Abstract
The p53 tumor suppressor is regulated by post-translational modification, including ubiquitination, phosphorylation and acetylation. It has previously been shown that the ubiquitin ligase Mdm2 also promotes the conjugation of Nedd8, a ubiquitin-like protein, to p53, inhibiting its transcriptional activity. We report the identification of FBXO11, a member of the F-box protein family and a component of the Skp1·Cullin1·F-box (SCF) complex, as a new p53-interacting protein. We show that FBXO11 promotes the neddylation of p53 both in vitro and in vivo. In addition to the C-terminal lysine residues, FBXO11 can also promote Nedd8 conjugation to Lys-320 and Lys-321, and neddylation of p53 leads to suppression of p53 function. This is consistent with recent studies showing that a lysine to arginine mutation at Lys-320 significantly enhances p53 function, although Lys-320 was originally identified as an acetylation site involving PCAF-mediated activation of p53. Our study provides an example of an F-box protein acting as an adaptor protein that can mediate the neddylation of a non-cullin substrate.
The p53 tumor suppressor has been dubbed the “guardian of the genome” because of its central role in the cellular response to stress. p53 functions as a transcription factor that is maintained at low levels in unstressed cells. In response to DNA damage, oncogene activation, hypoxia and oxidative damage, p53 levels and transcriptional activity are significantly enhanced, and the protein is able to initiate an anti-proliferative and pro-apoptotic program that prevents the multiplication of damaged and potentially pre-cancerous cells (1, 2). Although the mechanism of p53 activation is not fully understood, p53 is thought to be regulated primarily by post-translational modification, including ubiquitination, phosphorylation, and acetylation (3). In particular, Mdm2, a RING domain protein that is up-regulated in some tumors, is an important negative regulator of p53 function. Mdm2 binds directly to p53 and promotes its ubiquitination on six C-terminal lysines, thereby targeting it to the 26 S proteasome for degradation (4, 5). Following DNA damage, p53 is phosphorylated by Chk1 and Chk2 on serine and threonine residues within the Mdm2-binding region, resulting in a disruption of the Mdm2-p53 interaction and in an increase in p53 levels (6–8). This mechanism accounts in part for the activation of p53 in response to DNA damage.
It has recently been reported that in addition to its ubiquitin ligase activity, Mdm2 also promotes the conjugation of Nedd8 to p53 as part of its inhibitory function (9). Nedd8 is a small ubiquitin-like protein with 53% identity to ubiquitin (10) that is conserved from yeast to mammals, and the function of which was first described in yeast as the ortholog Rub1 (11, 12). Nedd8 is linked covalently through its C-terminal carboxyl group to the ε-amino group of lysine residues on target proteins in a ubiquitin-like manner. Nedd8 is activated by APPBP1-Uba3, a heterodimeric E1 enzyme, and conjugated to lysine by E2 enzyme Ubc12. The best-characterized substrates of Nedd8 conjugation are proteins of the cullin family. In particular, Cullin1 (or its yeast ortholog Cdc53), a component of the Skp1·Cullin1·F-box·Roc1 (SCF)3 complex, was shown to be neddylated, and its modification was shown to be important for the ubiquitin ligase function of some SCF complexes (12–14). Roc1, the RING domain-containing subunit within the SCF complex, functions as the E3 ligase in the Cullin1 neddylation reaction (15, 16). More recently, it has been reported that the ubiquitin ligase c-Cbl also promotes the neddylation of its target protein epidermal growth factor receptor (EGFR) (17). Unlike ubiquitination, neddylation does not directly promote the proteasomal degradation of target proteins, and although Nedd8 contains internal lysines, it does not form polyprotein chains like ubiquitin. Several lines of evidence point to the importance of the neddylation pathway in cell cycle control and viability. A temperature-sensitive mutation in SMC1, the hamster homolog of E1 component APP-BP1, results in severe cell cycle defects at nonpermissive temperature in the ts41 cell line (18), while deletion of the Uba3 gene in mice causes early embryonic lethality (19). Xirodimas et al. (9) found that Mdm2 promotes the neddylation of p53 on three of the six C-terminal lysines that are normally ubiquitinated, and that this neddylation of p53 inhibits its transcriptional activity. We report the identification of a new Nedd8 ligase for p53, an F-box domain protein called FBXO11, and show that this protein inhibits p53 activity without affecting its stability.
EXPERIMENTAL PROCEDURES
p53 Complex Purification
The epitope tagging strategy for the isolation of nuclear p53 protein complexes from human cells was performed essentially as described previously (20–22). To obtain the FLAG-HA-p53-expressing cell line, H1299 cells were transfected with pCIN4-FLAG-HA-p53(R175H) and selected for 2 weeks on 1 mg/ml G418 (Invitrogen), until p53-expressing clones were obtained. To prevent the degradation of nuclear p53, H1299/FLAG-HA-p53(R175H) cells were treated with the proteasome inhibitor MG132 (50 μM, Sigma) for 6 h before harvesting. For nuclear extract preparation, cells were incubated in buffer A (10 mM HEPES, pH 7.9, 10 mM NaCl, 0.1 mM EDTA, 1 mM DTT, and fresh protease inhibitor cocktail (Sigma)) for 15 min, Nonidet P-40 (Calbiochem) was added to 0.2%, and the samples were centrifuged for 5 min at 3000 rpm. Pellets were vortexed for 15 min in buffer C (20 mM HEPES, pH 7.9, 400 mM NaCl, 1 mM EDTA, 1 mM DTT, and fresh protease inhibitor), centrifuged for 30 min at 25,000 rpm, and filtered with 0.45-μm syringe filters (Nalgene). Supernatants were diluted with buffer D (20 mM HEPES, pH 7.9, 1 mM EDTA, fresh protease inhibitor) to the 200 mM final NaCl concentration and used as nuclear extracts for M2 immunoprecipitation using α-FLAG M2 affinity beads (Sigma) overnight at 4 °C. Binding proteins were eluted from the column with FLAG peptide (Sigma) at 4 °C overnight. M2 eluates were then subjected to a second step of affinity purification on α-HA beads (Sigma) at 4 °C overnight. p53 nuclear complexes were then eluted with HA peptide (Sigma) for 4 h at room temperature and resolved on a 4–12% gradient SDS-polyacrylamide gel (Invitrogen) followed by colloidal blue (Gelcode, Pierce) or silver staining. Specific bands were cut out from the gel and subjected to peptide sequencing by mass spectrometry.
