Abstract
We use a novel technique that allows for closed recirculation of vector genomes in the cardiac circulation using cardiopulmonary bypass, referred to here as molecular cardiac surgery with recirculating delivery (MCARD). We demonstrate that this platform technology is highly efficient in isolating the heart from the systemic circulation in vivo. Using MCARD, we compare the relative efficacy of single-stranded (ss) adeno-associated virus (AAV)6, ssAAV9 and self-complimentary (sc)AAV6-encoding enhanced green fluorescent protein, driven by the constitutive cytomegalovirus promoter to transduce the ovine myocardium in situ. MCARD allows for the unprecedented delivery of up to 48 green fluorescent protein genome copies per cell globally in the sheep left ventricular (LV) myocardium. We demonstrate that scAAV6-mediated MCARD delivery results in global, cardiac-specific LV gene expression in the ovine heart and provides for considerably more robust and cardiac-specific gene delivery than other available delivery techniques such as intramuscular injection or intracoronary injection; thus, representing a potential, clinically translatable platform for heart failure gene therapy.
Keywords: AAV vectors, molecular cardiac surgery, cardiac gene therapy
INTRODUCTION
In all, 20% of 40-year-old Americans will develop heart failure,1 with a 70–80% 8-year mortality and annual US cost of 37 billion dollars,2 inflicting a considerable burden on the current health-care infrastructure. Targeted genetic manipulation of cardiac function may offer a novel therapeutic strategy for these and other patients with chronic heart failure. However, long-term expression of therapeutic transgenes in a significant percentage of left ventricular (LV) cardiac myocytes is a likely prerequisite for clinically efficacious gene therapy for global LV dysfunction. Although this prerequisite has been met in murine and rodent models using systemic intravenous injection of single-stranded (ss) adeno-associated virus (AAV)6 (Gregorevic et al.3), self-complimentary (sc)AAV8 (Wang et al.4) and ssAAV9 (Inagaki et al.5) encoding reporter genes, equivalent results have never been reproduced in large adult animals, critically limiting the potential for successful clinical application. The non-scalability of simple delivery methods such as intravenous injection and the inefficiency and significant collateral organ gene delivery of both intracoronary (IC) and intramuscular (IM) injection6–8 underscore the need for a novel, highly efficient, global, cardiac organ-specific platform for cardiac gene delivery that is clinically translatable. Our hypothesis is that a novel integration of established cardiac surgical techniques, incorporating cardiopulmonary bypass (CPB) to isolate the heart in situ, retrograde (venous-to-arterial) high pressure recirculation of vector in the coronary circulation,9 vascular endothelial growth factor to increase endothelial permeability,9 coupled with selected AAV sero-types encoding the enhanced green fluorescent protein (EGFP) reporter gene, will allow, for the first time, the clear and unequivocal demonstration of robust cardiac gene expression in an ovine model with minimal collateral gene expression that is highly predictive of similar long-term efficacy in humans with congestive heart failure. Further, this study aims to compare the relative efficacy of ssAAV6, ssAAV9 and scAAV6-encoding EGFP, driven by the constitutive cytomegalovirus (CMV) promoter.
RESULTS
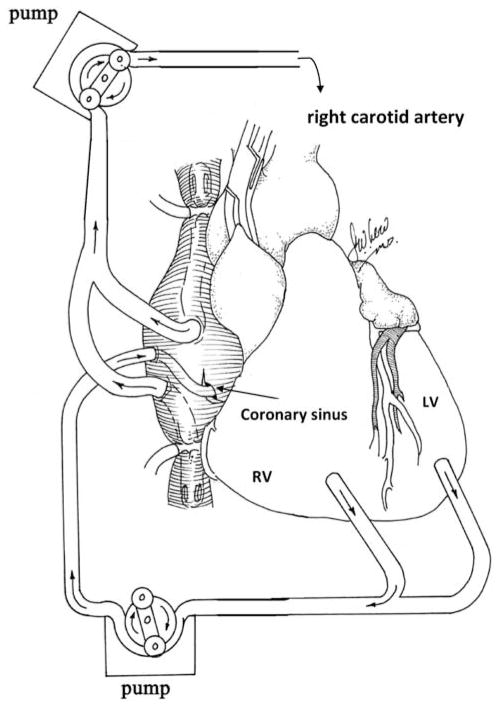
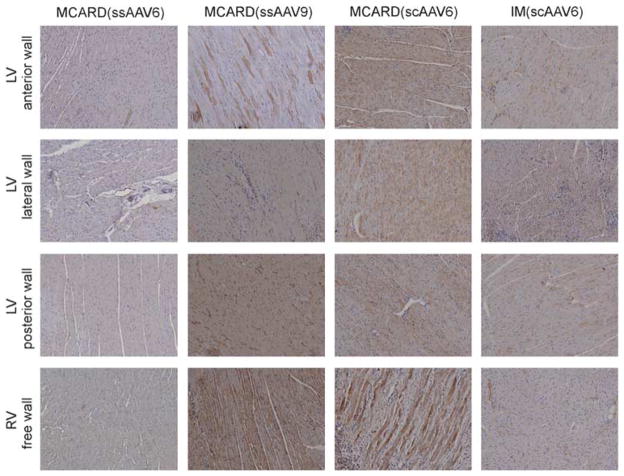
