amino acid sequence alignment of the G2 boxes of indicated RHO GTPases, frame highlights RAC1-I33 residue.
(D) RAC1 and CDC42 bind p110β in a GTP-dependent manner. Purified recombinant, GST-tagged and GDP/GTPγS-loaded RHO subfamily GTPases were incubated with lysates from COS7 cells expressing FLAG-p110β/p85.
(E) RAC-I33 is critical for p110β binding. Purified recombinant wild-type RAC1 and a RAC1-I33D mutant were compared for GTP-dependent binding to purified recombinant p110β/p85 (50 nM). Representative experiment and quantification of two independent experiments are shown (mean and SEM).
(F) RHO family GTPases do not bind p110α. 11 selected RHO family GTPases were expressed in E. coli, purified on glutathione agarose beads and loaded with GDP/GTPγS in vitro. HRAS was included as positive control. Pulldown was made from a lysate of transfected COS7 cells expressing FLAG-p110α/p85.
(G) RAC and CDC42 activate p110β wild-type but not p110β–RBD-DM in transfected cells. Constitutively active mutants of RAC1 and CDC42 (Myc-tagged) were expressed in COS7 cells along with empty vector, FLAG-p110β/p85 or FLAG-p110β-RBD-DM/p85.
(H) RAC1 and CDC42, but not RHOA activate p110β in transfected cells. Constitutively active mutants of RAC1, CDC42 and RHOA (Myc-tagged) were expressed in COS7 cells along with empty vector or FLAG-p110β/p85. Cells were serum-starved, and protein lysates were made for western blot analysis.
(I) RAC and CDC42 fail to stimulate p110α lipid kinase activity in vitro. Representative lipid kinase assay assessing the effect of increasing concentrations of purified recombinant GTPγS-loaded RAC1 and CDC42 on the activity of purified recombinant p110α/p85 protein complexes.
(J) RAC and CDC42 fail to activate p110α, p110γ and p110δ in transfected cells. Constitutively active mutants of RAC1 and CDC42 (Myc-tagged) were expressed in COS7 cells along with empty vector, FLAG-tagged p110α/p85, p110γ/p101 or p110δ/p85. Cells were serum-starved, and protein lysates were made for western blot analysis.
p110β Is a Direct RAC/CDC42 Target Protein
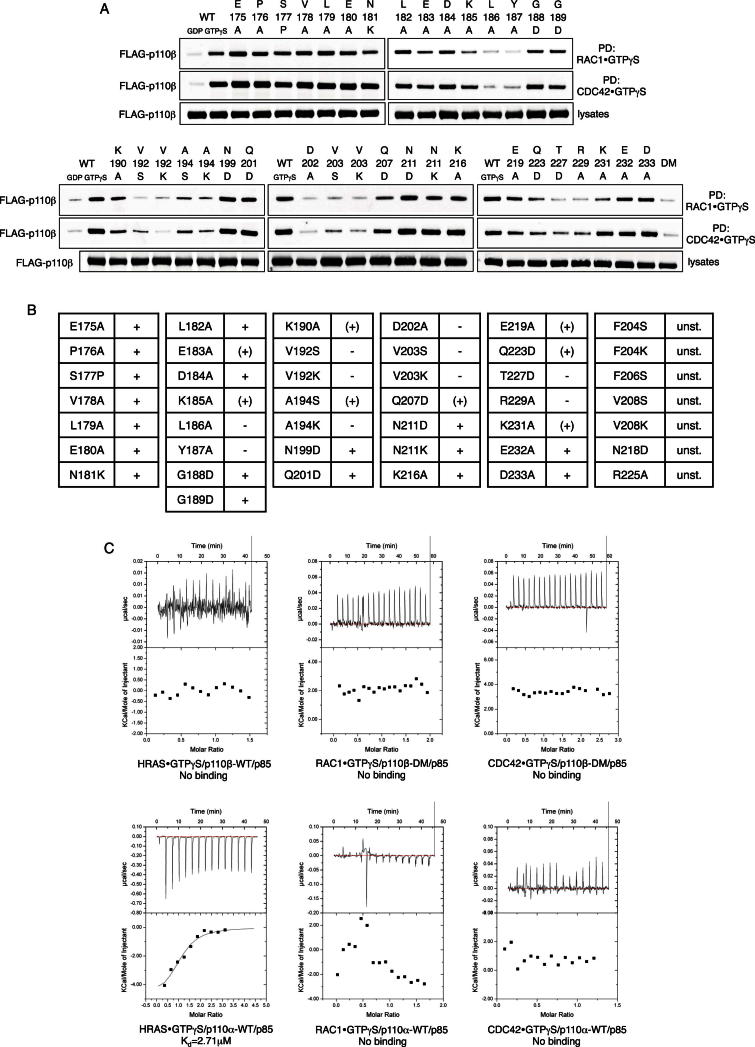
When comparing DIRAS with RAS, an obvious difference is the substitution of Asp33 within the G2 box of RAS with Ile37 in DIRAS (Figure 2D). This substitution is relevant to PI3K binding because an HRAS-D33I mutant showed attenuated binding to p110α and DIRAS1-I37D showed reduced binding to p110β, even though exchange of this residue did not enable RAS binding to p110β or DIRAS binding to p110α (Figure S2A), pointing to additional, G2-box-independent determinants of PI3K isoform specificity. Several members of the RHO subfamily of small GTPases harbor a hydrophobic isoleucine or valine residue in this position (Figure S2C), which prompted us to test p110β for binding to representative RHO family GTPases (Figure S2D). Surprisingly, p110β bound to both RAC1 and CDC42 in a GTP-dependent manner. Weaker binding to RHOG and minimal binding to RHOA was also observed (Figure 2E). Importantly, RAC1, CDC42, RHOG, and RHOA preparations bound similar amounts of GTP, indicating proper folding and functionality (Figure 2E, right lower graph), and a RAC1-I33D mutant showed reduced binding to p110β (Figure S2E), confirming a key role of this residue in GTPase binding to p110β.
The RAC1/CDC42-p110β interaction was isoform specific because neither RAC1 nor CDC42 significantly bound non-β isoforms under parallel conditions (Figures 2F and S2F). Strikingly, GTPγS-loaded RAC1 or CDC42 strongly stimulated p110β lipid kinase activity in vitro (Figure 2G), alone and in cooperation with a phosphotyrosine peptide (pY740), or when p110β was complexed with a less inhibitory, truncated p85 (Δp85 schematic in Figure 3C). Stimulation of p110β by active RAC1 and CDC42 was dose dependent (Figure 2H). Coexpression of constitutively active RAC1 or CDC42 (Figure S2G, lanes 4–6), but not RHOA (Figure S2H), along with p110β/p85 in COS7 cells strongly elevated cellular phospho-AKT and PIP3 levels (Figure 2I), indicating that both GTPases activate p110β in transfected cells. PIP3 levels were further enhanced by coexpression of Gβ1/Gγ2 subunits or by myristoylation of p110β (Figure 2I). In contrast, GTPγS-loaded RAC1/CDC42 did not stimulate p110α in vitro (Figure S2I), nor did V12-RAC1/CDC42 cooperate with p110α, p110γ, or p110δ to elevate cellular phospho-AKT levels (Figure S2J). Taken together, these data show that the RHO family GTPases RAC1 and CDC42 bind to p110β in an isoform-specific manner and potently and directly stimulate its lipid kinase activity.
Figure 3.
RAC and CDC42 Are Interactors of the p110β RAS-Binding Domain
(A) RAC1 and CDC42 do not bind p110β-RBD-DM in vitro. Purified recombinant p85/p110β (WT) and p85/p110β-RBD-DM (DM) protein complexes, at the indicated concentrations, were incubated with GST-tagged, GDP/GTPγS-loaded RAC1 and CDC42.
(B) RAC1 and CDC42 do not stimulate p110β-RBD-DM lipid kinase activity. Purified recombinant GTPγS-loaded RAC1/CDC42 (1 μM) were added to purified recombinant p85/p110β-WT or p85/p110β-RBD-DM in lipid kinase assays (n = 2; mean with SD). Data are part of experiment shown in Figure 2G.
(C) The N terminus of p85 is not required for p110β binding to RAC1/CDC42. Purified recombinant p110β/p85 protein complexes were incubated with GST-tagged, GDP/GTPγS-loaded RAC1, CDC42 and RAB5. Lanes 1/2: p85/p110β-WT; lanes 3/4: Δp85/p110β-WT; lanes 5/6: p85/p110β-RBD-DM. Δp85 is detailed in the schematic.
(D) Single RBD point mutations disrupt p110β binding to RAC1/CDC42. Amino acid alignment of type I PI3K RBDs with secondary structure elements of p110β (H, helix; E, β sheet) and color coding to illustrate effect of residue mutation on p110β binding to RAC1/CDC42. Green, unaltered binding; red, reduced/abolished binding; blue, unstable protein; yellow, p110β-RBD-DM residues; black, not mutated (Figures S3A and S3B for pull-down assays and table).
(E) Thermodynamic characterization of the RAC1/CDC42-p110β interaction by ITC. Binding of purified recombinant GTPγS-loaded RAC1 and CDC42 to purified recombinant p110β/Δ85 in solution was studied. Top: differential power over time; bottom: thermal energy (H) over molar ratio.
(F) Table summarizes results from ITC experiments. Numbers represent Kd values determined in independent experiments. n.b., no binding.
See also Figure S3.
RAC and CDC42 Are RBD Interactors of p110β
We next aimed to confirm that RAC and CDC42 are RBD interactors of p110β. Purified recombinant wild-type p110β/p85 bound to RAC1 and CDC42 in a concentration-dependent manner, whereas p110β-RBD-DM/p85 complexes showed no binding (Figure 3A). Similarly, RBD mutant p110β was not stimulated by active RAC1 or CDC42 in vitro (Figure 3B) or in cells (Figure S2E, lanes 7–9). To test whether the BCR homology domain (BHD) on p85, which had previously been shown to bind RAC and CDC42 (Bokoch et al., 1996; Zheng et al., 1994), is required for RAC/CDC42 binding to p110β, we truncated p85 (Δp85 schematic in Figure 3C) and probed for binding of p110β/Δp85 to RAC1 and CDC42 in vitro. Intriguingly, binding was unaffected by removal of the BHD but completely disrupted when full-length p85 was in complex with RBD mutant p110β, strongly arguing for the RBD as the RAC/CDC42-binding site. To further corroborate these findings, we generated 43 single point mutations covering 37 residues across the p110β RBD and assayed these mutants for binding to RAC1 and CDC42 (Figures 3D, S3A, and S3B). Of those, 17 mutations of 14 RBD residues affected binding to both GTPases without affecting p110β protein stability. Several of these residues were part of the RBDβ1 and β2 sheets or the loop adjacent to the RBDα1 helix (Zhang et al., 2011), areas known to be important for RAS binding in non-β isoforms (Pacold et al., 2000). Finally, we employed isothermal titration calorimetry (ITC) to study thermodynamics of the RAC1/CDC42-p110β interaction. In solution, RAC1⋅GTPγS bound to p110β/Δp85 with a molar ratio close to 1 and an average Kd of 1.42 μM, whereas the affinity measured for CDC42⋅GTPγS was 3.1 μM (Figures 3E and 3F). Similar affinities have been reported for the RAS-p110α and RAS-p110γ interactions (Pacold et al., 2000; Rodriguez-Viciana et al., 1996), indicating that RAC1 and CDC42 are plausible RBD interactors of p110β. No binding was observed between GTPγS-loaded RAC1/CDC42 and p110α or p110β-RBD-DM, respectively (Figure S3C).
Figure S3.
RAC and CDC42 Are Interactors of the p110β RAS-Binding Domain, Related to Figure 3
(A) Single point mutations across the p110β RBD disrupt binding to RAC and CDC42. 43 single point mutations of 37 RBD residues were introduced by site-directed mutagenesis. Mutants were expressed in COS7 cells, along with p85, and lysates were incubated with GST-tagged, GTPγS-loaded RAC1 and CDC42. Bound p110β was detected by western blot for FLAG. Mutants with reduced protein expression levels were deemed unstable and excluded from experiments.
(B) Table summarizing results from GST pulldown studies shown in (A). Point mutations made are listed along with their impact on binding to RAC1 and CDC42 (+, no effect on binding; (+), reduced binding; −, complete or near complete loss of binding; unst., unstable protein).