FBXO11 Cloning and Antibody Production
Full-length FBXO11 cDNA was amplified by PCR from Marathon-Ready HeLa cDNA (Clontech, BD Biosciences), which is tagged with an adaptor at the 5′-end and allows for the performance of 5′-RACE (rapid amplification of cDNA ends), and cloned into p3XFLAG-CMV-10 expression vector (Sigma). The expression plasmid for FBXO11(del-F-box) was produced by replacing the sequence corresponding to amino acids 152–197 with an XbaI site and cloning the fragment into p3XFLAG vector.
To prepare the FBXO11 antiserum, the DNA sequence corresponding to the last 110 amino acids of FBXO11 (residues 817–927) was amplified by PCR and subcloned into pGEX-2T (Promega). The GST-tagged epitope, GST-FBXO11 (817–927), was purified from Escherichia coli Rosetta(DE3)pLysS cells (Novagen) using GST-bind resin (Novagen) and sent to Covance for rabbit antiserum production. For antibody purification, the epitope sequence was cloned into pET14b Histagged bacterial expression plasmid, and His-FBXO11 (817–927) was purified from Rosetta cells using nickel-agarose beads (Qiagen). The antigen was then coupled to agarose beads using AminoLink Plus Immobilization Kit (Pierce), and α-FBXO11 was affinity-purified following the manufacturer’s protocol.
Purification of FBXO11 Nuclear Complex
HEK 293 cells were transfected with plasmid expressing FLAG-FBXO11 or FLAG-FBXO11(del-F-box) using the calcium phosphate method. Nuclear extract was prepared as described above in “p53 Complex Purification” and subjected to immunoprecipitation using M2 beads. Protein was eluted from the M2 column using FLAG peptide in BC100 buffer (20 mM Tris-Cl, pH 7.9, 100 mM NaCl, 0.2 mM EDTA, 20% glycerol) containing 0.2% Triton X-100 and fresh protease inhibitor for 2 h at 4 °C. A sample of the complex was run on an SDS-polyacrylamide gel, and Skp1, Cullin1, and Roc1 were detected by Western blot using antibodies α-Skp1 P19 (Santa Cruz Biotechnology), α-Cullin1 AS97 (Santa Cruz Biotechnology), and α-Roc1 (Zymed Laboratories Inc.).
GST-pulldown Assay
GST and GST-p53 were purified as described previously (23). 35S-Labeled FBXO11 was prepared by in vitro translation using the TNT reticulocyte lysate system (Promega). 3 μg of GST protein was pre-incubated with in vitro translated FBXO11 for 1.5 h at 4 °C in BC100 buffer containing 0.1% Triton X-100 and 1% bovine serum albumin. GST beads were then added, and the solution was incubated for another 1.5 h at 4 °C. The beads were washed, and bound protein was eluted for 1 h at 4 °C in BC100 buffer containing 0.1% Triton X-100 and 20 mM reduced glutathione (Sigma) and separated on an SDS-polyacrylamide gel for detection by autoradiography.
Co-immunoprecipitation Assay
HCT116 cells were lysed in BC100 buffer containing 0.2% Triton X-100 and fresh protease inhibitor. Total protein was quantitated using protein assay reagent (Bio-Rad). Lysate containing 1 mg of total protein was incubated for 1 h at 4 °C with 1 μg of normal rabbit IgG (Santa Cruz Biotechnology) or 1 μg of α-FBXO11, and A/G PLUS-Agarose beads (Santa Cruz Biotechnology) were added for overnight incubation, also at 4 °C. The beads were washed, and bound protein was eluted by boiling in SDS sample buffer. FBXO11 and p53 were detected by Western blot using α-FBXO11 and α-p53 DO-1 antibodies (Santa Cruz Biotechnology).
Effect of FBXO11 Overexpression on p53 Stability
H1299 cells were plated on 100-mm dishes and transfected using the calcium phosphate method with 1 μg of GFP expression plasmid, 1 μg of p53 expression plasmid (pCIN4-FLAG-p53), and an increasing amount of FBXO11 expression plasmid (2, 4, 8, and 16 μg of p3XFLAG-FBXO11) as indicated. Cells were harvested after 24 h, lysed in FLAG lysis buffer (50 mM Tris-Cl, pH 7.9, 137 mM NaCl, 1% Triton X-100, 10% glycerol, 1 mM NaF, 0.1% Sarkosyl, 1 mM EDTA, and 1 mM Na3VO4) containing fresh protease inhibitor and 1 mM DTT, and total protein was quantitated. FBXO11, p53, and GFP were detected by Western blot.