A schematic diagram of the molecular cardiac surgery with recirculating delivery (MCARD), perfusion circuit appears in Figure 1. The key components of this approach include isolation of the heart from the systemic circulation in vivo and recirculation of vector through the cardiac circulation for a total of 30 min. The vector was allowed to dwell in the heart for 10 min and recirculated via the coronary sinus for 20 min. Flow rates averaged approximately 80 cc min−1 during recirculation with a circuit volume of 300 cc and a delivery pressure of 80 mm Hg. Total CPB time averaged <2 h. After gene delivery, residual vector is washed from the coronary circulation to minimize systemic gene transfer. Direct fluorescence photography of whole cross-sections of the LV demonstrate robust green fluorescent protein (GFP) expression in the LV in the MCARD/scAAV6 group, less robust expression in the MCARD/ssAAV9 and MCARD/ssAAV6 groups, respectively and inhomogeneous expression in the IM/scAAV6 group (Figure 2). GFP immunostaining confirms that the majority of myocytes are transduced in representative sections of the LV in the MCARD/scAAV6 group only (Figure 3). Representative high-power photomicrographs further confirm the myocyte specificity of the transduced cells and that nearly 100% of cardiomyocytes in the LV anterior wall are transduced in the MCARD/scAAV6 group (Figure 4). GFP immunostaining also reveals minimal hepatic gene expression in the MCARD/scAAV6 group, while IM injection results in substantial hepatic gene expression (Figure 5). Western blotting reveals intense GFP expression in the LV, less intense but significant expression in the right atrium, and nearly undetectable hepatic expression in the MCARD/scAAV6 group. In contrast, in the IM injection group there is minimal GFP expression in the LV and RA, and significant hepatic GFP expression (Figure 6). Quantitative densitometric analysis of the western blot data confirms these qualitative findings and reveals that the higher expression of GFP in the liver in the IM group (0.21±0.05 (IM) vs 0.00±0.00 (MCARD)) is statistically significant (P<0.05). Quantitative PCR (Figure 7) reveals that the biodistribution of genome copies (GCs) in the MCARD groups is highest in the LV and low or undetectable in extracardiac tissues. Assuming that there are 8.14 pg DNA per ovine cell,10 the number of genome copies per cell in the LV using the scAAV6 transgene ranges from 1 gc per cell (lateral and posterior walls) to >3 gc per cell in the anterior wall. With ssAAV9 it ranged from 1 gc per cell in the lateral wall to >4 gc per cell in the posterior wall and with ssAAV6, the range was from 37 gc per cell (posterior wall) to 48 gc per cell (anterior wall). Over all serotypes, the ratio of vector genome concentration in the LV heart to liver (LV/liver) in the three MCARD groups ranged from 6 to 193 (6≤LV/liver ratio≤193). Using an interferon-γ Elispot assay, there was evidence of a T-cell-mediated immune response to the AAV6 capsid in one IM (scAAV6) group animal but none in the MCARD groups (scAAV6, ssAAV6 or ssAAV9), confirming that IM injection results in a more robust T-cell-mediated immune response against the AAV capsid than intravenous injection.11 None of the sheep in our series had preexisting immunity to the AAV6 or AAV9 capsid although preexisting immunity to GFP occurred in several animals.
Figure 1.
MCARD: high efficiency isolation of the heart in situ using CPB with true recirculating gene delivery globally to the myocardium retrograde via the coronary sinus.
Figure 2.
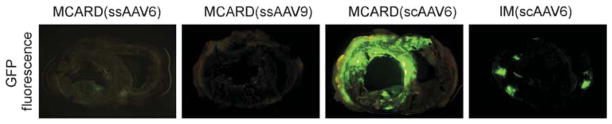
GFP fluorescence of whole mount cross-sections of the ovine heart taken at the mid-papillary level including both LV and RV demonstrating robust LV GFP fluorescence in the MCARD/scAAV6 group only.
Figure 3.
Representative GFP immunohistochemical sections displayed for MCARD and IM groups. Myocardial gene expression is most robust in the MCARD/scAAV6 group.
Figure 4.
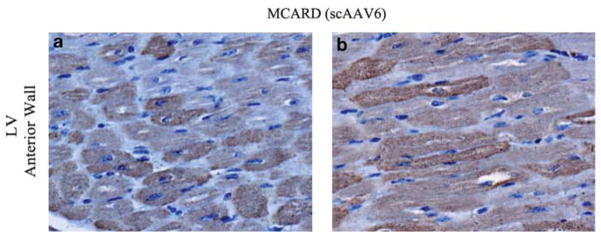
Representative high-power immunohistochemical transverse (a) and longitudinal (b) cross-sections displayed for the anterior wall for the MCARD/scAAV6 demonstrating that nearly 100% of cardiomyocytes are transduced unequivocally.
Figure 5.
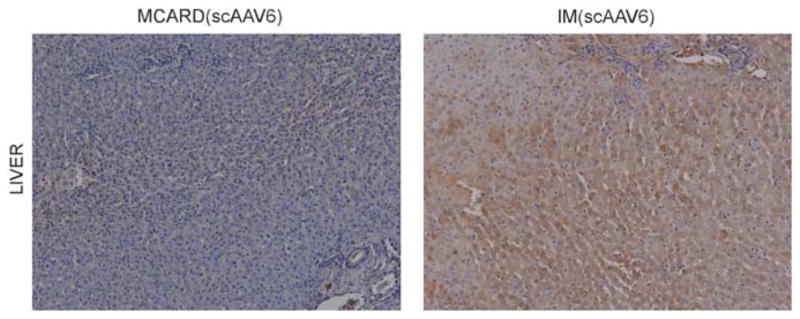
Representative GFP immunohistochemical sections displayed from liver for MCARD and IM groups (scAAV6) revealing significant collateral organ gene expression in the IM group but not in the MCARD group.