(C) Panel of representative ITC experiments investigating the thermodynamics of the interaction between indicated GTPγS-loaded small GTPases and recombinant p110/p85 complexes in solution. Top: the differential power recorded directly over time; bottom: thermal energy (H) over molar ratio.
p110β-RBD-DM Mice Show Signs of Reduced PI3K Activity
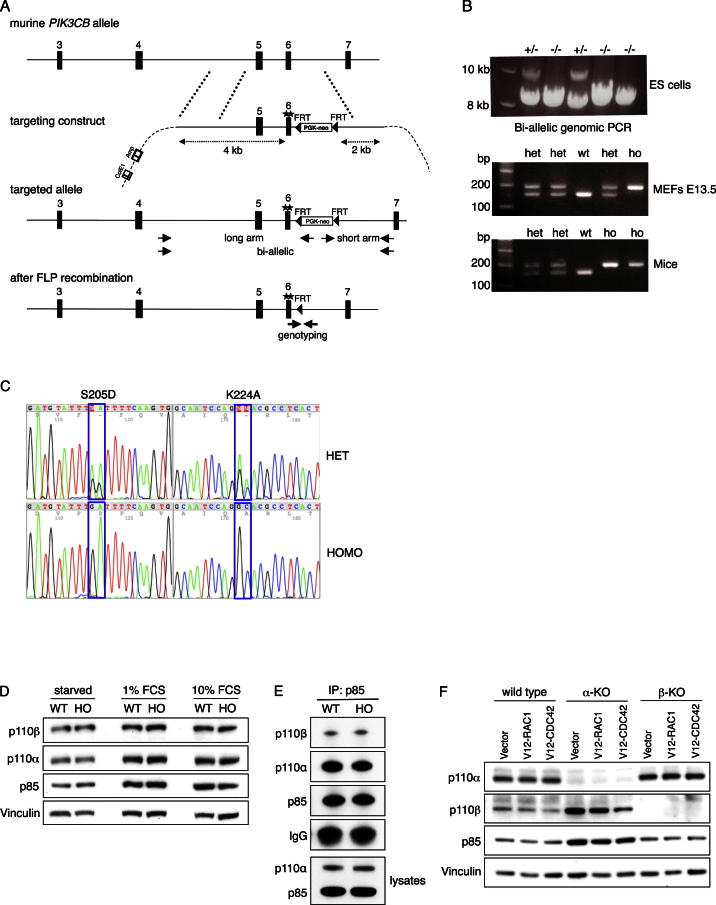
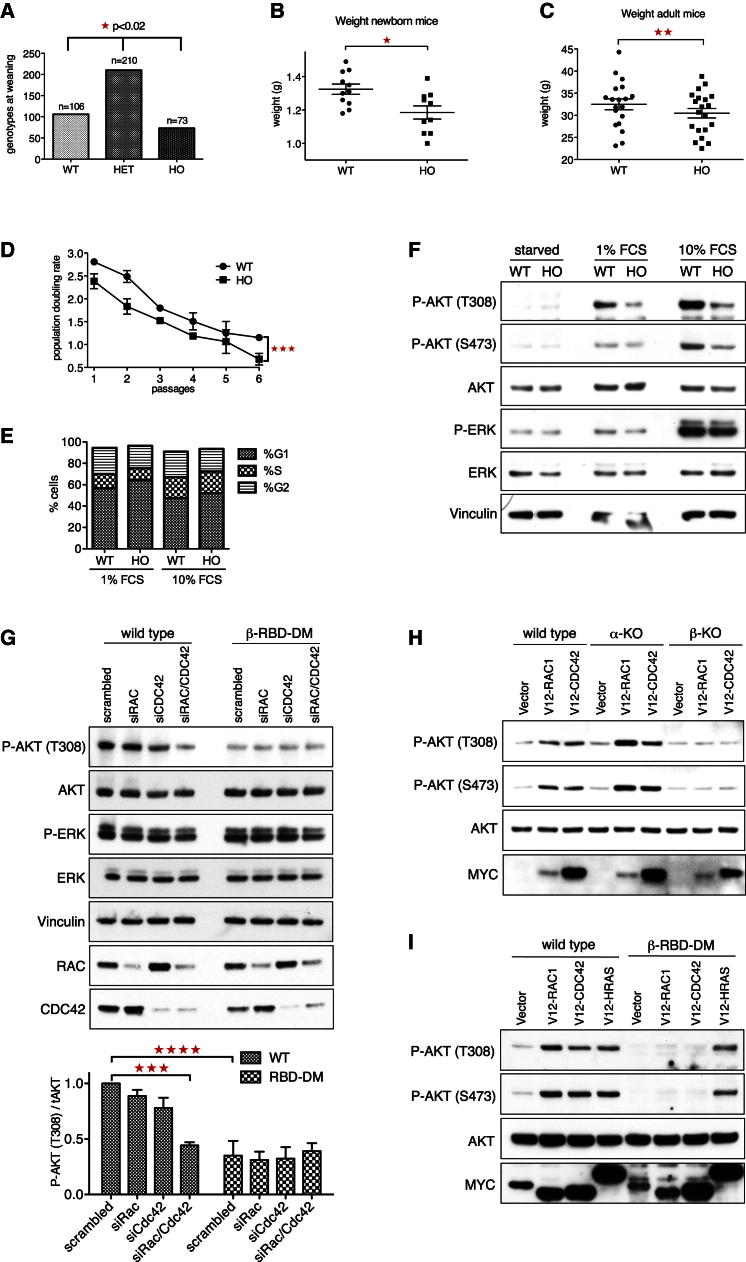
To study the role of interactor binding to the p110β RBD for PI3K signaling in vivo, we generated mice harboring the two p110β RBD point mutations (S205D/K224A) within their germline. Homologous recombination in embryonic stem (ES) cells was employed to replace exon 6 of the murine Pik3cb gene (Figure S4A), and germline transmission was achieved by eight-cell embryo injection (Figures S4B and S4C). p110β-RBD-DM mice were viable and fertile, although numbers of homozygous animals at the time of biopsy (around day 14) were moderately reduced (73 where 105 were expected; p < 0.02; Figure 4A), indicating incomplete lethality for undetermined reasons. Newborn homozygous p110β-RBD-DM pups were smaller than their wild-type littermates (Figure 4B). The size difference in adult mice was subtle but remained significant when same-sex litter- and cage-mates were compared (Figure 4C). MEFs homozygous for the p110β RBD mutation proliferated at a significantly slower rate than their wild-type counterparts (Figure 4D), which was reflected by a higher percentage of cells in G1 (1% fetal calf serum [FCS]: 56.7% ± 0.32% versus 64.2% ± 0.53%, n = 4, p < 0.001; 10% FCS: 47.5% ± 3.0% versus 52.2% ± 3.9%, n = 4, p < 0.05), fewer cells in G2 (1% FCS: 24.5% ± 1.4% versus 21.0% ± 1.9%, n = 4, p < 0.05; 10% FCS: 23.9% ± 1.4% versus 21.1% ± 1.6%, n = 4, p < 0.05) and fewer cells in S phase for 1% FCS (1% FCS: 13.3% ± 1.0% versus 11.0% ± 169%, n = 4, p < 0.05) (Figure 4E). Moreover, p110β-RBD-DM MEFs showed lower steady-state phospho-AKT levels (Figure 4F), suggesting that stimulatory signaling to p110β via its RBD contributes to PI3K activity in vivo. Expression levels of p110β, p110α, and p85 were indistinguishable among the genotypes (Figure S4D), and the stoichiometry of p110 subunit binding to p85 was undisturbed (Figure S4E).
Figure S4.
RAC/CDC42 Binding to p110β Regulates PI3K Activity In Vivo, Related to Figure 4
(A) Targeting strategy to replace exon 6 of the murine Pik3cb gene, encoding the p110β catalytic subunit. An FRT-flanked neomycin selection cassette was inserted into murine genomic DNA provided by a BAC clone, immediately downstream of Pik3cb exon 6, using Red/ET recombination technology (Genebridges). Selection cassette and flanking arms of genomic DNA were sub-cloned and mutations (★) introduced by site-directed mutagenesis. Arrows indicate priming sites for ES cell screening and genotyping. The neomycin selection cassette was later removed by crosses of heterozygous p110β-RBD-DM mice with FLPe mice.
(B) ES cell screening and genotyping. Homologous recombination in ES cells was confirmed by long genomic PCR bridging the entire targeting region (top, targeted allele 10kb, wild-type allele 8.5kb). After FLP recombination, MEFs (middle) and mice (bottom) were genotyped using primer pairs flanking the remaining FRT adjacent to exon 6.
(C) Genomic sequencing confirms presence of mutations. Genomic DNA from heterozygous and homozygous p110β-RBD-DM mice was isolated and exon 6 was PCR amplified and sequenced using standard techniques.
(D) Normal expression of type I PI3K subunits in p110β-RBD-DM MEFs. Wild-type and homozygous p110β-RBD-DM MEFs were maintained in 0%, 1% and 10% FCS, respectively. Lysates were made for western blot analysis.
(E) Undisturbed stoichiometry of type I PI3K subunits in p110β-RBD-DM MEFs. p85 protein was immunoprecipitated from whole-cell lysates made from wild-type and p110β-RBD-DM MEFs. The coprecipitation of p110α and p110β along with p85 was assessed by western blot.
(F) Deletion of p110α and p110β in conditional knockout MEFs. Immortalized p110αlox/lox;CreER+/− and p110βlox/lox;CreER+/− MEFs were treated with 4-hydroxytamoxifen (1 μM) for 3 consecutive days, prior to use in experiments.
Figure 4.
RAC/CDC42 Binding to p110β Regulates PI3K Activity In Vivo
(A) Reduced numbers of homozygous p110β-RBD-DM mice. Offspring from HET × HET crosses was genotyped at 2 weeks of age (n = 389; p < 0.02, chi-square analysis).
(B) Newborn p110β-RBD-DM pups are smaller. Newborn pups from HET × HET crosses were collected on the morning of birth, weighed, and genotyped (n = 31; mean ±SEM; p = 0.011, t test).
(C) Adult p110β-RBD-DM mice are smaller than their wild-type littermates. Weights of adult homozygous p110β-RBD-DM mice (12–30 weeks old) were compared to same-cage wild-type littermates (n = 39; mean ±SEM; paired t test).
(D) Reduced proliferation of p110β-RBD-DM MEFs. Early passage primary MEFs were grown in culture following a modified 3T3 protocol (n = 2 per genotype). Population doubling rate was calculated as PDR = (mean ±SEM; p < 0.001, two-way ANOVA).
(E) Accumulation of p110β-RBD-DM cells in G1. Cell-cycle profiles of early passage wild-type and homozygous p110β-RBD-DM primary MEFs growing in 1% or 10% FCS (n = 4, means; SEM in the Results section).
(F) p110β-RBD-DM MEFs show reduced steady-state phospho-AKT levels. Wild-type and p110β-RBD-DM MEFs were maintained in cell culture medium supplemented with 0%, 1%, or 10% FCS and harvested for western blot analysis.
(G) RAC1 and CDC42 cooperatively sustain phospho-AKT levels in wild-type MEFs. Wild-type and p110β-RBD-DM MEFs were transfected with scrambled duplex or gene-specific siRNA pools targeting RAC1, CDC42, or both and harvested for western blot analysis 30 hr after transfection. Graph shows phospho-AKT normalized to total AKT (n = 3; mean with SEM, one-way ANOVA).
(H) RAC1 and CDC42 activate p110β in vivo. Myc-V12-RAC1 and Myc-V12-CDC42 were nucleofected into immortalized wild-type, p110α-knockout, and p110β-knockout MEFs, along with a kinase-dead AKT reporter construct. The next day, cells were serum starved and harvested for western blot.
(I) RAC and CDC42 fail to activate PI3K in p110β-RBD-DM cells. Myc-V12-RAC1, Myc-V12-CDC42, or Myc-V12-HRAS were nucleofected into wild-type and p110β-RBD-DM MEFs as described in (H).
See also Figure S4.
RAC and CDC42 Regulate p110β In Vivo
To determine whether RAC and CDC42 are upstream activators of p110β in vivo, we transfected wild-type and p110β-RBD-DM MEFs with small interfering RNA (siRNA) pools (Dharmacon On-target plus) targeting these GTPases (Figure 4G). Although single knockdowns had only minor effects, combined knockdown of RAC1 and CDC42 significantly lowered phospho-AKT levels in wild-type, but not in p110β-RBD-DM cells, closing the gap in steady-state phospho-AKT levels between the genotypes and suggesting that endogenous RAC1 and CDC42 cooperatively activate p110β via its RBD. We next acutely expressed constitutively active mutants of RAC1 and CDC42 in wild-type, p110α-, and p110β-knockout MEFs (Figures 4H and S4F). Both RAC1 and CDC42 increased steady-state phospho-AKT levels in wild-type and p110α-deleted cells but not in p110β-knockout MEFs. Moreover, expression of V12-RAC1 and V12-CDC42 failed to elevate phospho-AKT levels in p110β-RBD-DM MEFs, whereas V12-HRAS did so in both wild-type and p110β-RBD-DM MEFs (Figure 4I). Taken together, these findings indicate that RAC1 and CDC42 activate PI3K in living cells by isoform-specific regulation of p110β through its RBD.