In Vivo Ubiquitination and Neddylation Assays
The p53 lysine to arginine mutants were created using QuikChange site-directed mutagenesis kit (Stratagene) following the manufacturer’s protocol. H1299 cells plated on 100-mm dishes were transfected using the calcium phosphate method with 8 μg of pCIN4-His-HA-Ub or pCIN4-His-HA-Nedd8, 2 μg of pCIN4-FLAG-p53, and 10 μg of p3XFLAG-FBXO11 or 2 μg of Mdm2 plasmid, as indicated. Cells were harvested after 24 h, and 10% of the cell suspension was kept for input and lysed in FLAG lysis buffer. The remaining cells were lysed in 1 ml of buffer I (6 M guanidinium-HCl, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris-Cl, pH 8.0, 0.2% Triton X-100, 10 mM β-mercaptoethanol, and 5 mM imidazole). 50 μl of nickel-agarose beads were added, and lysates were rotated for 2 h at room temperature. The beads were then washed for 5 min at room temperature 1× in 1 ml of buffer I, 1× in 1 ml of buffer II (8 M urea, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris-Cl, pH 8.0, 0.2% Triton X-100, 10 mM β-mercaptoethanol, and 5 mM imidazole), and 3× in 1 ml of buffer III (same composition as buffer II except pH 6.3). Bound proteins were eluted in 50 μl of 2× SDS sample buffer containing 500 mM imidazole and 150 mM DTT for 30 min at room temperature and resolved on an SDS-polyacrylamide gel. Neddylated p53 species were detected by Western blot using p53-specific DO-1 antibody (neddylation assay) or FL-393 antibody (Santa Cruz Biotechnology, ubiquitination assay).
In Vitro Neddylation Assay
His-p53 and GST-Mdm2 were purified as described previously (23). SCFFBXO11 nuclear complex was purified from an H1299 stable line expressing FLAG-FBXO11 following the protocol described above in “Purification of FBXO11 Nuclear Complex.” For the in vitro p53 neddylation assay, 10 ng of His-p53 was incubated with 2 μg of Nedd8, 10 ng of E1 (APPBP1-Uba3), 200 ng of E2 (UbcH12) (all from BostonBiochem) and SCFFBXO11 complex (~10 ng of FLAG-FBXO11) in a total reaction volume of 10 μl (40 mM Tris-Cl, pH 7.4, 5 mM MgCl2, 2 mM ATP, 2 mM DTT) at 37 °C for 2 h. The reaction was stopped by the addition of SDS sample buffer, and the samples were run on an SDS-polyacrylamide gel. p53 was detected by Western blot using DO-1 antibody.
Luciferase Assay
The pCIN4-FLAG-HA-p53-Nedd8 plasmid was constructed by fusing PCR-amplified and digested Nedd8 to the C terminus of p53. Mouse embryonic fibroblast (p53−/−, Mdm2−/−) double knock-out cells were transfected with 3 μg of p21-luciferase reporter plasmid, 1 μg of pRL-TK control vector, and the indicated amounts of pCIN4-FLAG-HA-p53 or pCIN4-FLAG-HA-p53-Nedd8 plasmid using the calcium phosphate method in 6-well plates. Lysis and measurements were carried out using the Dual-Luciferase Reporter Assay System (Promega) following the manufacturer’s protocol.
Immunofluorescence Staining
H1299 cells were plated on glass coverslips in 6-well plates and transfected with 0.5 μg of pCIN4-FLAG-HA-p53 or pCIN4-FLAG-HA-p53-Nedd8 plasmid using the calcium phosphate method. Cells were stained with α-HA antibody (Roche Applied Science) as described previously (21).
RNAi Knockdown of FBXO11
GFP (NNGCUACCUGUUC-CAUGGCCA) and FBXO11 (1, NNUAGUCCAUACCAACU-UCGU; 2, NNGAAUCAGGUCCUGGUGCAC; 3, NNCCG-GAGACCCAGGCGAGUG) siRNA oligos were purchased from Dharmacon. U2OS and H1299 cells were plated at 80 × 103 cells per well in a 12-well plate in Dulbecco’s modified Eagle’s medium containing 10% fetal calf serum and transfected the following day using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s protocol. The transfection was repeated after 24 h for a second round, and cells were harvested after 48 h and lysed in FLAG lysis buffer containing fresh protease inhibitor. Samples were run on a 12% SDS-polyacrylamide gel, and FBXO11, p53, p21, and β-actin were detected by Western blot using α-FBXO11, DO-1, C-19 (Santa Cruz Biotechnology), and AC-15 (Sigma) antibodies, respectively.
RESULTS
Purification of p53 Nuclear Complex
We used a biochemical purification strategy to identify p53-interacting factors in vivo. To this end, we generated a derivative of the p53-null human lung carcinoma H1299 cell line that stably expresses a double-tagged human p53 mutant (arginine 175 to histidine) containing N-terminal FLAG and HA epitopes (FLAG-HA-p53(R175H)) (Fig. 1A). We used transcriptionally inactive mutant p53 in the construct because wild-type p53 strongly induces apoptosis in transfected cells, making it difficult to generate a stable cell line.
FIGURE 1. Identification of FBXO11 in the p53 nuclear complex.
A, structure of the FLAG-HA-p53(R175H) construct used in the purification of p53-interacting proteins. TAD, transactivation domain; DBD, DNA binding domain. B, silver staining of the SDS-polyacrylamide gel containing protein standard (lane 1), control eluate from M2/HA IP of H1299 cell nuclear extract (lane 2), and p53 nuclear complex obtained by M2/HA immunoprecipitation of H1299/FLAG-HA-p53(R175H) stable cell line nuclear extract (lane 3). Cells used for purification were incubated for 6 h with proteasome inhibitor (25 μM MG132 and 25 μM MG101). Specific p53-interacting protein bands were analyzed by mass spectrometry, and FBXO11 peptide sequences are presented.
To isolate protein complexes containing p53, nuclear extracts from FLAG-HA-p53-expressing H1299 cells and from control cells (parental H1299) were first subjected to two rounds of immunoaffinity purification on M2- (α-FLAG) and HA-affinity columns. The eluted proteins were then fractionated on an SDS-polyacrylamide gel and visualized by colloidal-blue and silver staining (Fig. 1B). As expected, we identified Mdm2, a known E3 ligase for p53, as a specific component of the complex.