Figure 6.
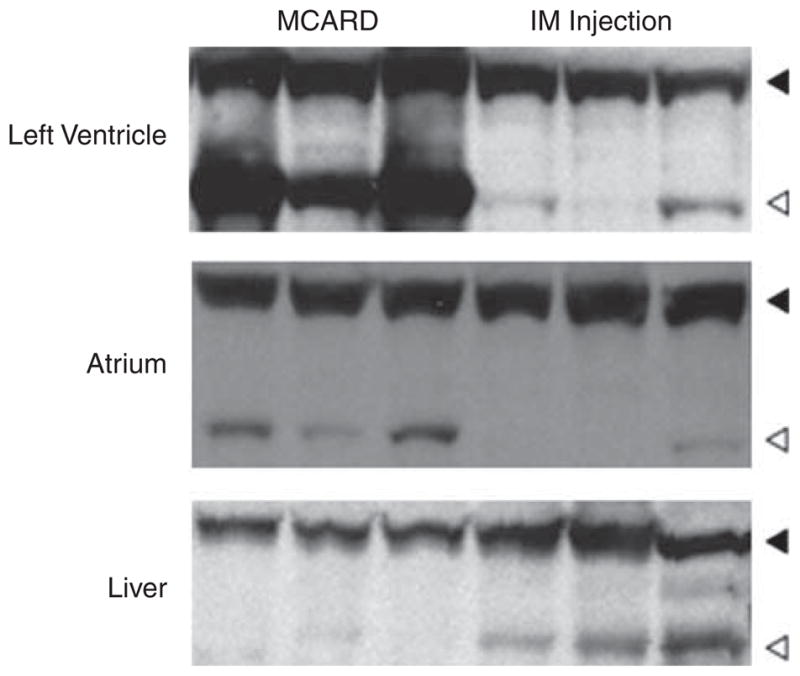
Western blots from MCARD and IM groups using the scAAV6 vector derived from the LV (anterior wall), left atrium and liver showing robust cardiac-specific GFP gene expression in the MCARD group with less robust LV gene expression in the IM group. There is significant hepatic expression only in the IM group.
Figure 7.
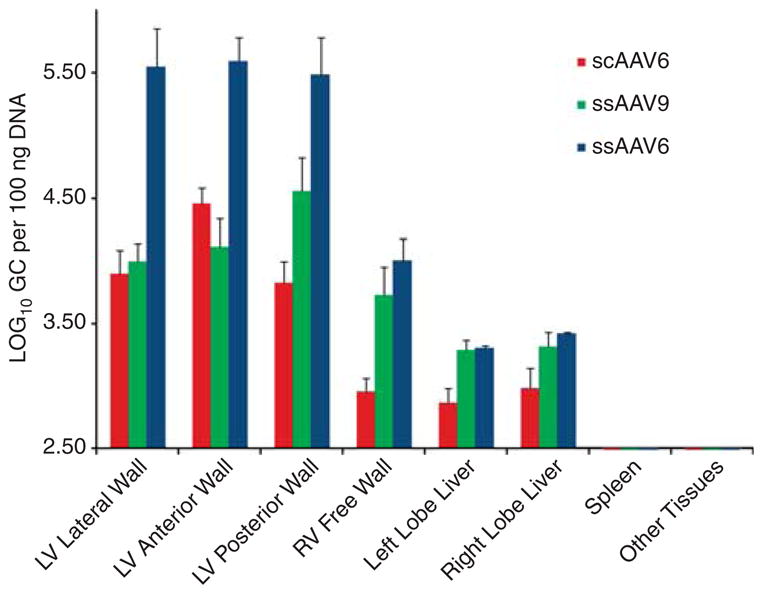
Biodistribution by vector serotype using MCARD delivery platform showing one to two orders of magnitude higher biodistribution of vector genomes in the LV then in the liver regardless of vector serotype (ssAAV6, ssAAV9 and scAAV6).
Sheep in this cohort received postoperative management reminiscent of that used to manage post-CPB patients. Routine laboratory values (complete blood count, liver function test, and so on) were obtained to assess physiologic function preoperatively and postoperatively. The mean values after the MCARD procedure were generally within normal limits for all parameters. There was no significant difference noted between baseline and post-surgical values, Table 1. We performed an electrocardiogram in all animals and analyzed the electrocardiogram for signs of myocardial ischemia (identified by T-wave changes, ST-segment displacement and the appearance of Q-waves) and conduction system abnormalities as well as common cardiac arrhythmias. The MCARD procedure was not associated with electrocardiogram signs of ischemia or other significant electrocardiogram abnormalities in any of the animals studied. We did not routinely perform echocardiography on the animals in this study but rather performed it selectively when clinically indicated. In all cases, echocardiography revealed normal postoperative LV chamber sizes and normal valvular and systolic function.
Table 1.