Activation of p110β Downstream of GPCRs Requires an Intact RBD
To study whether the p110β RBD is required for coupling p110β to GPCRs, we stimulated wild-type and p110β-RBD-DM MEFs with the lipid growth factors and GPCR agonists LPA and S1P. LPA and S1P dose-dependently induced both AKT and ERK phosphorylation in wild-type cells (Figures 5A and S5A). These responses were sensitive to pertussis toxin (Figure S5B), confirming GPCR involvement. In p110β-RBD-DM MEFs, LPA- and S1P-induced phosphorylation of AKT was strongly diminished, whereas ERK phosphorylation was undisturbed (Figures 5A and S5A). Also, in time course experiments, AKT phosphorylation was more transient when the p110β RBD was mutated (Figure 5B and not shown). In contrast, p110β-RBD-DM MEFs responded normally to epidermal growth factor (EGF), platelet-derived growth factor (PDGF), and insulin in dose-response (Figure S5C) and time course experiments (data not shown). Notably, in p110β-knockout cells, AKT phosphorylation in response to LPA was completely abolished (Figure 5C), indicating that the RBD is essential for much but not all p110β activation downstream of GPCRs.
Figure 5.
RAC Activates p110β Downstream of GPCRs
(A) LPA-induced AKT phosphorylation is attenuated in p110β-RBD-DM MEFs. Primary wild-type and homozygous p110β-RBD-DM MEFs were serum starved and stimulated with LPA for 5 min.
(B) LPA-induced AKT phosphorylation is more transient in p110β-RBD-DM MEFs. Primary wild-type and p110β-RBD-DM MEFs were serum starved and stimulated with LPA (10 μM) for indicated time periods.
(C) LPA-induced AKT phosphorylation is entirely dependent on p110β. Immortalized wild-type, p110α-knockout, p110β-knockout, and p110β-RBD-DM MEFs were serum starved and stimulated with indicated doses of LPA for 5 min.
(D) LPA-/S1P-induced AKT phosphorylation requires RAC. Immortalized wild-type MEFs were transfected with scrambled duplex or gene-specific siRNA pools targeting RAC1, CDC42 or both. Serum-starved cells were stimulated with LPA (10 μM) or S1P (1 μM) for 5 min.
(E) LPA-/S1P-induced AKT phosphorylation requires RAC activity.
Immortalized wild-type MEFs were serum starved, preincubated with EHT1864 for 30 min, and stimulated with LPA (10 μM) or S1P (1 μM) for 5 min.
(F) LPA induces rapid and transient activation of RAC. Serum-starved wild-type and p110β-RBD-DM MEFs were stimulated with LPA (10 μM) for indicated periods of time. RAC⋅GTP and total RAC were measured as described in the Extended Experimental Procedures (n = 3; mean with SEM).
See also Figure S5.
Figure S5.
RAC Activates p110β Downstream of G Protein-Coupled Receptors, Related to Figure 5
(A) S1P-induced AKT phosphorylation is attenuated in p110β-RBD-DM MEFs. Primary wild-type and homozygous p110β-RBD-DM MEFs were serum-starved and stimulated with sphingosine 1-phosphate (S1P) at indicated doses. Cells were harvested for western blot after 5 min.
(B) LPA/S1P-induced phosphorylation of AKT and ERK is sensitive to PTX. Immortalized wild-type MEFs were serum starved overnight and stimulated with EGF, LPA or S1P for 5 min. Pertussis toxin (PTX, 200 ng/ml) was added 16 prior to stimulation where indicated.
(C) Normal receptor tyrosine kinase signaling in p110β-RBD-DM MEFs. Primary wild-type and p110β-RBD-DM MEFs were serum-starved and stimulated with EGF, PDGF or insulin at indicated doses. Cells were harvested for western blot after 5 min.
(D) Deconvolution of RAC1 and CDC42 siRNA pools. Immortalized wild-type MEFs were transfected with scrambled duplex or gene-specific individual siRNA oligonucleotides targeting RAC1 or CDC42. 48 hr after transfection, serum-starved cells were stimulated with LPA (10 μM) for 5 min.
(E) RAC and CDC42 are not required for AKT phosphorylation induced by receptor tyrosine kinase agonists. Immortalized wild-type MEFs were transfected with scrambled duplex or gene-specific siRNA pools targeting RAC1 or CDC42. Serum-starved cells were stimulated with EGF (10 ng/ml), PDGF (10 ng/ml) or insulin (5 μg/ml) for 5 min, before they were harvested for western blot.
(F) AKT phosphorylation downstream of receptor tyrosine kinases does not require RAC activity. Immortalized wild-type MEFs were serum-starved and incubated with EHT1864 at the indicated doses for 30 min, before they were stimulated with EGF (10 ng/ml), PDGF (10 ng/ml) or insulin (5 μg/ml) for 5 min.
Activation of p110β Downstream of GPCRs Requires RAC
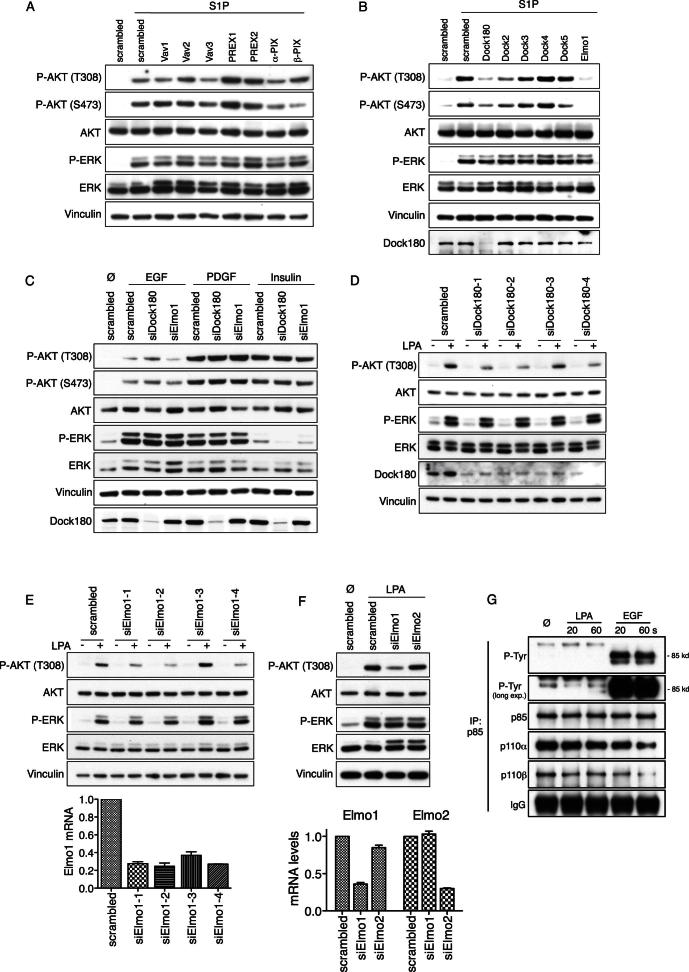
To test whether the identified p110β RBD interactors are required for linking p110β to GPCRs, we knocked down RAC1 and CDC42 in wild-type MEFs. Knockdown of RAC1 strongly impacted LPA/S1P-induced AKT phosphorylation, knockdown of CDC42 had only minor effects, and combination knockdown of both RAC1 and CDC42 had little additional effect compared to RAC1 knockdown alone (Figure 5D). Deconvolution experiments using single siRNA oligonucleotides confirmed the leading role of RAC1 in this pathway (Figure S5D). Neither RAC1 nor CDC42 knockdown affected LPA/S1P-induced phosphorylation of ERK or activation of either pathway induced by tyrosine kinase receptor agonists (EGF, PDGF, and insulin; Figure S5E). Similarly, EHT1864, a direct inhibitor of RAC but not CDC42 activation, dose-dependently inhibited AKT phosphorylation induced by LPA/S1P (Figure 5E), but not EGF, PDGF, or insulin (Figure S5F). Therefore, acute loss or inhibition of RAC phenocopied the signaling defect observed in p110β-RBD-DM MEFs. In line with this, RAC was activated very rapidly upon LPA stimulation, reaching its peak activity within 20 s (Figure 5F).
Dock180/Elmo1 Activates RAC Downstream of GPCRs and Upstream of p110β
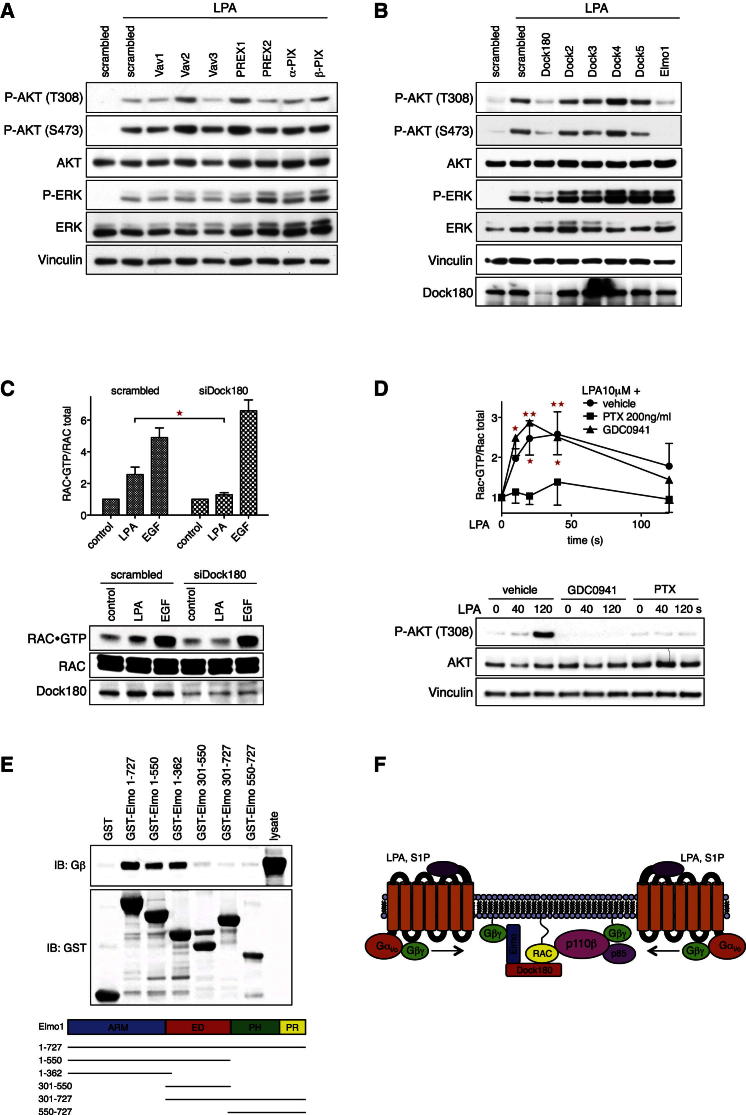
To provide further mechanistic insight into the GPCR-RAC-p110β pathway, we performed a small candidate siRNA screen to identify the guanine nucleotide exchange factor (RAC-GEF) involved. Transfection of wild-type MEFs with siRNA pools targeting the Dbl family RAC-GEFs Vav1-3, PREX1/2, and α-/β-PIX had no clear effect on LPA/S1P-induced AKT phosphorylation (Figures 6A and S6A). In contrast, knockdown of the Dock family RAC-GEF Dock180 or its adaptor protein Elmo1 interfered with AKT phosphorylation induced by LPA and S1P (Figure 6B and S6B), but not by EGF, PDGF, and insulin (Figure S6C). Specificity of results was confirmed in deconvolution experiments using individual siRNA oligonucleotides targeting Dock180 and Elmo1 (Figures S6D and S6E). Knockdown of Elmo2 had no effect (Figure S6F). Moreover, knockdown of Dock180 abolished LPA- but not EGF-induced RAC activation (Figure 6C), firmly placing Dock180/Elmo1 downstream of the LPA receptor and upstream of RAC and p110β. The DHR-1 domain of Dock180 has been shown to bind PIP3 (Côté et al., 2005), raising the possibility of a PIP3-driven feedback loop, in which RAC would be upstream and downstream of p110β. However, whereas sensitive to pertussis toxin, LPA-induced RAC activation was entirely insensitive to PI3K inhibition by GDC0941, a pan type I PI3K inhibitor, placing all detectable RAC activation upstream of p110β (Figure 6D). Recently, Dictyostelium ElmoE has been reported to be a direct Gβγ effector (Yan et al., 2012). We thus probed Gβγ subunits for direct binding to purified recombinant Elmo1 (Figure 6E) and found Gβγ to strongly bind to full-length Elmo and several N-terminal fragments. This altogether suggests a model in which Dock180 is recruited downstream of GPCRs, possibly through binding of Gβγ to the N terminus of Elmo1. At the same time, p110β is directly recruited by Gβγ. RAC is activated in proximity to p110β and binds to the RBD to fully activate p110β lipid kinase activity (schematic in Figure 6F). We could not detect any tyrosine phosphorylation on p85 in response to LPA (Figure S6G), and the tyrosine kinase inhibitors dasatinib, erlotinib, and PP2 (all at 1 μM) had no effect on the signaling pathway studied (data not shown).