Identification of FBXO11 as a p53-interacting Protein
Peptide sequencing of a major band of about 120 kDa by mass spectrometry identified three peptide sequences that matched a single translation product of a cDNA clone in the GenBank™ data base (accession number NM_025133). This clone encodes a major fragment of FBXO11, a little known member of the F-box protein family (24, 25). By rapid amplification of cDNA ends (RACE) and homology alignment with partial cDNA sequences in the data base, we cloned the full-length cDNA for human FBXO11 (Fig. 2A, Genbank™ accession number AY827075). The N terminus of FBXO11 contains the ~40 amino acid F-box motif common to all F-box protein family members (Fig. 2A). FBXO11 also contains a zinc finger-like domain called a UBR box at the C terminus. This domain was previously identified as the substrate-binding region of N-recognins, a family of ubiquitin ligases that recognize proteins through N-terminal signals and target them for degradation in a proteolytic pathway called the N-end rule pathway (26). The central region of FBXO11 contains a large domain with homology to the bacterial periplasmic protein NosD, a component of the nitrous oxide reductase complex (27). Overlapping with the NosD domain are three small CASH (carbohydrate-binding proteins and sugar hydrolases) domains of unclear function (28).
FIGURE 2. FBXO11 is a component of the SCF complex.
A, structure of FBXO11. B, FLAG-FBXO11 transfected into HEK 293 cells co-immunoprecipitates endogenous Cullin1, Skp1, and Roc1 (lane 2), as detected by Western blot, whereas FLAG-FBXO11(del-F-box) does not (lane 3).
To confirm the physical interaction between p53 and FBXO11, we first tested the binding of the two proteins in vitro in a GST-pulldown assay (Fig. 3A). 35S-Labeled in vitro translated FBXO11 bound to immobilized GST-p53 (Fig. 3A, lane 3) but not to GST alone (lane 2). We also investigated the interaction between endogenous p53 and FBXO11 protein in a co-immunoprecipitation assay (Fig. 3B). Total cellular extract from human colorectal carcinoma HCT116 cells was immunoprecipitated with α-FBXO11 or with normal rabbit IgG. As expected, Western blot analysis revealed that α-FBXO11 precipitated endogenous FBXO11 as well as endogenous p53 (Fig. 3B, lane 3), whereas the IgG control did not (lane 2).
FIGURE 3. FBXO11 interacts with p53 in vitro and in vivo.
A, GST (lane 2) and GST-p53 (lane 3) were used in a GST-pulldown assay with in vitro translated 35S-labeled FBXO11. B, HCT116 cellular extract was used for immunoprecipitation using normal rabbit IgG (lane 2) or anti-FBXO11 antibody (lane 3), and endogenous p53 protein was detected in the eluate.
FBXO11 Is a Component of the SCF Complex
F-box family proteins are known to interact through their F-box domain with Skp1 (29), a linker protein that couples the F-box protein to Cullin1 and Roc1, forming a complex known as the SCF ubiquitin ligase complex (30). Several members of the F-box protein family have been shown to bind to specific substrate proteins and to recuit them to the modular SCF complex, thus promoting their ubiquitination and degradation (31–37). Given that FBXO11 is a member of the F-box protein family, we investigated whether it is a component of the SCF complex in vivo. We transfected HEK 293 cells with plasmid expressing FLAG-tagged FBXO11, FLAG-tagged FBXO11 lacking the F-box domain (del-F-box), or empty vector and precipitated the expressed protein with M2 (α-FLAG) antibody from nuclear extract (Fig. 2B). FLAG-FBXO11 co-immunoprecipitated endogenous Skp1, Cullin1, and Roc1 (Fig. 2B, lane 2), whereas FLAG-FBXO11(del-F-box) (lane 3) and the empty control (lane 1) did not.
FBXO11 Does Not Regulate p53 Stability
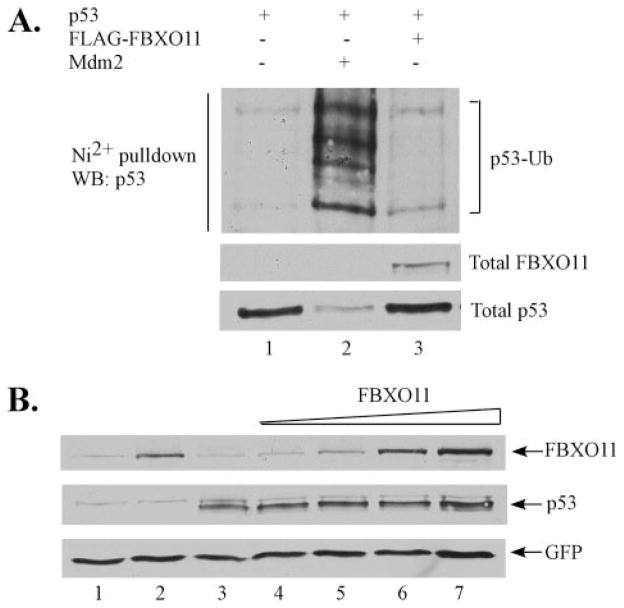
Because a major function of F-box proteins involves the ubiquitination and degradation of their target proteins, we first sought to determine whether FBXO11 ubiquitinates p53 and affects its stability. We found that FBXO11 does not promote the ubiquitination of p53 in an in vivo assay (Fig. 4A). We transfected H1299 cells with p53 plasmid as well as a vector expressing His-tagged ubiquitin, alone (Fig. 4A, lane 1), with Mdm2 plasmid (lane 2), or with FBXO11 plasmid (lane 3). His-ubiquitin-conjugated proteins were purified using nickel-agarose beads, and ubiquitin-modified p53 was detected by Western blot using p53-specific antibody. As expected, Mdm2 was able to promote the ubiquitination of p53 but FBXO11 was not. We also found that overexpression of FBXO11 in H1299 cells does not promote the degradation of p53 (Fig. 4B). H1299 cells were transfected with p53 alone (Fig. 4B, lane 3) and with increasing amounts of FBXO11 plasmid (lanes 4–7). The increase in FBXO11 had no effect on p53 protein levels. In addition, RNAi knockdown of FBXO11 in U2OS cells did not stabilize p53 compared with control knockdown using GFP siRNA (Fig. 7, upper panel).