Laboratory testing results: pre- and post-gene delivery
| Parameter | Cohort values
|
Reference | |
|---|---|---|---|
| Preoperative | 2–3 weeks postoperative | ||
| Hemoglobin (g dl−1) | 11.0±2.2 | 10.5±2.1 | 9–14 |
| Total white blood cells (103 μl) | 6.7±2.0 | 8.0±1.5 | 4–13 |
| Platelet count (103 μl) | 367.4±156.0 | 389±140.1 | 250–750 |
| Neutrophils (%) | 36.7±6.3 | 39.3±11.1 | NA |
| Lymphocytes (%) | 49.7±9.3 | 53.3±12.4 | NA |
| ALT/SGPT (U l−1) | 32.4±8.9 | 36.6±12.9 | 26–34 |
| AST/SGOT (U l−1) | 279.6±50.7 | 298.0±59.6 | 254–360 |
| Total bilirubin (mg dl−1) | 0.2±0.3 | 0.2±0.1 | 0.1–0.5 |
| Creatinine (mg dl−1) | 1.0±0.4 | 1.1±0.7 | 1.2–1.9 |
| Creatine phosphokinase (U l−1) | 119.7±48.8 | 120.9±39.4 | 100–547 |
Abbreviations: ALT/SGPT, alanine aminotransferase; AST/SGOT, aspartate aminotransferase, NA, not applicable.
DISCUSSION
MCARD used to deliver scAAV6-encoding EGFP results in robust, global LV gene expression, most intense in the anterior and lateral wall. Gene expression in the right ventricular (RV) is less consistent than in the LV. This pattern of cardiac gene expression is closely correlated with the anatomical pattern of coronary venous drainage, because the RV venous drainage enters into the proximal coronary sinus. We confirm the observation that scAAV has a substantially higher transduction efficiency than an equal titer/serotype of ssAAV.12 Although AAV9 has been touted as a ‘silver bullet’ for cardiac gene therapy,13 using MCARD we found substantially less cardiac gene expression using ssAAV9 compared with scAAV6, (scAAV6 ≫ ssAAV9>ssAAV6; Figures 2 and 3). In fact, the limited gene expression we observed in the ssAAV6 and ssAAV9 groups, in spite of up to 48 genome copies per cell in the LV, makes it unlikely that ssAAV6, ssAAV1 or ssAAV9 will result in robust cardiac gene expression in humans using the far less efficient and less cardiac-specific IC delivery method currently being utilized in ongoing clinical trials.14 Indeed, in contrast to IC injection where the concentration of GC in the liver is 100 times higher than in the heart,6 we found the inverse with MCARD (6≤ LV/liver ratio ≤193), representing a 600- to >19 000-fold increase in geographical specificity (Figure 7). Moreover, using IC injection of AAV, the biodistribution of only a small fraction of a genome copy per cell (<0.1) has been demonstrated in large animal myocardium7 indicating that using the IC delivery route, a paracrine mechanism would certainly be required as >90% of myocytes would never express the transgene.
In contrast to MCARD, despite using a 10-fold lower vector dose (1013 GC), IM injection caused significantly higher hepatic gene expression. In the IM group, 1013 GC was the highest IM dose allowable, using our AAV prep (~5×1012 GC/cc) to observe the upper limit of 10–15% (injectate volume)/(subtended myocardial segment volume), utilized in clinical trials15 to avoid myocardial edema. Higher IM doses would likely only have caused a more intense T-cell-mediated immune response and greater extracardiac gene expression because additional IM volume injection is likely to spill into the systemic circulation.16,17 Moreover, as MCARD uniquely isolates the heart and removes the AAV capsid from the circulation after delivery, there is no apparent maximum safe vector dose. Thus, even if the AAV prep were concentrated further, allowing for a higher IM dose at least 10 to 100 times as many vector genomes can be introduced into the MCARD circuit as could be delivered safely IM making the disparate dose comparison 1013 GC (IM) vs 1014 GC (MCARD) reasonable. MCARD limits interaction of capsid with antigen-presenting cells, minimizing the potential for a T-lymphocyte-mediated response. The gene delivery technology platform presented here remains theoretically superior to intravenous injection even using novel cardiotropic serotypes (‘biological nanoparticles’) coupled with a cardiac-specific promoter, although promising results have recently been demonstrated in mice, they have not yet been confirmed in larger animals.18 Indeed, such novel biological nano-particles too can be coupled with the MCARD delivery platform to provide additional advantages of higher efficiency and regional specificity. Furthermore, the cardiac-specific promoter, although limiting the expression of the transgene in organs other than the heart, will not limit the delivery of viral capsids to distant organs with its attendant potential to induce an immune response and cause organ damage. Thus, the MCARD delivery platform and novel biological nanoparticle engineering are complimentary and synergistic ways of ‘skinning the same cat’. Novel biological nanoparticles may also allow us to offer therapy to larger groups of patients by avoiding the problem of preexisting immunity against existing AAV serotypes. Our bias is that without an efficient gene delivery platform such as the one presented here, regardless of the AAV vector serotype utilized, successful gene therapy for human heart failure will likely remain an elusive goal. We were not able to achieve high enough titers of scAAV9 for comparison with scAAV6 but we hypothesize that MCARD/scAAV9 may have even greater therapeutic efficacy.