Figure 6.
Dock180/Elmo1 Activate RAC Downstream of GPCRs and Upstream of p110β
(A) siRNA pools targeting Dbl family RAC-GEFs fail to affect LPA-induced AKT phosphorylation. Immortalized wild-type MEFs were transfected with scrambled duplex or gene-specific siRNA pools targeting indicated Dbl family RAC-GEFs. A total of 48 hr after transfection, serum-starved cells were stimulated with LPA (10 μM) for 5 min.
(B) Dock180 and Elmo1 are essential for LPA-induced AKT phosphorylation. Immortalized wild-type MEFs were transfected with scrambled duplex or gene-specific siRNA pools targeting indicated Dock family RAC-GEFs. Then 48 hr after transfection, serum-starved cells were stimulated with LPA (10 μM) for 5 min.
(C) Dock180 is essential for LPA-induced RAC activation. Immortalized wild-type MEFs, transfected with scrambled duplex or Dock180-specific siRNA pools, were stimulated with LPA (10 μM) or EGF (10 ng/ml) for 20 s and active RAC was quantified (n = 4; mean with SEM; t test; bottom: a representative experiment).
(D) LPA-induced RAC activation is PI3K independent. Immortalized MEFs were preincubated with PTX (200 ng/ml, 16 hr) or GDC0941 (10 μM, 1 hr) and stimulated with LPA (10 μM) for the indicated time periods (n = 4; mean with SEM; one-way ANOVA; bottom: representative lysates).
(E) Gβγ subunits directly bind to the N terminus of Elmo1. GST-tagged full-length Elmo1 and fragments as shown (schematic) were incubated with lysates from COS7 cells expressing Gβ2 and Gγ1. Bound Gβ was detected by western blot.
(F) Model of GPCR-induced p110β activation. See text for details.
See also Figure S6.
Figure S6.
Dock180/Elmo1 Activate RAC Downstream of GPCRs and Upstream of p110β, Related to Figure 6
(A) siRNA pools targeting Dbl family RAC-GEFs fail to affect S1P-induced AKT phosphorylation. Immortalized wild-type MEFs were transfected with scrambled duplex or gene-specific siRNA pools targeting indicated Dbl family RAC-GEFs. Forty-eight hours after transfection, serum-starved cells were stimulated with S1P (1 μM) for 5 min.
(B) Dock180 and Elmo1 are essential for S1P-induced AKT phosphorylation. Immortalized wild-type MEFs were transfected with scrambled duplex or gene-specific siRNA pools targeting indicated Dock family RAC-GEFs. Forty-eight hours after transfection, serum-starved cells were stimulated with S1P (1 μM) for 5 min.
(C) Knockdown of Dock180 or Elmo1 does not affect signaling downstream of receptor tyrosine kinases. Immortalized wild-type MEFs were transfected with scrambled duplex or siRNA pools targeting Dock180 or Elmo1. Cells were serum starved and stimulated with EGF (10 ng/ml), PDGF (10 ng/ml) or insulin (5 μg/ml) for 5 min.
(D) Deconvolution of Dock180 siRNA pool. Immortalized wild-type MEFs were transfected with scrambled duplex or gene-specific individual siRNA oligonucleotides targeting Dock180. Forty-eight hours after transfection, serum-starved cells were stimulated with LPA (10 μM) for 5 min.
(E) Deconvolution of Elmo1 siRNA pool. Immortalized wild-type MEFs were transfected with scrambled duplex or gene-specific single siRNA oligonucleotides targeting Elmo1. Forty-eight hours after transfection, serum-starved cells were stimulated with LPA (10 μM) for 5 min. RNA was extracted in parallel experiments and Elmo1 mRNA levels were compared by qPCR (mean with SEM).
(F) Knockdown of Elmo1 but not Elmo2 affects LPA-induced AKT phosphorylation. Immortalized wild-type MEFs were transfected with scrambled duplex or gene-specific siRNA pools targeting Elmo1 or Elmo2, and serum-starved cells were stimulated with LPA for 5 min. Elmo1/Elmo2 mRNA levels were assessed by qPCR (mean with SEM).
(G) Rapid p85 tyrosine phosphorylation upon EGF but not LPA stimulation. Immortalized wild-type MEFs were serum-starved and stimulated with EGF (10 ng/ml) or LPA (10 μM) for 20 and 60 s, before cell lysates were made, and p85 immunoprecipitates were analyzed by western blot as shown.
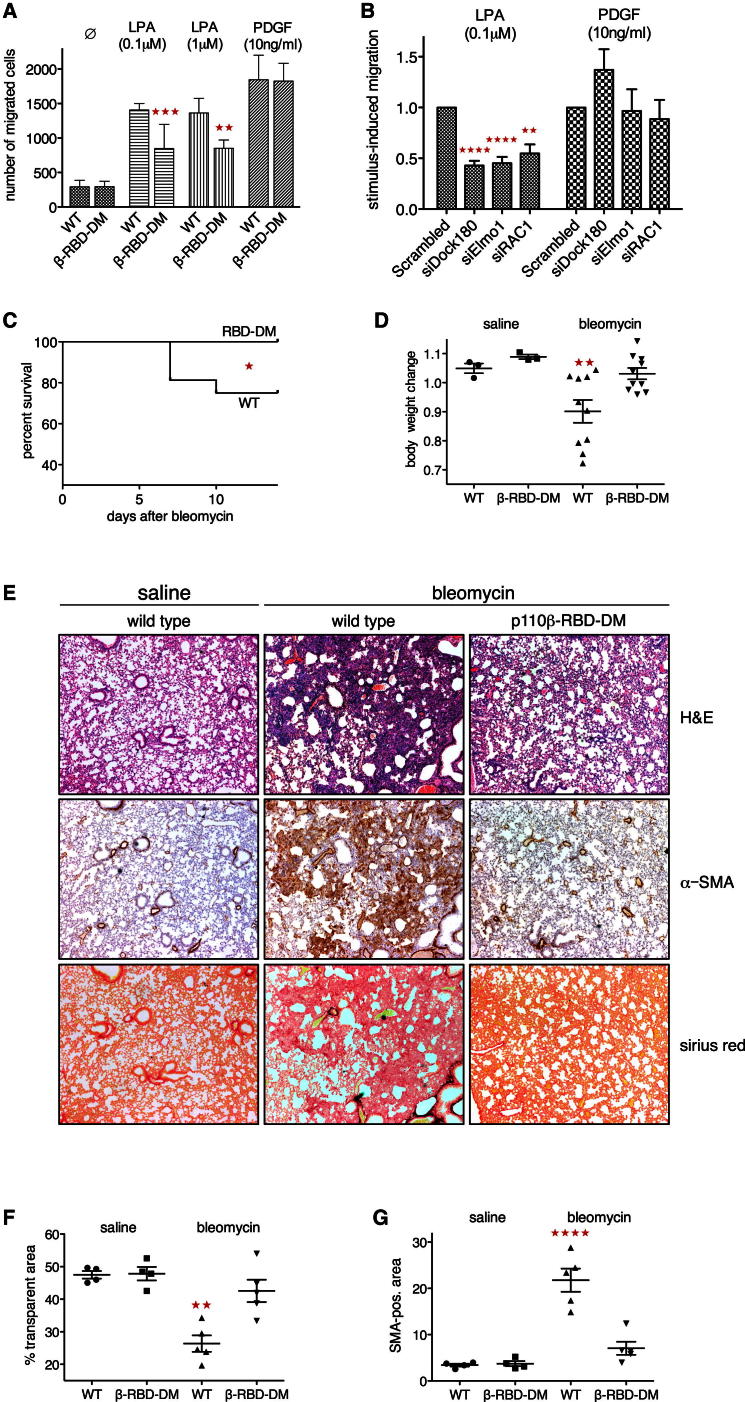
Disruption of the LPA-Dock180/Elmo1-RAC-p110β Axis Affects Fibroblast Chemotaxis
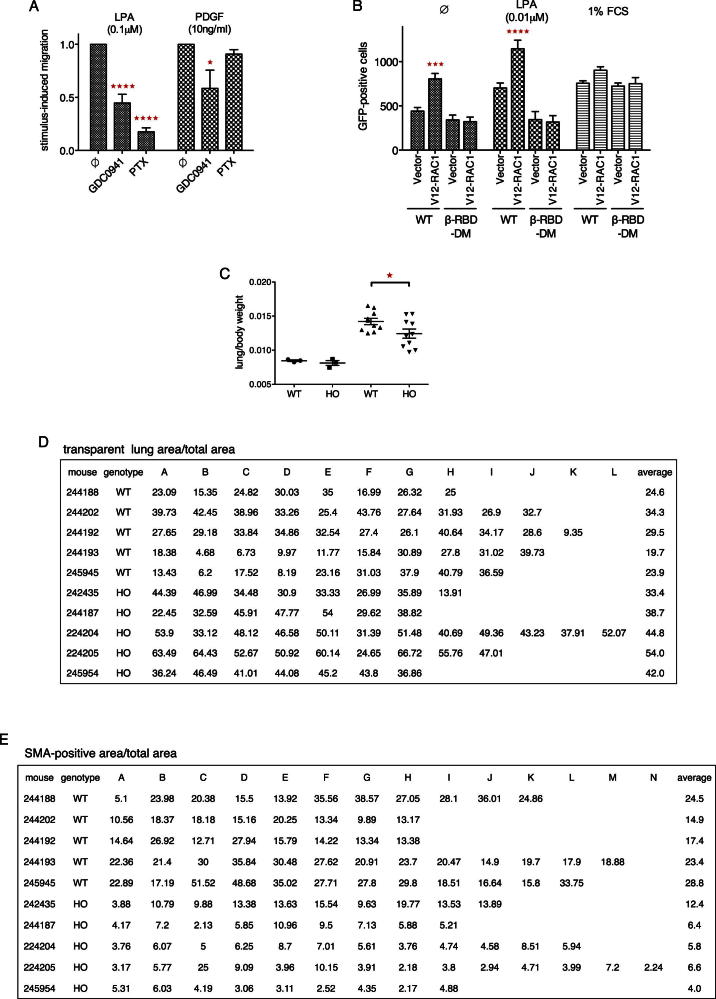
To assess the functional importance of the Dock180/Elmo1-RAC-p110β signaling axis for fibroblast chemotaxis, we employed transwell filter assays to study migration in gradients of LPA and PDGF. p110β-RBD-DM MEFs showed significantly reduced migration in LPA but not PDGF gradients (Figure 7A). Similarly, wild-type MEFs transfected with siRNA pools targeting Dock180, Elmo1, or RAC1 showed defective migration in gradients of LPA but not PDGF (Figure 7B), pointing to the specificity of this pathway for GPCR-induced chemotaxis. In contrast, PI3K activity was required for normal migration in either gradient, because pretreatment of wild-type cells with GDC0941 strongly affected migration toward LPA and PDGF, whereas pretreatment of cells with pertussis toxin selectively blocked migration in LPA gradients (Figure S7A).
Figure 7.
p110β-RBD-DM Mice Are Protected from Bleomycin-Induced Lung Fibrosis
(A) p110β-RBD-DM fibroblasts show reduced migration in gradients of LPA. Migration of wild-type and p110β-RBD-DM MEFs in gradients of LPA and PDGF was assessed in transwell filter assays (n = 3; mean with SD; one-way ANOVA).
(B) Dock180, Elmo1, and RAC1 are required for fibroblast migration in gradients of LPA. Immortalized wild-type MEFs were transfected with scrambled duplex or gene-specific siRNA pools targeting Dock180, Elmo1, or RAC1. Migration in gradients of LPA and PDGF was assessed in transwell filter assays and cell numbers were normalized to control conditions (n = 4; mean with SEM; one-way ANOVA).
(C) p110β-RBD-DM mice are protected against death from bleomycin-induced lung damage. Wild-type and homozygous p110β-RBD-DM mice were treated with a single intratracheal dose of bleomycin (1.25 U/kg) and observed for 14 days (n = 16 mice per genotype; Mantel-Cox test).
(D) p110β-RBD-DM mice are protected against weight loss following bleomycin instillation. Wild-type and p110β-RBD-DM mice received a single intratracheal dose of saline (n = 3 per genotype) or bleomycin (n = 10 per genotype) and weights were taken 14 days later (mean ±SEM; one-way ANOVA).
(E) p110β-RBD-DM mice are protected from bleomycin-induced lung fibrosis. Representative lung areas from wild-type and homozygous p110β-RBD-DM mice 14 days after treatment with intratracheal bleomycin (×4 magnification). Top: H&E; middle: IHC for α-SMA; bottom: Sirius red.
(F) p110β-RBD-DM mice are protected against loss of transparent lung areas following bleomycin instillation. Lungs were analyzed by H&E 14 days after bleomycin challenge. Multiple nonoverlapping areas of representative sections from each lung were photographed and transparent (white) areas were quantified using Nikon NIS elements software (mean ±SEM; one-way ANOVA; see Figure S7C for raw data).