FIGURE 4. FBXO11 does not promote the in vivo ubiquitination or degradation of p53.
A, H1299 cells were transfected with His-Ub, p53, Mdm2, and FBXO11 plasmid as indicated. Ubiquitinated proteins were purified using nickel-agarose beads and run on an SDS gel. Ubiquitinated p53 was detected by Western blot using p53-specific antibody. B, FBXO11 overexpression has no effect on p53 stability. H1299 cells were transfected with p53 (lanes 3–7), FBXO11 (lanes 2, 4 –7), and GFP (lanes 1–7) expression plasmids, and the level of expressed protein was detected by Western blot.
FIGURE 7. FBXO11 inhibits p53 transcriptional activity.
siRNA knockdown of FBXO11 using three different oligos (upper panel, lanes 2, 3, and 4) in U2OS results in an increase in p21 levels compared with control GFP siRNA (lane 1), whereas p53 levels are unaffected. Knockdown of FBXO11 in p53-null H1299 cells (lower panel, lanes 5 and 6) has no effect on p21 levels.
FBXO11 Promotes the Neddylation of p53 in Vivo and in Vitro
Roc1, the ubiquitin ligase subunit of the SCF complex, has been shown to promote the neddylation of cullins (15, 16), and it has previously been suggested that p53 function is regulated by conjugation to Nedd8 (9). We therefore decided to investigate whether FBXO11 can function as a Nedd8 E3 ligase for p53. FBXO11 was able to promote the neddylation of p53 in an in vivo neddylation experiment (Fig. 5A, lower panel). H1299 cells were transfected with p53 plasmid as well as a vector expressing His-tagged Nedd8, alone (Fig. 5A, lane 1) or with FBXO11 plasmid (lane 2). As for the in vivo ubiquitination assay described above, His-Nedd8-conjugated proteins were purified using nickel-agarose beads, and Nedd8-modified p53 was detected by Western blot using p53-specific antibody. His-Nedd8-conjugated p53 appears with the addition of FBXO11 plasmid (Fig. 5A, lane 2).
FIGURE 5. FBXO11 promotes the neddylation of p53 in vivo and in vitro.
A, upper panel, scheme of p53 lysine to arginine mutants used in this experiment. TAD, transactivation domain; DBD, DNA binding domain; NLS, nuclear localization signal; RD, C-terminal regulatory domain. Lower panel, H1299 cells were transfected with His-Nedd8, p53, and FBXO11 constructs as indicated. Neddylated proteins were purified using nickel-agarose beads and run on an SDS gel. Neddylated p53 was detected by Western blot using p53-specific antibody. p53 with lysine to arginine mutations on the six C-terminal lysines (6KR) has decreased neddylation by FBXO11 (lane 4), while p53 with two additional mutations (Lys-320 and Lys-321, 8KR) is not neddylated by FBXO11 (lane 6). B, SCFFBXO11 complex purified from a FLAG-FBXO11-expressing H1299 stable line promotes the neddylation of p53 in an in vitro neddylation reaction.
We also used nuclear FBXO11 complex (SCFFBXO11) purified from a FLAG-FBXO11-expressing H1299 stable line in an in vitro neddylation assay (Fig. 5B). Included in the in vitro reaction are the Nedd8 E1 enzyme, a heterodimer of APP-BP1 and Uba3, the Nedd8 E2 enzyme Ubc12, purified Nedd8, His-p53, and the potential E3 ligase, SCFFBXO11. The FBXO11 complex was able to promote the neddylation of p53 under these conditions (Fig. 5B, lane 2).
p53 is ubiquitinated on six C-terminal lysines, Lys-370, -372, -373, -381, -382, and -386 (4, 5), while Mdm2-mediated neddylation was shown to be limited to three of those residues, Lys-370, -372, and -373 (9). We used the p53 lysine mutants 6KR and 8KR (Fig. 5A, scheme in upper panel) in the in vivo neddylation experiment previously described to determine which sites are targeted by FBXO11. While there is a decrease in the neddylation of p53 with lysine to arginine mutations in the six C-terminal lysines (6KR; Fig. 5A, lane 4), neddylation of p53 is lost when two additional lysines, Lys-320 and Lys-321, are also mutated (8KR; lane 6).