In summary, we believe that the data presented represent the most convincing demonstration, to date, of robust AAV-mediated gene expression in the majority of cardiac myocytes in situ in a large animal model, resulting from the unique coupling of a highly efficient myocardial delivery platform with an optimized vector system. These findings suggest that scAAV6-mediated vector transfer is more efficient for large animal cardiac myocyte transduction than ssAAV6 or ssAAV9. The MCARD vector-mediated gene delivery platform allows for highly specific cardiac genome biodistribution, enhancing both efficacy and safety. Clinically, the MCARD approach, coupled with an appropriate therapeutic transgene, would be applicable to patients with end stage heart failure, initially in those who would undergo cardiac surgery for other reasons, such as valve repair or replacement or ventricular assist device placement. Although the procedure is currently performed using a median sternotomy for proof of principle, it could be readily performed through a small anterior thoracotomy using minimally invasive or robotic surgical techniques.
MATERIALS AND METHODS
Via median sternotomy, 1014 GC of scAAV6.CMV.EGFP (n=3), ssAAV6. CMV.EGFP (n=2) and ssAAV9.CMV.EGFP (n=2) was introduced into the ovine myocardium in male sheep (30–50 kg) in retrograde fashion via the coronary sinus, using CPB with complete isolation of the cardiac circuit. The vector was delivered at 80 mm Hg pressure, allowed to dwell for 10 min and recirculated for 20 min. All vector was flushed from the coronary circuit before weaning from CPB. Herein, we refer to this technique as MCARD (detailed below). Using a left anterior thoracotomy, control animals received 1013 GC of scAAV6.CMV.EGFP via IM injection, using a grid, spacing each injection 1 cm from its nearest neighbor covering the accessible LV wall (n=3). Necropsy was performed at 4 weeks. Specimens were retrieved from: LV lateral wall, posterior wall, anterior wall; the RV free wall, the right atrium, left atrium, interventricular septum, lung, liver, diaphragm, spleen, kidney, testes, triceps, intercostal and quadriceps muscles. Vector biodistribution and gene expression were assessed by quantitative PCR, western blotting, direct fluorescence and GFP immunohistochemistry.
Animal use and surgical protocol
All animals received humane care in compliance with the NIH and institutional guidelines established by the Institutional Animal Care and Use Committee of the University of Pennsylvania. Ten sheep (~1-year old) weighing 30–50 kg were the basis for the results presented here.
Preoperatively, each animal was prepared and draped and placed in the partial right lateral decubitus, elevated at 20°. The induction of anesthesia included ketamine, valium and diazepam. A cuffed, 7F endotracheal tube was inserted. Maintenance of anesthesia included fentanyl (total dose 15 mg kg−1) with the addition of isoflurane. Following induction, external defibrillation pads were placed. Invasive monitoring included the use of a 16G arterial catheter placed in the right femoral artery and a 5F triple-lumen central venous catheter inserted through the right internal jugular vein. The animal was then positioned in the supine position, prepped and draped to allow for a median sternotomy incision.
Details of the MCARD cardiac surgical technique
Initially, purse string sutures are placed in the right atrial appendage (for superior vena cava cannulation), the superior vena cava is doubly snared with 0-silk suture, umbilical tapes are placed around the aorta and main pulmonary artery, purse string sutures (4-0 prolene) are placed in the proximal ascending aorta, right atrium adjacent to the coronary sinus and right atrium adjacent to the inferior vena cava. Heparin is administered (130 U kg−1), and additional doses administered to keep the activated clotting time>400 s. The right carotid artery is cannulated using a 14F wire-wrapped cannula for systemic perfusion. The aortic root vent, superior vena cava cannula and retrograde cardioplegia catheter are then placed. Partial CPB is initiated. The inferior vena cava is cannulated; full CPB is initiated; the right and the left azygous (hemiazygous) veins are ligated; the inferior vena cava is snared with a double loop of 0-silk suture; a snare is place around the middle cardiac vein; vent cannulas are placed into the LV cavity through the apex and into the RV through the outflow tract. The LV, RV and aortic root vents are connected via a Y connector to the venous limb of the cardiac venous return circuit. The arterial limb of the cardiac circuit is connected to the coronary sinus catheter. The cardiac circuit is primed with Plegisol (Hospira, Inc., Lake Forest, IL, USA), total priming volume approximately 100 cc. The aorta is cross clamped. Cold (4 °C) crystalloid cardioplegia (Plegisol, 300 cc) is delivered antegrade. The heart is isolated by tightening the superior vena cava and inferior vena cava snares, cross-clamping the pulmonary artery and tightening the snare around the middle cardiac vein. All excess volume and air is removed from the cardiac circuit until the heart is fully decompressed. Cardiac circuit flow is initiated briefly (~15 s) until the coronary sinus pressure equals 80 mm Hg. Flow was about 60–100 cc min−1 average. The virus solution consisting of 1014 vg of ssAAV6.CMV.EGFP, ssAAV9.CMV.EGFP or scAAV6.CMV.EGFP along with 60 μg of vascular endothelial growth factor165 in a total of 20–35 cc phosphate-buffered saline is injected into the retrograde catheter and allowed to dwell for 10 min. The intravascular space is completely saturated with vector solution. After the 10-min dwell period, recirculation commences for 20 min, with coronary sinus pressure of 80 mm Hg, and flow adjusted to maintain a constant coronary sinus pressure. The coronary circuit is then flushed antegrade with 1000 cc of Hespan to wash out residual vector and vascular endothelial growth factor. The washout solution is discarded and not reinfused. The aortic cross clamp is removed and flow restored. Re-warming is initiated and all cannulas are removed once the heart is contracting well. Epinephrine is administered at 10–20 ml min−1 (1–2 mcg min−1). The animal is weaned from bypass after 20 min of reperfusion. Once off bypass, protamine is given and inotropic agents are discontinued, typically within 15 min. Bilateral chest thoracostomy tubes are placed, and sternal incisions are closed with steel wires in standard fashion.