(G) p110β-RBD-DM mice accumulate fewer activated lung fibroblasts following bleomycin instillation. Lungs were analyzed by immunohistochemistry for smooth muscle antigen (α-SMA) 14 days after bleomycin challenge. Multiple nonoverlapping areas of representative sections from each lung were photographed and SMA-positive (brown) areas were quantified using Nikon NIS elements software (mean ±SEM; one-way ANOVA; see Figure S7D for raw data).
See also Figure S7.
Figure S7.
p110β-RBD-DM Mice Are Protected from Bleomycin-Induced Lung Fibrosis, Related to Figure 7
(A) Fibroblasts migration in gradients of LPA and PDGF requires PI3K activity. Serum-starved immortalized wild-type MEFs were seeded onto fibronectin-coated membrane inserts and placed into 24-well plates containing serum-free medium and the indicated chemoattractants. Where indicated, GDC0941 (5 μM) or PTX (200 ng/ml) were added 30 min and 16 hr prior to seeding, respectively, and were present throughout experiments. After 6 hr, migrated cells were semiautomatically counted and numbers were normalized to control conditions (n = 3; mean with SEM; repeated-measures ANOVA).
(B) Constitutively active RAC1 promotes migration of wild-type but not p110β-RBD-DM fibroblasts. Migration of transfected wild-type and p110β-RBD-DM MEFs in starvation medium and gradients of LPA (10 nM) and FCS (1%) were assessed in transwell filter assays (n = 6 from 2 independent experiments; mean with SEM; 1way ANOVA).
(C) p110β-RBD-DM mice show less increase in lung weight after bleomycin challenge. Age- and sex-matched wild-type and p110β-RBD-DM mice received saline (n = 3 per genotype) or bleomycin (n = 10 per genotype) via intratracheal instillation and were culled 14 days later (mean ± SEM; t test).
(D) Morphometric quantification of transparent lung areas after bleomycin challenge. Wild-type and p110β-RBD-DM mice received a single dose bleomycin via intratracheal instillation. After two weeks, lungs were fixed, sectioned and stained with H&E. As many as possible nonoverlapping areas (A–L), avoiding artifacts from interlobes and major bronchi/vessels, of representative sections from each lung were photographed at low (4×) magnification and transparent (white) areas within each image were quantified using Nikon NIS elements software.
(E) Morphometric quantification of α-SMA-positive areas after bleomycin challenge. Wild-type and p110β-RBD-DM mice received a single dose bleomycin via intratracheal instillation. After two weeks, lungs were fixed, sectioned and stained with immunohistochemistry for smooth muscle antigen (α-SMA). As many as possible nonoverlapping areas (A–N), avoiding artifacts from bronchi and blood vessels, of representative sections from each lung were photographed at low (4×) magnification and positive (brown) areas within each image were quantified using Nikon NIS elements software.
In agreement with a key role of RAC upstream of p110β in fibroblast migration, acute expression of constitutively active RAC (V12-RAC1) stimulated migration of wild-type but not p110β-RBD-DM MEFs in the absence of chemoattractant and in the presence of a low concentration of LPA (10 nM) in the lower chamber, whereas migration toward 1% FCS was largely unaffected (Figure S7B).
p110β-RBD-DM Mice Are Protected from Bleomycin-Induced Lung Fibrosis
LPA has been identified as important fibroblast chemoattractant in bleomycin-induced lung fibrosis, a well-studied mouse model of human fibrotic lung disease, and was found to be elevated in patients with idiopathic lung fibrosis (Tager et al., 2008). We therefore wondered whether the disruption of p110β activation by LPA in p110β-RBD-DM mice would be sufficient to affect experimental lung fibrosis. Following a single intratracheal application of bleomycin, a quarter of all wild-type animals died or had to be culled according to local animal welfare regulations, whereas all p110β-RBD-DM mice survived (Figure 7C). Also, wild-type but not p110β-RBD-DM mice significantly lost body weight (Figure 7D) upon bleomycin treatment. Lung weights increased in both groups, but to a significantly lesser extent in p110β-RBD-DM mice (Figure S7C). Histology of lungs 14 days after bleomycin challenge revealed extended areas of fibrotic changes in wild-type animals (Figure 7E, hematoxylin and eosin staining [H&E], top), characterized by accumulation of activated, smooth muscle antigen-positive fibroblasts (Figure 7E, middle) and deposition of crosslinked collagen fibers (Figure 7E, Sirius red, bottom). Changes in p110β-RBD-DM mice were milder, with some mice showing almost normal lungs and others showing more limited areas of fibrosis. Morphometric analysis of multiple nonoverlapping lung areas confirmed the differences between the genotypes: transparent lung areas were significantly reduced (Figures 7F and S7D) and SMA-positive areas significantly increased (Figure 7G and S7E) in wild-type but not p110β-RBD-DM mice when compared to saline controls.
Discussion
RAS Proteins Do Not Regulate the Ubiquitous p110β Isoform
In this study, we show that, in contrast to what has been widely presumed, RAS is not a general regulator of type I PI3Ks. We find that out of the two ubiquitously expressed PI3K isoforms, only p110α is regulated by RAS, whereas p110β is a direct RAC and CDC42 target protein, indicating that key members of the pivotal RAS and RHO families of small G proteins directly regulate type I PI3Ks, with each family controlling their own distinct ubiquitous p110 isoform.
That p110β proved unable to physically and functionally interact with RAS is unexpected given the presence of a moderately conserved PI3K-type RBD in all four type I PI3K p110 catalytic subunits (Pacold et al., 2000). Comparison between the published structures of the four RBDs in their interactor-free states reveals little pointing to the distinct interactor specificity of p110β (R. Chaleil and P. Bates, personal communication). p110β is not only unable to interact with RAS under conditions readily revealing the transient, low-affinity interactions of RAS with other isoforms, but it also has an entirely distinct RAS superfamily GTPase interactor profile with a subfamily switch from RAS to RHO at the core of it, making any, for whatever reason, undetectable interaction with RAS unlikely. RBDs, classified on grounds of a ubiquitin fold structure with interactor specificities distinct from RAS, are not uncommon, as exemplified by the human formin FHOD1, a RAC interactor (Schulte et al., 2008), or the N terminus of Elmo1, shown to bind RHOG and the ARF family member ARL4A (Patel et al., 2011).
RAC and CDC42 Are Isoform-Specific RBD Interactors of p110β
Our biochemical experimentation identifies RAC1 and CDC42 as RBD interactors of p110β. An association of PI3K with RAC and CDC42 was first noticed nearly 20 years ago (Tolias et al., 1995), but was attributed to RAC/CDC42 binding to the amino-terminal BHD on p85, which has sequence homology to RHO-GAP domains (Bokoch et al., 1996; Zheng et al., 1994). These studies left the epistasis of information transfer between RAC/CDC42 and PI3K unclear. In retrospect, all functional data from these studies can be explained by the presence of p110β in the cell lysates and PI3K preparations used. Only the reported binding of RAC and CDC42 to recombinant p85 remains puzzling. We did not study monomeric p85, which has not been found in living cells (Geering et al., 2007), but our biochemical data strongly argue against an involvement of p85 in the RAC/CDC42-p110β interaction, because (1) RAC1 and CDC42 do not interact with p110α/p85 or p110δ/p85, (2) RAC1 and CDC42 bind normally to p110β in the absence of the p85 BH domain, and (3) RBD point mutations abrogate RAC1 and CDC42 binding to p110β in complex with full-length p85. A body of literature has accumulated identifying RAC or CDC42 as essential upstream activators of PI3K in various systems (Keely et al., 1997; Srinivasan et al., 2003; Weiner et al., 2002), but straightforward analysis of the relationship between RHO family GTPases and PI3K has been difficult, mainly because RAC and CDC42 also act downstream of PI3K, activated through PIP3-dependent GEFs (Welch et al., 2003). A very recent study using microscopy-based assays in transfected cells revisited the interaction of small GTPases and PI3K, confirming that both active RAS and RHO family GTPases can activate PI3K in living cells. However, based on experimentation exclusively with a p110α reporter construct, the authors interpreted PI3K regulation by RHO family members as indirect (Yang et al., 2012).
RAC/CDC42 Binding to p110β Controls PI3K Activity In Vivo
Our findings in MEFs suggest that RAC1 and CDC42 cooperatively control steady-state PI3K activity in living cells by isoform-specific activation of p110β and that this requires the p110β RBD. A model in which p110β provides basal, low-level PI3K activity has been proposed in the context of insulin signaling (Knight et al., 2006), and data showing PTEN-loss-driven prostate cancers to be entirely dependent on p110β, as well as metabolic findings in p110β kinase-dead mice, have been interpreted in the same way (Ciraolo et al., 2008; Jia et al., 2008). Our data point to RAC1 and CDC42 as drivers of such a basal, p110β-controlled activity, an idea consistent with a previous report finding that ectopic expression of wild-type but not RBD mutant p110β is sufficient to transform chicken embryo fibroblasts (Kang et al., 2006). It will be interesting to explore the oncogenic potential of the RAC/CDC42-p110β interaction in the setting of PTEN loss and also in the context of the recently discovered activating mutations in RAC (Hodis et al., 2012; Krauthammer et al., 2012) in human melanomas, where p110β could be an important downstream target.
GPCRs Activate p110β through its RBD
A firm body of in vivo evidence (Ciraolo et al., 2008; Guillermet-Guibert et al., 2008; Jia et al., 2008) has established p110β as a GPCR-regulated PI3K isoform. We find that mutation of the p110β RBD strongly attenuates p110β activation downstream of GPCRs, highlighting the importance of the RBD for p110β key signaling functions. The residual p110β activity in p110β-RBD-DM MEFs argues for a second, RBD-independent activation route, for which direct binding of Gβγ to p110β (Dbouk et al., 2012) is the most obvious candidate. A puzzling question is whether a cooperative effect of RBD interactor and Gβγ binding to p110β can suffice to fully activate p110β, or if additional phosphotyrosine input is required to overcome the strong inhibition of lipid kinase activity imposed by p85 (Zhang et al., 2011). Transactivation of receptor tyrosine kinases has been suggested to activate p110β downstream of GPCRs (Yart et al., 2002). Although we found no effect of tyrosine kinase inhibitors and no p85 tyrosine phosphorylation in response to LPA in support of such a mechanism in MEFs, there is evidence for cooperative GPCR and phosphotyrosine signaling to p110β in leukocytes (Kulkarni et al., 2011), and such a scenario appears possible in thrombocytes, where p110β is activated by integrins, ITAM-bearing receptors, and GPCRs (Martin et al., 2010).
Dock180/Elmo1 Couples GPCRs to RAC and p110β
RAC1 is essential for p110β activation downstream of the GPCRs for LPA and S1P, and the RAC-GEF Dock180/Elmo1 is upstream of both RAC and p110β in this pathway. The Dock/Elmo-RAC signaling axis is a highly conserved pathway controlling RAC-dependent key functions such as actin remodeling, migration, and phagocytosis (Côté et al., 2005). Recent findings in Dictyostelium have directly linked Gβγ subunits from GPCRs to Dock/Elmo-RAC and the cytoskeleton (Yan et al., 2012). Reminiscent of Dictyostelium ElmoE, we find the N terminus of human Elmo1 to directly bind Gβγ subunits, suggesting conservation of this pathway in mammals. In line with this, PI3K activity is not required for RAC activation by LPA, placing p110β entirely downstream of RAC in fibroblasts, which contrasts with a proposed PIP3-driven feedback loop controlling RAC activity upstream and downstream of PI3K in leukocytes (Weiner et al., 2002). Although PI3K is not required to activate RAC, it is still essential for fibroblast migration in gradients of LPA, indicating that PIP3-regulated pathways distinct from RAC activation contribute to GPCR-driven chemotaxis. Importantly, whereas our findings identify Dock180/Elmo1 downstream of LPA/S1P in MEFs, our experiments cannot rule out involvement of other RAC-GEFs within this pathway. We also cannot directly prove that the role of Dock180/Elmo1 in controlling fibroblast migration is exclusively and directly through activation of RAC upstream of p110β. Finally, different GPCRs and other cell types may signal through RAC-GEFs other than Dock180/Elmo1, and further studies will be required to determine whether the Dock180/Elmo1-RAC1-p110β axis is a fixed signaling module or just one example of how RAC/CDC42 is activated upstream of p110β.
Overall, the picture that emerges from these studies is one in which p110β regulation by GPCRs operates through a two-track signaling pathway with direct and indirect input into p110β. The direct route involves Gβγ interaction with p110β, whereas the indirect route goes through stimulation of RAC via Dock180/Elmo1, possibly also recruited through Gβγ (see Figure 6F). Such two-track wiring is reminiscent of receptor tyrosine kinase regulation of p110α via p85 interaction with tyrosine-phosphorylated receptor or adaptor protein and Grb2-Sos-RAS-p110α interaction. It can be speculated that this signaling logic might provide an improved ability to amplify a weak signal input or might increase the possibility for fine-tuning the signal through crosstalk of other pathways onto components such as Dock180/Elmo1, RAC itself, or RAC-GAPs terminating RAC activity. Another interesting issue is whether such Gβγ-Elmo1 and Gβγ-p110β interactions are mutually exclusive or can occur in a heterotrimeric complex. At present, we cannot distinguish between a model in which one Gβγ protein heterodimer binds directly to both Elmo1 and p110β simultaneously and one in which Elmo and p110β are engaged by two different Gβγ protein heterodimers (as shown in Figure 6F).