Fusion of Nedd8 to the C Terminus of p53 Inhibits Its Activity
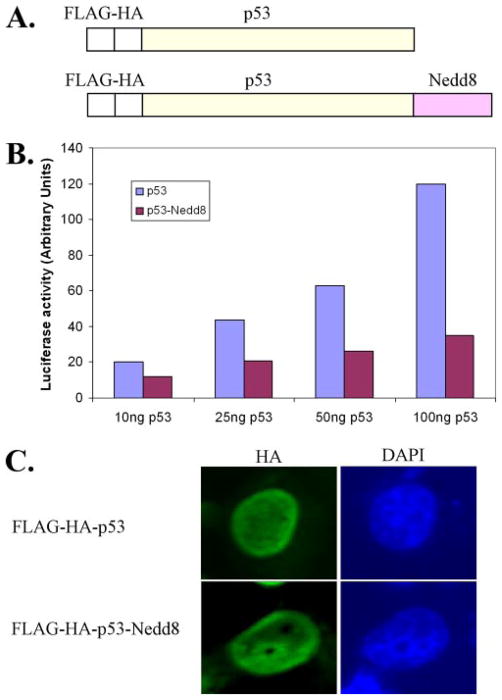
Given the difficulty of detecting neddylated p53 in vivo (9), we used an artificial system to study the effect of C-terminal Nedd8 conjugation to p53. We cloned a p53-Nedd8 construct by fusing Nedd8 to the C terminus of the p53 coding sequence (Fig. 6A), a method that was previously employed with ubiquitin to mimic protein monoubiquitination (38, 39). In a luciferase reporter assay using the endogenous promoter for the p21 gene (p21-luciferase), the p53-Nedd8 construct had decreased transactivation function compared to wild-type p53 (Fig. 6B). This is in agreement with previous data suggesting that Nedd8 conjugation to the C terminus of p53 is inhibitory (9). Because fusion of ubiquitin to the C terminus of p53 shifts the localization of p53 from the nucleus to the cytoplasm (38), we verified the subcellular localization of the p53-Nedd8 fusion protein (Fig. 6C). We transfected wild-type p53 and p53-Nedd8 plasmids into H1299 cells and carried out immunofluorescence staining to detect the expressed protein. We found that the p53-Nedd8 fusion protein, like wild-type p53, is predominantly nuclear, suggesting that Nedd8 conjugation to the C terminus inhibits p53 function without altering its subcellular localization.
FIGURE 6. Fusion of Nedd8 to the C terminus of p53 inhibits its activity.
A, schematic representation of the wild-type p53 and p53-Nedd8 constructs used in this experiment. B, activation of a p21-luciferase reporter by wild-type p53 and p53-Nedd8. The luciferase reporter construct was transfected into mouse embryonic fibroblast (p53−/−, Mdm2−/−) cells along with the indicated amounts of p53 plasmid, and luciferase activity was measured. C, sub-cellular localization of wild-type p53 and p53-Nedd8 protein. H1299 cells were transfected with expression plasmids for both constructs, and the expressed proteins were detected by immunofluorescence staining. Cells were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) to visualize nuclei.
FBXO11 Regulates the Transcriptional Activity of p53
It has previously been reported that Mdm2-mediated neddylation of p53 inhibits its transcriptional activity (9), and we showed that FBXO11 acts as a Nedd8 ligase for p53. To determine whether FBXO11 inhibits p53 activity, we carried out an RNAi knockdown experiment of FBXO11 in p53-positive U2OS cells (Fig. 7). Cells transfected with three different FBXO11 siRNAs (Fig. 7, lanes 2, 3, and 4) displayed enhanced levels of p21, a main p53 transcriptional target, compared to cells transfected with control GFP siRNA (lane 1), whereas p53 level was unchanged, indicating that FBXO11 inhibits p53 function without affecting its stability. In p53-null H1299 cells, siRNA knockdown of FBXO11 had no effect on the low p21 levels (Fig. 7, lower panel, lanes 5 and 6).
DISCUSSION
Mdm2 has been established as a key regulator of p53 activity. Knockout of Mdm2 in mice results in loss of p53 inhibition and p53-dependent embryonic death that can be rescued by knockout of Trp53 (40, 41). Mdm2 inhibits p53 primarily through the ubiquitin proteasome pathway. Mdm2 ubiquitinates p53 on six C-terminal lysines and targets it to the proteasome for degradation (4, 5). Interestingly, Mdm2 was also shown to have Nedd8 ligase activity for p53, and the neddylation of p53 was shown to be inhibitory, although the significance of this pathway in vivo is not yet clear (9). Studies on Mdm2 have shed light on important aspects of p53 regulation, but Mdm2-independent pathways are also known to be involved in activating and repressing p53. A number of proteins, including MdmX, COP1, Pirh2, and ARF-BP1 were found to be involved in the ubiquiti-nation and degradation of p53 (42–45), while several other proteins are known to regulate p53 activity through phosphorylation and acetylation (3).
In this report, we show that FBXO11, a novel p53-interacting protein that is a component of the SCF complex, can promote the neddylation of p53 and is an inhibitor of p53 transcriptional activity. FBXO11, a component of the SCF complex, was able to promote Nedd8 conjugation to p53 both in vivo and in vitro. FBXO11 does not promote the ubiquitination of p53 in vivo, and it does not affect p53 stability both in overexpression and in siRNA knockdown experiments.
We show that a p53-Nedd8 fusion protein, used to mimic Nedd8 conjugation to the C terminus of p53, has decreased transcription activation function compared with wild-type p53, in accordance with previous data suggesting that Nedd8 conjugation to p53 is inhibitory (9). While we expect that Nedd8 fusion to the C terminus of p53 approximates the effect of Nedd8 conjugation to lysines at the extreme C terminus of the protein, as previously reported for ubiquitin fusion proteins (38, 39), we recognize that this is an artificial system that may not fully recapitulate the effect of p53 neddylation by FBXO11 in vivo. In particular, Nedd8 C-terminal fusion does not approximate Nedd8 conjugation to Lys-320 and Lys-321, two internal lysines located within a nuclear localization signal, as discussed below, and may not have a similar effect on p53 sub-cellular localization as FBXO11-mediated neddylation. We also show that FBXO11 is an inhibitor of p53 function, as knockdown of FBXO11 using three different siRNA oligos results in an increase in the level of the p53 target p21, without affecting p53 stability.