Statistical methods
Comparisons between the MCARD/scAAV6 and IM/scAAV6 group, quantitative western blot intensities were performed using unpaired Student’s t-tests implemented using Microsoft Excel software (Microsoft, Malvern, PA, USA). Values presented in the text represent mean±s.e.m. One-way analysis of variance tests were used to survey anatomical specimens, respectively (that is, LV lateral wall, liver, and so on), vetting the hypothesis that all groups contained the same level of expression, where P<0.05 indicated significant difference.
Vector design and production
Ss and sc recombinant AAV-6 vectors, encoding EGFP gene under the control of the human CMV immediate early enhancer–promoter with a splice donor–acceptor sequence and polyadenylation signal from the human β-globin gene are produced. Human embryonic kidney 293 cells (American Type Culture Collection, Manassas, VA, USA) are seeded at a density of 1.5×107 cells per plate in 150 mm Corning tissue culture dishes the day before transfection. For each plate; 12 μg of XX680 plasmid (that is, the adenoviral backbone/components), 10 μg of pXR6 plasmid (that is, the plasmid containing AAV2-rep and AAV6-cap) and 6 μg of the inverted terminal repeat-containing plasmid pTRUFR19 for production of single-stranded vectors or pTR-CMV-GFP20 for production of self complementary vectors are used for polyethylenimine (Poly-sciences, Inc., Warrington, PA, USA) mediated transfection.21 Between 200 and 400 plates are used for the production of 1×1014 DNase resistant particles. Three days post-transfection, cells are rinsed from the plates and pelleted by centrifuged at 1500 r.p.m. then frozen at −80 °C. For 10 plates of cells the pellet is re-suspended in 5 ml of 25 mM Tris HCl pH8.5, on ice. The cells are then sonicated at 26% amplitude for 25×1 s bursts (Brandson Digital Sonifier, Danbury, CT, USA) on ice. The lysate is treated with 50 μl of leupeptin (10 mg ml−1) and 25 μl of DNase I (10 U μl−1; Roche, Branchburg, NJ, USA) for 30–60 min at 37 °C. The lysate is then centrifugation at 5000 r.p.m. for 10 min (Sorvall Legend RT, Arlington, MA, USA). The clarified lysate is then layered onto an iodixanol gradient as described by Zolotuhkin et al.,22 and centrifuged for 1 h at 70 000 r.p.m. in an SW-70.1 rotor (Beckman, Brea, CA, USA). The recombinant virus is removed from the gradient by needle puncture between the 60 and 40% iodixanol interface. The virus is further purified by FPLC (AKTA FPLC; GE Health Care, Waukesha, WI, USA) using HiTrap QHP columns (GE Health Care). AAV is applied to the column, washed with 10 column volumes of 25 mM Tris HCl pH8.5 and eluted in 25 mM Tris HCl pH8.5 with a linear 25 mM to 1 M NaCl gradient. Flow speed of the pump is set to 1 ml min−1 for all column steps. Elute is collected and dialyzed against 1× phosphate-buffered saline supplemented with 5% sorbitol. DNase-resistant virus particle number is determined by performing slot blot hybridization of purified vector genomes and by sybergreen real-time PCR (HotStart-IT SYBR Green quantitative PCR, USB Corp., Cleveland, OH, USA).
Forward primer 5′-ACGTAAACGGCCACAAGTTC-3′and reverse primer 5′-AAGTCGTGCTGCTTCATGTG-3′. All AAV stocks are frozen at −80 °C in single aliquots of 1×1014 particles and thawed during the surgical procedure.
Quantitative real-time PCR
Real-time PCR was performed using the MyiQ Real-Time PCR Detection System and analyzed using the MyiQ software package (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Briefly, the DNA from blood was isolated by the High Pure Nucleic Acid Kit (Roche Applied Science, Branford, CT, USA) and the DNA from tissue was purified with Qiagen Blood and Tissue DNeasy kit (Qiagen, Valencia, CA, USA) following the protocols. Absorbance at 260 nm (A260) was measured for each DNA sample using the NanoDrop (ND-1000) spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). A total amount of 100 ng template DNA per reaction was then taken for quantitative PCR assay.
Quantitative PCR analysis was performed in optical 96-well plates using iQ SYBRGreen Supermix (Bio-Rad Laboratories, Inc.) to monitor double-stranded DNA synthesis. All samples were analyzed in duplicate or triplicate in volume of 25 μl. Each reaction consisted of 12.5 μl 2× SYBRGreen Supermix, 1 μl of specific primer mix and 100 ng of DNA template.
A 10-fold dilution series of pTRUFR GFP (5 μl containing 1.45×108 copies to 1.45×103 copies) was used as the external quantitation standard. The copy number was calculated based on the molecular weight of pTRUFR GFP.
PCR for standard curve and experimental samples were performed together in the same run. The MyiQ software extrapolated sample concentration from the standard curve. Reactions were performed using the default two-step amplification plus melting curve protocol with GFP-specific forward 5′-TATA TCATGGCCGACAAGCA-3′ and reverse 5′-GAACTCCAGCAGGACCATGT-3′ primers.