See Supplemental Information online for the Extended Discussion, including information on other GTPase interactors of p110β, the phenotype of p110β-RBD mutant mice, and resistance of these mice to bleomycin-induced lung fibrosis.
Extended Discussion.
Other GTPase Interactors of p110β
The only small GTPase previously found to interact with p110β is RAB5. p110β/p85 copurified with RAB5 in a study searching for RAB5 interactors (Christoforidis et al., 1999) and RAB5 was identified in a yeast two hybrid screen searching for p110β interactors (Kurosu and Katada, 2001). p110β appears to associate with RAB5 in the early endocytic pathway, where it has been proposed to indirectly contribute to PtdIns(3)P formation (Shin et al., 2005), and RAB5 and p110β have further been suggested to be part of the autophagy-promoting Vps34–Vps15–Beclin1–Atg14L complex (Dou et al., 2010). The RAB5-binding site on p110β remains to be determined, yet several findings argue against RAB5 binding to p110β through the RBD: (1) binding of RAB5 is entirely unaffected by p110β RBD mutations (Figure 3C); (2) RAB5 does not appear to stimulate p110β (Figure 1C, (Christoforidis et al., 1999; Rodriguez-Viciana et al., 2004); (3) the affinity between RAB5 and p110β in solution appears to be significantly higher (data not shown) than for typical PI3K RBD interactors, suggesting a more stable interaction, allowing to effectively localize p110β to RAB5-positve subcellular compartments.
Out of the murine 34 members of the RAS subfamily of small GTPases, only DIRAS-1 and DIRAS-2 showed binding to p110β (Figures S1C and S2B). Human DIRAS-3, also termed ARHI or NOEY2, a tumor suppressor in ovarian cancer and putative autophagy regulator (Lu et al., 2008), showed no interaction (data not shown). DIRAS-1 and -2 are relatively understudied RAS subfamily GTPases with low endogenous GTPase activity, whose expression appears to be limited to brain and heart (Ellis et al., 2002; Kontani et al., 2002). And whereas the relatedness to RAS and the inability to bind to RBD mutant p110β (Figure S2B) argues for binding to the RBD in vitro, the absence of any detectable stimulatory effect on p110β, the limited tissue distribution and growth-inhibitory effects make the role of DIRAS proteins as RBD interactors of p110β in vivo unclear, and further studies will be dedicated to this subject.
p110β-RBD-DM Mice
Direct analysis of signaling complexes regulating type I PI3K isoforms in living cells is notoriously difficult due to the transient, low-affinity nature of these interactions and their instability in solution. We therefore generated p110β-RBD-DM mice as a tool to study the impact of interactor binding to p110β for PI3K signaling in vivo. p110β-RBD-DM pups and mice are slightly smaller than wild-type littermates, and are born in slightly sub-Mendelian ratios, both features reported—to an overall more severe extent—from p110β knockout and p110β kinase-dead knock in mice (β-KD) (Ciraolo et al., 2010; Guillermet-Guibert et al., 2008; Kulkarni et al., 2011). A straightforward comparison between RBD mutant mice and these models is, however, complicated not only by different genetic backgrounds, but also by the different genetics of these models. Complete loss of p110β catalytic subunit will inadvertently lead to some disturbance of subunit composition and/or recruitment to signaling complexes, an artifact noticeable in recently published work (Utermark et al., 2012). Also one of the β-KD models (Ciraolo et al., 2008) has been found to have reduced p110β expression, both in embryos and adult tissues, placing these animals between knockout and other β-KD mice. A milder phenotype of p110β-RBD-DM mice is after all not unexpected, given that the RBD mutations do not disrupt basal p110β activity or RBD-independent activation routes, and some p110β activity might be enough for normal development and life under controlled conditions. We also saw moderately reduced proliferation of p110β-RBD-DM primary MEFs, indicating that p110β regulation through its RBD interactors is essential for normal cell proliferation, which appears to contradict findings from β-KD MEFs and reconstituted p110β-KO MEFs suggesting that p110β lipid kinase activity is not required for proliferation. (Ciraolo et al., 2008; Jia et al., 2008). Although no role of p110β lipid kinase activity for cell proliferation appears surprising in light of the reported growth defect in these mice, the discrepancies between these studies could well be down to technical reasons, such as the use of short term assays against the 3T3 protocol employed in our study and – at least for reconstituted KO cells - the use of immortalized and repeatedly manipulated cells against the use of early passage primary p110β-RBD-DM MEFs.
p110β-RBD-DM Mice Are Resistant to Bleomycin-Induced Lung Fibrosis
Fibroblasts are motile cells with proposed key roles in the development and progression of a wide range of diseases, including cancer and fibrosis (Wynn and Ramalingam, 2012). LPA has been identified as a critical fibroblast chemoattractant in experimental lung fibrosis, and LPA1 receptor knockout mice are protected in this model (Tager et al., 2008). Our findings establish p110β as critical downstream target of LPA in vivo. By inference, the protection of p110β-RBD-DM mice against bleomycin-induced lung fibrosis, together with the reduced migration of p110β-RBD-DM fibroblasts in LPA-gradients, suggests that p110β activation downstream of LPA might be critical for fibroblast chemotaxis to sites of tissue damage in vivo. However, the RAC-p110β axis might also be required for other fibroblast-specific functions involved in fibrogenesis such as secretion and remodelling of extracellular matrix components. Further studies will be required to decipher the precise role of p110β in the development of fibrosis in the lung and potentially other organ systems, and to learn whether a similar mechanism exists to recruit fibroblasts to sites of primary tumors and metastases, processes that could then be targeted by isoform-specific p110β inhibitors or strategies specifically disrupting the Dock180/Elmo1-RAC-p110β signaling axis.
Experimental Procedures
Detailed procedures are described in the Extended Experimental Procedures.
Extended Experimental Procedures.
Reagents and Antibodies
Pertussis toxin, EHT1864, sphingosine 1-phosphate and NSC23766 were from Tocris Bioscience; lysophosphatidic acid, GDP, GTPγS, insulin, bleomycin and protease inhibitors were from Sigma Aldrich; EGF and PDGF were from PeproTech. Antibodies to p110α, p110β, p85, phospho-AKT (T308), phospho-AKT (S473), total AKT, phospho-ERK, total ERK and P-Tyr were from Cell Signaling Technology. Antibodies to Dock180 were from Santa Cruz Biotechnologies, antibodies to FLAG, α-tubulin and vinculin from Sigma Aldrich. Monoclonal antibody to Myc and GST tags were made in house.
Plasmids and Cloning
cDNAs for p110α, p110β, p110γ and p110δ were PCR amplified from a murine cDNA library introducing an N-terminal FLAG or Myr-FLAG tag (containing a myristoylation signal). Murine p85, Δp85, Gβ1 and Gγ2 were amplified untagged, and all were cloned into pSG5 and fully sequenced. RBD mutations were introduced by site-directed mutagenesis and final constructs were fully sequenced. RAS and RHO subfamily GTPases as well as RAB5 were PCR-amplified from murine cDNA, cloned into pGEX-2T and sequenced. RAB5, DIRAS1 and DIRAS2 were subcloned into pcDNA3 introducing an N-terminal Myc tag and mutations were generated by site-directed mutagenesis. pEF-V12-RAC, pEF-V12-CDC42 and pEF-V14-RHOA were gifts from Erik Sahai, pcDNA-Myc-V12-HRAS, -KRAS and -NRAS were from UMR cDNA Resource Center. Human Elmo1 and fragments were PCR amplified from cDNA, cloned into pGEX-2T and sequenced.
Protein Purification
To produce purified recombinant GTPases on beads, cDNAs in pGEX-2T were transformed into BL21 E. coli (Stratagene). Protein expression was induced by addition of isopropyl β-D–thiogalactoside (IPTG, 0.1 mM) overnight at 16°C. Bacterial lysates were made and GST-fusion proteins were recovered on glutathione agarose beads. To produce soluble untagged GTPases, cDNAs encoding RAC1, CDC42, HRAS and DIRAS1 were subcloned into pGEX-6P and GTPases were cleaved off GST tag and beads by overnight incubation with HRV 3C protease (in-house made) at 4°C. When required, soluble GTPases were loaded with nucleotide in solution by addition of GDP or GTPγS (20× molar excess) and EDTA (5 mM final concentration), followed by incubation at 30°C for 30 min and addition of MgCl2 to a final concentration of 10 mM on ice. Loaded GTPases were gel-filtrated, concentrated (all at 4°C) and snap-frozen.
To express full-length GST-p110/p85 protein complexes, cDNAs were subcloned into pBacPAK-His3-GST, preserving the GST tag for p110 and removing it for p85/Δp85. High 5 (Life Technologies) insect cells were co–infected with baculoviruses encoding the respective PI3K subunits, lysed after 72 hr, and complexes were recovered on glutathione agarose, evaluated by gel electrophoresis and either used for pull down experiments on beads, or cleaved off overnight with HRV 3C protease, gel-filtrated and snap frozen.
For production of recombinant Gβγ, cDNAs for murine Gβ2 and Gγ1 were subcloned into pBacPAK-His3-TEV, removing the HIS tag for Gβ and preserving it for Gγ. Gβγ complexes were purified from High 5 cell lysates on NTA agarose, gel-filtrated and snap frozen.
Protein-Binding Assays
For GST pulldown assays, approximately 10 μg of GST fusion protein were immobilized on 20 μl glutathione sepharose beads. Immobilized GTPases were loaded with nucleotide by adding 50 μl of GTPase loading buffer (20 mM Tris-HCl pH 7.4, 5 mM EDTA, 25 mM NaCl) containing GDP or GTPγS at 2 mM. After 20 min at 37°C, MgCl2 was added to a final concentration of 10 mM on ice. Proteins on beads were incubated with either 1 ml of cell lysate made from a confluent 60 mm dish of transfected COS7 cells, or with recombinant purified protein diluted in GST-binding buffer (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM MgCl2, 1% Triton X-100, 5 mg/ml BSA). After 1 hr, beads were washed and proteins solubilized by boiling in LDS sample buffer (Life Science Technologies).
[3H]-GTP Uptake of Small GTPases
To compare GTP uptake across different preparations of purified recombinant GTPases, each sample was loaded on beads with 2 mM GTP containing 1 μCi [3H]-GTP, following the procedure detailed above. Unbound GTP was removed by rigorous washes, GTPases on beads were resuspended in scintillation fluid, and counts per minutes were recorded in a scintillation counter over a GST only background. Data are presented as counts per minute normalized to protein amounts, estimated by western blot for GST and quantification on the Li-Cor Odyssey system.
Lipid Kinase Assays
Purified recombinant, untagged and soluble p110/p85 protein complexes were incubated in kinase assay buffer (50 mM HEPES, [pH 7.4], 50 mM NaCl, 5 mM MgCl2, 0.03% CHAPS, 2 mM DTT, 25 μM ATP) with 4 μM PI(4,5)P2-diC8 as substrate for 1 hr at 25°C. Reactions were terminated by adding 6 mM EDTA, and PI(3,4,5)P3 was quantified using a commercial competitive ELISA (Echelon Biosciences). For some reactions, recombinant Gβ2γ1 dimers were added a 1 μM or a PDGF receptor-derived recombinant phospho-tyrosine peptide (pY740: DGG(pY)MDMSKDE) was present at 10 μM.
Lipid Extraction and PIP3 Quantification
For quantification of cellular PI(3,4,5)P3-levels, acidic lipids were extracted from overnight serum-starved COS7 cells following a standard lipid extraction protocol. In brief, cells were lysed in ice-cold trichloroacetic acid (TCA, 0.5 M). Pellets were washed in 5%TCA/1 mM EDTA and neutral lipids were extracted with MeOH:CHCl3 (2:1) and discarded. Acidic lipids were extracted with MeOH:CHCl3:HCl (80:40:1), recovered by phase-split, dried and resuspended. PI(3,4,5)P3-levels were quantified by competitive ELISA (Echelon Biosciences).
Isothermal Titration Calorimetry
Purified recombinant soluble PI3K protein complexes and soluble nucleotide-loaded GTPases were gel-filtrated into ITC buffer (phosphate-buffered saline [pH 7.4], 5 mM MgCl2, 1 mM Tris(2-carboxyethyl)phosphine hydrochloride) at 4°C. PI3K was loaded into the cell of a MicroCal iTC200 microcalorimeter at concentrations of approximately 20 μM. GTPases were loaded into the syringe at 200 μM. In a typical experiment, 16 injections of GTPase into the cell were recorded at 15°C, and relevant thermodynamic parameters were analyzed and calculated using the instrument’s software (Origin). GTPase injections into buffer only were done to determine heats of dilution.