While Mdm2 was reported to promote the neddylation of p53 on three lysine residues in the C-terminal regulatory domain of p53, we found that FBXO11 also neddylates p53 on two lysines, Lys-320 and Lys-321, within a nuclear localization signal that spans residues 300–323. This raises the possibility that neddylation by FBXO11 may affect p53 localization, sequestering p53 away from its nuclear site of action, but this issue is yet to be addressed. Interestingly, a recent report showed that thymocytes, epithelial, and retinal cells and E1A/Ras-expressing mouse embryonic fibroblasts from mice carrying a homozygous lysine 317 to arginine mutation, the site corresponding to human lysine 320, undergo increased p53-dependent apoptosis in response to DNA damage compared with wild-type cells, whereas p53 level remains unchanged (46). This data suggests that modification at lysine 320 is important for the repression of p53 activity in vivo, and it would be interesting to see whether FBXO11-mediated neddylation has a role in this repression.
The observation that a RING finger-containing complex is able to promote Nedd8 conjugation of p53 is in line with other evidence showing that RING finger proteins, including Mdm2 and c-Cbl, can act as Nedd8 E3 ligases (9, 17). Roc1, the RING domain-containing subunit of SCF, was in fact the first protein shown to have Nedd8 ligase activity in the Cullin1 neddylation reaction. It is somewhat surprising, however, that FBXO11 can bind p53 without promoting its ubiquitination or degradation, since SCF complexes are generally thought to be involved in the ubiquitination and proteasomal targeting of their substrates. Our data suggests that the SCF complex can also promote nondegradative modifications of target proteins, and we give the first example of an SCF protein promoting Nedd8 conjugation to its substrate. This finding raises the question of whether F-box proteins can promote the neddylation of additional non-cullin substrates. It would be interesting to see whether other F-box proteins, some of which have already been shown to ubiquitinate their targets and affect their stability, can also promote Nedd8 conjugation, and whether neddylation has a role in the regulation of substrate degradation.
It is unclear how the modular SCF complex, which comprises three constant subunits, Skp1, Cullin1, Roc1, and a variable substrate-binding F-box protein could distinguish between Nedd8 and ubiquitin conjugation. FBXO11 is a member of the F-box protein sub-family lacking a distinct unifying domain (the O in FBXO11 stands for other) (24). The two other F-box sub-families are FBXW, for proteins containing WD-40 repeat domains that are involved in phosphoprotein binding (47), and FBXL, for proteins containing leucine-rich repeat (LRR) domains, which also mediate protein-protein interactions (48, 49). A few F-box proteins have been well studied; among them Skp2 (FBXL1), β-Trcp (FBXW1), Cdc4 (FBXW7), and structural studies suggest that the SCF complex maintains a C-shape structure with the substrate-binding domain of the F-box protein at one end and the Roc1/E2 subunits at the other (50, 51). Although Cullin1 acts as a scaffold, maintaining the overall rigid structure of the complex, the variable F-box protein could provide flexibility, particularly through the linker region between the F-box and substrate-binding domains, in the positioning of the substrate relative to Roc1 and the E2 enzyme, which could allow for differential recruitment of Nedd8 versus ubiquitin-conjugating machinery.
p53 was previously shown to be neddylated and inhibited by Mdm2. We report here the identification of a novel Nedd8 ligase for p53, FBXO11, a component of the SCF complex, and show that FBXO11 is an inhibitor of p53 transcriptional activity. Although the role of Nedd8 modification in the regulation of p53 in vivo is not yet understood, our results raise the possibility that Nedd8 conjugation may be involved in the regulation of p53 function under some conditions. FBXO11 was initially identified as Vit1, a gene located on chromosome 2p21 and down-regulated in vitiligo, a skin disorder characterized by loss of melanocytes (52). It would be interesting to determine whether the p53 Nedd8-conjugation pathway has a role in this or in similar disease processes.
Footnotes
This work was supported in part by grants from the NCI, National Institutes of Health (to W. G.).
The abbreviations used are: SCF, Skp1·Cullin1·F-box·Roc1; HA, hemagglutinin; DTT, dithiothreitol; GST, glutathione S-transferase; E1, activating enzyme; E2, carrier protein; E3, protein isopeptide ligase; siRNA, small interfering RNA; GFP, green fluorescent protein.
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-Bank™/EBI Data Bank with accession number(s) AY827075.