The reaction conditions were: 95 °C for 5 min; 40 cycles at 95 °C for 15 s and 60 °C for 45 s. The melt curve protocol was performed after the amplification and consisted of 1 min at 55 °C followed by 80 cycles with 0.5 °C increments in each step. Threshold values for threshold cycle (Ct) determination were generated automatically by the MyiQ software.
Western blotting
Frozen tissues (heart, liver, kidney, lung, diaphragm, muscles and testis) were disrupted by Omni TH Tissue Homogenizer in RIPA lysis buffer (Cell Signaling Technology, Danvers, MA, USA) supplemented with phenylmethylsulfonyl fluoride, protease inhibitor cocktail and sodium orthovanadate. Tissue debris were removed by centrifugation at 10 000×g for 10 min. Resulting protein extract concentrations were quantified by using the Bradford method (Bio-Rad Laboratories, Inc.) according to the manufacturer’s protocol. A total of 65 μg of each protein sample were loaded in one well and resolved in a NuPAGE 12%-BT gel (Invitrogen, Carlsbad, CA, USA). For western blot analysis proteins were electro-transferred to PVDF membranes (Bio-Rad Laboratories, Inc.) at 40 V and 4 °C overnight in a XCell II Blot Module (Invitrogen). Immunodetection was revealed by using a polyclonal anti-EGFP (Invitrogen) and anti-glyceraldehyde 3-phosphate dehydrogenase antibody (Sigma, Ronkonkoma, NJ, USA) with Invitrogen Western Breeze Chromogenic Kit. Expression level approximations were taken by comparing the band intensity of the GFP protein to a housekeeping glyceraldehyde 3-phosphate dehydrogenase gene product by quantitative densitometric analysis.
Whole mount fluorescence photography
The whole heart (LV and RV) images displayed in Figure 1a were direct and unmodified photographs of mid papillary muscle whole heart cross-sections with the specimens illuminated with the GFP excitation frequency coupled with a band pass filter to select out light in the GFP fluorescence range.
Immunological studies
Blood sample collection and PBMC isolation
The blood samples were collected through venipuncture using EDTA as anticoagulant and sent to the Immunology Core at the Gene Therapy Program of the University of Pennsylvania. The peripheral blood mononuclear cells (PBMC) were isolated using Ficoll–Paque gradient centrifugation (GE-Amersham Biosciences, Waukesha, WI, USA) and cryopreserved in liquid nitrogen. The day before the assay, PBMC samples were recovered from cryopreservation and cultured in complete R10 medium (RPMI 1640 with 10% fetal bovine serum) at 37 °C overnight before being enumerated for ELISpot assay (see below).
Interferon-γ ELISPOT assay
Interferon-γ ELISPOT assays were performed using the bovine/ovine/equine interferon-γ ELISPOT kit (Mabtech, Nacka Strand, Sweden). PBMCs were added at the density of 2×105 cells per well and stimulated for 18–20 h at 37 °C with peptide libraries derived from the AAV6 or AAV9 VP1 protein or EGFP protein at the concentration of 2 μg ml−1. Medium alone served as a negative control, and phytohemagglutinin as a positive control. The peptide libraries were synthesized as 15-mers with 10-amino-acid overlap with the proceeding peptide (Mimotopes, Clayton, Australia). The AAV6 or 9-VP1 peptide library was divided into three pools such that pool 1A contains the first 50 peptides of AAV VP1, pool 1B contains peptides 51–100 and pool 1C contains peptides 101–146. Spots were counted with an ELISPOT reader (AID, San Diego, CA, USA). Peptide-specific cells were represented as spot-forming cells per 106 PBMCs. A positive response to any peptide pools is arbitrarily defined as three times over background (medium alone control) and the response is over 55 spot-forming cells per 106 PBMCs.
Acknowledgments
This work was sponsored in part by the National Heart Lung and Blood Institute (1-R01-HL083078-01A2), CR Bridges, PI; we would like to acknowledge the Gene Therapy Resource Program (GTRP) of the National Heart, Lung and Blood Institute, National Institutes of Health for providing a subset of the gene vectors used in this study and the Immunology Core of the Gene Therapy Program at the University of Pennsylvania, supported by NIH P30-DK047757 (PI: James Wilson). The authors have no proprietary disclosures to report. We thank the animal care staff and technicians for excellent care and handling of the animals in the Biomedical Research Building Vivarium-University of Pennsylvania.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Lloyd-Jones DM, Larson MG, Leip EP, Beiser A, D’Agostino RB, Kannel WB, et al. Lifetime risk for developing congestive heart failure: the Framingham Heart Study. Circulation. 2002;106:3068–3072. doi: 10.1161/01.cir.0000039105.49749.6f. [DOI] [PubMed] [Google Scholar]
- 2.Lloyd-Jones DM, Adams R, Carnethon M, De Simone J, Ferguson TB, Flegal K, et al. Heart Disease and Stroke Statistics- 2009 update. A report from the American Heart Association statistics committee and stroke statistics subcommittee. Circulation. 2009;119:480–486. doi: 10.1161/CIRCULATIONAHA.108.191259. [DOI] [PubMed] [Google Scholar]