Generation of p110β-RBD-DM Mice
A gene targeting vector was designed to replace exon 6 of the murine Pik3cb gene, encoding the p110β catalytic subunit, with an engineered version bearing two single point mutations leading to the exchange of two RBD key residues (S205D, K224A). An FRT-flanked neomycin selection cassette (Genebridges) was PCR-amplified and inserted into murine genomic DNA immediately downstream of Pik3cb exon 6 in a murine BAC clone, using Red/ET recombination technology (Genebridges). Selection cassette and flanking arms of genomic DNA (arms of homology, 4kb upstream and 2kb downstream of exon 6, respectively) were subcloned into a targeting vector backbone taken from PGKneolox2DTA.2 (Phil Soriano), again by using Red/ET-based recombination in E. coli. Plasmid DNA was isolated and mutations were introduced by site-directed mutagenesis (Quikchange XL, Stratagene). The targeting construct was fully sequenced, linearized, and electroporated into C57BL/6 ES cells. Neomycin-resistant ES cell clones were screened for homologous recombination by genomic PCR bridging the short arm of the construct and confirmed by PCRs bridging the long arm of homology and the entire targeting region (biallelic PCR). Takara LA-Taq DNA polymerase was used for amplification of long genomic fragments. Several correctly targeted ES clones were injected into blastocysts or 8-cell embryos and implanted into pseudopregnant foster mothers following standard techniques. Offspring of chimera was genotyped for germline transmission. Two independent founder lines were maintained, and mutations were confirmed by genomic sequencing. The neomycin selection cassette was removed by crosses of heterozygous p110β-RBD-DM mice with FLPe deleter mice, leaving a single FRT site in intron 6/7, which was used for PCR genotyping. Genotyping was later outsourced to an automated genotyping provider (Transnetyx).
Mouse Embryonic Fibroblasts
To generate wild-type and homozygous p110β-RBD-DM MEFs, E13.5 mouse embryos from intercrosses of heterozygous p110β-RBD-DM mice were collected, minced and trypsinized. Heads were separated for genotyping. MEFs from each four wild-type and homozygous embryos were made and used throughout this study. For immortalization, MEFs were transduced with SV40 large T antigen. To generate p110α and p110β knockout MEFs, mice harboring conditional floxed alleles for p110α (Zhao et al., 2006) or p110β (Jia et al., 2008) were crossbred with Rosa26CreER(T2) mice to generate p110αlox/lox;CreER+/− and p110βlox/lox;CreER+/− embryos. Conditional MEFs made from these embryos were immortalized, expanded and treated with 4-hydroxytamoxifen (1 μM) for 3 consecutive days, prior to use in experiments. Deletion of p110α or p110β was assessed by western blot.
Cell Culture, Plasmid, and siRNA Transfections
COS7 cells and mouse embryonic fibroblasts (MEFs) were maintained in DMEM, supplemented with 10% fetal calf serum (FCS). For serum starvation, MEFs were washed twice with sterile phosphate buffered saline (PBS) and incubated in serum-free DMEM, supplemented with 0.5% fatty acid-free bovine serum albumin (BSA, Sigma Aldrich). Plasmids were transfected into COS7 cells with Lipofectamine 2000 (Life Science Technologies), following the manufacturer’s protocol. MEFs were plasmid transfected by electroporation using the Amaxa Nucleofector system.
siRNA was transfected into immortalized MEFs using Dharmafect 4 (Dharmacon), following manufacturer’s protocols. siGENOME siRNA pools were used to target RAC-GEFs, ON-TARGETplus pools for RAC1 and CDC42, and Non-Targeting Pool 2 for control transfections (all from Dharmacon). siRNA sequences are listed in Table S1.
Immunoprecipitation and Western Blotting
Western blotting was performed following standard methods. In brief, cells were washed twice in ice-cold PBS, before they were lysed directly in preheated LDS sample buffer (Life Science Technologies). Samples were heated to 98°C for 3 min, run on Nupage Bis-Tris gels (Life Science Technologies) and transferred to PVDF membranes (Millipore). Immunodetection of proteins was performed either by conventional HRP-based chemiluminescence or by fluorescence-based detection using the Li-Cor Odyssey system and therefore recommended protocols and reagents.
For immunoprecipitation of p85, MEFs were lysed in IP buffer (20 mM Tris-HCl [pH 7.4], 150 mmol NaCl, 5 mmol MgCl2, 1% Triton X-100, protease inhibitor cocktail [Sigma Aldrich], 10 mM β-glycerol phosphate, 10 mM NaF, 10 mM sodium pyrophosphate), and lysates were cleared by centrifugation. Anti-p85 antibody was precoupled to protein A sepharose beads (Sigma Aldrich) and was incubated with lysates for 2 hr. Beads were washed, boiled in LDS sample buffer and analyzed by western blot. Lysates were run in parallel.
Quantitative Real-Time PCR
To compare Elmo1/Elmo2 mRNA abundance after siRNA transfections, total RNA was extracted from transfected immortalized wild-type MEFs (QIAGEN RNA mini kit), and 1 μg of total mRNA was reverse-transcribed into cDNA. Quantitative real-time PCR (qPCR) was performed following standard protocols, Elmo1/2 mRNA levels were normalized to beta-actin and changes estimated using the ΔΔCt method. Primers used were QIAGEN Quantitect Primer assays for Elmo1, Elmo2 and beta-actin.
Growth Factor and Lysophospholipid Signaling Assays
For growth factor and lysophospholipid stimulation experiments, early passage primary MEFs were seeded at 3 × 105 cells in 6 cm cell culture dishes, starved in serum-free medium, supplemented with 0.5% BSA, overnight and stimulated as desired. Cells were rapidly washed in ice-cold PBS and directly lysed in preheated LDS sample buffer, prior to analysis by western blot.
Proliferation Assays
To study proliferation of MEFs in culture, early passage primary MEFs (P2) from each two wild-type and homozygous p110β-RBD-DM embryos, derived from HET x HET crosses, were seeded at a defined number, grown in full serum for 3 days, trypsinized, counted in triplicate and reseeded at original numbers (modified 3T3 protocol). Each such cycle represents one passage for analysis. Population doubling rate was calculated as . For cell cycle analysis, primary MEFs growing in 1% or 10% FCS were digested off, fixed in 70% ethanol, stained with propidium iodide, and analyzed by flow cytometry.
Migration Assays
For migration assays, serum-starved immortalized MEFs were seeded onto 8 μM pore size cell culture inserts, coated with 10 μg/ml fibronectin. Inserts were placed into wells containing 500 μl serum-free DMEM, supplemented with 0.5% fatty-acid-free BSA and chemoattractant as required. Cells were allowed to migrate for 6 hr, before they were fixed in 4% paraformaldehyde for 10 min. Nonmigrated cells were scraped off using cotton wool tips, and membranes were recovered and mounted onto microscopic slides in VECTASHIELD HardSet mounting medium with DAPI (Vector laboratories). Several nonoverlapping low-magnification fields were photographed on a NIKON fluorescence microscope, and cells were semiautomatically counted using the object count function of NIKON NIS elements. For studies with siRNA-transfected MEFs, cells were transfected 48 hr prior to migration experiment. To assess migration of plasmid transfected MEFs, cells were cotransfected with pmaxGFP and migrated GFP-positive cells were counted.
RAC Activation Assay
For RAC activation assays, the GST-tagged active RAC and CDC42-binding domain of PAK1 (GST-PAK-PBD) was expressed in E. coli and purified on glutathione agarose beads (10 μg protein on 20 μl bead volume per reaction). Immortalized wild-type and p110β-RBD-DM MEFs were seeded on 150 mm tissue culture dishes, grown to 50% confluency, serum-starved overnight, inhibitor-treated as desired and stimulated one at a time. For siRNA experiments, cells were harvested 48 hr after transfection and following an overnight starvation. For harvest, cells were rapidly washed twice in ice-cold PBS and lysed in 1 ml GST lysis buffer (20 mM Tris-HCl [pH 7.4], 150 mmol NaCl, 5 mmol MgCl2, 1% Triton X-100, 1 mM DTT, protease inhibitors, 10 mM β-glycerol phosphate, 10 mM NaF). Cells were scraped off and lysates were cleared at full speed for 2 min in a cooled microcentrifuge. Supernatants were snap frozen in liquid nitrogen. Once all dishes were harvested, lysates were carefully thawed and incubated with GST-PAK-PBD for 1 hr. Beads were washed several times and bound RAC was identified and quantified by western blot (Li-Cor Odyssey). Lysates were analyzed in parallel and RAC-GTP/total RAC ratios were determined.
Histology and Morphometric Analysis
Lungs were processed for histology after fixation in neutral-buffered formalin. Frontal sections through central levels of each lung were stained with H&E, Sirius Red or immunohistochemistry (IHC) for smooth muscle actin (anti-α-SMA, DAKO) following standard protocols. To quantify fibrotic changes, as many as possible nonoverlapping low-magnification (4×) microscopic fields from representative sections of each lung stained with H&E, were photographed, avoiding artifacts from interlobe spaces and major bronchi and blood vessels. The transparent area of each section, made up mainly by alveolar spaces and small airways was semiautomatically quantified using NIKON NIS elements. Areas stained positive for α-SMA were quantified in a parallel fashion.
Isothermal Titration Calorimetry
Purified recombinant soluble PI3K protein complexes were loaded into the cell of a MicroCal iTC200 microcalorimeter at concentrations of approximately 20 μM. Nucleotide-loaded GTPases were loaded into the syringe at 200 μM. In a typical experiment, 16 injections of GTPase into the cell were recorded at 15°C, and relevant thermodynamic parameters were analyzed and calculated using the instrument’s software (Origin).
Bleomycin-Induced Lung Fibrosis
All animal experimentation was carried out in compliance with UK Home Office animal welfare regulations. Age- and sex-matched wild-type and homozygous p110β-RBD-DM mice from intercrosses of heterozygous p110β-RBD-DM animals were used to study bleomycin-induced lung fibrosis. Mice (8–12 weeks old) were anaesthetized with isoflurane, and bleomycin in sterile normal saline (1.25 U/kg) or saline alone (50 μl) was given by intratracheal instillation. After 14 days, mice were culled and lungs were recovered for further analysis.
Acknowledgments
We thank Jean Zhao for p110α-loxP and p110β-loxP mice, Erik Sahai for plasmids, Raphael Chaleil and Paul Bates for their expertise on structure issues, Pablo Rodriguez-Viciana for discussion, and Miriam Molina-Arcas and David Hancock for help and advice. We thank LRI Core Technology Facilities, including Transgenic Services, Biological Resources, Peptide Synthesis, Equipment Park, and Cell Services.
Published: May 23, 2013
Footnotes
Supplemental Information includes Extended Discussion, Extended Experimental Procedures, seven figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2013.04.031.