References
- 1.Levine AJ. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Lane D, Levine AJ. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.Brooks CL, Gu W. Curr Opin Cell Biol. 2003;15:164–171. doi: 10.1016/s0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- 4.Nakamura S, Roth JA, Mukhopadhyay T. Mol Cell Biol. 2000;20:9391–9398. doi: 10.1128/mcb.20.24.9391-9398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez MS, Desterro JM, Lain S, Lane DP, Hay RT. Mol Cell Biol. 2000;20:8458–8467. doi: 10.1128/mcb.20.22.8458-8467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chehab NH, Malikzay A, Appel M, Halazonetis TD. Genes Dev. 2000;14:278–288. [PMC free article] [PubMed] [Google Scholar]
- 7.Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge SJ, Mak TW. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 8.Shieh SY, Ahn J, Tamai K, Taya Y, Prives C. Genes Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- 9.Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP. Cell. 2004;118:83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 10.Kamitani T, Kito K, Nguyen HP, Yeh ET. J Biol Chem. 1997;272:28557–28562. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- 11.Liakopoulos D, Doenges G, Matuschewski K, Jentsch S. EMBO J. 1998;17:2208–2214. doi: 10.1093/emboj/17.8.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lammer D, Mathias N, Laplaza JM, Jiang W, Liu Y, Callis J, Goebl M, Estelle M. Genes Dev. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Read MA, Brownell JE, Gladysheva TB, Hottelet M, Parent LA, Coggins MB, Pierce JW, Podust VN, Luo RS, Chau V, Palombella VJ. Mol Cell Biol. 2000;20:2326–2333. doi: 10.1128/mcb.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu K, Chen A, Pan ZQ. J Biol Chem. 2000;275:32317–32324. doi: 10.1074/jbc.M004847200. [DOI] [PubMed] [Google Scholar]
- 15.Kamura T, Conrad MN, Yan Q, Conaway RC, Conaway JW. Genes Dev. 1999;13:2928–2933. doi: 10.1101/gad.13.22.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morimoto M, Nishida T, Nagayama Y, Yasuda H. Biochem Biophys Res Commun. 2003;301:392–398. doi: 10.1016/s0006-291x(02)03051-6. [DOI] [PubMed] [Google Scholar]
- 17.Oved S, Mosesson Y, Zwang Y, Santonico E, Shtiegman K, Marmor MD, Kochupurakkal BS, Katz M, Lavi S, Cesareni G, Yarden Y. J Biol Chem. 2006;281:21640–21651. doi: 10.1074/jbc.M513034200. [DOI] [PubMed] [Google Scholar]
- 18.Handeli S, Weintraub H. Cell. 1992;71:599–611. doi: 10.1016/0092-8674(92)90594-3. [DOI] [PubMed] [Google Scholar]
- 19.Tateishi K, Omata M, Tanaka K, Chiba T. J Cell Biol. 2001;155:571–579. doi: 10.1083/jcb.200104035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo J, Su F, Chen D, Shiloh A, Gu W. Nature. 2000;408:377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 21.Nikolaev AY, Li M, Puskas N, Qin J, Gu W. Cell. 2003;112:29–40. doi: 10.1016/s0092-8674(02)01255-2. [DOI] [PubMed] [Google Scholar]
- 22.Nikolaev AY, Papanikolaou NA, Li M, Qin J, Gu W. Biochem Biophys Res Commun. 2004;323:1216–1222. doi: 10.1016/j.bbrc.2004.08.227. [DOI] [PubMed] [Google Scholar]
- 23.Li M, Luo J, Brooks CL, Gu W. J Biol Chem. 2002;277:50607–50611. doi: 10.1074/jbc.C200578200. [DOI] [PubMed] [Google Scholar]
- 24.Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kipreos ET, Pagano M. Genome Biol. 2000;1:REVIEWS3002. doi: 10.1186/gb-2000-1-5-reviews3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tasaki T, Mulder LC, Iwamatsu A, Lee MJ, Davydov IV, Varshavsky A, Muesing M, Kwon YT. Mol Cell Biol. 2005;25:7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zumft WG, Viebrock-Sambale A, Braun C. Eur J Biochem. 1990;192:591–599. doi: 10.1111/j.1432-1033.1990.tb19265.x. [DOI] [PubMed] [Google Scholar]
- 28.Ciccarelli FD, Copley RR, Doerks T, Russell RB, Bork P. Trends Biochem Sci. 2002;27:59–62. doi: 10.1016/s0968-0004(01)02046-1. [DOI] [PubMed] [Google Scholar]
- 29.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 30.Willems AR, Schwab M, Tyers M. Biochim Biophys Acta. 2004;1695:133–170. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 31.Carrano AC, Eytan E, Hershko A, Pagano M. Nat Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- 32.Sutterluty H, Chatelain E, Marti A, Wirbelauer C, Senften M, Muller U, Krek W. Nat Cell Biol. 1999;1:207–214. doi: 10.1038/12027. [DOI] [PubMed] [Google Scholar]
- 33.Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 34.Wu G, Lyapina S, Das I, Li J, Gurney M, Pauley A, Chui I, Deshaies RJ, Kitajewski J. Mol Cell Biol. 2001;21:7403–7415. doi: 10.1128/MCB.21.21.7403-7415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI. EMBO J. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hatakeyama S, Kitagawa M, Nakayama K, Shirane M, Matsumoto M, Hattori K, Higashi H, Nakano H, Okumura K, Onoe K, Good RA, Nakayama K. Proc Natl Acad Sci U S A. 1999;96:3859–3863. doi: 10.1073/pnas.96.7.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW. Genes Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 39.Terrell J, Shih S, Dunn R, Hicke L. Mol Cell. 1998;1:193–202. doi: 10.1016/s1097-2765(00)80020-9. [DOI] [PubMed] [Google Scholar]
- 40.Jones SN, Roe AE, Donehower LA, Bradley A. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 41.Montes de Oca Luna R, Wagner DS, Lozano G. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 42.Chen D, Kon N, Li M, Zhang W, Qin J, Gu W. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 43.Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, O’Rourke K, Koeppen H, Dixit VM. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 44.Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, Parant JM, Lozano G, Hakem R, Benchimol S. Cell. 2003;112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 45.Marine JC, Jochemsen AG. Biochem Biophys Res Commun. 2005;331:750–760. doi: 10.1016/j.bbrc.2005.03.151. [DOI] [PubMed] [Google Scholar]
- 46.Chao C, Wu Z, Mazur SJ, Borges H, Rossi M, Lin T, Wang JY, Anderson CW, Appella E, Xu Y. Mol Cell Biol. 2006;26:6859–6869. doi: 10.1128/MCB.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith TF, Gaitatzes C, Saxena K, Neer EJ. Trends Biochem Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- 48.Enkhbayar P, Kamiya M, Osaki M, Matsumoto T, Matsushima N. Proteins. 2004;54:394–403. doi: 10.1002/prot.10605. [DOI] [PubMed] [Google Scholar]
- 49.Kobe B, Kajava AV. Curr Opin Struct Biol. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 50.Cardozo T, Pagano M. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 51.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 52.Le Poole IC, Sarangarajan R, Zhao Y, Stennett LS, Brown TL, Sheth P, Miki T, Boissy RE. Pigment Cell Res. 2001;14:475–484. doi: 10.1034/j.1600-0749.2001.140608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]