- 3.Gregorevic P, Blankinship MJ, Allen JM, Crawford RW, Meuse L, Miller DG, et al. Systemic delivery of genes to striated muscles using adeno-associated viral vectors. Nat Med. 2004;10:828–834. doi: 10.1038/nm1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Z, Zhu T, Qiao C, Zhou L, Wang B, Zhang J, et al. Adeno-associated virus serotype 8 efficiently delivers genes to muscle and heart. Nat Biotechnol. 2005;23:321–328. doi: 10.1038/nbt1073. [DOI] [PubMed] [Google Scholar]
- 5.Inagaki K, Fuess S, Storm TA, Gibson GA, Mctiernan CF, Kay MA, et al. Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Mol Ther. 2006;14:45–53. doi: 10.1016/j.ymthe.2006.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bridges CR. ‘Recirculating cardiac delivery’ method of gene delivery should be called ‘non-recirculating’ method [letter] Gene Therapy. 2009;16:939–940. doi: 10.1038/gt.2009.35. [DOI] [PubMed] [Google Scholar]
- 7.Byrne MJ, Power JM, Preovolos A, Mariani JA, Hajjar RJ, Kaye DM. Recirculating cardiac delivery of AAV2/1SERCA2a improves myocardial function on an experimental model of heart failure in large animals. Gene Therapy. 2008;15:1550–1557. doi: 10.1038/gt.2008.120. [DOI] [PubMed] [Google Scholar]
- 8.French BA, Mazur W, Geske RS, Bolli R. Direct in vivo gene transfer into porcine myocardium using replication-deficient adenoviral vectors. Circulation. 1994;90:2414–2424. doi: 10.1161/01.cir.90.5.2414. [DOI] [PubMed] [Google Scholar]
- 9.Bridges CR, Gopal K, Holt DE, Yarnall C, Cole S, Anderson RB, et al. Efficient myocyte gene delivery with complete cardiac surgical isolation in situ. J Thorac Cardiovasc Surg. 2005;130:1364–1370. doi: 10.1016/j.jtcvs.2005.07.035. [DOI] [PubMed] [Google Scholar]
- 10.Rodgers RJ, O’Shea JD, Bruce NW. Morphometric analysis of the ovine corpus luteum. J Anat. 1984;138:757–770. [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Figueredo J, Calcedo R, Wilson JM. Cross-presentation of adeno-associated virus serotype 2 capsids activates cytotoxic T cells but does not render hepatocytes effective cytolytic targets. Hum Gene Ther. 2007;18:185–194. doi: 10.1089/hum.2007.001. [DOI] [PubMed] [Google Scholar]
- 12.Gao GP, Lu Y, Sun X, Johnston J, Calcedo R, Grant R, et al. High-level transgene expression in nonhuman primate liver with novel adeno-associated virus serotypes containing self-complementary genomes. J Virol. 2006;80:6192–6194. doi: 10.1128/JVI.00526-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hajjar RJ, Samulski RJ. A Silver bullet to treat heart failure. Gene Therapy. 2006;13:997. doi: 10.1038/sj.gt.3302747. [DOI] [PubMed] [Google Scholar]
- 14.Hajjar RJ, Zsebo K, Deckelbaum L, Thompson C, Rudy J, Yaroshinsky A, et al. Design of a phase 1/2 trial of intracoronary administration of AAV1/SERCA2a in patients with heart failure. J Card Fail. 2008;14:355–367. doi: 10.1016/j.cardfail.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Rosengart TK, Lee LY, Patel SR, Sanborn TA, Parikh M, Bergman GW, et al. Angiogenesis gene therapy: phase 1 assessment of direct intramyocardial administration of an adenovirus vector expressing VEGF121 cDNA to individuals with clinically significant severe coronary artery disease. Circulation. 1999;100:468–474. doi: 10.1161/01.cir.100.5.468. [DOI] [PubMed] [Google Scholar]
- 16.Grossman PM, Han Z, Palasis M, Barry JJ, Lederman RJ. Incomplete retention after direct myocardial injection. Cathet Cardiovasc Intervent. 2002;55:392–397. doi: 10.1002/ccd.10136. [DOI] [PubMed] [Google Scholar]
- 17.Von Harsdorf R, Schott RJ, Shen YT, Vatner SF, Mahdavi V, Nadal-Ginard B. Gene injection into canine myocardium as a useful model for studying expression in the heart of large mammals. Circ Res. 1993;72:688–695. doi: 10.1161/01.res.72.3.688. [DOI] [PubMed] [Google Scholar]
- 18.Asokan A, Conway JC, Phillips JL, Li C, Hegge J, Sinnott R, et al. Reengineering a receptor footprint of adeno-associated virus enables selective and systemic gene transfer to muscle. Nat Biotechnol. 2010;28:79–82. doi: 10.1038/nbt.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zolotukhin S, Potter M, Hauswirth W, Guy J, Muzyczka N. T ‘humanized’ green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J Virol. 1996;70:4646–4654. doi: 10.1128/jvi.70.7.4646-4654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCarty D, Monahan P, Samulski RJ. Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Therapy. 2001;8:1248–1254. doi: 10.1038/sj.gt.3301514. [DOI] [PubMed] [Google Scholar]
- 21.Grieger J, Choi V, Samulski RJ. Production and characterization of adeno-associated viral vectors. Nat Protoc. 2006;1:1412–1428. doi: 10.1038/nprot.2006.207. [DOI] [PubMed] [Google Scholar]
- 22.Zolotuchin S, Byrne BJ, Mason E, Zolotuchin I, Potter M, Chestnut K, et al. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Therapy. 1999;6:973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]