Supplemental Information
References
- Bokoch G.M., Vlahos C.J., Wang Y., Knaus U.G., Traynor-Kaplan A.E. Rac GTPase interacts specifically with phosphatidylinositol 3-kinase. Biochem. J. 1996;315:775–779. doi: 10.1042/bj3150775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforidis S., Miaczynska M., Ashman K., Wilm M., Zhao L., Yip S.C., Waterfield M.D., Backer J.M., Zerial M. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat. Cell Biol. 1999;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- Ciraolo E., Iezzi M., Marone R., Marengo S., Curcio C., Costa C., Azzolino O., Gonella C., Rubinetto C., Wu H. Phosphoinositide 3-kinase p110beta activity: key role in metabolism and mammary gland cancer but not development. Sci. Signal. 2008;1:ra3. doi: 10.1126/scisignal.1161577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté J.F., Motoyama A.B., Bush J.A., Vuori K. A novel and evolutionarily conserved PtdIns(3,4,5)P3-binding domain is necessary for DOCK180 signalling. Nat. Cell Biol. 2005;7:797–807. doi: 10.1038/ncb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dbouk H.A., Vadas O., Shymanets A., Burke J.E., Salamon R.S., Khalil B.D., Barrett M.O., Waldo G.L., Surve C., Hsueh C. G protein-coupled receptor-mediated activation of p110β by Gβγ is required for cellular transformation and invasiveness. Sci. Signal. 2012;5:ra89. doi: 10.1126/scisignal.2003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funamoto S., Meili R., Lee S., Parry L., Firtel R.A. Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell. 2002;109:611–623. doi: 10.1016/s0092-8674(02)00755-9. [DOI] [PubMed] [Google Scholar]
- Geering B., Cutillas P.R., Nock G., Gharbi S.I., Vanhaesebroeck B. Class IA phosphoinositide 3-kinases are obligate p85-p110 heterodimers. Proc. Natl. Acad. Sci. USA. 2007;104:7809–7814. doi: 10.1073/pnas.0700373104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillermet-Guibert J., Bjorklof K., Salpekar A., Gonella C., Ramadani F., Bilancio A., Meek S., Smith A.J., Okkenhaug K., Vanhaesebroeck B. The p110beta isoform of phosphoinositide 3-kinase signals downstream of G protein-coupled receptors and is functionally redundant with p110gamma. Proc. Natl. Acad. Sci. USA. 2008;105:8292–8297. doi: 10.1073/pnas.0707761105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Ramjaun A.R., Haiko P., Wang Y., Warne P.H., Nicke B., Nye E., Stamp G., Alitalo K., Downward J. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–968. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- Hodis E., Watson I.R., Kryukov G.V., Arold S.T., Imielinski M., Theurillat J.P., Nickerson E., Auclair D., Li L., Place C. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S.P., Schoenwaelder S.M., Goncalves I., Nesbitt W.S., Yap C.L., Wright C.E., Kenche V., Anderson K.E., Dopheide S.M., Yuan Y. PI 3-kinase p110beta: a new target for antithrombotic therapy. Nat. Med. 2005;11:507–514. doi: 10.1038/nm1232. [DOI] [PubMed] [Google Scholar]
- Jia S., Liu Z., Zhang S., Liu P., Zhang L., Lee S.H., Zhang J., Signoretti S., Loda M., Roberts T.M., Zhao J.J. Essential roles of PI(3)K-p110beta in cell growth, metabolism and tumorigenesis. Nature. 2008;454:776–779. doi: 10.1038/nature07091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Denley A., Vanhaesebroeck B., Vogt P.K. Oncogenic transformation induced by the p110beta, -gamma, and -delta isoforms of class I phosphoinositide 3-kinase. Proc. Natl. Acad. Sci. USA. 2006;103:1289–1294. doi: 10.1073/pnas.0510772103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keely P.J., Westwick J.K., Whitehead I.P., Der C.J., Parise L.V. Cdc42 and Rac1 induce integrin-mediated cell motility and invasiveness through PI(3)K. Nature. 1997;390:632–636. doi: 10.1038/37656. [DOI] [PubMed] [Google Scholar]
- Knight Z.A., Gonzalez B., Feldman M.E., Zunder E.R., Goldenberg D.D., Williams O., Loewith R., Stokoe D., Balla A., Toth B. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauthammer M., Kong Y., Ha B.H., Evans P., Bacchiocchi A., McCusker J.P., Cheng E., Davis M.J., Goh G., Choi M. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat. Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni S., Sitaru C., Jakus Z., Anderson K.E., Damoulakis G., Davidson K., Hirose M., Juss J., Oxley D., Chessa T.A. PI3Kβ plays a critical role in neutrophil activation by immune complexes. Sci. Signal. 2011;4:ra23. doi: 10.1126/scisignal.2001617. [DOI] [PubMed] [Google Scholar]
- Kurosu H., Maehama T., Okada T., Yamamoto T., Hoshino S., Fukui Y., Ui M., Hazeki O., Katada T. Heterodimeric phosphoinositide 3-kinase consisting of p85 and p110beta is synergistically activated by the betagamma subunits of G proteins and phosphotyrosyl peptide. J. Biol. Chem. 1997;272:24252–24256. doi: 10.1074/jbc.272.39.24252. [DOI] [PubMed] [Google Scholar]
- Maier U., Babich A., Nürnberg B. Roles of non-catalytic subunits in gbetagamma-induced activation of class I phosphoinositide 3-kinase isoforms beta and gamma. J. Biol. Chem. 1999;274:29311–29317. doi: 10.1074/jbc.274.41.29311. [DOI] [PubMed] [Google Scholar]
- Martin V., Guillermet-Guibert J., Chicanne G., Cabou C., Jandrot-Perrus M., Plantavid M., Vanhaesebroeck B., Payrastre B., Gratacap M.P. Deletion of the p110beta isoform of phosphoinositide 3-kinase in platelets reveals its central role in Akt activation and thrombus formation in vitro and in vivo. Blood. 2010;115:2008–2013. doi: 10.1182/blood-2009-04-217224. [DOI] [PubMed] [Google Scholar]
- Orme M.H., Alrubaie S., Bradley G.L., Walker C.D., Leevers S.J. Input from Ras is required for maximal PI(3)K signalling in Drosophila. Nat. Cell Biol. 2006;8:1298–1302. doi: 10.1038/ncb1493. [DOI] [PubMed] [Google Scholar]
- Pacold M.E., Suire S., Perisic O., Lara-Gonzalez S., Davis C.T., Walker E.H., Hawkins P.T., Stephens L., Eccleston J.F., Williams R.L. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell. 2000;103:931–943. doi: 10.1016/s0092-8674(00)00196-3. [DOI] [PubMed] [Google Scholar]
- Patel M., Chiang T.C., Tran V., Lee F.J., Côté J.F. The Arf family GTPase Arl4A complexes with ELMO proteins to promote actin cytoskeleton remodeling and reveals a versatile Ras-binding domain in the ELMO proteins family. J. Biol. Chem. 2011;286:38969–38979. doi: 10.1074/jbc.M111.274191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P., Warne P.H., Dhand R., Vanhaesebroeck B., Gout I., Fry M.J., Waterfield M.D., Downward J. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P., Warne P.H., Vanhaesebroeck B., Waterfield M.D., Downward J. Activation of phosphoinositide 3-kinase by interaction with Ras and by point mutation. EMBO J. 1996;15:2442–2451. [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P., Sabatier C., McCormick F. Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol. Cell. Biol. 2004;24:4943–4954. doi: 10.1128/MCB.24.11.4943-4954.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte A., Stolp B., Schönichen A., Pylypenko O., Rak A., Fackler O.T., Geyer M. The human formin FHOD1 contains a bipartite structure of FH3 and GTPase-binding domains required for activation. Structure. 2008;16:1313–1323. doi: 10.1016/j.str.2008.06.008. [DOI] [PubMed] [Google Scholar]
- Srinivasan S., Wang F., Glavas S., Ott A., Hofmann F., Aktories K., Kalman D., Bourne H.R. Rac and Cdc42 play distinct roles in regulating PI(3,4,5)P3 and polarity during neutrophil chemotaxis. J. Cell Biol. 2003;160:375–385. doi: 10.1083/jcb.200208179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suire S., Condliffe A.M., Ferguson G.J., Ellson C.D., Guillou H., Davidson K., Welch H., Coadwell J., Turner M., Chilvers E.R. Gbetagammas and the Ras binding domain of p110gamma are both important regulators of PI(3)Kgamma signalling in neutrophils. Nat. Cell Biol. 2006;8:1303–1309. doi: 10.1038/ncb1494. [DOI] [PubMed] [Google Scholar]
- Tager A.M., LaCamera P., Shea B.S., Campanella G.S., Selman M., Zhao Z., Polosukhin V., Wain J., Karimi-Shah B.A., Kim N.D. The lysophosphatidic acid receptor LPA1 links pulmonary fibrosis to lung injury by mediating fibroblast recruitment and vascular leak. Nat. Med. 2008;14:45–54. doi: 10.1038/nm1685. [DOI] [PubMed] [Google Scholar]
- Tolias K.F., Cantley L.C., Carpenter C.L. Rho family GTPases bind to phosphoinositide kinases. J. Biol. Chem. 1995;270:17656–17659. doi: 10.1074/jbc.270.30.17656. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B., Welham M.J., Kotani K., Stein R., Warne P.H., Zvelebil M.J., Higashi K., Volinia S., Downward J., Waterfield M.D. P110delta, a novel phosphoinositide 3-kinase in leukocytes. Proc. Natl. Acad. Sci. USA. 1997;94:4330–4335. doi: 10.1073/pnas.94.9.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaesebroeck B., Guillermet-Guibert J., Graupera M., Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat. Rev. Mol. Cell Biol. 2010;11:329–341. doi: 10.1038/nrm2882. [DOI] [PubMed] [Google Scholar]
- Weiner O.D., Neilsen P.O., Prestwich G.D., Kirschner M.W., Cantley L.C., Bourne H.R. A PtdInsP(3)- and Rho GTPase-mediated positive feedback loop regulates neutrophil polarity. Nat. Cell Biol. 2002;4:509–513. doi: 10.1038/ncb811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch H.C., Coadwell W.J., Stephens L.R., Hawkins P.T. Phosphoinositide 3-kinase-dependent activation of Rac. FEBS Lett. 2003;546:93–97. doi: 10.1016/s0014-5793(03)00454-x. [DOI] [PubMed] [Google Scholar]
- Yan J., Mihaylov V., Xu X., Brzostowski J.A., Li H., Liu L., Veenstra T.D., Parent C.A., Jin T. A Gβγ effector, ElmoE, transduces GPCR signaling to the actin network during chemotaxis. Dev. Cell. 2012;22:92–103. doi: 10.1016/j.devcel.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.W., Shin M.G., Lee S., Kim J.R., Park W.S., Cho K.H., Meyer T., Do Heo W. Cooperative activation of PI3K by Ras and Rho family small GTPases. Mol. Cell. 2012;47:281–290. doi: 10.1016/j.molcel.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yart A., Chap H., Raynal P. Phosphoinositide 3-kinases in lysophosphatidic acid signaling: regulation and cross-talk with the Ras/mitogen-activated protein kinase pathway. Biochim. Biophys. Acta. 2002;1582:107–111. doi: 10.1016/s1388-1981(02)00144-0. [DOI] [PubMed] [Google Scholar]
- Zhang X., Vadas O., Perisic O., Anderson K.E., Clark J., Hawkins P.T., Stephens L.R., Williams R.L. Structure of lipid kinase p110β/p85β elucidates an unusual SH2-domain-mediated inhibitory mechanism. Mol. Cell. 2011;41:567–578. doi: 10.1016/j.molcel.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Bagrodia S., Cerione R.A. Activation of phosphoinositide 3-kinase activity by Cdc42Hs binding to p85. J. Biol. Chem. 1994;269:18727–18730. [PubMed] [Google Scholar]
Supplemental References
- Ciraolo E., Morello F., Hobbs R.M., Wolf F., Marone R., Iezzi M., Lu X., Mengozzi G., Altruda F., Sorba G. Essential role of the p110beta subunit of phosphoinositide 3-OH kinase in male fertility. Mol. Biol. Cell. 2010;21:704–711. doi: 10.1091/mbc.E09-08-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z., Chattopadhyay M., Pan J.A., Guerriero J.L., Jiang Y.P., Ballou L.M., Yue Z., Lin R.Z., Zong W.X. The class IA phosphatidylinositol 3-kinase p110-beta subunit is a positive regulator of autophagy. J. Cell Biol. 2010;191:827–843. doi: 10.1083/jcb.201006056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C.A., Vos M.D., Howell H., Vallecorsa T., Fults D.W., Clark G.J. Rig is a novel Ras-related protein and potential neural tumor suppressor. Proc. Natl. Acad. Sci. USA. 2002;99:9876–9881. doi: 10.1073/pnas.142193799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontani K., Tada M., Ogawa T., Okai T., Saito K., Araki Y., Katada T. Di-Ras, a distinct subgroup of ras family GTPases with unique biochemical properties. J. Biol. Chem. 2002;277:41070–41078. doi: 10.1074/jbc.M202150200. [DOI] [PubMed] [Google Scholar]
- Kurosu H., Katada T. Association of phosphatidylinositol 3-kinase composed of p110beta-catalytic and p85-regulatory subunits with the small GTPase Rab5. J. Biochem. 2001;130:73–78. doi: 10.1093/oxfordjournals.jbchem.a002964. [DOI] [PubMed] [Google Scholar]
- Lu Z., Luo R.Z., Lu Y., Zhang X., Yu Q., Khare S., Kondo S., Kondo Y., Yu Y., Mills G.B. The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells. J. Clin. Invest. 2008;118:3917–3929. doi: 10.1172/JCI35512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H.W., Hayashi M., Christoforidis S., Lacas-Gervais S., Hoepfner S., Wenk M.R., Modregger J., Uttenweiler-Joseph S., Wilm M., Nystuen A. An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J. Cell Biol. 2005;170:607–618. doi: 10.1083/jcb.200505128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utermark T., Rao T., Cheng H., Wang Q., Lee S.H., Wang Z.C., Iglehart J.D., Roberts T.M., Muller W.J., Zhao J.J. The p110α and p110β isoforms of PI3K play divergent roles in mammary gland development and tumorigenesis. Genes Dev. 2012;26:1573–1586. doi: 10.1101/gad.191973.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn T.A., Ramalingam T.R. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat. Med. 2012;18:1028–1040. doi: 10.1038/nm.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.J., Cheng H., Jia S., Wang L., Gjoerup O.V., Mikami A., Roberts T.M. The p110alpha isoform of PI3K is essential for proper growth factor signaling and oncogenic transformation. Proc. Natl. Acad. Sci. USA. 2006;103:16296–16300. doi: 10.1073/pnas.0607899103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.